REVIEW: Regulation of Zygotic Genome and Cellular Pluripotency
D. V. Onichtchouk1* and A. S. Voronina2
1University of Freiburg, Developmental Biology Unit, Biologie 1, 79194 Freiburg, Germany; E-mail: Daria.Onichtchouk@biologie.uni-freiburg.de2Research Center of Biotechnology RAS (Bach Institute of Biochemistry), 119071 Moscow, Russia; E-mail: voronina_a@mail.ru
* To whom correspondence should be addressed.
Received March 17, 2015; Revision received July 31, 2015
Events, manifesting transition from maternal to zygotic period of development are studied for more than 100 years, but underlying mechanisms are not yet clear. We provide a brief historical overview of development of concepts and explain the specific terminology used in the field. We further discuss differences and similarities between the zygotic genome activation and in vitro reprogramming process. Finally, we envision the future research directions within the field, where biochemical methods will play increasingly important role.
KEY WORDS: zebrafish, ZGA, MBT, pluripotency, Oct4, NanogDOI: 10.1134/S0006297915130088
Abbreviations and definitions: ESC, embryonic stem cells; IPSC, induced pluripotent stem cells; maternal transcription – transcription that occurs in the egg and stops in the first meiotic prophase; MBT, midblastula transition (a particular case of MZT, which occurs in several organisms. MBT is a moment of time, when ZGA, degradation of maternal transcripts, desynchronization of cell cycle, and locomotor activity of the cells start. The definition MBT is mostly applied to amphibians, fish, and Drosophila); MZT, maternal to zygotic transition (several events happening after fertilization until establishment of zygotic control over development including ZGA, degradation of maternal transcripts, desynchronization of cell cycle, and acquisition of cell motility. The term MZT can be applied to all metazoans); pluripotency – capacity of cells to differentiate along cell lineages contributing to all embryonic but not extraembryonic tissues. Early cells of vertebrate embryos before beginning of gastrulation, ESC, derived from mammals, birds, and fish and IPSC are pluripotent; totipotency – unrestricted capacity of cells to differentiate along cell lineages contributing to all embryonic and extraembryonic tissues. Zygote is totipotent by definition; early mammalian cells are also totipotent; ZGA, zygotic genome activation (start of zygotic transcription, which occurs in species-specific interval after fertilization).
Fertilized egg, or zygote, provides all gene products, which are
necessary to start embryonic development in multicellular animal. After
a while, control over development passes over to zygotic gene products.
Zygotic genome activation occurs in mammalian embryos in two (mouse) to
eight (cow) cell stage, and in the lower vertebrate embryos at blastula
stage, which consists of hundreds of actively dividing cells. Position
of each cell within the embryo defines to which cell type (skin,
muscles, nerves, etc.) the progeny of a given cell will belong. For
some time after zygotic genome activation, the destiny of the cells is
not yet fixed and can be reversed, when the cells are transplanted from
one to the other part of the embryo. This ability of a cell to develop
into any cell type of adult organism is called pluripotency. The
unrestricted developmental potential of the cell is stepwise restricted
during development: first, the cell is assigned to one of three germ
layers (ectoderm, endoderm or mesoderm), than to tissue-specific
precursors, and finally it differentiates to one cell type, or gives
rise to adult stem cell, capable of differentiating to two-three cell
types. Dozens of genes defining cellular fate during development have
been discovered during the last decades. Most of these genes encode
evolutionary conserved signaling molecules and transcription factors
involved in embryo patterning. Mutations of these genes are frequently
observed in human disease, including cancer. We have learned a lot
about genes, coordinating development in space; but in contrast to
that, we know relatively little about the genes controlling development
in time. The current review focuses on exactly the type of factors,
which activate zygotic genome and coordinate genetic programs in time,
during the transition from pluripotency to differentiated cells. For
the reader interested in related fields, we recommend recent in-depth
reviews [1-5] discussing
different types of pluripotent cells and [6-8] discussing pluripotent stem cell origins within
different taxons.
MATERNAL PERIOD OF DEVELOPMENT – EARLY OBSERVATIONS
At the end of the 19th and beginning of the 20th century, most contemporary biologists, who studied fish, amphibians, echinoderms, and mollusks, believed that pregastrula development is defined by information contained in the zygote [9]. One of the first attempts to define the earliest effects of sperm on development involved experimentation on sea urchins. Theodor Bovery, one of the authors of chromatin inheritance theory (1862-1915), summarized the studies of sea urchin hybridization in his last manuscript published in 1918 after his death [10]. Bovery distinguished nuclei-independent and nuclei-dependent periods of development. The first period, lasting from fertilization until beginning of gastrulation, depends on ooplasm rather than on chromatin of the egg or sperm. Indeed, enucleated eggs of sea urchin could be induced to develop by sperm of sea anemone, which belongs to different class of animals [11]. Nuclear substance of sea anemone sperm in this case is not participating in development, as evidenced by lack of sperm decondensation and sea urchin-like type of development. Development according to the maternal type even in cases when the nuclei had been removed from the eggs occurs after hybridization of close echinoderm species (Parechinus sp. and Paracentrotus sp.). When enucleated eggs of Sphaerechinus sea urchins were fertilized by sperm of relatively distant species (Parechinus or Paracentrotus), sperm chromosomes decondensed, and embryos developed by maternal type until gastrulation, and subsequently became developmentally arrested. These and similar experiments led Bovery to the conclusion that beginning of development is independent on nuclear function and is defined by egg cytoplasm. Conclusions from classical experiments of Bovery were confirmed by cross-hybridization experiments performed on frogs by Moore [12]. Rana pipiens eggs fertilized by sperm of close species (Rana palustris), first developed according to maternal type, while the influence of paternal genome became detectable later in development. Hybrids between more distant frog species (e.g. Rana pipiens eggs fertilized by Rana catesbeiana sperm) became arrested at gastrulation and died, as developmental programs encoded in maternal and paternal genomes differed too much to be fitted together. However, if X-rays destroyed the genome of Rana catesbeiana sperm, the eggs fertilized by this sperm developed as haploids until metamorphosis or even later [13]. Thus, even if non-specifically activated, the eggs can develop until gastrulation without requirement for nuclear material.
MORPHOGENETIC FUNCTION OF NUCLEI: WORK OF NEYFAKH
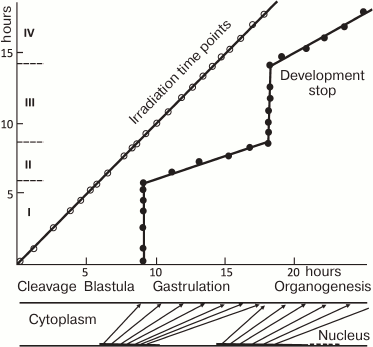
The theory growing from observations of the beginning of the 20th century was expanded and complemented by X-ray irradiation experiments on the teleost fish loach (Misgurnus fossilis) [14-18] performed by soviet scientist A. A. Neyfakh. When the fertilized eggs of loach were irradiated with a lethal dose of X-rays, they developed until late blastula, stopped, and died. This has shown that nuclei have no influence on initial development. In contrast, X-ray irradiation after midblastula led to progressively later developmental arrest (Fig. 1). This result implied that nuclear activity, or “morphogenetic function of nuclei” as formulated by Neyfakh [15], starts approximately at the midblastula stage. “Morphogenetic function of nuclei” appearing within the period between mid- and late blastula (2.5 h in loach) parallels desynchronization of cell cycles due to interphase lengthening and defines the whole gastrulation process, which lasts 9 h. Neyfakh determined the period from fertilization of the egg till beginning of morphogenetic function of nuclei in fish, echinoderms, mollusks, and amphibians [15, 16]. What switches “morphogenetic function of nuclei” on? Rott and Sheveleva [19] suggested that “morphogenetic function of nuclei” could be triggered by certain threshold ratio between nucleus and cytoplasm. The early cleavages of embryo do not change total volume: the volume of each cell is reduced two times during each cleavage cycle. Thus, absolute DNA content of a single cell does not change, but the ratio of DNA to cytoplasmic components constantly grows. One can speculate that rapid cell divisions are possible until this ratio reaches a certain level. If this is so, slowing the cell cycle should start in haploid embryos one cell division later. This was indeed confirmed experimentally [19].
