REVIEW: Diversity of Potassium Channel Ligands: Focus on Scorpion Toxins
A. I. Kuzmenkov*, E. V. Grishin, and A. A. Vassilevski*
Shemyakin–Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, 117997 Moscow, Russia; E-mail: aleksey.kuzmenkov@gmail.com; avas@ibch.ru* To whom correspondence should be addressed.
Received June 16, 2015; Revision received July 21, 2015
Potassium (K+) channels are a widespread superfamily of integral membrane proteins that mediate selective transport of K+ ions through the cell membrane. They have been found in all living organisms from bacteria to higher multicellular animals, including humans. Not surprisingly, K+ channels bind ligands of different nature, such as metal ions, low molecular mass compounds, venom-derived peptides, and antibodies. Functionally these substances can be K+ channel pore blockers or modulators. Representatives of the first group occlude the channel pore, like a cork in a bottle, while the second group of ligands alters the operation of channels without physically blocking the ion current. A rich source of K+ channel ligands is venom of different animals: snakes, sea anemones, cone snails, bees, spiders, and scorpions. More than a half of the known K+ channel ligands of polypeptide nature are scorpion toxins (KTx), all of which are pore blockers. These compounds have become an indispensable molecular tool for the study of K+ channel structure and function. A recent special interest is the possibility of toxin application as drugs to treat diseases involving K+ channels or related to their dysfunction (channelopathies).
KEY WORDS: ion channel, potassium channel, ligand, pore blocker, modulator, venom, scorpion, toxinDOI: 10.1134/S0006297915130118
Abbreviations: 4-AP, 4-aminopyridine; BKCa, large-conductance calcium-activated K+ channels; CSα/α, cysteine-stabilized helix-loop-helix; CSα/β, cysteine-stabilized α-helix β-sheet; ICK, inhibitor cystine knot; IKCa, intermediate-conductance calcium-activated K+ channels; K+ channels, potassium channels; K2P, two-pore-domain K+ channels; Kir, inward-rectifier K+ channels; KTx, polypeptide K+ channel blockers from scorpion venom; Kv, voltage-gated K+ channels; RP-HPLC, reversed-phase high-performance liquid chromatography; SKCa, small-conductance calcium-activated K+ channels; TEA, tetraethylammonium; TM, transmembrane (segment).
Potassium ion (K+) channels comprise a superfamily of
integral membrane proteins that provide passive selective transport of
K+ across the cell membrane. K+ channels have
been found in all living organisms from bacteria to higher
multicellular animals, including humans. Two major functions of
K+ channels are maintaining the resting membrane potential
and forming the action potential in electrically excitable cells. It is
difficult to overestimate the role of these proteins in numerous
physiological processes, such as ion transport, nerve signal
transmission, cell communication and proliferation, neurohumoral
regulation, and immune response. This makes study of the structure,
mechanism of action, and modulation of K+ channels one of
the most important tasks of modern bioorganic chemistry. Moreover,
multiple studies have demonstrated that K+ channels are
involved in the development of various pathological states, which makes
these channels promising pharmacological targets.
Traditionally, studies of K+ channels include identification of novel channel ligands and subsequent use of these ligands as specific tools to elucidate the mechanisms of channel action and to search for new members of the superfamily. Based on the mechanism of action, K+ channel ligands can be classified as either pore blockers or modulators. Channel blockers “plug” the pore, similarly to the cork in a bottle, whereas modulators affect channel functions without mechanically preventing ion flow through the channel. Scorpion venoms are the major source of K+ channel ligands. These venoms are complex mixtures of tens or even hundreds of compounds, predominantly, short polypeptides. Interestingly, all known K+ channel ligands isolated from scorpion venoms are pore blockers.
About 250 K+ channel ligands from scorpion venom have been identified by now, which supposedly represent only 0.5% of total number of potentially active compounds. Further studies will broaden our understanding of structural and functional properties of K+ channels and enable us to find highly selective ligands with potential pharmacological applications.
K+ CHANNEL SUPERFAMILY
K+ channels have been found in all living organisms. In humans, 78 genes code for major (α-) subunits of these transmembrane proteins (see below). It is now commonly believed that K+ channels appeared around the time of the origin of life on Earth, a hypothesis that is supported by identifications of more than 200 eukaryotic K+ channel-related channel-like proteins in Archaea and bacteria [1].
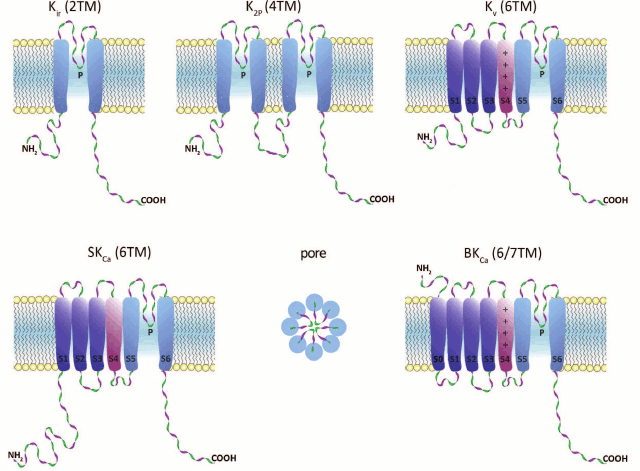
K+ channels are composed of α- and β-subunits. The structure and major functions of a channel are determined by the α-subunits, whereas β-subunits affect the channel kinetics. Human K+ channels can be subdivided into five structural and functional groups based on the structure of their α-subunits (Fig. 1).
Fig. 1. K+ channel families (topological diagrams). Center of bottom row, schematic representation of the channel pore formed by pore regions of four individual subunits. TM, transmembrane segment; P, pore loop; S0-S6, TM segments; S4, voltage sensor (in Kv and BKCa).
1. Inward-rectifier K+ channels (Kir) are homo- or heterotetrameric complexes in which each subunit consists of two transmembrane (TM) segments with a pore region (P) between them. Their function is regulated by nucleotides (ATP, ADP), phosphorylation, G proteins, and phosphatidylinositol 4,5-bisphosphate. In humans, Kir channels are encoded by 15 different genes [2, 3].
2. Two-pore-domain K+ channels (K2P) contain four TM segments, and their α-subunits undergo dimerization upon channel formation. These channels are regulated by a broad array of factors: pH, temperature, and cell membrane tension. In humans, 15 genes coding for K2P channels have been found [4, 5].
3. α-Subunits of voltage-gated (voltage-dependent) K+ channels (Kv) consist of six TM segments (S1-S6) with one pore region (P) located between S5 and S6. Four α-subunits form a complete channel. An important structural feature of Kv channels is the presence of the voltage-sensing domain (VSD) formed by four TM segments (S1-S4). S4 contains regularly positioned positively charged amino acid residues that play the role of a voltage sensor. Voltage-gated K+ channels are the most abundant group of potassium channels (40 genes for Kv channels have been identified in humans) [6].
4. Calcium-activated intermediate- and small-conductance (IKCa and SKCa, respectively) potassium channels are formed by subunits consisting of six TM segments (S1-S6) with the pore region (P) located between S5 and S6, similarly to those of Kv channels. However, in KCa channels, the S4 segment is almost insensitive to changes in potential; the channels are activated by Ca2+ via a calmodulin-mediated mechanism. In humans, proteins of this family are encoded by four genes [7, 8].
5. Ca2+-activated large-conductance channels (BKCa) are encoded by four slo genes in humans. Two members of this family contain seven TM segments. An interesting property of these channels is that they can be activated not only by changes in potential, but by a number of ions (Ca2+, Na+, or Cl–, depending on the channel isoform) [9].
K+ channels of higher plants can be subdivided into three groups: Shaker-like channels, tandem-pore K+ channels (TPK), and Kir-like channels [10]. Similarly to Kv channels, Shaker-like channel subunits are composed of six TM segments and are activated by changes in membrane potential. TPK and Kir-like channel subunits have four and two TM segments and are related to human K2P and Kir channels, respectively [11, 12].
The smallest K+ channel α-subunit (94 amino acid residues) was found in PBCV-1 virus parasitizing green algae of the Chlorella genus [13]. In prokaryotes, K+ channel subunits usually have either two (MthK, KirB, and KcsA) or six (Kch, KvAP, and Mlo1) TM segments. Quite often, these channels also contain additional Ca2+- or nucleotide-binding cytoplasmic domains [14]. Unusual topology of the channel-forming α-subunit (S1-S2-S3-S4-S5-P-S6-S7-P-S8) was found in fungi; this structure seems to be exclusive to Fungi, and not present in other kingdoms [15]. Interestingly, the genome of infusoria Paramecium tetraurelia contains 298 genes for K+ channels, which is 3.8 times more than in the human genome [14].
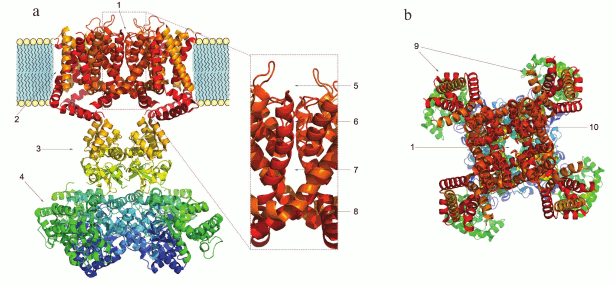
Domain structure of K+ channels. The membrane part of a Kv channel α-subunit consists of two regions (Fig. 2): the pore region and the voltage-sensing region. The pore region is formed by two transmembrane segments (S5-S6) connected by the pore loop (P). The voltage-sensing region is composed of four transmembrane segments (S1-S4). The mature Kv channel consists of the pore domain, formed by four pore regions of four different α-subunits, four voltage-sensing domains, and four cytoplasmic domains [16].
Fig. 2. 3D structure of the voltage-gated Kv1.2 channel (PDB ID: 2A79): a) side view; b) extracellular view. 1) Pore; 2) homotetramer of membrane parts of α-subunits; 3) α-subunit cytoplasmic domain; 4) β-subunits; 5) external vestibule; 6) selectivity filter; 7) internal cavity; 8) inner gate; 9) voltage-sensing domains; 10) pore domain.
The structure and the mechanism of action of the Kv channel pore domain are similar to those of voltage-gated Na+ and Ca2+ channels. In the open state, a Kv channel can conduct 106-108 K+ ions per second [17, 18]. Kv channels are one of the most diverse family of membrane proteins [19], but its members have a highly conserved seven amino acid sequence TTVGYGD [20] that forms the channel selectivity filter. The filter is responsible for the highly selective transport of K+, but not, for example, Na+, even though the Na+ ion radius is only 0.4 Å smaller. This high selectivity is provided by the presence of oxygen atoms inside the pore that coordinate K+ ions lacking the hydration shell (the radius of a Na+ ion is too small to form stable coordination bonds). Oxygen atoms of the polypeptide chain carbonyls stabilize dehydrated K+ ions from four different directions and “substitute” the hydration shell during ion passage through the pore [21]. The crystal structure of a bacterial KcsA channel obtained in the laboratory of MacKinnon [22] proved the earlier hypothesis of Hodgkin who studied K+ flux in squid giant axons and in 1955 suggested that ions pass the channel one after another [23]. When transported into the cell, K+ ions enter the external vestibule, lose their hydration shell, pass through the selectivity filter, get hydrated in the channel cavity, and exit through the intracellular gate. K+ ions can also be transported in the opposite direction [24].
The voltage-sensing domain is essential for the Kv channel response to membrane potential changes. The process involves conversion of the electric field energy accumulated on the membrane into mechanical energy, which is then transferred to the pore domain and causes its conformational changes and pore opening. The S4 segment serves as a voltage sensor due to the high content of positively charged amino acid residues [25]. Translocation of the voltage sensor necessary for the pore opening requires physical interaction between certain amino acid residues of the transmembrane S6 fragment and the juxtamembrane “linker” helix that connects the S4 and S5 segments [26, 27]. The cytoplasmic domain also plays an important role in the channel activity regulation (e.g. in N-type inactivation (see below) and some cases of α-subunit tetramerization [28]).
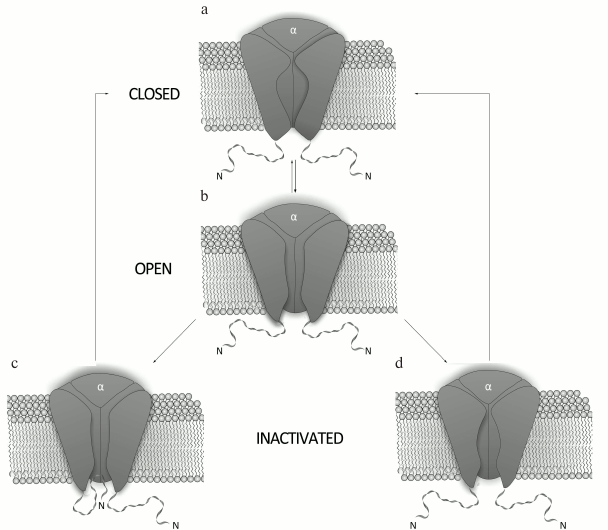
Mechanism of K+ channel activity. The mechanism of Kv channel function is based on conformational changes of the channel protein (Fig. 3). Kv channels are closed at resting potential and do not conduct ions. An increase in the membrane potential (depolarization) affects charged amino acid residues of the voltage sensor and can cause channel opening [29, 30]. The transmembrane S4 segment responds to the changes in the membrane potential, which causes conformational changes in the voltage-sensing domain followed by changes in the pore domain [31]. Due to an extremely high sensitivity of the voltage sensor, the channel can respond even to slight potential changes [32]. The pore opens only when all four voltage sensors are activated [33]; the open channel conducts ions until it enters an inactivation phase.
Fig. 3. Mechanism of action of a voltage-gated K+ channel: a) closed state; b) open (activated) state; c) N-type inactivation; d) C-type inactivation.
There are two mechanisms for Kv channel inactivation: the N-type and the C-type. During N-type inactivation, the channel stays in the open conformation, but the pore is blocked from the membrane inner side by the N-terminal fragment of the α-subunit [34]. The removal of this fragment abolishes channel inactivation via the N-type mechanism, while its addition to the system in a form of a synthetic peptide restores inactivation [35, 36]. The C-type inactivation occurs at a slower rate and does not involve the N-terminus of the channel cytoplasmic domain [37]. Structural elements responsible for the C-type inactivation are located in the external vestibule of the selectivity filter [38] and in the vicinity of the S6 segment N-terminus [39]. The C-type inactivation is caused by structural changes in the channel external vestibule and by narrowing of the channel mouth, which result in blockade of the ion flux [40]. Decrease in the membrane potential to resting state values (repolarization) causes channel transition from inactivation into the closed state.
K+ CHANNEL LIGANDS
K+ channel ligands can be divided into several groups, the three major being metal ions [41], small organic molecules [42], and polypeptide toxins [43]. K+ channel ligands can be either pore blockers or modulators. Pore blockers prevent ion flow through the channel by binding to the pore region [44]. Modulators do not directly block the ion flow, but rather affect the channel functional properties by interacting with other regions of the channel protein (e.g. voltage-sensing domain or auxiliary β-subunit) (Fig. 4) [45, 46].
Fig. 4. Regions of ligand interaction with K+ channels: 1) blockers of the pore outer part; 2) blockers of the pore inner part; 3, 4) modulators acting from the outer and inner side of the membrane, respectively; 5) inactivation inhibitors.
In addition to the major ligand groups, isoform-specific antibodies have been obtained for a number of K+ channels that are able to selectively block the ion flow through the corresponding channels [47]. K+ channel ligands were identified in parasitic worm secretions [48]. Defensins from various organisms have also been shown to affect K+ flux [49, 50].
Most K+ channel isoforms can be nonselectively inhibited by Cs+, Ba2+, Cd2+, Pb2+, Co2+, Ni2+, and other metal ions in millimolar concentrations [51]. These ions were widely used in early studies of K+ channel function [52] but later substituted by ligands with higher affinity and selectivity [53].
Small Organic Molecules
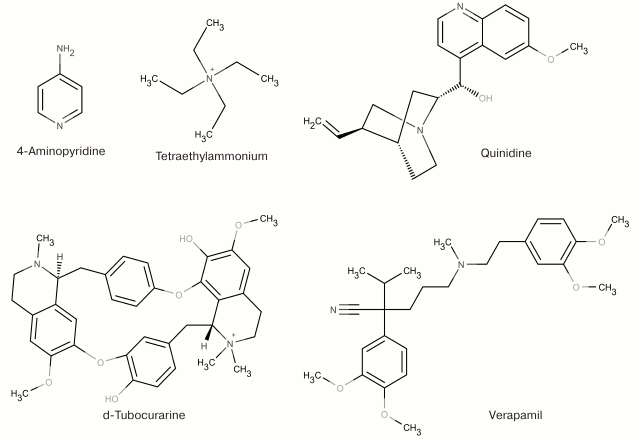
At the dawn of the ion flux studies, the two most widely used agents for pharmacological identification of K+ channels were 4-aminopyridine (4-AP) and tetraethylammonium (TEA). This was due to the fact that they both inhibit (in millimolar concentrations) a majority of K+ channels without producing any visible effects on Na+ and Ca2+ channels [54, 55]. These two compounds remain the only low molecular mass ligands for some K+ channels [53]. Other organic molecules have been identified that are capable of binding to the pore inner part and modulating various K+ channels (quinidine, d-tubocurarine, and verapamil) [56-58]. Some of the low molecular mass ligands of K+ channels are shown in Fig. 5.