Fig. 1. Dependence of stage of stopping the development of loach embryos from the moment of exposure (from [16] with modifications). Loach embryos were irradiated with 40 krad at different time points, which led to developmental arrest later. The irradiation time points are shown on Y (vertical) axis, and the stage when development stopped are shown on the X (horizontal) axis. X- and Y-axes, hours of normal development at 21.5°C. Roman numerals on the Y-axis, periods the morphogenetic activity of the nuclei. During periods II and IV core morphogenetic function of nuclei is critical, during periods I and III it is dispensable for development. Below the X-axis is a scheme of dependence of development of the embryo (“cytoplasm”, upper line) on morphogenetic functions of nuclei (“nuclei”, lower line). Periods of nuclear activity (corresponding to II and IV on the Y-axis) are marked in bold. Arrows connect the points of morphogenetic function of the nuclei with the stages it controls.
Spirin and coworkers studied the molecular basis for morphogenetic nuclear function and showed that the majority of mRNA synthesized at the gastrula stage is inactive and stored in the form of ribonucleoproteins called informosomes. This finding led to creation of the concept of “masking” of mRNA [20, 21]. Masking and demasking of mRNA play an important role in the development of embryos. It has been suggested that periodic demasking of specific mRNAs is the cause of the periodicity of the morphogenetic function of nuclei observed by Neyfakh. It became clear that in addition to transcription, there is another level of gene regulation in embryonic development – the level of translational regulation. This was confirmed by many works of the last decades [22]. Especially, the role of translational regulation of the genes involved in cell cycle was studied [23].
MIDBLASTULA TRANSITION (MBT)
The work of Neyfakh and Neyfakh’s laboratory laid the scientific grounds for widely used “midblastula transition” definition. Unfortunately, these works are cited only in papers or reviews of scientists of Russian origin. The reason for this forgetfulness was an artificial isolation of soviet scientist Neyfakh from the world scientific community (see [18] for details). French scientists Signoret and Lefreschne formulated the Midblastula Transition (MBT) concept in 1970s. The concept became popular after John Gerhardt’s review in 1980 [24].
Usually, the term “midblastula transition” (MBT) is applied to organisms, starting their embryonic life from series of rapid synchronous cleavages: amphibians, fish, and Drosophila. During MBT in the embryos of these animals several events, like lengthening and desynchronization of the cell cycle, beginning of cell locomotor activity, maternal RNA degradation, and start of zygotic transcription (ZGA, Zygotic Gene Activation) occur at the same time. In other animals, MBT does not exist. For example, ZGA in mice starts at the two-cell stage, while maternal RNA degradation starts after fertilization. Sea urchin embryos are transcriptionally active from the moment of fertilization, and transcription reaches its maximum at the 16-32-cell stage. In general, the processes manifesting the transmission of developmental control from maternal to zygotic genes (Maternal to Zygotic transition, MZT) are similar in metazoans. The definition of MBT can be applied to embryos in which these events occur simultaneously, while a broader MZT definition is used in other cases [25-27].
Cell cycle analysis of early synchronous phase of axolotl development demonstrated that G1 phase is missing and in each blastomere DNA replication begins at telophase [28]. At MBT, after 10-11 cell divisions, G1 appears, and its duration varies from cell to cell [29]. Appearance of G1 may point out that certain factors necessary to enter S phase become exhausted. MBT studies in the anuran amphibian Xenopus [30-32] and teleosts Danio rerio [32] and Fundulus [33] have shown that in parallel to cell cycle lengthening, embryonic cells become motile and transcriptionally active. In 2013, Collart and coworkers suggested a possible MBT mechanism: they showed that during Xenopus laevis MBT, the levels of four replication factors drop and become critical for replication start [34]. This causes lengthening of the cell cycle and beginning of transcription in the embryo.
Drosophila embryos start their development with a series of fast mitoses, similarly to fish and amphibians, but the cellular membrane between replicating nuclei is not formed. During the first nine cycles, which last 10 min each, dividing nuclei are located in the center of the egg. During the 10th cycle, nuclei migrate to the periphery (cortex) of the egg and form syncytial blastoderm persisting until the 13th cell cycle. Mitotic cycles 10-13-lengthen from 10 to 25 min. At the 14th cycle, which lasts 60 min, cellular membrane appears and separates the nuclei (this process is called cellularization), mitoses become asynchronous, and the cells start to move. The cell cycle duration in Drosophila is controlled by nuclear-cytoplasmic (N/C) ratio: haploid embryos compensate the lack of nuclear material with additional cleavage at the syncytial blastoderm stage. In contrast, artificial increase in the N/C ratio leads to skipping of one cell cycle at the blastoderm stage. As in fish and amphibians, cell cycle lengthening at Drosophila MBT coincides with regulated degradation of maternal RNAs and ZGA. However, in contrast to cell cycle, early gene activity in Drosophila is controlled not by one, but by at least three different mechanisms [35, 36]. Transcription of 127 out of 300 early genes is not controlled by N/C ratio but depends on time after fertilization. Transcription of other 88 genes depends on N/C ratio, and many of the other 85 early genes are switching on before MBT at cycles 11/12 using additional mechanisms [36].
Transcriptional events at MBT can be subdivided to two parts: maternal RNA degradation and ZGA [25].
MATERNAL RNA DEGRADATION
Transcript destabilization is accomplished by at least two pathways: the “maternal pathway” encoded exclusively in maternal genome, and the “zygotic pathway”. The difference between maternal and zygotic degradation pathways was demonstrated in mice [37-39], Danio rerio [40, 41], and C. elegans [42]. Traditionally, shortening of polyA tail is considered as the first step of mRNA degradation. However, in contrast to other animals, fertilization-induced deadenylation of maternal transcripts in Xenopus does not lead to their degradation prior to ZGA [43-45]. Maternal mRNA degradation in Drosophila is achieved by combination of maternal and zygotic mechanisms. The maternal mechanism is responsible for destabilization of 20%, and zygotic mechanism of 15% of transcripts [46, 47]. Egg activation triggers maternal mechanism by switching on the synthesis of RNA-binding protein Smaug from the maternally stored mRNA templates [46]. Smaug specifically binds to SMG-recognition elements on target mRNAs, leading to their deadenylation and subsequent degradation. Mechanisms of the zygotic mRNA degradation pathway in Drosophila remain unclear. Zygotic gene activator Zelda (Zld) may play a role in this process as degradation rates of many maternal mRNAs are changed in Zelda mutants [48]. Zygotically expressed miRNA mir430 in Danio is switched on by zygotic gene activator Nanog and is responsible for the degradation of hundreds of maternal mRNAs [49, 50]. Degradation of a separate set of mRNA, different from mir430 targets, is impaired in Danio rerio MZspg mutants lacking transcription factor Pou5f3 [51].
ZYGOTIC GENOME ACTIVATION
During zygotic genome activation (ZGA), 12-15% of Danio rerio, Drosophila, and mouse genes start to be rapidly transcribed [25]. Apart of general mechanisms like N/C ratio, ZGA is controlled by sequence-specific transcription factors: proteins that can recognize and bind to specific DNA sequences. In Drosophila, the earliest genes are activated by single maternally provided transcription factor Zelda (Zld). Zelda binds to specific TAGteam sequences on the regulatory regions of early genes to activate their transcription [48, 52, 53]. During the cell cycles following MBT, Zelda coordinates gene expression in time and space [54-56]. It is believed that Zelda keeps open chromatin structure, facilitating the local binding of other transcription factors [57, 58]. Thus, Zelda coordinates ZGA and post-ZGA events in Drosophila.
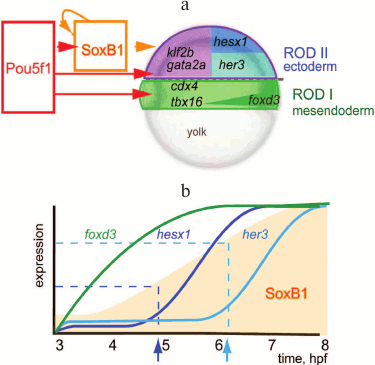
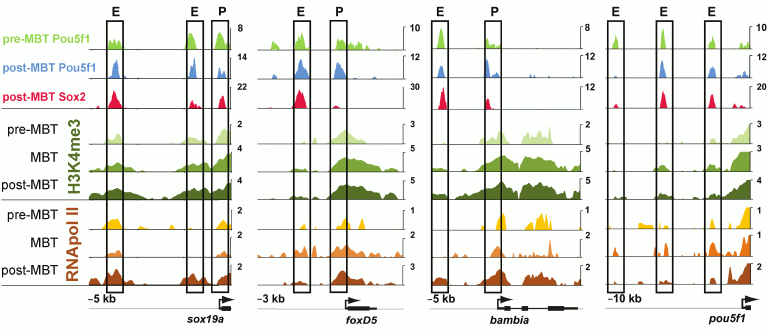
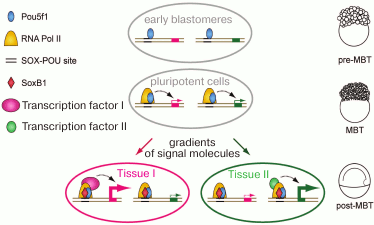
In Danio rerio, three transcription factors activate the zygotic genome: Pou5f3 (previously called Pou5f1), genes of the SoxB1 group, and Nanog [50, 59]. After ZGA, these factors activate tissue-specific genes [51, 60] (Fig. 2). Transcription in Danio rerio starts after the first 10 cell cycles. In MZspg mutants, devoid of maternal and zygotic functions of Pou5f3, expression of 595 zygotic genes is reduced twofold or more [51]. Pou5f3- and SoxB1-binding genomic regions are located in close proximity to developmental regulatory genes including Wnt, Nodal, BMP, and FGF signaling components and transcription factors controlling lineage specification in the embryo. Like Zelda in Drosophila, Pou5f3 and SoxB1 activate the earliest Danio rerio genes [59]. Pou5f3 and Nanog binding regions at and after MBT are enriched in active chromatin marks such as H3K4me3 and RNApolII [59, 61] (Fig. 3). RNApolII and chromatin-modifying factors do not have their own DNA-binding specificity, and it is possible that Pou5f3 recruits these factors to DNA. Binding regions of zygotic genome activators contain specific binding motifs TAGteam in Drosophila and SOX-POU in Danio and are usually bound by many other transcription factors, including tissue specific [48, 53, 56, 58, 59, 62] (see also Fig. 4). It is believed that Pou5f1/3, SoxB1, and Nanog, like Zelda, change local chromatin structure and facilitate binding of other factors to their binding sites. The mechanism of these changes is only starting to be revealed and can be different between Drosophila and vertebrates [63, 64].
Fig. 2. Model of gene-regulatory network at early gastrula stage of the Danio rerio embryo (modified from [60], printed with kind permission from John Wiley and Sons). a) Schematic representation of Danio rerio embryo in the early gastrula stage. Pou5f1 (Pou5f3) is present in all cells of the embryo; SoxB1 is present throughout ectoderm (violet outline). Tissue-specific transcription repressors, cdx4, foxD3, and tbx16, are activated by Pou5f3 and expressed in mesendoderm (green). Pou5f3 together with SoxB1 activates tissue-specific transcriptional repressors gata2a, klf2b, her3, and hesx1: gata2a and klf2b are expressed in the epithelial domain of the ectoderm (violet); her3 and hesx1 are expressed in the neural domain of the ectoderm (dark and light blue). b) Expression of Pou5f3 and SoxB1 target genes in time; hpf – hours post-fertilization; the vertical axis represents the amount of mRNA in the embryo [51]. Pou5f3 is present at the same concentration, SoxB1 concentration grows with time (orange outline). Foxd3 is switched on in the earliest time point, as it requires only Pou5f3 for its expression. To activate her3 and hesx1, different threshold concentrations of SoxB1 (indicated by dark and light blue arrows, respectively) in addition to Pou5f3 are necessary.
Fig. 3. Regulatory regions of genes associated early Pou5f1 and Sox2 contain marks of active chromatin ([59], published with permission from AAAS). Pre- and post-MBT Pou5f1, post-MBT Sox2, Pol II binding, and H3K4me3 modifications at four loci. ChIP-seq data for Pou5f1 and Sox2 are shown in reads per base. For H3K4me3 and Pol II, log2 Ma2C signal is shown for stages indicated; E, enhancer; P, promoter; X-axis indicates distance (kb) from Transcription Start Site.
Fig. 4. A hypothetical model for activation of the embryo genome (ZGA) and subsequent event (modified from [59], reproduced with the permission of AAAS). Before ZGA, Pou5f1 binds to the SOX-POU “priming sites” and attracts Pol II to ensure robust activation at ZGA. After ZGA, SoxB1 proteins co-occupy SOX-POU sites; both factors cooperate with developmental regulators to ensure precise transcriptional timing and proper level of expression.
PLURIPOTENCY PERIOD IN DEVELOPMENT
Totipotency and pluripotency are temporal cell states during development of the multicellular organisms, which pass necessary one-cell stage during their life cycle. Totipotency is unrestricted capacity of cell to develop all embryonic and extraembryonic cells of the organism. Zygote and early mammalian blastomeres are totipotent [65]. Pluripotency is the ability of cell to differentiate into all embryonic tissues. Cells of early vertebrate embryos at least until the beginning of gastrulation are pluripotent, as shown by transplantation experiments in mice, chick, fish, and frogs [66-70]. Notably, the duration of the pluripotency period greatly varies between different vertebrates: from 6 h after fertilization in Danio to five or more days in mammals.