Fig. 5. Nonselective blockers of K+ channels.
TEA and structurally similar molecules can block K+ channels from either extracellular or cytoplasmic sides of the membrane [59]. Other hydrophobic cations (d-tubocurarine and verapamil) also “plug” the inner part of the pore with their ammonium or amine groups and block the ion flux (Fig. 6) [60]. Different mechanism of action was proposed for 4-AP that has molecular properties similar to those of the hydrophobic cations, interacts with the channel in the open state, competes with TEA for the binding site on the open channel, and stays bound after channel transition into the closed state. However, the small size of the 4-AP molecule prevents it from plugging the pore. According to recent findings, the site for 4-AP binding is located in a space between the TM S6 fragments [61]. When the channel opens, 4-AP approaches the binding site, binds to the channel, and stabilizes the channel closed state, thereby preventing the ion flux [62].
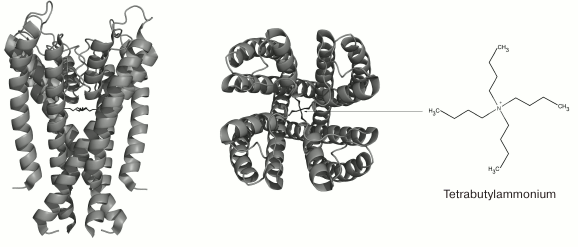
Fig. 6. 3D-Structure of tetrabutylammonium complex with the KcsA channel (in the closed state) (PDB ID: 2JK5): side and extracellular views.
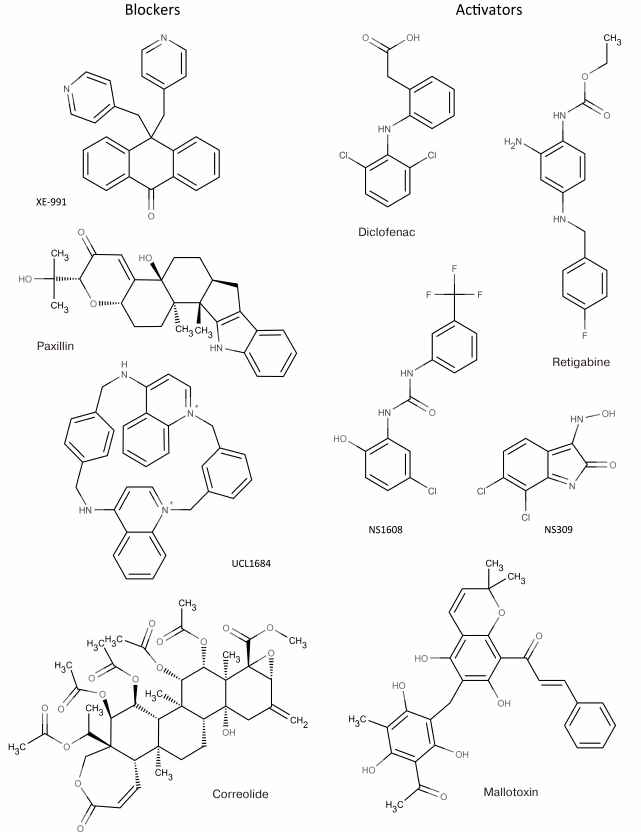
In addition to nonselective K+ channel ligands, a significant number of highly selective activators for K+ channels have been found. These compounds were originally developed as drug prototypes, and some of them are already used in clinics. Table 1 shows the most “popular” blockers and activators of K+ channels; structures of some of these compounds are shown in Fig. 7.
Table 1. The best known blockers and
activators of K+ channels (according to the International
Union of Basic and Clinical Pharmacology, IUPHAR) [2, 4, 6, 7]
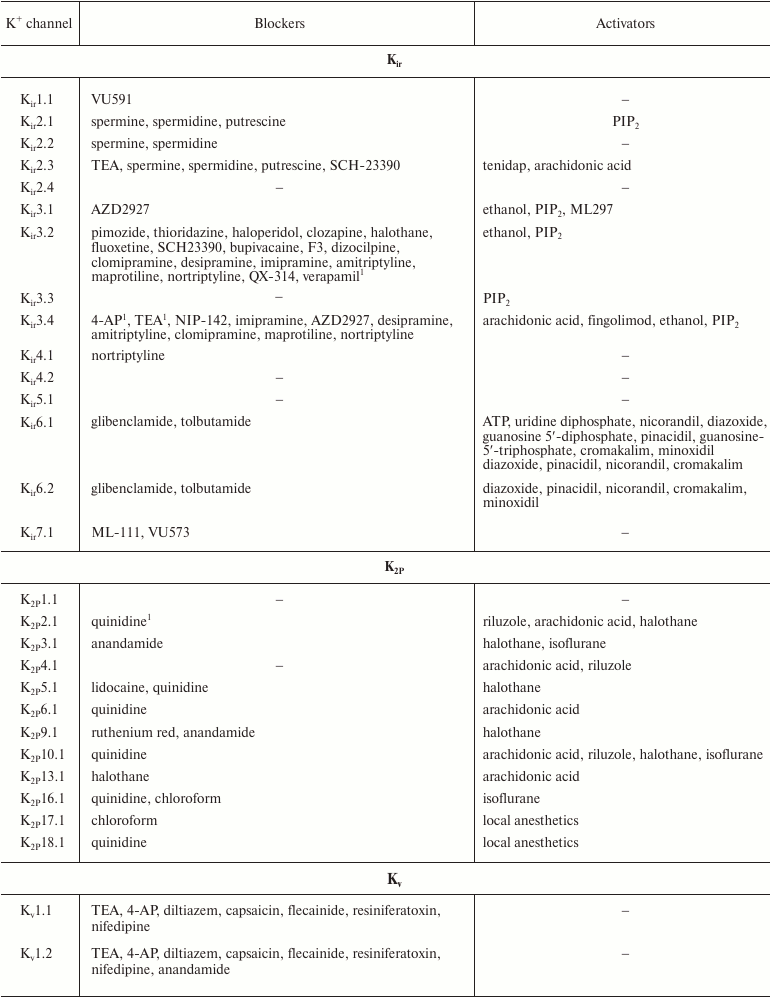
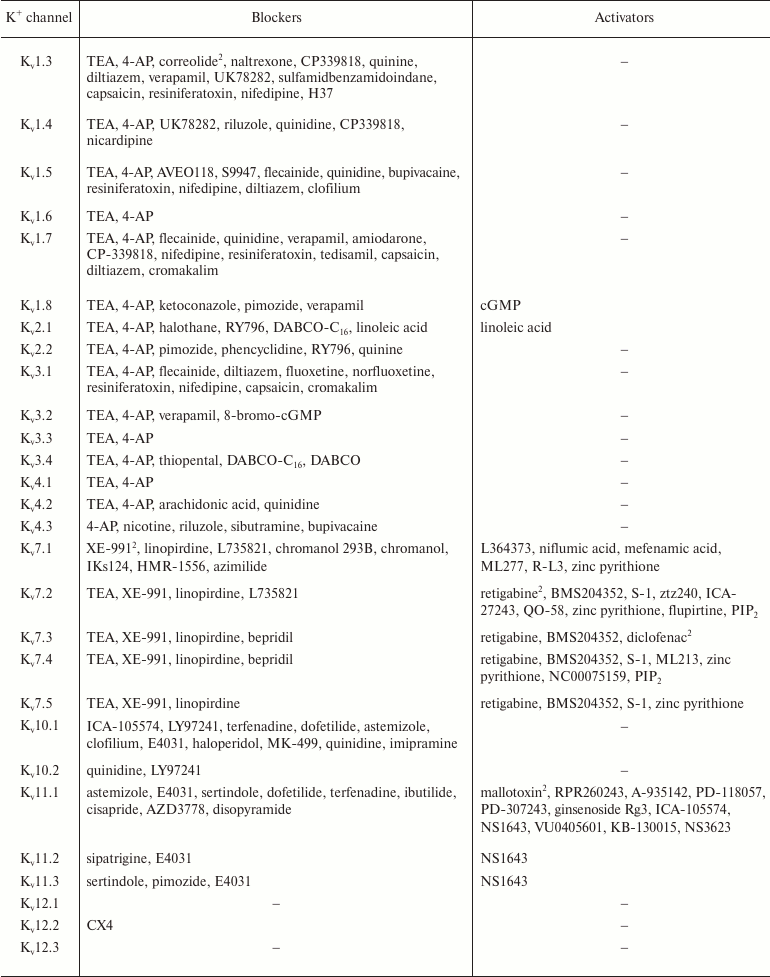
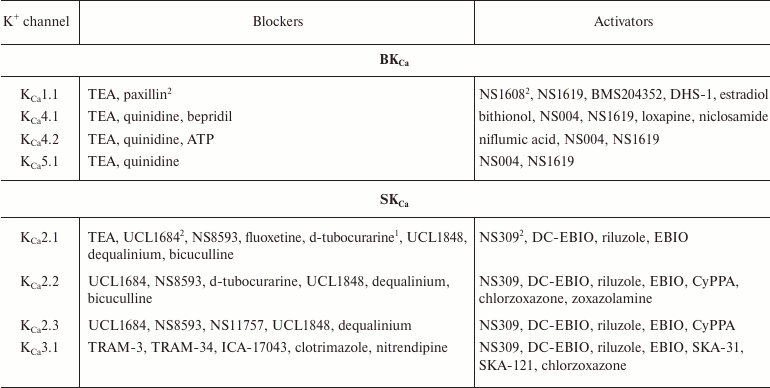
1 Nonselective blockers shown in Fig. 5.
2 Blockers and activators shown in Fig. 7.
Fig. 7. Low molecular mass blockers and activators of K+ channels.
Polypeptide Toxins
A great number of polypeptide ligands of K+ channels have been found in the venoms from bees, snakes, sea anemones, cone snails, spiders, and scorpions. These compounds, often called toxins, are synthesized in the animal venom glands as precursors and then processed into active molecules [63, 64].
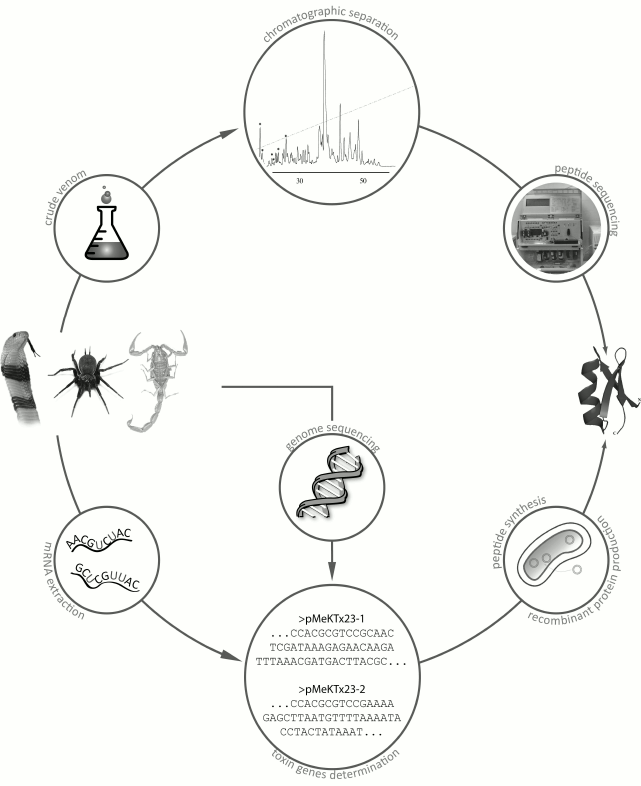
Although the number of known venomous animals is huge, only a few of their venoms have been studied, and even fewer have been characterized in details. The lack of information is mostly because venoms are complex mixtures of multiple components, and their comprehensive characterization might take several decades [65, 66]. There are two main strategies for studying animal venoms (Fig. 8). The first, more traditional strategy, involves purification of individual components directly from the venom by various chromatographic methods [67]. The second, more recent strategy, originated due to the rapid development of DNA sequencing techniques and involves construction and analysis of cDNA libraries from mRNAs isolated from the animal venom glands [68, 69].
Fig. 8. Main strategies for identification of active peptides in animal venoms. The first strategy is purification of active components from crude venom; the second strategy is identification of venom components by construction of cDNA libraries from mRNA of the venom glands with subsequent production of compounds of interest by chemical synthesis or by biosynthesis in recombinant systems.
The first strategy requires sufficient amounts of crude venom that could be obtained, for example, by electrostimulation of venom glands. The crude venom is then subjected to a series of chromatographic procedures, usually starting with size-exclusion chromatography followed by several rounds of ion-exchange or reversed-phase high-performance liquid chromatography (RP-HPLC). Then the purified components are sequenced by the Edman degradation method [70].
Most often, the main active compounds of animal venoms are peptides. Their amino acid sequences can be determined by isolating total mRNA from the venom gland, reverse transcribing this mRNA into corresponding cDNA, and cDNA sequencing with subsequent analysis of the resulting cDNA library or individual sequences [71]. Amino acid sequences can be deduced by in silico translation, and the corresponding peptides can be produced by chemical synthesis or in a recombinant system [72]. Total sequencing of genomes of all venomous animals might provide much valuable information; however, only very few entire genomes of venomous species have been deciphered so far [73-76].
Other traditional approaches to identification and characterization of K+ channel ligands are electrophysiological measurements (methods of patch clamp and two-electrode voltage clamp) [77, 78] and radioligand binding assay [79, 80]. Electrophysiological studies are traditionally conducted in artificial membranes, eukaryotic cells, and Xenopus laevis oocytes expressing K+ channel genes of interest. Radioligand binding assay uses radioactively labeled channel ligands, and the activity of a particular compound is determined from the substitution of the labeled ligand in its complex with the channel by the studied compound. Recently, new methods have been developed based on the use of fluorescently labeled ligands [81] (e.g. fluorescence-activated cell sorting [82] and single-molecule fluorescence microscopy [83]), that have largely replaced radioligand binding assay [83].
Below, we describe groups of animals whose venoms contain K+ channel ligands, in the following order: vertebrates, sea invertebrates, and terrestrial invertebrates.
Snakes. More than 2650 species of snakes have been described; 500 of these species are considered venomous [84]. The most dangerous snakes belong to the families Viperidae (vipers and rattlesnakes) and Elapidae (cobras and kraits) [85]. Snake venoms contain a broad array of biologically active compounds: enzymes, polypeptides with anticoagulant, hypotensive, and other activities, various organic molecules (e.g. nucleotides) [86], and polypeptide toxins that are capable of interactions with membrane proteins, including K+ channels [87]. K+ channel blockers, named dendrotoxins, have been found in the venom of snakes belonging to the Dendroaspis genus (mambas) (Fig. 9a) [88]. Their molecules consist of 57-60 amino acid residues (Table 2), six of which are cysteines that form the Kunitz-type fold with C1-C6, C2-C4, and C3-C5 disulfide bond formation pattern [89]. Dendrotoxins undergo a single posttranslational modification – cyclization of the N-terminal glutamine into pyroglutamate.
Table 2. K+ channel ligands from
snake venom

Notes: ID, accession number in the UniProt database; Z, pyroglutamate
residue; 1, active toward Kv channels (here and below,
figures indicate activity toward corresponding channels). Cysteine
residues are shown in bold.
Fig. 9. Spatial structures of K+ channel ligands. a) I-DTX (PDB ID: 1DEM), a Kv channel blocker from Dendroaspis polylepis polylepis snake venom; b) ShK (PDB ID: 1ROO), a high-affinity K+ channel blocker from Stichodactyla helianthus sea anemone venom; c) PVIIA (PDB ID: 1AV3), a Kv channel blocker from Conus purpurascens mollusk venom; d) tertiapin (PDB ID: 1TER), a K+ channel peptide ligand from Apis mellifera bee venom; e) hanatoxin (PDB ID: 1D1H) from Grammostola rosea Chilean bird-eating spider venom; f) noxiustoxin (PDB ID: 1SXM) from Centruroides noxius scorpion venom.
Dendrotoxins are selective toward Kv channels (Kv1.1, Kv1.2, and Kv1.6) and act at concentrations ranging from pico- to nanomolar and above [90, 91].