GENETIC CONTROL OF PLURIPOTENCY IN EMBRYONIC STEM CELLS
Pluripotent Embryonic Stem Cells (ESC) and Induced Pluripotent Stem Cells (IPSC) confer great potential for use in regenerative medicine. For derivation of the first (ESC) in vitro cultures, morula stage embryos were used [71, 72]. Another pluripotent cell type, Epiblast Stem Cells (EpiSCs), was derived from later developmental stage [73]. ES and EpiS cells maintain pluripotency and can indefinitely proliferate in culture. Changing of culture conditions leads to in vitro differentiation towards various cell lineages. When transplanted into blastocyst stage mouse embryos, donor cells mixed with host cells and contributed to development of chimeric mice. High transcription level and “open” chromatin is a characteristic feature of ES cells [74]. Notably, the transcriptomes of mouse and human ES cells are quite divergent [75, 76]: biological characteristics of human ESC are closer to mouse EpiSC, than to mouse ESC [77-80]. ESC and EpiSCs need different culture conditions and activity of different signaling pathways for their propagation in a Petri dish [3]. All pluripotent cell types, including ESC and EpiSC, express homologs of ZGA activators, which play a central role in pluripotency maintenance: “pluripotency transcription factors” Pou5f1/Oct4, Sox2, and Nanog. These key transcription factors control the expression of several thousand genes in ESC [62, 75, 81]. Oct4, Sox2, and Nanog usually bind to enhancers, marked by presence of transcriptional co-activator p300 [62, 81]. Transcription of the nearby genes usually depends on the presence of one or two of the pluripotency transcription factors [5]. In mammalian ESC, Oct4–Sox2 complex binds to specific SOX-POU sequences [62, 76, 81]. SOX-POU sequences bind Nanog, Oct4, Sox2, Klf2/4, and downstream components of the LIF, BMP [81], and Wnt [82] pathways. Thus, SOX-POU modules integrate many signaling pathways and transcription factors to coordinate the signals of pluripotency and differentiation and balance them. Recent data are pointing out that the genetic regulatory network (GRN) regulated by “pluripotency transcription factors” in mammalian embryos is not necessarily the same as in in vitro cell cultures [83-85]. ESC were successfully derived not only from mammals, but also from chicken [86] and teleost fish medaka (Oryzias latipes) [87-89].
INDUCED PLURIPOTENT STEM CELLS (IPSC)
The central role in of “pluripotency transcription factors” in establishment and maintenance of pluripotency was confirmed by the discovery that forced expression of Oct4(Pou5f1), Sox2, Klf4, and c-Myc (OSKM) can reprogram somatic mammalian cells, fibroblasts, to Induced Pluripotent Stem Cells (IPSC) [90]. During the reprogramming process, epigenetic differences between differentiated and pluripotent cells are wiped out and some percentage of fibroblasts turns to ES-like cells [91, 92]. Reprogramming can be achieved not only by using Oct4, Sox2, Klf4, and c-Myc mixture, but also by alternative combinations of factors employing Nanog, Lin28, ESRRB, NR5A2, and other genes [3, 93, 94], which are not exclusively limited to genes expressed in ES cells or early embryonic stages [95, 96]. Currently, it is impossible to predict which new combination of transcription factors will induce reprogramming, although this question is of great theoretical and practical importance.
Capacity to be reprogrammed is not an exclusive propensity of mammalian cells. Rossello and coauthors [97] demonstrated that forced expression of mammalian OKSM mix could reprogram somatic cells of birds, fish, and even Drosophila. Thus, one can assume that some features of pluripotency GRNs were present in the common ancestor living about 550 million years ago.
EVOLUTION OF EARLY GENE REGULATORY NETWORKS IN VERTEBRATES
Recent molecular data support the “hourglass model”, which postulates maximum variability between different organisms of certain taxa at the beginning of development and in the adult, and maximum similarity between them in the middle of embryogenesis, at the so-called “phylotypic stage” [98-103]. Indeed, comparative studies between different mammalian species point to the high plasticity of early GRNs, defining pluripotency and zygotic genome activation [104, 105]. Gene expression analysis comparing mice and chick embryos even led to the conclusion that the early pluripotency network evolved de novo in mammals [106]. However, in spite of high variability of early GRNs, several observations, such as similarity of chromatin state in ESC and early Danio rerio embryos [107, 108], or presence of Pou5f1/3, Nanog (except Xenopus), and SoxB1 homologs in early development of vertebrates point to the conserved mechanisms controlling pluripotency. A critical role of Pou5f1/3, SoxB1, and Nanog homologs for early development is confirmed by pleiotropic lethal phenotypes of null-mutants for these genes in mice, Danio rerio, and Xenopus (except Nanog) [60, 109-113]. Although the phenotypic manifestations of Pou5f1/3 or Nanog mutations differ from species to species, in most cases it is possible to rescue these deficiencies with homologs of Pou5f1/3 and Nanog from evolutionarily distant species, which points to functional conservation of these factors [51, 114-119]. Analysis of early GRNs led Erwin and Davidson to the concept of “kernels”, central components of GRNs, which have an important developmental role, control the large number of downstream target genes, and are therefore less prone to evolutionary change than the other network components [120]. The “pluripotency transcription factors” Pou5f1/3, SoxB1, and Nanog match well to this theory, as kernels of the earliest zygotic GRN.
We have briefly outlined the current state of knowledge in one of the most rapidly developing parts of modern biology, concerning pluripotency in development. This field appeared from the merge of classical model organism developmental biology and stem cell biology. Molecular biological methods allowed identification of the genes important for stem cell survival and maintenance. In parallel, epigenetic studies led to discovery of specific histone modifications characteristic to both early embryos and embryonic stem cells, and to characterization of main transcription factor binding. However, in spite of a large body of data, molecular mechanisms controlling the balance between pluripotency and differentiation in stem cells are not yet understood completely. This understanding would require integrating the gene expression and epigenetic data with functional activity of encoded proteins, metabolites produced by cells, and cellular phenotypes changing in time during differentiation. Modern biochemical methods will allow making further steps in this direction. Thus, use of quantitative mass-spectroscopy methods enabled the characterization of various protein complexes being formed by Pou5f1/Oct4 in ES cells [121, 122]. Studies of ES cell metabolic products demonstrated that oxidative state of cells changes during differentiation, and direction and speed of cellular differentiation can be changed by experimentally perturbing some metabolic pathways [123]. Quantitative biochemical methods such as SILAC, which allows tracing time-resolved dynamics of total protein synthesis in cells, have great potential, as they can be applied both to cultured cells and to whole developing organisms.
Development of an organism from a single cell follows a strict schedule and is tightly controlled in time and space. Understanding the principles of this control plays a key role for ongoing attempts to grow tissues and organs from Embryonic Stem Cells. Therefore, the answer to the central question of biology – what are the rules that make an embryo from a single cell – has not only theoretical, but a great practical significance for the future development of new fields of medicine, such as regenerative therapies.
We are grateful to Vladimir Korzh and Yulia Kraus for critical reading of our manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (grant ON86/3-1 to D.V.O.) and by the Russian Foundation for Basic Research (grant No. 14-54-12008 to A.S.V.).
REFERENCES
1.Adachi, K., and Scholer, H. R. (2012) Directing
reprogramming to pluripotency by transcription factors, Curr. Opin.
Gen. Dev., 22, 416-422.
2.Chambers, I., and Tomlinson, S. R. (2009) The
transcriptional foundation of pluripotency, Development,
136, 2311-2322.
3.Hanna, J. H., Saha, K., and Jaenisch, R. (2010)
Pluripotency and cellular reprogramming: facts, hypotheses, unresolved
issues, Cell, 143, 508-525.
4.Sterneckert, J., Hoing, S., and Scholer, H. R.
(2012) Concise review: Oct4 and more: the reprogramming
expressway, Stem Cells, 30, 15-21.
5.Young, R. A. (2011) Control of the embryonic stem
cell state, Cell, 144, 940-954.
6.Bosch, T. C. (2009) Hydra and the evolution of stem
cells, BioEssays, 31, 478-486.
7.Bosch, T. C., Anton-Erxleben, F., Hemmrich, G., and
Khalturin, K. (2010) The hydra polyp: nothing but an active stem cell
community, Dev. Growth Differ., 52, 15-25.
8.Tanaka, E. M., and Reddien, P. W. (2011) The
cellular basis for animal regeneration, Dev. Cell, 21,
172-185.
9.Conklin, E. G. (1908) The mechanism of heredity,
Science, 27, 89-99.
10.Boveri, T. (1918) Zwei Fehlerquellen bei
Merogonieversuchen und die Entwicklungsfaehigkeit merogonischer und
partiellermerogonischer Seeigelbastarde, Arch. Entw. Mech.,
44, 417-471.
11.Godlevski, E. (1906) Untersuchungen ueber die
Bastardierung der Echiniden- und Crinoidenfamilie, Arch. Entw.
Mech., 20, 579-643.
12.Moore, J. (1941) Developmental rate of hybrid
frogs, J. Exp. Zool., 86, 405-422.
13.Rugh, R., and Exner, F. (1940) Developmental
effects resulting from exposure to X-rays. 11. Development of leopard
frog eggs activated by bullfrog sperm, Proc. Am. Phil. Soc.,
83, 607-619.
14.Neyfakh, A. A. (1959) X-ray inactivation of
nuclei as method for studying their function in the early development
of fishes, J. Embryol. Exp. Morphol., 7, 173-192.
15.Neyfakh, A. A. (1964) Radiation investigation of
nucleo–cytoplasmic interrelations in morphogenesis and
biochemical differentiation, Nature, 201, 880-884.
16.Neyfakh, A. A. (1961) Comparative study with the
use of radiation of developmental function of nuclei in the development
of animals, Zh. Obshch. Biol., 22, 42-57.
17.Korzh, V. (2009) Before maternal-zygotic
transition… there was morphogenetic function of nuclei,
Zebrafish, 6, 295-302.
18.VP%5BAuthor%5D&cauthor=true&cauthor_uid=20429375
Korzh, V. P., and Minin, A. A. (2010) A short history of loach or why
remember morphogenetic function nuclei? The 50th anniversary of A. A.
Neyfakh’s discovery of the morphogenetic function of the nucleus,
Ontogenez, 41, 150-158.
19.Rott, N. N., and Sheveleva, G. A. (1968) Changes
in the rate of cell divisions in the course of early development of
diploid and haploid loach embryos, J. Embryol. Exp. Morphol.,
20, 141-150.
20.Spirin, A. S. (1966) “Masked” forms
of mRNA, Curr. Top Dev. Biol., 1, 1-38.
21.Spirin, A. S. (1969) Informosomes, Eur. J.
Biochem., 10, 20-35.
22.Voronina, A. S. (2002) Translational regulation
in early development of eukaryotes, Mol. Biol. (Moscow), 36,
956-969.
23.Kronja, I., and Orr-Weaver, T. L. (2011)
Translational regulation of the cell cycle: when, where, how and why?
Phil. Trans. R. Soc., 366, 3638-3652.
24.Gerhart, J. (1980) Mechanisms regulating pattern
formation in the amphibian egg and early embryo, in: Biological
Regulation and Development (Goldberg, R., ed.) Plenum Press, New
York, pp. 133-315.
25.Tadros, W., and Lipshitz, H. D. (2009) The
maternal-to-zygotic transition: a play in two acts, Development,
136, 3033-3042.
26.Yasuda, G. K., and Schubiger, G. (1992) Temporal
regulation in the early embryo: is MBT too good to be true? Trends
Genet., 8, 124-127.
27.Langley, A. R., Smith, J. C., Stemple, D. L., and
Harvey, S. A. (2014) New insights into the maternal to zygotic
transition, Development, 141, 3834-3841.
28.Signoret, J., and Lefresne, J. (1974)
Determination, by tritiated thymidine incorporation, of the stages of
cellular cycle in the axolotl germ in synchronous period of
segmentation, C. R. Hebd. Seances Acad. Sci., 279,
1189-1191.
29.Lefresne, J., Andeol, Y., and Signoret, J. (1998)
Evidence for introduction of a variable G1 phase at the midblastula
transition during early development in axolotl, Dev. Growth
Differ., 40, 497-508.
30.Newport, J., and Kirschner, M. (1982) A major
developmental transition in early Xenopus embryos: I.
Characterization and timing of cellular changes at the midblastula
stage, Cell, 30, 675-686.
31.Newport, J., and Kirschner, M. (1982) A major
developmental transition in early Xenopus embryos: II. Control
of the onset of transcription, Cell, 30, 687-696.
32.Kane, D. A., and Kimmel, C. B. (1993) The
zebrafish midblastula transition, Development, 119,
447-456.
33.Trinkaus, J. P. (1992) The midblastula
transition, the YSL transition and the onset of gastrulation in
Fundulus, Development, Suppl. S, 116, 75-80.
34.Collart, C., Allen, G. E., Bradshaw, C. R.,
Smith, J. C., and Zegerman, P. (2013) Titration of four replication
factors is essential for the Xenopus laevis midblastula
transition, Science, 341, 893-896.
35.Edgar, B. A., and Schubiger, G. (1986) Parameters
controlling transcriptional activation during early Drosophila
development, Cell, 44, 871-877.
36.Lu, X., Li, J. M., Elemento, O., Tavazoie, S.,
and Wieschaus, E. F. (2009) Coupling of zygotic transcription to
mitotic control at the Drosophila midblastula transition,
Development, 136, 2101-2110.
37.Piko, L., and Clegg, K. B. (1982) Quantitative
changes in total RNA, total poly(A), and ribosomes in early mouse
embryos, Dev. Biol., 89, 362-378.
38.Alizadeh, Z., Kageyama, S., and Aoki, F. (2005)
Degradation of maternal mRNA in mouse embryos: selective degradation of
specific mRNAs after fertilization, Mol. Reprod. Dev.,
72, 281-290.
39.Hamatani, T., Carter, M. G., Sharov, A. A., and
Ko, M. S. (2004) Dynamics of global gene expression changes during
mouse preimplantation development, Dev. Cell, 6,
117-131.
40.Ferg, M., Sanges, R., Gehrig, J., Kiss, J.,
Bauer, M., Lovas, A., Szabo, M., Yang, L., Straehle, U., Pankratz, M.
J., Olasz, F., Stupka, E., and Muller, F. (2007) The TATA-binding
protein regulates maternal mRNA degradation and differential zygotic
transcription in zebrafish, EMBO J., 26, 3945-3956.
41.Aanes, H., Winata, C. L., Lin, C. H., Chen, J.
P., Srinivasan, K. G., Lee, S. G., Lim, A. Y., Hajan, H. S., Collas,
P., Bourque, G., Gong, Z., Korzh, V., Alestrom, P., and Mathavan, S.
(2011) Zebrafish mRNA sequencing deciphers novelties in transcriptome
dynamics during maternal to zygotic transition, Genome Res.,
21, 1328-1338.
42.Baugh, L. R., Hill, A. A., Slonim, D. K., Brown,
E. L., and Hunter, C. P. (2003) Composition and dynamics of the
Caenorhabditis elegans early embryonic transcriptome,
Development, 130, 889-900.
43.Paillard, L., Omilli, F., Legagneux, V., Bassez,
T., Maniey, D., and Osborne, H. B. (1998) EDEN and EDEN-BP, a
cis-element and an associated factor that mediate
sequence-specific mRNA deadenylation in Xenopus embryos, EMBO
J., 17, 278-287.
44.Audic, Y., Omilli, F., and Osborne, H. B. (1997)
Post-fertilization deadenylation of mRNAs in Xenopus
laevis embryos is sufficient to cause their degradation at the
blastula stage, Mol. Cell. Biol., 17, 209-218.
45.Voeltz, G. K., and Steitz, J. A. (1998) AUUUA
sequences direct mRNA deadenylation uncoupled from decay during
Xenopus early development, Mol. Cell. Biol., 18,
7537-7545.