Sea anemones. Sea anemones (Actiniaria) belong to the coral polyps (Anthozoa). Most sea anemones are solitary sedentary organisms inhabiting solid sea bottom surface [92].
Similarly to snake venoms, crude venoms of sea anemones are complex mixtures of numerous compounds: actinoporins (~20 kDa) that can form pores in lipid bilayer [93], ligands of voltage-gated Na+ channels [94], K+ channel blockers [95], and polypeptide ligands of acid-sensing ion channels [96] and thermoreceptors [97].
More than 25 polypeptide ligands of K+ channels have been identified in sea anemone venom [98]. All of these can be subdivided into three structurally different groups [99]. The first group includes AeK, BgK, HmK, and ShK polypeptides. These polypeptides are composed of 34-37 amino acid residues. Six of the residues are cysteines that form C1-C6, C2-C4, and C3-C5 disulfide bonds [95, 100-102], promoting formation of the toxin spatial structure as a combination of α- and/or 310-helices (Fig. 9b) [85]. The second group includes AsKC1 and APEKTx1 molecules (58-65 amino acid residues) with Kunitz-type fold [99]. Their six cysteine residues form C1-C6, C2-C4, and C3-C5 disulfide bonds [103, 104]. Toxins of the third group (BDS-I, Am II, and APETx1) consist of 42-48 residues and display different disulfide bond formation pattern: C1-C5, C2-C4, and C3-C6 [105-107]. The spatial structure of these toxins is represented by several β-strands [85]. Amino acid sequences of some K+ channel blockers from sea anemones are shown in Table 3. Some active forms of the polypeptides undergo posttranslational modifications, such as conversion of proline into 4-hydroxyproline or amidation of the C-terminal amino acid residue [106]. Polypeptides from sea anemones mostly modulate Kv channels (including Kv11 and Kv3.4) [105, 107] and IKCa channels [108] in pico- and nanomolar concentrations.
Table 3. K+ channel ligands from
sea anemone venoms
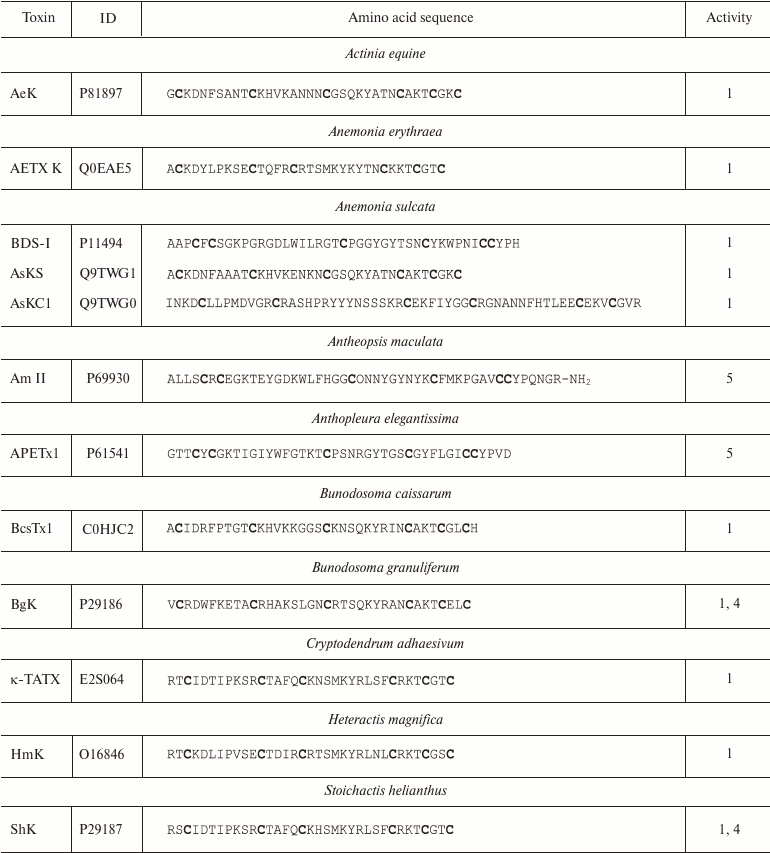
Notes: 1, 4, and 5, active toward Kv (except
Kv11), IKCa, and Kv11 channels,
respectively; O, 4-hydroxyproline residue; -NH2, amidation.
Cysteine residues are shown in bold.
Cone snails. Cone snails are predatory sea mollusks from the Conidae family that includes about 600 species [109]. The major weapons of these snails are short disulfide bond-containing peptides named conopeptides [110].
Conopeptides interacting with K+ channels (κ-conotoxins) are extremely diverse and can be classified into several groups according to their primary structure [111]. Amino acid sequences of some κ-conotoxins are shown in Table 4. κA-Conotoxins (Ac4.2, MIVA, PIVF, SmIVA, SIVA, and other) are peptides of 24-37 amino acid residues. Six of these residues are cysteines that comprise the CC-C-C-C-C motif. The pattern of formed disulfide bonds can be either C1-C5, C2-C3, C4-C6 or C1-C3, C2-C5, C4-C6 [112-114]. κO-Conotoxins (e.g. PVIIA) are short (~27 amino acid residues) peptides with six cysteine residues, whose distribution in the amino acid sequence (C-C-CC-C-C) differs from that in κA-conotoxins. The disulfide bond formation pattern is C1-C4, C2-C5, and C3-C6 (Fig. 9c) [115, 116]. κM-Conotoxins (RIIIJ and RIIIK) are composed of 24-25 amino acids; the number of cysteine residues and the pattern of disulfide bond formation in these polypeptides are the same as in κO-conotoxins; however, the distribution of six cysteine residues in the amino acid sequence is different: CC-C-C-CC [117, 118]. Another group, κI-conotoxins (BeTx, SrXIA, and ViTx), includes molecules composed of ~30 amino acid residues with eight cysteine residues (C-C-CC-CC-C-C) and varying patterns of disulfide bond formation [119-122]. Cone snail venom also contains κL-conotoxins (vil14a) and κJ-conotoxins (α/κ-pl14a) – small (~25 amino acid residues) peptides with two disulfide bonds. They have similar cysteine residue distribution (C-C-C-C), but different patterns of disulfide bond formation: C1-C4, C2-C3 in κL-conotoxins and C1-C3, C2-C4 in κJ-conotoxins [123, 124]. Two toxins should also be mentioned that have an unusual structure: conkunitzin-S1 (Conk-S1), a 60 amino acid peptide with the Kunitz-type fold and only two disulfide bonds (C1-C4, C2-C3) [125], and contryphan-Vn (Con-Vn), a short peptide of nine amino acids with one disulfide bond [126]. These variations in the cysteine residue distribution and disulfide bonds patterns give origin to a wide variety of the κ-conotoxin spatial structures, such as inhibitor cystine knot (ICK, see below), as in PVIIA [116], Kunitz-type fold [127], or even helix-loop-helix motif [123].
Table 4. Conotoxins, ligands of
K+ channels

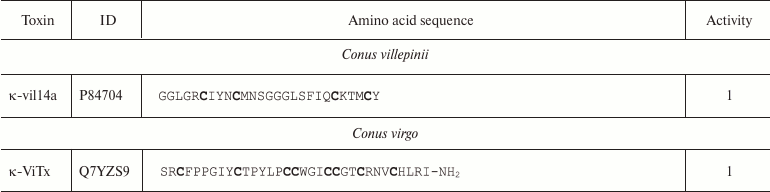
Notes: 1 and 2, active toward Kv (except Kv11) and
BKCa channels, respectively; O, 4-hydroxyproline residue; Z,
pyroglutamate residue; E, 4-carboxyglutamate residue; T
and S, O-glycosylated threonine and serine residues,
respectively; WD, D-tryptophan residue; -NH2,
amidation. Cysteine residues are shown in bold.
Molecules of κ-conotoxins and other peptides from the cone snail venom are often subjected to posttranslational modifications that are rarely seen in other organisms [128]. In addition to typical modifications (cyclization of N-terminal glutamine, hydroxyproline formation, C-terminal amidation), other types of the polypeptide chain modification are observed, such as 4-carboxyglutamate formation, serine and threonine O-glycosylation [129], and incorporation of D-amino acids (e.g. D-tryptophan) in the polypeptide chain [130]. The major targets of κ-conotoxins are Kv channels; however, some can affect the function of BKCa channels [111].
Bees. Bee venom has been extensively characterized, and its components, such as melittin, secapin, apamin, and tertiapin, have become classical instruments in biochemical and physiological studies (Fig. 9d) [131]. Apamin and tertiapin, short peptides of 18 and 21 amino acid residues, respectively, affect various K+ channels (Table 5) [132, 133]. Their molecules undergo posttranslational C-terminal amidation. Four cysteine residues form two disulfide bonds (C1-C3 and C2-C4), and together with an amidated C-terminal residue, generate a spatial structure with a short β-turn and an α-helix [134-136]. Apamin preferentially binds to SKCa channels [137, 138], while tertiapin mostly affects BKCa [139] and Kir channels [140, 141].
Table 5. Short polypeptide ligands of
K+ channels from bee venom

Notes: 2, 3, and 6, active toward BKCa, SKCa, and
Kir channels, respectively; -NH2, amidation.
Cysteine residues are shown in bold.
Spiders. More than 45,000 species of spiders are known today [142]. However, venoms of only about 100 species have been studied [143], and just a few of these have been characterized in details [144]. Spider venoms are complex mixtures of biologically active compounds that can be divided into three groups: low molecular mass (1 kDa) components, peptides (1-10 kDa), and high molecular mass (>10 kDa) compounds (enzymes and neurotoxins) [143]. K+ channel ligands in spider venoms are peptides that inhibit mostly channel activation via interaction with the voltage sensor [145]. This interaction is preceded by the binding of the toxin to the membrane due to the presence of hydrophobic amino acid residues on the molecule surface [146]. Spider venom toxins consist of ~30-40 amino acid residues (Table 6) with the C1-C4, C2-C5, C3-C6 pattern of disulfide bond formation [147-150]. Their spatial structure is represented by the inhibitor cystine knot (ICK) fold characterized by the presence of β-hairpin and characteristic “knot”-like (hence the name) structure in which the third disulfide bond (C3-C6) penetrates the ring formed by the two other disulfide bonds and amino acid residues of the polypeptide chain between them (Fig. 9e).
Table 6. K+ channel ligands from
spider venom
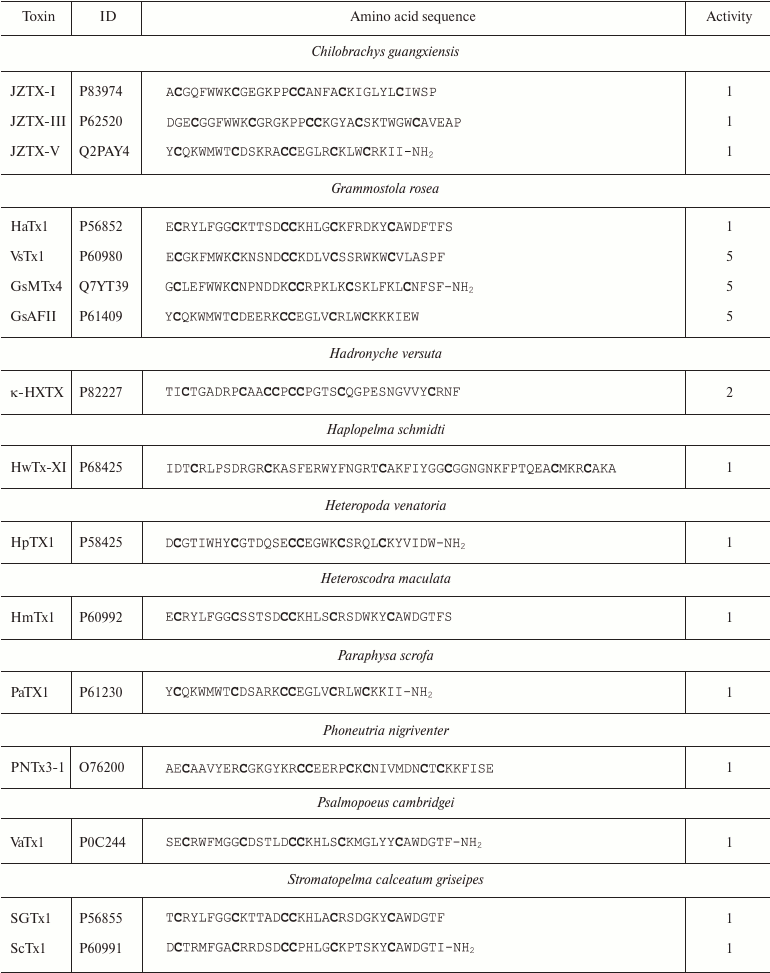

Notes: 1, 2, and 5, active toward Kv, BKCa, and
Kv11 channels, respectively; -NH2, amidation.
Cysteine residues are shown in bold.
Some spider venom polypeptides (κ-HXTX and PNTx3-1) have an additional fourth disulfide bond, but it does not noticeably affect the spatial structure of the molecule [151]. Spider venoms also contain K+ ligands with the Kunitz-type fold, which act as weak pore blockers [152].
Spider toxins mostly interact with Kv channels: hanatoxin (HaTx1) is specific against Kv2.1 [147]; HpTx1 and PaTx1 bind to the Kv4 channels [149, 153]; ScTx1 and HmTx1 bind to the Kv2 and Kv4 channels [154]. Some ligands of voltage-gated Na+ and Ca2+ channels, e.g. JZTX-I and VsTx1, can also inhibit K+ channels [155-157].
Scorpions. An extremely important role in understanding the mechanism of action and physiological role of K+ channels belongs to polypeptide channel blockers from scorpion venom (KTx). The application of these molecules in K+ channel studies started over 30 years ago, when the first short peptide, noxioustoxin (Fig. 9f), was purified from the venom of the scorpion Centruroides noxius [158] and found to affect potassium flux in the squid giant axon [159]. Isolation of charybdotoxin (ChTX) from the venom of the scorpion Leiurus quinquestriatus hebraeus [160] and its use for modeling ligand–receptor interactions (toxin binding to channels) helped to achieve a deeper understanding of the structure–functional properties of K+ channels [161, 162]. Later, an array of isoform-specific selective blockers of K+ channels were identified in the venoms of various scorpion species. Such peptides as iberiotoxin (IbTX) [163], margatoxin (MgTX) [164], kaliotoxin [165], and agitoxin-2 (AgTx2) [166] have become classical molecular instruments for studying K+ channels [167].
Structural properties of K+ channel blockers from scorpion venom. The UniProt database includes ~250 K+ channel blockers from scorpion venoms (KTx). All of them are small polypeptides of ~20-75 amino acid residues (Mw ~ 2400-8500 Da) with two to four S–S bridges [168, 169]. These toxins are synthesized as precursors with the N-terminal signal peptide that is cleaved off in the process of toxin maturation [170]. Sometimes, KTx precursors contain a propeptide fragment of unknown function immediately after the signal peptide that is also cleaved off during processing [171]. Some peptides undergo posttranslational modification, such as conversion of the N-terminal glutamine into pyroglutamate [172], C-terminal amidation of the C-terminal glycine, or removal of C-terminal positively charged residues (lysine, arginine) [173].
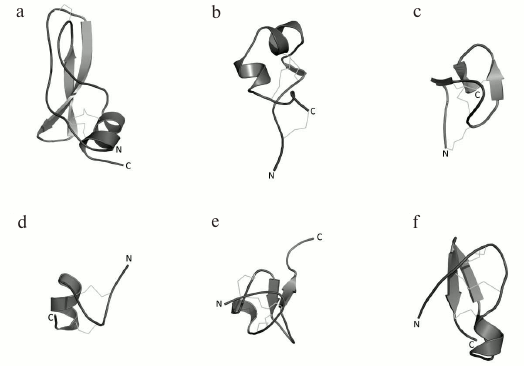
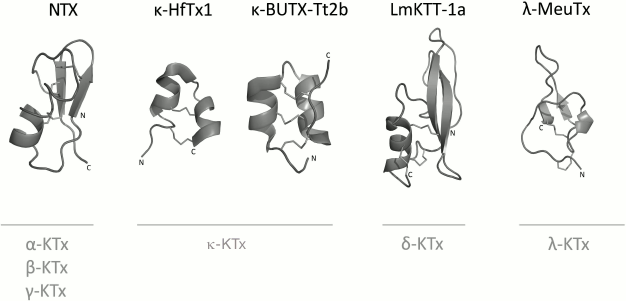
K+ channel blockers from scorpion venoms can be classified into five groups according to their structure (see Fig. 10 for representatives of different structural classes).