46.Tadros, W., Westwood, J. T., and Lipshitz, H. D.
(2007) Titration of four replication factors is essential for the
Xenopus laevis midblastula transition, Dev. Cell,
12, 847-849.
47.De Renzis, S., Elemento, O., Tavazoie, S., and
Wieschaus, E. F. (2007) Unmasking activation of the zygotic genome
using chromosomal deletions in the Drosophila embryo, PLoS
Biol., 5, e117.
48.Liang, H. L., Nien, C. Y., Liu, H. Y., Metzstein,
M. M., Kirov, N., and Rushlow, C. (2008) The zinc-finger protein Zelda
is a key activator of the early zygotic genome in Drosophila,
Nature, 456, 400-403.
49.Giraldez, A. J., Mishima, Y., Rihel, J., Grocock,
R. J., Van Dongen, S., Inoue, K., Enright, A. J., and Schier, A. F.
(2006) Zebrafish MiR-430 promotes deadenylation and clearance of
maternal mRNAs, Science, 312, 75-79.
50.Lee, M. T., Bonneau, A. R., Takacs, C. M.,
Bazzini, A. A., Divito, K. R., Fleming, E. S., and Giraldez, A. J.
(2013) Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during
the maternal-to-zygotic transition, Nature, 503,
360-364.
51.Onichtchouk, D., Geier, F., Polok, B.,
Messerschmidt, D. M., Mossner, R., Wendik, B., Song, S., Taylor, V.,
Timmer, J., and Driever, W. (2010) Zebrafish Pou5f1-dependent
transcriptional networks in temporal control of early development,
Mol. Syst. Biol., 6, 354.
52.Farley, E., and Levine, M. (2012) HOT DNAs: a
novel class of developmental enhancers, Genes Dev., 26,
873-876.
53.Ten Bosch, J. R., Benavides, J. A., and Cline, T.
W. (2006) The TAGteam DNA motif controls the timing of
Drosophila pre-blastoderm transcription, Development,
133, 1967-1977.
54.Kanodia, J. S., Liang, H. L., Kim, Y., Lim, B.,
Zhan, M., Lu, H., Rushlow, C. A., and Shvartsman, S. Y. (2012) Pattern
formation by graded and uniform signals in the early Drosophila
embryo, Biophys. J., 102, 427-433.
55.Nien, C. Y., Liang, H. L., Butcher, S., Sun, Y.,
Fu, S., Gocha, T., Kirov, N., Manak, J. R., and Rushlow, C. (2011)
Temporal coordination of gene networks by Zelda in the early
Drosophila embryo, PLoS Genet., 7, e1002339.
56.Satija, R., and Bradley, R. K. (2012) The TAGteam
motif facilitates binding of 21 sequence-specific transcription factors
in the Drosophila embryo, Genome Res., 22,
656-665.
57.Kvon, E. Z., Stampfel, G., Yanez-Cuna, J. O.,
Dickson, B. J., and Stark, A. (2012) HOT regions function as patterned
developmental enhancers and have a distinct cis-regulatory
signature, Genes Dev., 26, 908-913.
58.Nien, C. Y., Liang, H. L., Butcher, S., Sun, Y.,
Fu, S., Gocha, T., Kirov, N., Manak, J. R., and Rushlow, C. (2011)
Temporal coordination of gene networks by Zelda in the early
Drosophila embryo, PLoS Genet., 7, e1002339.
59.Leichsenring, M., Maes, J., Mossner, R., Driever,
W., and Onichtchouk, D. (2013) Pou5f1 transcription factor controls
zygotic gene activation in vertebrates, Science, 341,
1005-1009.
60.Onichtchouk, D. (2012) Pou5f1/oct4 in
pluripotency control: insights from zebrafish, Genesis,
50, 75-85.
61.Bogdanovic, O., Fernandez-Minan, A., Tena, J. J.,
De la Calle-Mustienes, E., Hidalgo, C., Van Kruysbergen, I., Van
Heeringen, S. J., Veenstra, G. J., and Gomez-Skarmeta, J. L. (2012)
Dynamics of enhancer chromatin signatures mark the transition from
pluripotency to cell specification during embryogenesis, Genome
Res., 22, 2043-2053.
62.Kim, J., Chu, J., Shen, X., Wang, J., and Orkin,
S. H. (2008) An extended transcriptional network for pluripotency of
embryonic stem cells, Cell, 132, 1049-1061.
63.Foo, S. M., Sun, Y., Lim, B., Ziukaite, R.,
O’Brien, K., Nien, C. Y., Kirov, N., Shvartsman, S. Y., and
Rushlow, C. A. (2014) Zelda potentiates morphogen activity by
increasing chromatin accessibility, Curr. Biol., 24,
1341-1346.
64.Li, X. Y., Harrison, M. M., Villalta, J. E.,
Kaplan, T., and Eisen, M. B. (2014) Establishment of regions of genomic
activity during the Drosophila maternal to zygotic transition,
eLife, 3, e03737.
65.Burton, A., and Torres-Padilla, M. E. (2014)
Chromatin dynamics in the regulation of cell fate allocation during
early embryogenesis, Nat. Rev. Mol. Cell Biol., 15,
723-734.
66.Lawson, K. A., Meneses, J. J., and Pedersen, R.
A. (1991) Clonal analysis of epiblast fate during germ layer formation
in the mouse embryo, Development, 113, 891-911.
67.Garcia-Martinez, V., and Schoenwolf, G. C. (1992)
Positional control of mesoderm movement and fate during avian
gastrulation and neurulation, Dev. Dyn., 193,
249-256.
68.Ho, R. K., and Kimmel, C. B. (1993) Commitment of
cell fate in the early zebrafish embryo, Science, 261,
109-111.
69.Domingo, C., and Keller, R. (2000) Cells remain
competent to respond to mesoderm-inducing signals present during
gastrulation in Xenopus laevis, Dev. Biol., 225,
226-240.
70.Okabayashi, K., and Asashima, M. (2003) Tissue
generation from amphibian animal caps, Curr. Opin. Genet. Dev.,
13, 502-507.
71.Evans, M. J., and Kaufman, M. H. (1981)
Establishment in culture of pluripotent cells from mouse embryos,
Nature, 292, 154-156.
72.Thomson, J. A., Itskovitz-Eldor, J., Shapiro, S.
S., Waknitz, M. A., Swiergiel, J. J., Marshall, V. S., and Jones, J. M.
(1998) Embryonic stem cell lines derived from human blastocysts,
Science, 282, 1145-1147.
73.Tesar, P. J., Chenoweth, J. G., Brook, F. A.,
Davies, T. J., Evans, E. P., Mack, D. L., Gardner, R. L., and McKay, R.
D. (2007) New cell lines from mouse epiblast share defining features
with human embryonic stem cells, Nature, 448,
196-199.
74.Mikkelsen, T. S., Ku, M., Jaffe, D. B., Issac,
B., Lieberman, E., Giannoukos, G., Alvarez, P., Brockman, W., Kim, T.
K., Koche, R. P., Lee, W., Mendenhall, E., O’Donovan, A.,
Presser, A., Russ, C., Xie, X., Meissner, A., Wernig, M., Jaenisch, R.,
Nusbaum, C., Lander, E. S., and Bernstein, B. E. (2007) Genome-wide
maps of chromatin state in pluripotent and lineage-committed cells,
Nature, 448, 553-560.
75.Boyer, L. A., Lee, T. I., Cole, M. F., Johnstone,
S. E., Levine, S. S., Zucker, J. P., Guenther, M. G., Kumar, R. M.,
Murray, H. L., Jenner, R. G., Gifford, D. K., Melton, D. A., Jaenisch,
R., and Young, R. A. (2005) Core transcriptional regulatory circuitry
in human embryonic stem cells, Cell, 122, 947-956.
76.Loh, Y. H., Wu, Q., Chew, J. L., Vega, V. B.,
Zhang, W., Chen, X., Bourque, G., George, J., Leong, B., Liu, J., Wong,
K. Y., Sung, K. W., Lee, C. W., Zhao, X. D., Chiu, K. P., Lipovich, L.,
Kuznetsov, V. A., Robson, P., Stanton, L. W., Wei, C. L., Ruan, Y.,
Lim, B., and Ng, H. H. (2006) The Oct4 and Nanog transcription network
regulates pluripotency in mouse embryonic stem cells, Nat.
Genet., 38, 431-440.
77.Tesar, P. J., Chenoweth, J. G., Brook, F. A.,
Davies, T. J., Evans, E. P., Mack, D. L., Gardner, R. L., and McKay, R.
D. (2007) New cell lines from mouse epiblast share defining features
with human embryonic stem cells, Nature, 448,
196-199.
78.Chenoweth, J. G., McKay, R. D., and Tesar, P. J.
(2010) Epiblast stem cells contribute new insight into pluripotency and
gastrulation, Dev. Growth Differ., 52, 293-301.
79.Chenoweth, J. G., and Tesar, P. J. (2010)
Isolation and maintenance of mouse epiblast stem cells, Methods Mol.
Biol., 636, 25-44.
80.Gafni, O., Weinberger, L., Mansour, A. A., Manor,
Y. S., Chomsky, E., Ben-Yosef, D., Kalma, Y., Viukov, S., Maza, I.,
Zviran, A., Rais, Y., Shipony, Z., Mukamel, Z., Krupalnik, V., Zerbib,
M., Geula, S., Caspi, I., Schneir, D., Shwartz, T., Gilad, S.,
Amann-Zalcenstein, D., Benjamin, S., Amit, I., Tanay, A., Massarwa, R.,
Novershtern, N., and Hanna, J. H. (2013) Derivation of novel human
ground state naive pluripotent stem cells, Nature, 504,
282-286.
81.Chen, X., Xu, H., Yuan, P., Fang, F., Huss, M.,
Vega, V. B., Wong, E., Orlov, Y. L., Zhang, W., Jiang, J., Loh, Y. H.,
Yeo, H. C., Yeo, Z. X., Narang, V., Govindarajan, K. R., Leong, B.,
Shahab, A., Ruan, Y., Bourque, G., Sung, W. K., Clarke, N. D., Wei, C.
L., and Ng, H. H. (2008) Integration of external signaling pathways
with the core transcriptional network in embryonic stem cells,
Cell, 133, 1106-1117.
82.Cole, M. F., Johnstone, S. E., Newman, J. J.,
Kagey, M. H., and Young, R. A. (2008) Tcf3 is an integral component of
the core regulatory circuitry of embryonic stem cells, Genes
Dev., 22, 746-755.
83.Frum, T., Halbisen, M. A., Wang, C., Amiri, H.,
Robson, P., and Ralston, A. (2013) Oct4 cell-autonomously promotes
primitive endoderm development in the mouse blastocyst, Dev.
Cell, 25, 610-622.
84.Le Bin, G. C., Munoz-Descalzo, S., Kurowski, A.,
Leitch, H., Lou, X., Mansfield, W., Etienne-Dumeau, C., Grabole, N.,
Mulas, C., Niwa, H., Hadjantonakis, A. K., and Nichols, J. (2014) Oct4
is required for lineage priming in the developing inner cell mass of
the mouse blastocyst, Development, 141, 1001-1010.
85.Wu, G., Han, D., Gong, Y., Sebastiano, V.,
Gentile, L., Singhal, N., Adachi, K., Fischedick, G., Ortmeier, C.,
Sinn, M., Radstaak, M., Tomilin, A., and Scholer, H. R. (2013)
Establishment of totipotency does not depend on Oct4A, Nat. Cell
Biol., 15, 1089-1097.
86.Pain, B., Clark, M. E., Shen, M., Nakazawa, H.,
Sakurai, M., Samarut, J., and Etches, R. J. (1996) Long-term in
vitro culture and characterization of avian embryonic stem cells
with multiple morphogenetic potentialities, Development,
122, 2339-2348.
87.Hong, N., He, B. P., Schartl, M., and Hong, Y.
(2013) Medaka embryonic stem cells are capable of generating entire
organs and embryo-like miniatures, Stem Cells Dev.,
22, 750-757.
88.Hong, Y., Winkler, C., and Schartl, M. (1996)
Pluripotency and differentiation of embryonic stem cell lines from the
medakafish (Oryzias latipes), Mech. Dev.,
60, 33-44.
89.Hong, Y., Winkler, C., and Schartl, M. (1998)
Production of medakafish chimeras from a stable embryonic stem cell
line, Proc. Natl. Acad. Sci. USA, 95, 3679-3684.
90.Takahashi, K., and Yamanaka, S. (2006) Induction
of pluripotent stem cells from mouse embryonic and adult fibroblast
cultures by defined factors, Cell, 126, 663-676.
91.Maherali, N., Sridharan, R., Xie, W., Utikal, J.,
Eminli, S., Arnold, K., Stadtfeld, M., Yachechko, R., Tchieu, J.,
Jaenisch, R., Plath, K., and Hochedlinger, K. (2007) Directly
reprogrammed fibroblasts show global epigenetic remodeling and
widespread tissue contribution, Cell Stem Cell, 1,
55-70.
92.Pijnappel, W. W., Esch, D., Baltissen, M. P., Wu,
G., Mischerikow, N., Bergsma, A. J., Van der Wal, E., Han, D. W.,
Bruch, H., Moritz, S., Lijnzaad, P., Altelaar, A. F., Sameith, K.,
Zaehres, H., Heck, A. J., Holstege, F. C., Scholer, H. R., and Timmers,
H. T. (2013) A central role for TFIID in the pluripotent transcription
circuitry, Nature, 495, 516-519.
93.Ichida, J. K., Blanchard, J., Lam, K., Son, E.
Y., Chung, J. E., Egli, D., Loh, K. M., Carter, A. C., Di Giorgio, F.
P., Koszka, K., Huangfu, D., Akutsu, H., Liu, D. R., Rubin, L. L., and
Eggan, K. (2009) A small-molecule inhibitor of tgf- signaling replaces
sox2 in reprogramming by inducing Nanog, Cell Stem Cell,
5, 491-503.
94.Yu, J., Vodyanik, M. A., Smuga-Otto, K.,
Antosiewicz-Bourget, J., Frane, J. L., Tian, S., Nie, J., Jonsdottir,
G. A., Ruotti, V., Stewart, R., Slukvin, I. I., and Thomson, J. A.
(2007) Induced pluripotent stem cell lines derived from human somatic
cells, Science, 318, 1917-1920.
95.Montserrat, N., Nivet, E., Sancho-Martinez, I.,
Hishida, T., Kumar, S., Miquel, L., Cortina, C., Hishida, Y., Xia, Y.,
Esteban, C. R., and Izpisua Belmonte, J. C. (2013) Reprogramming of
human fibroblasts to pluripotency with lineage specifiers, Cell Stem
Cell, 13, 341-350.
96.Shu, J., Wu, C., Wu, Y., Li, Z., Shao, S., Zhao,
W., Tang, X., Yang, H., Shen, L., Zuo, X., Yang, W., Shi, Y., Chi, X.,
Zhang, H., Gao, G., Shu, Y., Yuan, K., He, W., Tang, C., Zhao, Y., and
Deng, H. (2013) Induction of pluripotency in mouse somatic cells with
lineage specifiers, Cell, 153, 963-975.