Fig. 10. Examples of structural folds in different groups of KTx: CSα/β in NTX; CSα/α with two disulfide bonds in κ-HfTx1; CSα/α with three disulfide bonds in κ-BUTX-Tt2b; Kunitz-type fold in LmKTT-1a; ICK in λ-MeuTx.
1. Peptides with CSα/β (cysteine-stabilized α-helix-β-sheet) fold. This type of peptide structure is the most represented fold in KTx; it is also present in Na+ channel ligands and a group of chlorotoxin-like peptides [174-176].
2. KTx with two parallel α-helices connected with two disulfide bonds: CSα/α 2(C-C) [168].
3. Peptides with amino acid sequences similar to those of peptides from the first group, but with different spatial structure, CSα/α (cysteine-stabilized helix-loop-helix), due to an alternative pattern of disulfide bond formation [177].
4. KTx with the Kunitz-type fold characteristic of serine protease inhibitors [178].
5. KTx with the inhibitor cystine knot (ICK) fold that is present in polypeptides from various groups of living organisms (plants, fungi, and invertebrates) [179].
Table 7 shows the relative occurrence of different structural folds in known KTx (according to the UniProt database). It is evident that the CSα/β fold dominates among the KTx structures.
Table 7. Occurrence of different structural
folds in KTx (total number of toxins analyzed, 247)
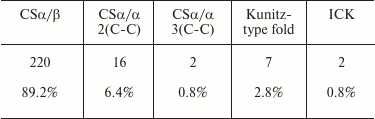
KTx with CSα/β fold. As mentioned above, CSα/β is the most widely represented type of fold in scorpion venom toxins. It is characterized by the presence of two conserved motifs, CXXXC and CXC (where C is cysteine and X is any amino acid residue), in the amino acid sequence. Based on structural and functional properties, KTx with CSα/β fold can be classified into three families: α-KTx, β-KTx, and γ-KTx [180].
The α-KTx family includes ~27 subfamilies (based on similarity of their primary structures) [181]. More than 165 polypeptides are now known to belong to this family, and this number continues to grow. α-KTx consist of ~20-40 amino acid residues, six or eight being cysteines that form three or four disulfide bonds. The majority of toxins in this family have been isolated from the venoms of scorpions of the Buthidae family. α-KTx have also been identified in the venoms of scorpions from other families (Chactidae, Euscorpiidae, Vaejovidae, Caraboctonidae, Hemiscorpiidae, and Scorpionidae) [182-184].
β-KTx are larger than α-KTx (~45-75 amino acid residues) due to the presence of an additional linear N-terminal fragment. The C-terminal fragments of these toxins contain six cysteines that form three disulfide bonds. Most of β-KTx (22 peptides) have been found in scorpions from the Buthidae family; two β-KTx were discovered in the transcriptome and venom of Hadrurus gertschi from the family Caraboctonidae [169, 185]. It is believed that, in addition to the blocking effect on K+ channels, β-KTx also possess cytolytic activity that is determined by the presence of the N-terminal linear fragment [186, 187].
The structure of γ-KTx is similar to that of α-KTx. They are composed of ~35-45 amino acid residues and have three or four disulfide bonds. This group is distinguished by its selective activity toward the only type of Kv channels – Kv11. At present, 29 γ-KTx are known; all of them have been identified in the venom or the venom gland transcriptome of scorpions from the Buthidae family [188, 189].
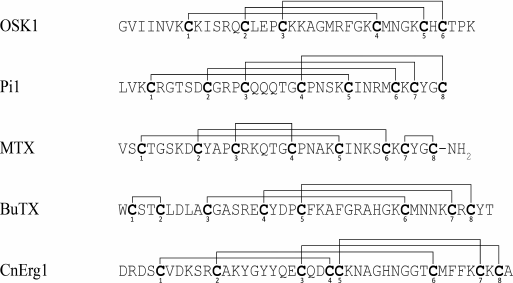
As mentioned above, the number of disulfide bonds in KTx with CSα/β fold varies from three to four. The pattern of the disulfide bond formation can differ in different molecules (Fig. 11), but it does not affect the fold of the toxins [190-194].
Fig. 11. Disulfide bond formation pattern in KTx with CSα/β fold.
KTx with CSα/α fold and two disulfide bonds. Peptides of this group are also called κ-KTx (similarly to the group 3 toxins that have CSα/α fold and three disulfide bonds). They are weak inhibitors of K+ channels and have been found only in the venom of scorpions of the Scorpionidae and Hemiscorpiidae families. κ-KTx consist of ~20-25 amino acid residues, four of which are cysteine residues that form two intramolecular disulfide bonds [168]. Cysteine residues are located within the conserved motifs C1XXXC2 and C3XXXC4 (where C is cysteine residue, X is any amino acid residue), and form the C1-C4 and C2-C3 disulfide bonds (Fig. 12). The spatial structure of these toxins is represented by two parallel α-helices.
Fig. 12. Disulfide bond formation pattern in κ-KTx (κ-hefutoxin).
KTx with CSα/α fold and three disulfide bonds. Recently, a new type of fold was found in a group of scorpion venom toxins. Even if these peptides have the traditional positioning of cysteine residues typically found in the majority of CSα/β toxins, the patter of disulfide bond formation is C1-C5, C2-C4, C3-C6 (Fig. 13) instead of the classical C1-C4, C2-C5, C3-C6. The spatial structure of these toxins is represented by two α-helices connected by a loop. Peptides of this group are conventionally called κ-KTx (similarly to KTx with the CSα/α fold and two disulfide bonds) [195].
Fig. 13. Disulfide bond formation pattern in CSα/α-fold peptides with cysteine residue distribution characteristic of CSα/β-fold toxins.
Only two short peptides (28-29 amino acid residues) with this type of structural fold have been found so far. They were isolated from the venoms of related scorpions Tityus serrulatus and Tityus trivittatus of the Buthidae family [177].
KTx with Kunitz-type fold. KTx with this type of fold can possess a dual activity. Almost all of them inhibit proteolytic enzymes (e.g. trypsin) in nanomolar concentration, and some of the toxins block Kv channels. Seven different toxins with such properties have been identified so far: six from the venom of Buthidae family scorpions and one from the venom of Caraboctonidae scorpions [178]. KTx with the Kunitz-type fold consist of ~60-70 residues, six or eight of them are cysteines that form three or four intramolecular disulfide bonds with the C1-C5, C2-C3, C4-C6 or C1-C7, C2-C4, C3-C5, C6-C8 patterns (Fig. 14). The Kunitz-type fold is represented by two antiparallel β-strands and two or, more often, one helical regions [196]. Toxins of this group are named δ-KTx.
Fig. 14. Disulfide bond formation pattern in K+ channel blockers with the Kunitz-type fold.
KTx with the inhibitor cystine knot fold. Ten peptides with the inhibitor cystine knot (ICK) fold have been found in scorpion venom; only two of them are active toward K+ channels. The structure of these peptides (37 amino acid residues) is represented by a β-sheet formed by three antiparallel β-strands, and a 310-helix [197]. Two disulfide bonds (C1-C4 and C2-C5) and atoms of the polypeptide chain form a ring that is penetrated by the third disulfide bond C3-C6 (Fig. 15). In addition to weak Kv channel-blocking properties, toxins of this group also affect the activity of Ca2+ channels [198].
Fig. 15. Disulfide bond formation pattern in KTx with the ICK fold (λ-MeuTx).
Variety of K+ channel blockers from scorpion venom. As mentioned above, ~250 KTx have been identified in scorpions. Table 8 shows one representative of each KTx subfamily (the toxins are placed under the Latin names of the corresponding scorpion species, and the species names are arranged alphabetically). All K+ channels ligands from scorpion venom are pore blockers that act at concentrations ranging from pM to µM and above. The most active Kv1.3 channel blocker Vm24 from scorpion Vaejovis mexicanus smithi is characterized by complex dissociation constant (Kd) of 2.9 pM [199]. Some of the toxins selectively inhibit specific isoforms of Kv (e.g. BeKm-1 and OSK2) [188, 200] or BKCa channels (IbTX) [163]. No selective blockers for the Kir and K2P channels have been identified in scorpion venom so far [53].
Table 8. KTx subfamilies
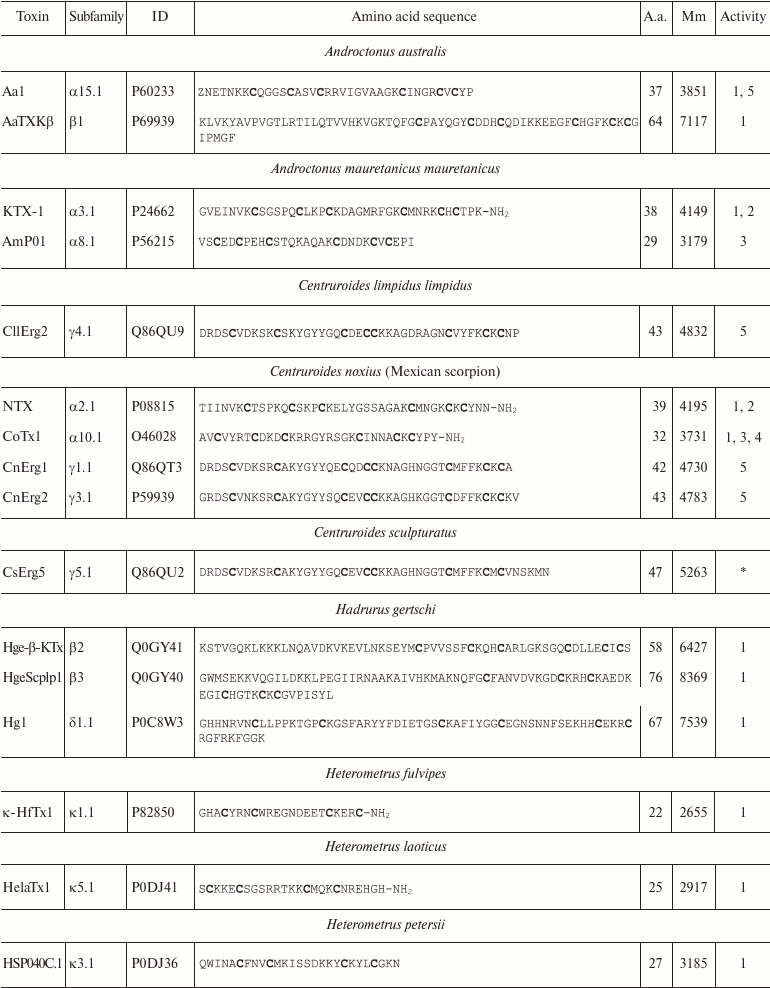
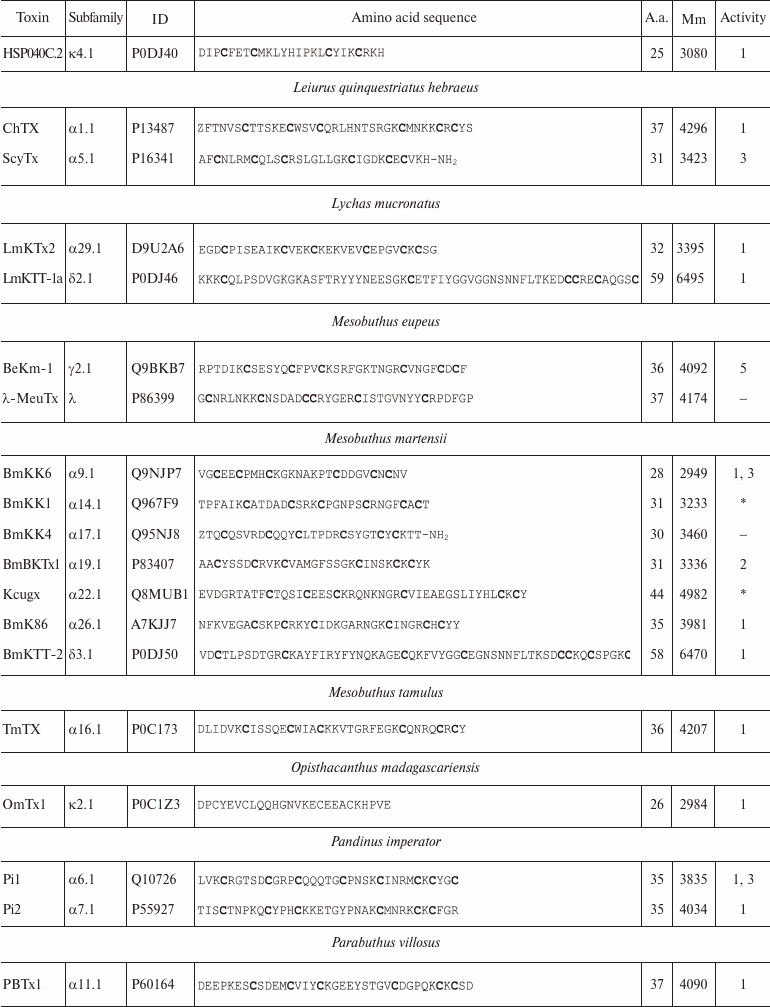
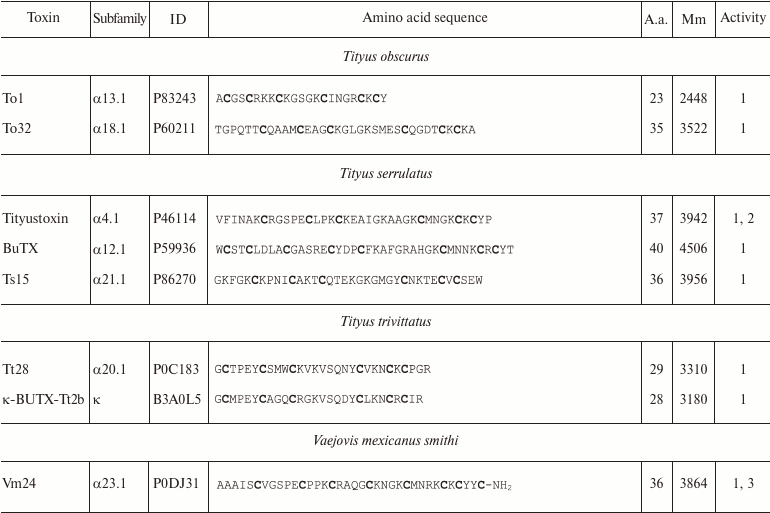
Notes: The table shows one representative of each subfamily. α,
β, and γ, toxins with CSα/β-fold; κ, toxins
with CSα/α fold with two or three disulfide bonds; δ,
peptides with Kunitz-type fold; λ, peptides with ICK fold.
Activity toward: 1, Kv channels (except Kv11
channels); 2, BKCa channels; 3, SKCa channels; 4,
IKCa channels, 5, Kv11 channels; –, no data
available; *, amino acid sequence was determined from cDNA sequencing,
no data on the activity available. Posttranslational modifications: Z,
pyroglutamate residue, -NH2, amidation; a.a., amino acids;
Mm, molecular mass. Cysteine residues are shown in bold.
KTx interaction with K+ channels. K+ channel ligands can be either pore blockers or modulators (see above). Interestingly, unlike in spiders, K+ channel modulators are not typical for scorpion venoms; all KTx are pore blockers.
Since the known KTx predominantly affect Kv channels and, to a lesser extent, BKCa and SKCa channels, and the most active peptides of this group have CSα/β fold, the model for the interaction of the toxin with the channel (ligand–receptor binding) was based on data obtained for this type of toxins [201-203].
Early studies of α-KTx suggested that the channel pore region is highly conserved and has a similar structure in all groups of K+ channels [204]. Later, this hypothesis received robust experimental confirmation: first, spatial structures of several channels were resolved [22, 205, 206]; second, a hybrid channel based on the K+ channel KscA from the bacterium Streptomyces lividans was obtained, and it was blocked by some α-KTx (the wild-type channels are insensitive to the toxins) [207, 208].
Some of the toxins were found to interact with the selectivity filter in the pore region of Kv and BKCa channels via the β-hairpin amino acid residues [209-211]. Other blockers interact with the lower part of the SKCa channel vestibule via amino acid residues of the α-helix, but not of the β-hairpin [212-214]. Moreover, some γ-KTx interact with Kv11 channels by binding to a site located at a considerable distance from the selectivity filter due to the presence of elongated extracellular loops in the channel molecule [215, 216].
Such broad structural diversity of peptides capable of interacting with K+ channels suggests existence of key molecular determinants responsible for the ligand binding to the receptor. Several studies demonstrated that mutations of certain amino acid residues in both the channels and the toxins could considerably change KTx affinity and selectivity.
Many KTx contain the so-called “functional dyad” [217] that is involved in ligand–receptor interaction and is responsible for K+ flux blockade. This dyad is composed of two highly conserved amino acid residues, the first being lysine, and the second being tyrosine, phenylalanine [213, 217], or leucine [218]. According to the functional dyad model, the lysine side chain enters the pore, where a ring of negatively charged aspartate or glutamate residues surrounds it (each aspartate or glutamate residue belonging to one of the four α-subunits of the K+ channel tetramer). Substitution of this lysine residue with another amino acid considerably decreases toxin affinity [219, 220]. The aromatic or aliphatic residue of the dyad might interact with a tyrosine or tryptophan residue of one of the channel α-subunits [221-223].