97.Rossello, R. A., Chen, C. C., Dai, R., Howard, J.
T., Hochgeschwender, U., and Jarvis, E. D. (2013) Mammalian genes
induce partially reprogrammed pluripotent stem cells in non-mammalian
vertebrate and invertebrate species, eLife, 2,
e00036.
98.Duboule, D. (1994) Temporal colinearity and the
phylotypic progression: a basis for the stability of a vertebrate
Bauplan and the evolution of morphologies through heterochrony,
Development (Suppl.), 135-142.
99.Raff, R. A. (1996) The Shape of Life: Genes,
Development and the Evolution of Animal Form, Chicago Press.
100.Sander, K. (1976) Specification of the basic
body plan in insect embryogenesis, Adv. Insect Physiol.,
12, 125-238.
101.Domazet-Loso, T., and Tautz, D. (2010) A
phylogenetically based transcriptome age index mirrors ontogenetic
divergence patterns, Nature, 468, 815-818.
102.Kalinka, A. T., and Tomancak, P. (2012) The
evolution of early animal embryos: conservation or divergence?
Trends Ecol. Evol., 27, 385-393.
103.Piasecka, B., Lichocki, P., Moretti, S.,
Bergmann, S., and Robinson-Rechavi, M. (2013) The hourglass and the
early conservation models – co-existing patterns of developmental
constraints in vertebrates, PLoS Genet., 9, e1003476.
104.Kunarso, G., Chia, N. Y., Jeyakani, J., Hwang,
C., Lu, X., Chan, Y. S., Ng, H. H., and Bourque, G. (2010) Transposable
elements have rewired the core regulatory network of human embryonic
stem cells, Nat. Genet., 42, 631-634.
105.Xie, D., Chen, C. C., Ptaszek, L. M., Xiao, S.,
Cao, X., Fang, F., Ng, H. H., Lewin, H. A., Cowan, C., and Zhong, S.
(2010) Rewirable gene regulatory networks in the preimplantation
embryonic development of three mammalian species, Genome Res.,
20, 804-815.
106.Fernandez-Tresguerres, B., Canon, S., Rayon,
T., Pernaute, B., Crespo, M., Torroja, C., and Manzanares, M. (2010)
Evolution of the mammalian embryonic pluripotency gene regulatory
network, Proc. Natl. Acad. Sci. USA, 107,
19955-19960.
107.Vastenhouw, N. L., Zhang, Y., Woods, I. G.,
Imam, F., Regev, A., Liu, X. S., Rinn, J., and Schier, A. F. (2010)
Chromatin signature of embryonic pluripotency is established during
genome activation, Nature, 464, 922-926.
108.Lindeman, L. C., Andersen, I. S., Reiner, A.
H., Li, N., Aanes, H., Ostrup, O., Winata, C., Mathavan, S., Muller,
F., Alestrom, P., and Collas, P. (2011) Prepatterning of developmental
gene expression by modified histones before zygotic genome activation,
Dev. Cell, 21, 993-1004.
109.Lunde, K., Belting, H. G., and Driever, W.
(2004) Zebrafish pou5f1/pou2, homolog of mammalian Oct4, functions in
the endoderm specification cascade, Curr. Biol., 14,
48-55.
110.Okuda, Y., Ogura, E., Kondoh, H., and Kamachi,
Y. (2010) B1 SOX coordinate cell specification with patterning and
morphogenesis in the early zebrafish embryo, PLoS Genet.,
6, e1000936.
111.Reim, G., and Brand, M. (2006) Maternal control
of vertebrate dorsoventral axis formation and epiboly by the POU domain
protein Spg/Pou2/Oct4, Development, 133, 2757-2770.
112.Reim, G., Mizoguchi, T., Stainier, D. Y.,
Kikuchi, Y., and Brand, M. (2004) The POU domain protein spg
(pou2/Oct4) is essential for endoderm formation in cooperation with the
HMG domain protein casanova, Dev. Cell, 6, 91-101.
113.Xu, C., Fan, Z. P., Muller, P., Fogley, R.,
Dibiase, A., Trompouki, E., Unternaehrer, J., Xiong, F., Torregroza,
I., Evans, T., Megason, S. G., Daley, G. Q., Schier, A. F., Young, R.
A., and Zon, L. I. (2012) Nanog-like regulates endoderm formation
through the Mxtx2-Nodal pathway, Dev. Cell, 22,
625-638.
114.Morrison, G. M., and Brickman, J. M. (2006)
Conserved roles for Oct4 homologues in maintaining multipotency during
early vertebrate development, Development, 133,
2011-2022.
115.Lavial, F., Acloque, H., Bertocchini, F.,
Macleod, D. J., Boast, S., Bachelard, E., Montillet, G., Thenot, S.,
Sang, H. M., Stern, C. D., Samarut, J., and Pain, B. (2007) The Oct4
homologue PouV and Nanog regulate pluripotency in chicken embryonic
stem cells, Development, 134, 3549-3563.
116.Dixon, J. E., Allegrucci, C., Redwood, C.,
Kump, K., Bian, Y., Chatfield, J., Chen, Y. H., Sottile, V., Voss, S.
R., Alberio, R., and Johnson, A. D. (2010) Axolotl Nanog activity in
mouse embryonic stem cells demonstrates that ground state pluripotency
is conserved from urodele amphibians to mammals, Development,
137, 2973-2980.
117.Theunissen, T. W., Costa, Y., Radzisheuskaya,
A., Van Oosten, A. L., Lavial, F., Pain, B., Castro, L. F., and Silva,
J. C. (2011) Reprogramming capacity of Nanog is functionally conserved
in vertebrates and resides in a unique homeodomain, Development,
138, 4853-4865.
118.Theunissen, T. W., Van Oosten, A. L.,
Castelo-Branco, G., Hall, J., Smith, A., and Silva, J. C. (2011) Nanog
overcomes reprogramming barriers and induces pluripotency in minimal
conditions, Curr. Biol., 21, 65-71.
119.Tapia, N., Reinhardt, P., Duemmler, A., Wu, G.,
Arauzo-Bravo, M. J., Esch, D., Greber, B., Cojocaru, V., Rascon, C. A.,
Tazaki, A., Kump, K., Voss, R., Tanaka, E. M., and Scholer, H. R.
(2012) Reprogramming to pluripotency is an ancient trait of vertebrate
Oct4 and Pou2 proteins, Nat. Commun., 3, 1279.
120.Davidson, E. H., and Erwin, D. H. (2006) Gene
regulatory networks and the evolution of animal body plans,
Science, 311, 796-800.
121.Ding, J., Xu, H., Faiola, F., Ma’ayan,
A., and Wang, J. (2012) Oct4 links multiple epigenetic pathways to the
pluripotency network, Cell Res., 22, 155-167.
122.Van den Berg, D. L., Snoek, T., Mullin, N. P.,
Yates, A., Bezstarosti, K., Demmers, J., Chambers, I., and Poot, R. A.
(2010) An Oct4-centered protein interaction network in embryonic stem
cells, Cell Stem Cell, 6, 369-381.
123.Yanes, O., Clark, J., Wong, D. M., Patti, G.
J., Sanchez-Ruiz, A., Benton, H. P., Trauger, S. A., Desponts, C.,
Ding, S., and Siuzdak, G. (2010) Metabolic oxidation regulates
embryonic stem cell differentiation, Nat. Chem. Biol., 6,
411-417.