It should be noted that this model is far from a comprehensive description of the ligand–receptor interactions. Thus, some of the toxins lack the dyad but display high affinity toward specific K+ channel isoforms [224]. Moreover, a new model has been proposed that describes the importance of the “functional triad” (earlier identified Lys residue and two additional hydrophobic residues) for the toxin binding with the pore [225].
An alternative model of the “ring of basic residues” was suggested based on the results of molecular docking studies. Such ring is present in many Kv channel blockers, and its positively charged residues form salt bridges with the negatively charged residues of the channel α-subunit. In some toxins, the ring is formed by four lysine or arginine resides, but this number may vary in other toxins [221, 222].
The observed differences in the selectivity of various KTx toward K+ channel isoforms imply that neither the “functional dyad” nor the “ring of basic residues” model can comprehensively describe unique toxin–channel interactions. More and more data have been published that show that the determining role in the toxin binding to the channel does not belong exclusively to the residues of the dyad or the ring. For example, asparagine residue Asn30 of AgTx2 was found essential for toxin–channel complex stabilization [167]. Therefore, a third model was proposed that emphasizes the necessity for creation of functional maps of “complementary” surfaces of toxins and channels involved in close contact. In this model, each residue is given its “coefficient of importance”. Obviously, the dyad residues will have the maximum coefficient values [226].
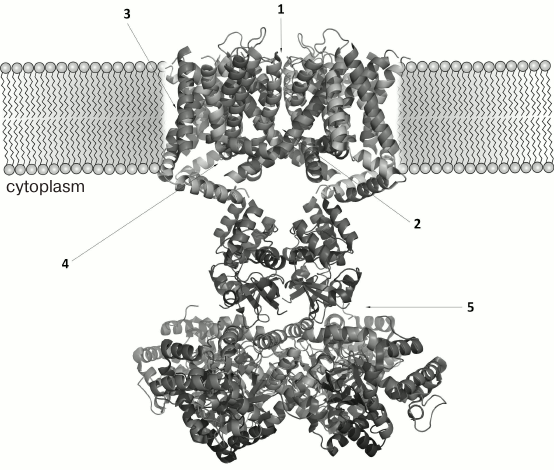
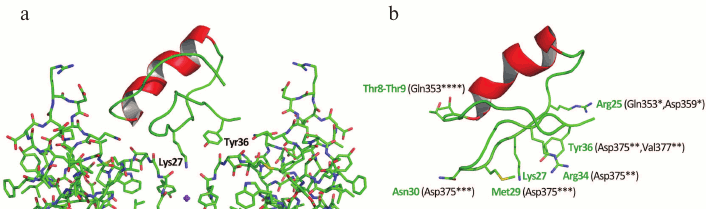
The three-dimensional structure of ChTX complex with a chimeric Kv1.2-Kv2.1 channel has been resolved. It demonstrated that the interaction of the toxin with the channel does not considerably change the structure of the channel selectivity filter (Fig. 16), and it confirmed the important role of functional dyad lysine residue (Lys27) in the interaction. Lys27 is surrounded by four carbonyl groups of tyrosine residues of four channel α-subunits. The α-helical part of the toxin molecule faces away, while the part of the toxin formed by a fragment of the β-hairpin faces toward the pore. A tyrosine residue (Tyr36) is positioned to pack simultaneously against Asp375 and Val377 of one subunit, and Met29 is able to pack against Asp375 of an adjacent subunit. The side chain of Arg25 is within close proximity of the channel Gln353, and the peptide backbone near Thr8-Thr9 is held against Gln353 of another subunit. There are also residues that might be involved in long-range electrostatic interactions: Arg34 and Arg25 of the toxin electrostatically interact with Asp375 and Asp359 of the channel, respectively. Asn30 is another residue that appears to be close enough to Asp375 to enable an H-bonding interaction.
Fig. 16. a) Spatial structure of chimeric Kv1.2-Kv2.1 channel complex with ChTX (PDB ID: 4JTA). The key lysine residue Lys27 and the aromatic residue (Tyr36) of the dyad are shown. The side chain of Lys27 enters the channel pore. b) Amino acid residues of ChTX that interact with the channel residues (in parentheses). Asterisks indicate to which one of the four α-subunits the residues belong.
The resolved structure of the ChTX complex with Kv1.2-Kv2.1 confirmed the earlier proposed models and explained the importance of certain residues (e.g. Lys27) in the interaction of the toxin with the channel. However, some questions remain that require further study. Thus, Tyr36 seems to participate in the complex formation due to the presence of its hydroxyl group and to the large size of its residue, but not because of its hydrophobic nature, as had been previously believed. To understand the molecular mechanisms of toxin selectivity, future studies should aim at resolving crystal structures of toxin complexes with channels from different families (Kv, BKCa, and SKCa).
PRACTICAL APPLICATIONS OF POLYPEPTIDE LIGANDS OF K+
CHANNELS
K+ channel polypeptide ligands from living organisms have always played an important role in the investigations of K+ channels. Early studies used toxins to purify the channels. Later, radioactive and fluorescently labeled toxin analogs were widely applied as molecular tools for investigating pharmacological properties of K+ channels and their localization [227, 228].
The role of K+ channels in cell excitability has been extensively studied; however, the importance of these channels in cell migration, proliferation, and angiogenesis has been demonstrated relatively recently [229]. The functional diversity and wide occurrence of K+ channels make this superfamily of membrane proteins an important pharmacological target in the development of new drugs (including polypeptides) [230].
Radioactive and fluorescently labeled toxin analogs. Toxins labeled with radioactive iodine 125I on Tyr/His residues [231, 232] or with tritium 3H [233] have been widely applied for pharmacological profiling of K+ channels, channel purification from tissues, and investigation of channel distribution and subunit composition [227, 234-236]. However, the use of radioactive labeled toxins poses a number of serious problems, such as difficulties in the utilization of radioactive waste, specific working conditions, and high cost of experiments [237].
In recent years, fluorescently labeled ligands have been increasingly used in K+ channel studies (Fig. 17). Most of them are synthesized by covalent attachment of fluorescent organic molecules to the amino groups of toxins (N-terminal or lysine side chain) [238] or to a cysteine residue introduced into the toxin sequence [239]. In addition, biotinylated derivatives of toxins are used in combination with fluorescently labeled streptavidin [240]. The most common fluorescent labels for toxins are Cy3, Cy5, Alexa488, Alexa546, and TAMRA [237].
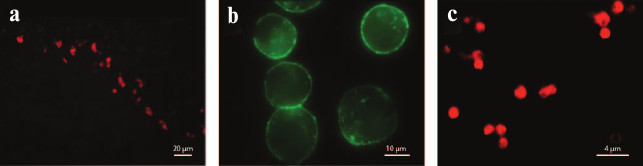
Fig. 17. K+ channel staining with fluorescently labeled toxins and their analogs. a) Rat cerebellum Kv channels stained with HgTX1-A19C-Alexa546 (from [239] with kind permission from Springer). b) HEK293 cells expressing BKCa channels treated with biotinylated IbTX and visualized with streptavidin-Alexa488 (from [240] with permission from Springer). c) Hybrid KcsA-Kv1.3 channels on the surface of spheroplasts [241] (with kind permission from K. S. Kudryashova).
Fluorescent toxin derivatives have been used to demonstrate the ligand–receptor interactions at the monomolecular level [83] and to identify subpopulations of cells expressing specific Kv isoforms by flow cytometry [82].
K+ channels as targets in treatment of various pathologies. Ion channels form the third largest group (after G protein-coupled receptors and kinases) of proteins involved in signal transduction in human cells [51]. At present, ~13% of all medicinal compounds sold on the pharmaceutical market affect ion channels [242], which puts these membrane proteins in second place of the most widely used pharmacological targets after G protein-coupled receptors [243]. Since K+ channels (78 genes in humans) are the largest superfamily of ion channels, it is not surprising that they are involved in pathogenesis of various diseases and, therefore, can be promising pharmacological targets [244].
The involvement of K+ channels in autoimmune diseases, such as multiple sclerosis, rheumatoid arthritis, psoriasis, and type I diabetes, has been extensively studied [245-247]. One of the major roles in the development of these disorders belongs to effector T-lymphocytes. T-lymphocyte proliferation and activation, as well as interleukin production by these cells, require high Ca2+ concentrations in the cytoplasm, which are achieved through the functioning of Ca2+ channels [248]. Low membrane potential is maintained by release of K+ via homotetrameric Kv1.3 and IKCa channels [249-251]. During the development of an autoimmune disorder, the biochemical profile of T-lymphocytes undergoes drastic changes that include hyperexpression of Kv1.3 channels [245]. Inhibition of K+ channels prevents K+ efflux from the cells, which, in turn, abolishes Ca2+ entry into the cytoplasm and suppresses proliferation of the effector T-lymphocytes and interleukin biosynthesis [252]. The functional activity of these cells is predominantly supported by Kv1.3 channels; therefore, inhibition of this isoform may be an efficient approach to autoimmune disease therapy. The membrane potential of other types of lymphocytes is maintained mostly by IKCa channels, and the functioning of those cells will not be affected [253].
It has long been observed that scorpion bites inhibit the development of multiple sclerosis [254]. Elucidation of molecular mechanism of this disease allowed proposing the use of K+ channel blockers from animal venoms as therapeutic agents in multiple sclerosis [230]. The most progress has been achieved in the development of a Kv1.3 channel blocker based on the ShK toxin from the venom of the sea anemone Stichodactyla helianthus (Fig. 9b) [95, 255]. A number of improved toxin derivatives have been synthesized [256], and one of them, ShK-186, has already passed stage I of clinical trials [257]. Scorpion toxins MgTX, OSK1, ChTX, and their derivatives were found to be promising candidates for the treatment of autoimmune disease in model systems [258-260].
Recently, an important role of Kv1.3 in asthma pathogenesis has been demonstrated [261]. Channel inhibition with ShK-186 provided an efficient treatment of the disease in model experiments [262]. In addition, combined inhibition of Kv1.3 and IKCa could be an approach to preventing tissue rejection in organ transplantation [263].
About 10 different types of K+ channels are known to be involved in the development of cardiovascular disorders. Kir2.1, Kir3.4, K2P17.1, Kv1.5, Kv4.3, Kv7.1, and Kv11.1 channels [264] were found to be major players in some types of arrhythmias (atrial fibrillation, Andersen syndrome, and long or short QT syndromes) [264]. In most cases, the dysfunction of a particular channel (channelopathy) results from mutations in the corresponding gene [265]. Therefore, selective modulators of these channels (e.g. BeKm-1) may be promising therapeutic agents in cardiovascular diseases [188].
As mentioned above, K+ channels are the major molecules responsible for the generation of the action potential and nerve signal transmission (see introductory part). Disturbances in the normal functioning of these membrane proteins are related to numerous neurophysiological disorders, such as seizures, epilepsy, and autism [266-268]. Similarly to arrhythmia, the reasons for abnormal channel activity can be mutations in the channel-encoding genes (e.g. neuronal Kv1.1, Kv1.2, Kv4.2, Kv7.2, and Kv7.3 channels). Therapy of neurophysiological disorders uses mostly low molecular mass K+ channels ligands [51, 53].
Abnormal expression of K+ channels has been described in many types of tumor cells [269]. Some isoforms (Kir1.1, Kir3.4, K2P5.1, K2P9.1, Kv1.1, Kv1.3, Kv1.5, Kv10.1, Kv10.2, Kv11.1, KCa1.1, KCa2.3, and KCa3.1) play an important role in cancer pathogenesis and, therefore, can be considered as targets for treatment of oncological diseases [229]. An even broader array of K+ channels is involved the chronic pain-associated processes [270], which make K+ channel activators promising candidates as analgesics [271].
CONCLUSION
K+ channels are a superfamily of integral membrane proteins responsible for passive transport of K+ across the cell membrane. Similarly to other ion channels, the activity of K+ channels can be modulated by various ligands. All ligands of K+ channels can be classified into several groups, the three major groups being metal ions, small organic molecules, and polypeptide toxins. In addition, specific antibodies have been obtained against many of K+ channel isoforms. K+ channel ligands can be either pore blockers or modulators.
About 400 polypeptide ligands of K+ channels have been identified in animal venoms; ~250 of them are scorpion venom toxins (KTx). All KTx are pore blockers that can be subdivided into five groups according to their structural and functional properties. Some of the toxins (such as ChTX, BeKm-1, OSK1, and apamin) have become classical molecular tools in studies of K+ channels. In recent years, K+ channels have been considered promising pharmacological targets in the therapy of autoimmune, respiratory, and neurodegenerative diseases, and K+ channel ligands have emerged as potential agents for treating these disorders.
Every year, new polypeptide ligands of K+ channels are added to the UniProt database because of high scientific interest in this type of compounds. However, further studies should focus not only on identification and purification of these molecules, but on their detailed structural and physiological characterization.
This work was supported by the Russian Science Foundation (project No. 14-14-01180).
REFERENCES
1.Gonzalez, C., Baez-Nieto, D., Valencia, I.,
Oyarzun, I., Rojas, P., Naranjo, D., and Latorre, R. (2012) K(+)
channels: function-structural overview, Compr. Physiol.,
2, 2087-2149.
2.Kubo, Y., Adelman, J. P., Clapham, D. E., Jan, L.
Y., Karschin, A., Kurachi, Y., Lazdunski, M., Nichols, C. G., Seino,
S., and Vandenberg, C. A. (2005) International Union of Pharmacology.
LIV. Nomenclature and molecular relationships of inwardly rectifying
potassium channels, Pharmacol. Rev., 57, 509-526.
3.Hibino, H., Inanobe, A., Furutani, K., Murakami,
S., Findlay, I., and Kurachi, Y. (2010) Inwardly rectifying potassium
channels: their structure, function, and physiological roles,
Physiol. Rev., 90, 291-366.
4.Goldstein, S. A. N., Bayliss, D. A., Kim, D.,
Lesage, F., Plant, L. D., and Rajan, S. (2005) International Union of
Pharmacology. LV. Nomenclature and molecular relationships of two-P
potassium channels, Pharmacol. Rev., 57, 527-540.
5.Plant, L. D., Rajan, S., and Goldstein, S. A. N.
(2005) K2P channels and their protein partners, Curr. Opin.
Neurobiol., 15, 326-333.
6.Gutman, G. A., Chandy, K. G., Grissmer, S.,
Lazdunski, M., McKinnon, D., Pardo, L. A., Robertson, G. A., Rudy, B.,
Sanguinetti, M. C., Stuhmer, W., and Wang, X. (2005) International
Union of Pharmacology. LIII. Nomenclature and molecular relationships
of voltage-gated potassium channels, Pharmacol. Rev., 57,
473-508.
7.Wei, A. D., Gutman, G. A., Aldrich, R., Chandy, K.
G., Grissmer, S., and Wulff, H. (2005) International Union of
Pharmacology. LII. Nomenclature and molecular relationships of
calcium-activated potassium channels, Pharmacol. Rev.,
57, 463-472.
8.Kohler, M., Hirschberg, B., Bond, C. T., Kinzie, J.
M., Marrion, N. V., Maylie, J., and Adelman, J. P. (1996)
Small-conductance, calcium-activated potassium channels from mammalian
brain, Science, 273, 1709-1714.
9.Salkoff, L., Butler, A., Ferreira, G., Santi, C.,
and Wei, A. (2006) High-conductance potassium channels of the SLO
family, Nat. Rev. Neurosci., 7, 921-931.
10.Wang, Y., and Wu, W.-H. (2013) Potassium
transport and signaling in higher plants, Annu. Rev. Plant
Biol., 64, 451-476.
11.Gambale, F., and Uozumi, N. (2006) Properties of
Shaker-type potassium channels in higher plants, J. Membr.
Biol., 210, 1-19.
12.Hedrich, R. (2012) Ion channels in plants,
Physiol. Rev., 92, 1777-1811.
13.Kang, M., Moroni, A., Gazzarrini, S.,
DiFrancesco, D., Thiel, G., Severino, M., and Van Etten, J. L. (2004)
Small potassium ion channel proteins encoded by chlorella viruses,
Proc. Natl. Acad. Sci. USA, 101, 5318-5324.
14.Loukin, S. H., Kuo, M. M.-C., Zhou, X.-L.,
Haynes, W. J., Kung, C., and Saimi, Y. (2005) Microbial K+
channels, J. Gen. Physiol., 125, 521-527.
15.Ketchum, K. A., Joiner, W. J., Sellers, A. J.,
Kaczmarek, L. K., and Goldstein, S. A. (1995) A new family of outwardly
rectifying potassium channel proteins with two pore domains in tandem,
Nature, 376, 690-695.
16.Long, S. B., Campbell, E. B., and Mackinnon, R.
(2005) Crystal structure of a mammalian voltage-dependent Shaker family
K+ channel, Science, 309, 897-903.
17.Grissmer, S., Nguyen, A. N., Aiyar, J., Hanson,
D. C., Mather, R. J., Gutman, G. A., Karmilowicz, M. J., Auperin, D.
D., and Chandy, K. G. (1994) Pharmacological characterization of five
cloned voltage-gated K+ channels, types Kv1.1, 1.2, 1.3,
1.5, and 3.1, stably expressed in mammalian cell lines, Mol.
Pharmacol., 45, 1227-1234.
18.Hess, P., and Tsien, R. W. (1984) Mechanism of
ion permeation through calcium channels, Nature, 309,
453-456.
19.Salkoff, L., Wei, A. D., Baban, B., Butler, A.,
Fawcett, G., Ferreira, G., and Santi, C. M. (2005) Potassium channels
in C. elegans, WormBook, pp. 1-15.
20.Heginbotham, L., Lu, Z., Abramson, T., and
MacKinnon, R. (1994) Mutations in the K+ channel signature
sequence, Biophys. J., 66, 1061-1067.
21.Bezanilla, F., and Armstrong, C. M. (1972)
Negative conductance caused by entry of sodium and cesium ions into the
potassium channels of squid axons, J. Gen. Physiol., 60,
588-608.
22.Doyle, D. A., Morais Cabral, J., Pfuetzner, R.
A., Kuo, A., Gulbis, J. M., Cohen, S. L., Chait, B. T., and MacKinnon,
R. (1998) The structure of the potassium channel: molecular basis of
K+ conduction and selectivity, Science, 280,
69-77.
23.Hodgkin, A. L., and Keynes, R. D. (1955) The
potassium permeability of a giant nerve fibre, J. Physiol.,
128, 61-88.
24.MacKinnon, R. (2003) Potassium channels, FEBS
Lett., 555, 62-65.
25.Aggarwal, S. K., and MacKinnon, R. (1996)
Contribution of the S4 segment to gating charge in the Shaker
K+ channel, Neuron, 16, 1169-1177.
26.Lu, Z., Klem, A. M., and Ramu, Y. (2001) Ion
conduction pore is conserved among potassium channels, Nature,
413, 809-813.
27.Lu, Z., Klem, A. M., and Ramu, Y. (2002) Coupling
between voltage sensors and activation gate in voltage-gated
K+ channels, J. Gen. Physiol., 120,
663-676.
28.Barros, F., Dominguez, P., and De la Pena, P.
(2012) Cytoplasmic domains and voltage-dependent potassium channel
gating, Front. Pharmacol., 3, 49.
29.Sigworth, F. J. (1994) Voltage gating of ion
channels, Q. Rev. Biophys., 27, 1-40.
30.Bezanilla, F. (2000) The voltage sensor in
voltage-dependent ion channels, Physiol. Rev., 80,
555-592.
31.Mannuzzu, L. M., Moronne, M. M., and Isacoff, E.
Y. (1996) Direct physical measure of conformational rearrangement
underlying potassium channel gating, Science, 271,
213-216.
32.Schoppa, N. E., McCormack, K., Tanouye, M. A.,
and Sigworth, F. J. (1992) The size of gating charge in wild-type and
mutant Shaker potassium channels, Science, 255,
1712-1715.
33.Zagotta, W. N., Hoshi, T., and Aldrich, R. W.
(1994) Shaker potassium channel gating. III. Evaluation of kinetic
models for activation, J. Gen. Physiol., 103,
321-362.
34.Demo, S. D., and Yellen, G. (1991) The
inactivation gate of the Shaker K+ channel behaves like an
open-channel blocker, Neuron, 7, 743-753.
35.Armstrong, C. M., and Bezanilla, F. (1977)
Inactivation of the sodium channel. II. Gating current experiments,
J. Gen. Physiol., 70, 567-590.
36.Zagotta, W. N., Hoshi, T., and Aldrich, R. W.
(1990) Restoration of inactivation in mutants of Shaker potassium
channels by a peptide derived from ShB, Science, 250,
568-571.
37.Hoshi, T., Zagotta, W. N., and Aldrich, R. W.
(1990) Biophysical and molecular mechanisms of Shaker potassium channel
inactivation, Science, 250, 533-538.
38.Oliva, C., Gonzalez, V., and Naranjo, D. (2005)
Slow inactivation in voltage gated potassium channels is insensitive to
the binding of pore occluding peptide toxins, Biophys. J.,
89, 1009-1019.
39.Hoshi, T., Zagotta, W. N., and Aldrich, R. W.
(1991) Two types of inactivation in Shaker K+ channels:
effects of alterations in the carboxy-terminal region, Neuron,
7, 547-556.
40.Choi, K. L., Aldrich, R. W., and Yellen, G.
(1991) Tetraethylammonium blockade distinguishes two inactivation
mechanisms in voltage-activated K+ channels, Proc. Natl.
Acad. Sci. USA, 88, 5092-5095.
41.Hagiwara, S., Miyazaki, S., and Rosenthal, N. P.
(1976) Potassium current and the effect of cesium on this current
during anomalous rectification of the egg cell membrane of a starfish,
J. Gen. Physiol., 67, 621-638.
42.Robertson, D. W., and Steinberg, M. I. (1990)
Potassium channel modulators: scientific applications and therapeutic
promise, J. Med. Chem., 33, 1529-1541.
43.Moczydlowski, E., Lucchesi, K., and Ravindran, A.
(1988) An emerging pharmacology of peptide toxins targeted against
potassium channels, J. Membr. Biol., 105, 95-111.
44.MacKinnon, R., and Miller, C. (1989) Mutant
potassium channels with altered binding of charybdotoxin, a
pore-blocking peptide inhibitor, Science, 245,
1382-1385.
45.Swartz, K. J. (2007) Tarantula toxins interacting
with voltage sensors in potassium channels, Toxicon, 49,
213-230.
46.Swartz, K. J., and MacKinnon, R. (1997) Hanatoxin
modifies the gating of a voltage-dependent K+ channel
through multiple binding sites, Neuron, 18, 665-673.
47.Gomez-Varela, D., Zwick-Wallasch, E., Knotgen,
H., Sanchez, A., Hettmann, T., Ossipov, D., Weseloh, R.,
Contreras-Jurado, C., Rothe, M., Stuhmer, W., and Pardo, L. A. (2007)
Monoclonal antibody blockade of the human Eag1 potassium channel
function exerts antitumor activity, Cancer Res., 67,
7343-7349.
48.Chhabra, S., Chang, S. C., Nguyen, H. M., Huq,
R., Tanner, M. R., Londono, L. M., Estrada, R., Dhawan, V., Chauhan,
S., Upadhyay, S. K., Gindin, M., Hotez, P. J., Valenzuela, J. G.,
Mohanty, B., Swarbrick, J. D., Wulff, H., Iadonato, S. P., Gutman, G.
A., Beeton, C., Pennington, M. W., Norton, R. S., and Chandy, K. G.
(2014) Kv1.3 channel-blocking immunomodulatory peptides from parasitic
worms: implications for autoimmune diseases, FASEB J.,
28, 3952-3964.
49.Xiang, F., Xie, Z., Feng, J., Yang, W., Cao, Z.,
Li, W., Chen, Z., and Wu, Y. (2015) Plectasin, first animal toxin-like
fungal defensin blocking potassium channels through recognizing channel
pore region, Toxins (Basel), 7, 34-42.
50.Yang, W., Feng, J., Xiang, F., Xie, Z., Zhang,
G., Sabatier, J.-M., Cao, Z., Li, W., Chen, Z., and Wu, Y. (2015)
Endogenous animal toxin-like human β-defensin 2 inhibits own K(+)
channels through interaction with channel extracellular pore region,
Cell. Mol. Life Sci., 72, 845-853.
51.Wulff, H., Castle, N. A., and Pardo, L. A. (2009)
Voltage-gated potassium channels as therapeutic targets, Nat. Rev.
Drug Discov., 8, 982-1001.
52.Hermann, A., and Gorman, A. L. (1979) Blockade of
voltage-dependent and Ca2+-dependent K+ current
components by internal Ba2+ in molluscan pacemaker neurons,
Experientia, 35, 229-231.
53.Wulff, H., and Zhorov, B. S. (2008) K+
channel modulators for the treatment of neurological disorders and
autoimmune diseases, Chem. Rev., 108, 1744-1773.
54.Shen, K. Z., Lagrutta, A., Davies, N. W.,
Standen, N. B., Adelman, J. P., and North, R. A. (1994)
Tetraethylammonium block of Slowpoke calcium-activated potassium
channels expressed in Xenopus oocytes: evidence for tetrameric
channel formation, Pflugers Arch., 426, 440-445.
55.Zheng, W., and Zhang, Y. P. (1986) Research
progress on a new potassium channel-blocking agent-4-aminopyridine,
Sheng Li Ke Xue Jin Zhan, 17, 254-258.
56.Illes, P., Norenberg, W., and Gebicke-Haerter, P.
J. (1996) Molecular mechanisms of microglial activation. B. Voltage-
and purinoceptor-operated channels in microglia, Neurochem.
Int., 29, 13-24.
57.Brown, D. A., and Selyanko, A. A. (1985) Membrane
currents underlying the cholinergic slow excitatory post-synaptic
potential in the rat sympathetic ganglion, J. Physiol.,
365, 365-387.
58.DeCoursey, T. E., Chandy, K. G., Gupta, S., and
Cahalan, M. D. (1985) Voltage-dependent ion channels in T-lymphocytes,
J. Neuroimmunol., 10, 71-95.
59.French, R. J., and Shoukimas, J. J. (1981)
Blockage of squid axon potassium conductance by internal
tetra-N-alkylammonium ions of various sizes, Biophys. J.,
34, 271-291.
60.Faraldo-Gomez, J. D., Kutluay, E., Jogini, V.,
Zhao, Y., Heginbotham, L., and Roux, B. (2007) Mechanism of
intracellular block of the KcsA K+ channel by
tetrabutylammonium: insights from X-ray crystallography,
electrophysiology and replica-exchange molecular dynamics simulations,
J. Mol. Biol., 365, 649-662.
61.Armstrong, C. M., and Loboda, A. (2001) A model
for 4-aminopyridine action on K channels: similarities to
tetraethylammonium ion action, Biophys. J., 81,
895-904.
62.Del Camino, D., Kanevsky, M., and Yellen, G.
(2005) Status of the intracellular gate in the activated-not-open state
of shaker K+ channels, J. Gen. Physiol., 126,
419-428.
63.Martin-Eauclaire, M. F., Ceard, B., Ribeiro, A.
M., Diniz, C. R., Rochat, H., and Bougis, P. E. (1992) Molecular
cloning and nucleotide sequence analysis of a cDNA encoding the main
beta-neurotoxin from the venom of the South American scorpion Tityus
serrulatus, FEBS Lett., 302, 220-222.
64.Vlasak, R., and Kreil, G. (1984) Nucleotide
sequence of cloned cDNAs coding for preprosecapin, a major product of
queen-bee venom glands, Eur. J. Biochem., 145,
279-282.
65.Mozhayeva, G. N., Naumov, A. P., Nosyreva, E. D.,
and Grishin, E. V. (1980) Potential-dependent interaction of toxin from
venom of the scorpion Buthus eupeus with sodium channels in
myelinated fibre: voltage clamp experiments, Biochim. Biophys.
Acta, 597, 587-602.
66.Zhu, L., Peigneur, S., Gao, B., Tytgat, J., and
Zhu, S. (2013) Two recombinant α-like scorpion toxins from
Mesobuthus eupeus with differential affinity toward insect and
mammalian Na(+) channels, Biochimie, 95, 1732-1740.
67.Rodriguez de la Vega, R. C., Schwartz, E. F., and
Possani, L. D. (2010) Mining on scorpion venom biodiversity,
Toxicon, 56, 1155-1161.
68.Quintero-Hernandez, V., Ortiz, E., Rendon-Anaya,
M., Schwartz, E. F., Becerril, B., Corzo, G., and Possani, L. D. (2011)
Scorpion and spider venom peptides: gene cloning and peptide
expression, Toxicon, 58, 644-663.
69.Kozlov, S., Malyavka, A., McCutchen, B., Lu, A.,
Schepers, E., Herrmann, R., and Grishin, E. (2005) A novel strategy for
the identification of toxin-like structures in spider venom,
Proteins, 59, 131-140.
70.Vassilevski, A. A., Kozlov, S. A., Egorov, T. A.,
and Grishin, E. V. (2010) Purification and characterization of
biologically active peptides from spider venoms, Methods Mol.
Biol., 615, 87-100.
71.Kuzmenkov, A. I., Vassilevski, A. A.,
Kudryashova, K. S., Nekrasova, O. V., Peigneur, S., Tytgat, J.,
Feofanov, A. V., Kirpichnikov, M. P., and Grishin, E. V. (2015)
Variability of potassium channel blockers in Mesobuthus eupeus
scorpion venom with focus on Kv1.1: an integrated transcriptomic and
proteomic study, J. Biol. Chem., 290, 12195-12209.
72.Shijin, Y., Hong, Y., Yibao, M., Zongyun, C.,
Han, S., Yingliang, W., Zhijian, C., and Wenxin, L. (2008)
Characterization of a new Kv1.3 channel-specific blocker, J123, from
the scorpion Buthus martensii Karsch, Peptides,
29, 1514-1520.
73.Weinstock, G. M. (2006) Insights into social
insects from the genome of the honeybee Apis mellifera,
Nature, 443, 931-949.
74.Sanggaard, K. W., Bechsgaard, J. S., Fang, X.,
Duan, J., Dyrlund, T. F., Gupta, V., Jiang, X., Cheng, L., Fan, D.,
Feng, Y., Han, L., Huang, Z., Wu, Z., Liao, L., Settepani, V.,
Thogersen, I. B., Vanthournout, B., Wang, T., Zhu, Y., Funch, P.,
Enghild, J. J., Schauser, L., Andersen, S. U., Villesen, P., Schierup,
M. H., Bilde, T., and Wang, J. (2014) Spider genomes provide insight
into composition and evolution of venom and silk, Nat. Commun.,
5, 3765.
75.Cao, Z., Yu, Y., Wu, Y., Hao, P., Di, Z., He, Y.,
Chen, Z., Yang, W., Shen, Z., He, X., Sheng, J., Xu, X., Pan, B., Feng,
J., Yang, X., Hong, W., Zhao, W., Li, Z., Huang, K., Li, T., Kong, Y.,
Liu, H., Jiang, D., Zhang, B., Hu, J., Hu, Y., Wang, B., Dai, J., Yuan,
B., Feng, Y., Huang, W., Xing, X., Zhao, G., Li, X., Li, Y., and Li, W.
(2013) The genome of Mesobuthus martensii reveals a unique
adaptation model of arthropods, Nat. Commun., 4,
2602.
76.Vonk, F. J., Casewell, N. R., Henkel, C. V.,
Heimberg, A. M., Jansen, H. J., McCleary, R. J. R., Kerkkamp, H. M.,
Vos, R. A., Guerreiro, I., Calvete, J. J., Wuster, W., Woods, A. E.,
Logan, J. M., Harrison, R. A., Castoe, T. A., De Koning, A. P. J.,
Pollock, D. D., Yandell, M., Calderon, D., Renjifo, C., Currier, R. B.,
Salgado, D., Pla, D., Sanz, L., Hyder, A. S., Ribeiro, J. M. C.,
Arntzen, J. W., Van den Thillart, G. E., Boetzer, M., Pirovano, W.,
Dirks, R. P., Spaink, H. P., Duboule, D., McGlinn, E., Kini, R. M., and
Richardson, M. K. (2013) The king cobra genome reveals dynamic gene
evolution and adaptation in the snake venom system, Proc. Natl.
Acad. Sci. USA, 110, 20651-20656.
77.Gao, B., Peigneur, S., Tytgat, J., and Zhu, S.
(2010) A potent potassium channel blocker from Mesobuthus eupeus
scorpion venom, Biochimie, 92, 1847-1853.
78.Chandy, K. G., Cahalan, M., Pennington, M.,
Norton, R. S., Wulff, H., and Gutman, G. A. (2001) Potassium channels
in T-lymphocytes: toxins to therapeutic immunosuppressants,
Toxicon, 39, 1269-1276.
79.Kharrat, R., Mansuelle, P., Sampieri, F., Crest,
M., Oughideni, R., Van Rietschoten, J., Martin-Eauclaire, M. F.,
Rochat, H., and El Ayeb, M. (1997) Maurotoxin, a four disulfide bridge
toxin from Scorpio maurus venom: purification, structure and
action on potassium channels, FEBS Lett., 406,
284-290.
80.Helms, L. M., Felix, J. P., Bugianesi, R. M.,
Garcia, M. L., Stevens, S., Leonard, R. J., Knaus, H. G., Koch, R.,
Wanner, S. G., Kaczorowski, G. J., and Slaughter, R. S. (1997)
Margatoxin binds to a homomultimer of K(V)1.3 channels in Jurkat cells.
Comparison with K(V)1.3 expressed in CHO cells, Biochemistry,
36, 3737-3744.
81.Nekrasova, O. V., Ignatova, A. A., Nazarova, A.
I., Feofanov, A. V., Korolkova, Y. V., Boldyreva, E. F., Tagvei, A. I.,
Grishin, E. V., Arseniev, A. S., and Kirpichnikov, M. P. (2009)
Recombinant Kv channels at the membrane of Escherichia
coli bind specifically agitoxin2, J. Neuroimmune
Pharmacol., 4, 83-91.
82.Beeton, C., Wulff, H., Singh, S., Botsko, S.,
Crossley, G., Gutman, G. A., Cahalan, M. D., Pennington, M., and
Chandy, K. G. (2003) A novel fluorescent toxin to detect and
investigate Kv1.3 channel up-regulation in chronically activated
T-lymphocytes, J. Biol. Chem., 278, 9928-9937.
83.Freudenthaler, G., Axmann, M., Schindler, H.,
Pragl, B., Knaus, H.-G., and Schutz, G. J. (2002) Ultrasensitive
pharmacological characterization of the voltage-gated potassium channel
K(V)1.3 studied by single-molecule fluorescence microscopy,
Histochem. Cell Biol., 117, 197-202.
84.Warrell, D. A. (2010) Snake bite, Lancet,
375, 77-88.
85.Mouhat, S., Andreotti, N., Jouirou, B., and
Sabatier, J.-M. (2008) Animal toxins acting on voltage-gated potassium
channels, Curr. Pharm. Des., 14, 2503-2518.
86.Doley, R., and Kini, R. M. (2009) Protein
complexes in snake venom, Cell. Mol. Life Sci., 66,
2851-2871.
87.Koh, D. C. I., Armugam, A., and Jeyaseelan, K.
(2006) Snake venom components and their applications in biomedicine,
Cell. Mol. Life Sci., 63, 3030-3041.
88.Harvey, A. L., and Anderson, A. J. (1985)
Dendrotoxins: snake toxins that block potassium channels and facilitate
neurotransmitter release, Pharmacol. Ther., 31,
33-55.
89.Berndt, K. D., Guntert, P., and Wuthrich, K.
(1993) Nuclear magnetic resonance solution structure of dendrotoxin K
from the venom of Dendroaspis polylepis polylepis, J. Mol.
Biol., 234, 735-750.
90.Strydom, D. J. (1973) Protease inhibitors as
snake venom toxins, Nat. New Biol., 243, 88-89.
91.Robertson, B., Owen, D., Stow, J., Butler, C.,
and Newland, C. (1996) Novel effects of dendrotoxin homologues on
subtypes of mammalian Kv1 potassium channels expressed in
Xenopus oocytes, FEBS Lett., 383, 26-30.
92.Daly, M., Fautin, D. G., and Cappola, V. A.
(2003) Systematics of the hexacorallia (Cnidaria: Anthozoa), Zool.
J. Linn. Soc., 139, 419-437.
93.Anderluh, G., and Macek, P. (2002) Cytolytic
peptide and protein toxins from sea anemones (Anthozoa: Actiniaria),
Toxicon, 40, 111-124.
94.Norton, R. S. (1991) Structure and
structure–function relationships of sea anemone proteins that
interact with the sodium channel, Toxicon, 29,
1051-1084.
95.Castaneda, O., Sotolongo, V., Amor, A. M.,
Stocklin, R., Anderson, A. J., Harvey, A. L., Engstrom, A., Wernstedt,
C., and Karlsson, E. (1995) Characterization of a potassium channel
toxin from the Caribbean Sea anemone Stichodactyla helianthus,
Toxicon, 33, 603-613.
96.Diochot, S., Baron, A., Rash, L. D., Deval, E.,
Escoubas, P., Scarzello, S., Salinas, M., and Lazdunski, M. (2004) A
new sea anemone peptide, APETx2, inhibits ASIC3, a major acid-sensitive
channel in sensory neurons, EMBO J., 23, 1516-1525.
97.Andreev, Y. A., Kozlov, S. A., Koshelev, S. G.,
Ivanova, E. A., Monastyrnaya, M. M., Kozlovskaya, E. P., and Grishin,
E. V. (2008) Analgesic compound from sea anemone Heteractis
crispa is the first polypeptide inhibitor of vanilloid receptor 1
(TRPV1), J. Biol. Chem., 283, 23914-23921.
98.Frazao, B., Vasconcelos, V., and Antunes, A.
(2012) Sea anemone (Cnidaria, Anthozoa, Actiniaria) toxins: an
overview, Mar. Drugs, 10, 1812-1851.
99.Honma, T., and Shiomi, K. (2005) Peptide toxins
in sea anemones: structural and functional aspects, Mar. Biotechnol.
(N.Y.), 8, 1-10.
100.Minagawa, S., Ishida, M., Nagashima, Y., and
Shiomi, K. (1998) Primary structure of a potassium channel toxin from
the sea anemone Actinia equine, FEBS Lett., 427,
149-151.
101.Cotton, J., Crest, M., Bouet, F., Alessandri,
N., Gola, M., Forest, E., Karlsson, E., Castaneda, O., Harvey, A. L.,
Vita, C., and Menez, A. (1997) A potassium-channel toxin from the sea
anemone Bunodosoma granulifera, an inhibitor for Kv1 channels.
Revision of the amino acid sequence, disulfide-bridge assignment,
chemical synthesis, and biological activity, Eur. J. Biochem.,
244, 192-202.
102.Gendeh, G. S., Young, L. C., De Medeiros, C.
L., Jeyaseelan, K., Harvey, A. L., and Chung, M. C. (1997) A new
potassium channel toxin from the sea anemone Heteractis
magnifica: isolation, cDNA cloning, and functional expression,
Biochemistry, 36, 11461-11471.
103.Peigneur, S., Billen, B., Derua, R., Waelkens,
E., Debaveye, S., Beress, L., and Tytgat, J. (2011) A bifunctional sea
anemone peptide with Kunitz type protease and potassium channel
inhibiting properties, Biochem. Pharmacol., 82,
81-90.
104.Schweitz, H., Bruhn, T., Guillemare, E.,
Moinier, D., Lancelin, J. M., Béress, L., and Lazdunski, M.
(1995) Kalicludines and kaliseptine. Two different classes of sea
anemone toxins for voltage sensitive K+ channels, J.
Biol. Chem., 270, 25121-25126.
105.Diochot, S., Schweitz, H., Beress, L., and
Lazdunski, M. (1998) Sea anemone peptides with a specific blocking
activity against the fast inactivating potassium channel Kv3.4, J.
Biol. Chem., 273, 6744-6749.
106.Honma, T., Hasegawa, Y., Ishida, M., Nagai, H.,
Nagashima, Y., and Shiomi, K. (2005) Isolation and molecular cloning of
novel peptide toxins from the sea anemone Antheopsis maculate,
Toxicon, 45, 33-41.
107.Diochot, S., Loret, E., Bruhn, T., Beress, L.,
and Lazdunski, M. (2003) APETx1, a new toxin from the sea anemone
Anthopleura elegantissima, blocks voltage-gated human
ether-a-go-go-related gene potassium channels, Mol. Pharmacol.,
64, 59-69.
108.Rauer, H., Pennington, M., Cahalan, M., and
Chandy, K. G. (1999) Structural conservation of the pores of
calcium-activated and voltage-gated potassium channels determined by a
sea anemone toxin, J. Biol. Chem., 274, 21885-21892.
109.Tucker, J. K., and Tenorio, M. J. (2009)
Systematic Classification of Recent and Fossil Conoidean
Gastropods, Conchbooks, Hackenheim, p. 133.
110.Livett, B. G., Gayler, K. R., and Khalil, Z.
(2004) Drugs from the sea: conopeptides as potential therapeutics,
Curr. Med. Chem., 11, 1715-1723.
111.Lewis, R. J., Dutertre, S., Vetter, I., and
Christie, M. J. (2012) Conus venom peptide pharmacology, Pharmacol.
Rev., 64, 259-298.
112.Teichert, R. W., Jacobsen, R., Terlau, H.,
Yoshikami, D., and Olivera, B. M. (2007) Discovery and characterization
of the short kappaA-conotoxins: a novel subfamily of excitatory
conotoxins, Toxicon, 49, 318-328.
113.Santos, A. D., McIntosh, J. M., Hillyard, D.
R., Cruz, L. J., and Olivera, B. M. (2004) The A-superfamily of
conotoxins: structural and functional divergence, J. Biol.
Chem., 279, 17596-17606.
114.Wang, C.-Z., Jiang, H., Ou, Z.-L., Chen, J.-S.,
and Chi, C.-W. (2003) cDNA cloning of two A-superfamily conotoxins from
Conus striatus, Toxicon, 42, 613-961.
115.Shon, K. J., Stocker, M., Terlau, H., Stuhmer,
W., Jacobsen, R., Walker, C., Grilley, M., Watkins, M., Hillyard, D.
R., Gray, W. R., and Olivera, B. M. (1998) κ-Conotoxin PVIIA is a
peptide inhibiting the shaker K+ channel, J. Biol.
Chem., 273, 33-38.
116.Savarin, P., Guenneugues, M., Gilquin, B.,
Lamthanh, H., Gasparini, S., Zinn-Justin, S., and Menez, A. (1998)
Three-dimensional structure of kappa-conotoxin PVIIA, a novel potassium
channel-blocking toxin from cone snails, Biochemistry,
37, 5407-5416.
117.Ferber, M., Sporning, A., Jeserich, G.,
DeLaCruz, R., Watkins, M., Olivera, B. M., and Terlau, H. (2003) A
novel conus peptide ligand for K+ channels, J. Biol.
Chem., 278, 2177-2183.
118.Al-Sabi, A., Lennartz, D., Ferber, M., Gulyas,
J., Rivier, J. E. F., Olivera, B. M., Carlomagno, T., and Terlau, H.
(2004) κM-conotoxin RIIIK, structural and functional novelty in a
K+ channel antagonist, Biochemistry, 43,
8625-8635.
119.Aguilar, M. B., Lopez-Vera, E., Heimer de la
Cotera, E. P., Falcon, A., Olivera, B. M., and Maillo, M. (2007)
I-conotoxins in vermivorous species of the West Atlantic: peptide sr11a
from Conus spurius, Peptides, 28, 18-23.
120.Fan, C.-X., Chen, X.-K., Zhang, C., Wang,
L.-X., Duan, K.-L., He, L.-L., Cao, Y., Liu, S.-Y., Zhong, M.-N.,
Ulens, C., Tytgat, J., Chen, J.-S., Chi, C.-W., and Zhou, Z. (2003) A
novel conotoxin from Conus betulinus, kappa-BtX, unique in
cysteine pattern and in function as a specific BK channel modulator,
J. Biol. Chem., 278, 12624-12633.
121.Aguilar, M. B., Perez-Reyes, L. I., Lopez, Z.,
De la Cotera, E. P. H., Falcon, A., Ayala, C., Galvan, M., Salvador,
C., and Escobar, L. I. (2010) Peptide sr11a from Conus
spurius is a novel peptide blocker for Kv1 potassium channels,
Peptides, 31, 1287-1291.
122.Kauferstein, S., Huys, I., Lamthanh, H.,
Stocklin, R., Sotto, F., Menez, A., Tytgat, J., and Mebs, D. (2003) A
novel conotoxin inhibiting vertebrate voltage-sensitive potassium
channels, Toxicon, 42, 43-52.
123.Moller, C., Rahmankhah, S., Lauer-Fields, J.,
Bubis, J., Fields, G. B., and Mari, F. (2005) A novel conotoxin
framework with a helix-loop-helix (Cs alpha/alpha) fold,
Biochemistry, 44, 15986-15996.
124.Imperial, J. S., Bansal, P. S., Alewood, P. F.,
Daly, N. L., Craik, D. J., Sporning, A., Terlau, H., Lopez-Vera, E.,
Bandyopadhyay, P. K., and Olivera, B. M. (2006) A novel conotoxin
inhibitor of Kv1.6 channel and nAChR subtypes defines a new superfamily
of conotoxins, Biochemistry, 45, 8331-8340.
125.Bayrhuber, M., Vijayan, V., Ferber, M., Graf,
R., Korukottu, J., Imperial, J., Garrett, J. E., Olivera, B. M.,
Terlau, H., Zweckstetter, M., and Becker, S. (2005) Conkunitzin-S1 is
the first member of a new Kunitz-type neurotoxin family. Structural and
functional characterization, J. Biol. Chem., 280,
23766-23770.
126.Massilia, G. R., Schinina, M. E., Ascenzi, P.,
and Polticelli, F. (2001) Contryphan-Vn: a novel peptide from the venom
of the Mediterranean snail Conus ventricosus, Biochem.
Biophys. Res. Commun., 288, 908-913.
127.Dy, C. Y., Buczek, P., Imperial, J. S., Bulaj,
G., and Horvath, M. P. (2006) Structure of conkunitzin-S1, a neurotoxin
and Kunitz-fold disulfide variant from cone snail, Acta Crystallogr.
D Biol. Crystallogr., 62, 980-990.
128.Terlau, H., and Olivera, B. M. (2004) Conus
venoms: a rich source of novel ion channel-targeted peptides,
Physiol. Rev., 84, 41-68.
129.Craig, A. G., Zafaralla, G., Cruz, L. J.,
Santos, A. D., Hillyard, D. R., Dykert, J., Rivier, J. E., Gray, W. R.,
Imperial, J., DelaCruz, R. G., Sporning, A., Terlau, H., West, P. J.,
Yoshikami, D., and Olivera, B. M. (1998) An O-glycosylated
neuroexcitatory conus peptide, Biochemistry, 37,
16019-16025.
130.Eliseo, T., Cicero, D. O., Romeo, C., Schinina,
M. E., Massilia, G. R., Polticelli, F., Ascenzi, P., and Paci, M.
(2004) Solution structure of the cyclic peptide contryphan-Vn, a
Ca2+-dependent K+ channel modulator,
Biopolymers, 74, 189-198.
131.Palma, M. S. (2013) Hymenoptera insect
peptides, in Handbook of Biologically Active Peptides, Academic
Press, San Diego, pp. 416-422.
132.Haux, P., Sawerthal, H., and Habermann, E.
(1967) Sequence analysis of bee venom neurotoxin (apamine) from its
tryptic and chymotryptic cleavage products, Hoppe Seylers Z.
Physiol. Chem., 348, 737-738.
133.Ovchinnikov, Y. A., Miroshnikov, A. I.,
Kudelin, A. B., Kostina, M. B., Boikov, V. A., Magazanik, L. G., and
Gotgilf, I. M (1980) Structure and presynaptic action of tertiapin
– a neurotoxin from the venom of honey bee Apis mellifera,
Bioorg. Khim., 6, 359-365.
134.Bystrov, V. F., Okhanov, V. V., Miroshnikov, A.
I., and Ovchinnikov, Y. A. (1980) Solution spatial structure of apamin
as derived from NMR study, FEBS Lett., 119, 113-117.
135.Xu, X., and Nelson, J. W. (1993) Solution
structure of tertiapin determined using nuclear magnetic resonance and
distance geometry, Proteins, 17, 124-137.
136.Pease, J. H., and Wemmer, D. E. (1988) Solution
structure of apamin determined by nuclear magnetic resonance and
distance geometry, Biochemistry, 27, 8491-8498.
137.Lazdunski, M. (1983) Apamin, a neurotoxin
specific for one class of Ca2+-dependent K+
channels, Cell Calcium, 4, 421-428.
138.Labbe-Jullie, C., Granier, C., Albericio, F.,
Defendini, M. L., Ceard, B., Rochat, H., and Van Rietschoten, J. (1991)
Binding and toxicity of apamin. Characterization of the active site,
Eur. J. Biochem., 196, 639-645.
139.Kanjhan, R., Coulson, E. J., Adams, D. J., and
Bellingham, M. C. (2005) Tertiapin-Q blocks recombinant and native
large conductance K+ channels in a use-dependent manner,
J. Pharmacol. Exp. Ther., 314, 1353-1361.
140.Jin, W., and Lu, Z. (1998) A novel
high-affinity inhibitor for inward-rectifier K+ channels,
Biochemistry, 37, 13291-13299.
141.Jin, W., Klem, A. M., Lewis, J. H., and Lu, Z.
(1999) Mechanisms of inward-rectifier K+ channel inhibition
by tertiapin-Q, Biochemistry, 38, 14294-14301.
142.NMBE – World Spider Catalog (2015) World
Spider Cat. Nat. Hist. Museum Bern (http://www.wsc.nmbe.ch) Accessed
January 29, 2015.
143.Vasilevskii, A. A., Kozlov, S. A., and Grishin,
E. V. (2009) Molecular diversity of spider venoms, Usp. Biol.
Khim., 49, 211-274.
144.Kuhn-Nentwig, L., and Nentwig, W. (2013) The
cytotoxic mode of action of the venom of Cupiennius salei
(Ctenidae), Spider Ecophysiol., 16, 217-228.
145.Wang, J. M., Roh, S. H., Kim, S., Lee, C. W.,
Kim, J. I., and Swartz, K. J. (2004) Molecular surface of tarantula
toxins interacting with voltage sensors in K(v) channels, J. Gen.
Physiol., 123, 455-467.
146.Lee, S.-Y., and MacKinnon, R. (2004) A
membrane-access mechanism of ion channel inhibition by voltage sensor
toxins from spider venom, Nature, 430, 232-235.
147.Swartz, K. J., and MacKinnon, R. (1995) An
inhibitor of the Kv2.1 potassium channel isolated from the venom of a
Chilean tarantula, Neuron, 15, 941-949.
148.Takahashi, H., Kim, J. I., Min, H. J., Sato,
K., Swartz, K. J., and Shimada, I. (2000) Solution structure of
hanatoxin1, a gating modifier of voltage-dependent K(+) channels:
common surface features of gating modifier toxins, J. Mol.
Biol., 297, 771-780.
149.Diochot, S., Drici, M. D., Moinier, D., Fink,
M., and Lazdunski, M. (1999) Effects of phrixotoxins on the Kv4 family
of potassium channels and implications for the role of Ito1 in cardiac
electrogenesis, Br. J. Pharmacol., 126, 251-263.
150.Chagot, B., Escoubas, P., Villegas, E.,
Bernard, C., Ferrat, G., Corzo, G., Lazdunski, M., and Darbon, H.
(2004) Solution structure of phrixotoxin 1, a specific peptide
inhibitor of Kv4 potassium channels from the venom of the theraphosid
spider Phrixotrichus auratus, Protein Sci., 13,
1197-1208.
151.Wang, X., Connor, M., Smith, R., Maciejewski,
M. W., Howden, M. E., Nicholson, G. M., Christie, M. J., and King, G.
F. (2000) Discovery and characterization of a family of insecticidal
neurotoxins with a rare vicinal disulfide bridge, Nat. Struct.
Biol., 7, 505-513.
152.Liang, S. (2004) An overview of peptide toxins
from the venom of the Chinese bird spider Selenocosmia huwena
Wang [=Ornithoctonus huwena Wang], Toxicon, 43,
575-585.
153.Sanguinetti, M. C., Johnson, J. H., Hammerland,
L. G., Kelbaugh, P. R., Volkmann, R. A., Saccomano, N. A., and Mueller,
A. L. (1997) Heteropodatoxins: peptides isolated from spider venom that
block Kv4.2 potassium channels, Mol. Pharmacol., 51,
491-498.
154.Escoubas, P., Diochot, S., Celerier, M.-L.,
Nakajima, T., and Lazdunski, M. (2002) Novel tarantula toxins for
subtypes of voltage-dependent potassium channels in the Kv2 and Kv4
subfamilies, Mol. Pharmacol., 62, 48-57.
155.Li-Smerin, Y., and Swartz, K. J. (1998) Gating
modifier toxins reveal a conserved structural motif in voltage-gated
Ca2+ and K+ channels, Proc. Natl. Acad. Sci.
USA, 95, 8585-8589.
156.Yuan, C., Yang, S., Liao, Z., and Liang, S.
(2007) Effects and mechanism of Chinese tarantula toxins on the Kv2.1
potassium channels, Biochem. Biophys. Res. Commun., 352,
799-804.
157.Bemporad, D., Sands, Z. A., Wee, C. L.,
Grottesi, A., and Sansom, M. S. P. (2006) Vstx1, a modifier of Kv
channel gating, localizes to the interfacial region of lipid bilayers,
Biochemistry, 45, 11844-11855.
158.Possani, L. D. (1982) The primary structure of
noxiustoxin: a K+ channel blocking peptide from the venom of
the scorpion Centruroides noxius Hoffmann, Carlsberg Res.
Commun., 47, 285-289.
159.Carbone, E., Wanke, E., Prestipino, G.,
Possani, L. D., and Maelicke, A. (1982) Selective blockage of
voltage-dependent K+ channels by a novel scorpion toxin,
Nature, 296, 90-91.
160.Miller, C., Moczydlowski, E., Latorre, R., and
Phillips, M. (1985) Charybdotoxin, a protein inhibitor of single
Ca2+-activated K+ channels from mammalian
skeletal muscle, Nature, 313, 316-318.
161.Sugg, E. E., Garcia, M. L., Reuben, J. P.,
Patchett, A. A., and Kaczorowski, G. J. (1990) Synthesis and structural
characterization of charybdotoxin, a potent peptidyl inhibitor of the
high conductance Ca(2+)-activated K+ channel, J. Biol.
Chem., 265, 18745-18745.
162.Miller, C. (1995) The charybdotoxin family of
K+ channel-blocking peptides, Neuron, 15,
5-10.
163.Galvez, A., Gimenez-Gallego, G., Reuben, J. P.,
Roy-Contancin, L., Feigenbaum, P., Kaczorowski, G. J., and Garcia, M.
L. (1990) Purification and characterization of a unique, potent,
peptidyl probe for the high conductance calcium-activated potassium
channel from venom of the scorpion Buthus tamulus, J. Biol.
Chem., 265, 11083-11090.
164.Garcia-Calvo, M., Leonard, R. J., Novick, J.,
Stevens, S. P., Schmalhofer, W., Kaczorowski, G. J., and Garcia, M. L.
(1993) Purification, characterization, and biosynthesis of margatoxin,
a component of Centruroides margaritatus venom that selectively
inhibits voltage-dependent potassium channels, J. Biol. Chem.,
268, 18866-18874.
165.Crest, M., Jacquet, G., Gola, M., Zerrouk, H.,
Benslimane, A., Rochat, H., Mansuelle, P., and Martin-Eauclaire, M. F.
(1992) Kaliotoxin, a novel peptidyl inhibitor of neuronal BK-type
Ca(2+)-activated K+ channels characterized from
Androctonus mauretanicus mauretanicus venom, J. Biol.
Chem., 267, 1640-1647.
166.Garcia, M. L., Garcia-Calvo, M., Hidalgo, P.,
Lee, A., and MacKinnon, R. (1994) Purification and characterization of
three inhibitors of voltage-dependent K+ channels from
Leiurus quinquestriatus var. hebraeus venom,
Biochemistry, 33, 6834-6839.
167.Gao, Y.-D., and Garcia, M. L. (2003)
Interaction of agitoxin2, charybdotoxin, and iberiotoxin with potassium
channels: selectivity between voltage-gated and Maxi-K channels,
Proteins, 52, 146-154.
168.Srinivasan, K. N., Sivaraja, V., Huys, I.,
Sasaki, T., Cheng, B., Kumar, T. K. S., Sato, K., Tytgat, J., Yu, C.,
San, B. C. C., Ranganathan, S., Bowie, H. J., Kini, R. M., and
Gopalakrishnakone, P. (2002) κ-Hefutoxin1, a novel toxin from the
scorpion Heterometrus fulvipes with unique structure and
function. Importance of the functional diad in potassium channel
selectivity, J. Biol. Chem., 277, 30040-30047.
169.Diego-Garcia, E., Schwartz, E. F.,
D’Suze, G., Gonzalez, S. A. R., Batista, C. V. F., Garcia, B. I.,
De la Vega, R. C. R., and Possani, L. D. (2007) Wide phylogenetic
distribution of scorpine and long-chain beta-KTx-like peptides in
scorpion venoms: identification of “orphan” components,
Peptides, 28, 31-37.
170.Zeng, X.-C., Zhang, L., Nie, Y., and Luo, X.
(2012) Identification and molecular characterization of three new
K+-channel specific toxins from the Chinese scorpion
Mesobuthus martensii Karsch revealing intronic number
polymorphism and alternative splicing in duplicated genes,
Peptides, 34, 311-323.
171.Diego-Garcia, E., Batista, C. V. F.,
Garcia-Gomez, B. I., Lucas, S., Candido, D. M., Gomez-Lagunas, F., and
Possani, L. D. (2005) The Brazilian scorpion Tityus costatus
Karsch: genes, peptides and function, Toxicon, 45,
273-283.
172.Pisciotta, M., Coronas, F. I., Bloch, C.,
Prestipino, G., and Possani, L. D. (2000) Fast K(+) currents from
cerebellum granular cells are completely blocked by a peptide purified
from Androctonus australis Garzoni scorpion venom, Biochim.
Biophys. Acta, 1468, 203-212.
173.Bagdany, M., Batista, C. V. F., Valdez-Cruz, N.
A., Somodi, S., Rodriguez de la Vega, R. C., Licea, A. F., Varga, Z.,
Gaspar, R., Possani, L. D., and Panyi, G. (2005) Anuroctoxin, a new
scorpion toxin of the alpha-KTx 6 subfamily, is highly selective for
Kv1.3 over IKCa1 ion channels of human T-lymphocytes, Mol.
Pharmacol., 67, 1034-1044.
174.Mouhat, S., Jouirou, B., Mosbah, A., De Waard,
M., and Sabatier, J.-M. (2004) Diversity of folds in animal toxins
acting on ion channels, Biochem. J., 378, 717-726.
175.Arzamasov, A. A., Vasilevskii, A. A., and
Grishin, E. V. (2014) Chlorotoxin and related peptides are short insect
toxins from scorpion venom, Bioorg. Khim., 40,
387-398.
176.Blanc, E., Sabatier, J. M., Kharrat, R.,
Meunier, S., El Ayeb, M., Van Rietschoten, J., and Darbon, H. (1997)
Solution structure of maurotoxin, a scorpion toxin from Scorpio
maurus, with high affinity for voltage-gated potassium channels,
Proteins, 29, 321-333.
177.Saucedo, A. L., Flores-Solis, D., Rodriguez de
la Vega, R. C., Ramirez-Cordero, B., Hernandez-Lopez, R., Cano-Sanchez,
P., Noriega Navarro, R., Garcia-Valdes, J., Coronas-Valderrama, F., De
Roodt, A., Brieba, L. G., Domingos Possani, L., and Del Rio-Portilla,
F. (2012) New tricks of an old pattern: structural versatility of
scorpion toxins with common cysteine spacing, J. Biol. Chem.,
287, 12321-12330.
178.Chen, Z.-Y., Hu, Y.-T., Yang, W.-S., He, Y.-W.,
Feng, J., Wang, B., Zhao, R.-M., Ding, J.-P., Cao, Z.-J., Li, W.-X.,
and Wu, Y.-L. (2012) Hg1, novel peptide inhibitor specific for Kv1.3
channels from first scorpion Kunitz-type potassium channel toxin
family, J. Biol. Chem., 287, 13813-13821.
179.Chen, Z., Hu, Y., Han, S., Yin, S., He, Y., Wu,
Y., Cao, Z., and Li, W. (2011) ImKTx1, a new Kv1.3 channel blocker with
a unique primary structure, J. Biochem. Mol. Toxicol.,
25, 244-251.
180.Tytgat, J., Chandy, K. G., Garcia, M. L.,
Gutman, G. A., Martin-Eauclaire, M. F., Van der Walt, J. J., and
Possani, L. D. (1999) A unified nomenclature for short-chain peptides
isolated from scorpion venoms: alpha-KTx molecular subfamilies,
Trends Pharmacol. Sci., 20, 444-447.
181.Diego-Garcia, E., Caliskan, F., and Tytgat, J.
(2014) The Mediterranean scorpion Mesobuthus gibbosus
(Scorpiones, Buthidae): transcriptome analysis and organization of the
genome encoding chlorotoxin-like peptides, BMC Genomics,
15, 295.
182.He, Y., Zhao, R., Di, Z., Li, Z., Xu, X., Hong,
W., Wu, Y., Zhao, H., Li, W., and Cao, Z. (2013) Molecular diversity of
Chaerilidae venom peptides reveals the dynamic evolution of scorpion
venom components from Buthidae to non-Buthidae, J. Proteomics,
89, 1-14.
183.Diego-Garcia, E., Peigneur, S., Debaveye, S.,
Gheldof, E., Tytgat, J., and Caliskan, F. (2013) Novel potassium
channel blocker venom peptides from Mesobuthus gibbosus
(Scorpiones: Buthidae), Toxicon, 61, 72-82.
184.Cologna, C. T., Peigneur, S., Rosa, J. C.,
Selistre-de-Araujo, H. S., Varanda, W. A., Tytgat, J., and Arantes, E.
C. (2011) Purification and characterization of Ts15, the first member
of a new α-KTX subfamily from the venom of the Brazilian scorpion
Tityus serrulatus, Toxicon, 58, 54-61.
185.Schwartz, E. F., Diego-Garcia, E., Rodriguez de
la Vega, R. C., and Possani, L. D. (2007) Transcriptome analysis of the
venom gland of the Mexican scorpion Hadrurus gertschi
(Arachnida: Scorpiones), BMC Genomics, 8, 119.
186.Zhu, S., Gao, B., Aumelas, A., Del Carmen
Rodriguez, M., Lanz-Mendoza, H., Peigneur, S., Diego-Garcia, E.,
Martin-Eauclaire, M.-F., Tytgat, J., and Possani, L. D. (2010)
MeuTXKbeta1, a scorpion venom-derived two-domain potassium channel
toxin-like peptide with cytolytic activity, Biochim. Biophys.
Acta, 1804, 872-883.
187.Diego-Garcia, E., Abdel-Mottaleb, Y., Schwartz,
E. F., De la Vega, R. C. R., Tytgat, J., and Possani, L. D. (2008)
Cytolytic and K+ channel blocking activities of beta-KTx and
scorpine-like peptides purified from scorpion venoms, Cell. Mol.
Life Sci., 65, 187-200.
188.Korolkova, Y. V., Kozlov, S. A., Lipkin, A. V.,
Pluzhnikov, K. A., Hadley, J. K., Filippov, A. K., Brown, D. A.,
Angelo, K., Strobaek, D., Jespersen, T., Olesen, S. P., Jensen, B. S.,
and Grishin, E. V. (2001) An ERG channel inhibitor from the scorpion
Buthus eupeus, J. Biol. Chem., 276, 9868-9876.
189.Corona, M., Gurrola, G. B., Merino, E.,
Cassulini, R. R., Valdez-Cruz, N. A., Garcia, B., Ramirez-Dominguez, M.
E., Coronas, F. I. V., Zamudio, F. Z., Wanke, E., and Possani, L. D.
(2002) A large number of novel Ergtoxin-like genes and ERG
K+-channels blocking peptides from scorpions of the genus
Centruroides, FEBS Lett., 532, 121-126.
190.Gairi, M., Romi, R., Fernandez, I., Rochat, H.,
Martin-Eauclaire, M. F., Van Rietschoten, J., Pons, M., and Giralt, E.
(1997) 3D structure of kaliotoxin: is residue 34 a key for channel
selectivity? J. Pept. Sci., 3, 314-319.
191.Torres, A. M., Bansal, P., Alewood, P. F.,
Bursill, J. A., Kuchel, P. W., and Vandenberg, J. I. (2003) Solution
structure of CnErg1 (Ergtoxin), a HERG specific scorpion toxin,
FEBS Lett., 539, 138-142.
192.M’Barek, S., Lopez-Gonzalez, I.,
Andreotti, N., Di Luccio, E., Visan, V., Grissmer, S., Judge, S., El
Ayeb, M., Darbon, H., Rochat, H., Sampieri, F., Beraud, E., Fajloun,
Z., De Waard, M., and Sabatier, J.-M. (2003) A maurotoxin with
constrained standard disulfide bridging: innovative strategy of
chemical synthesis, pharmacology, and docking on K+
channels, J. Biol. Chem., 278, 31095-31104.
193.Fajloun, Z., Mosbah, A., Carlier, E.,
Mansuelle, P., Sandoz, G., Fathallah, M., Di Luccio, E., Devaux, C.,
Rochat, H., Darbon, H., De Waard, M., and Sabatier, J. M. (2000)
Maurotoxin versus Pi1/HsTx1 scorpion toxins. Toward new insights in the
understanding of their distinct disulfide bridge patterns, J. Biol.
Chem., 275, 39394-39402.
194.Coronas, F. V., De Roodt, A. R., Portugal, T.
O., Zamudio, F. Z., Batista, C. V. F., Gomez-Lagunas, F., and Possani,
L. D. (2003) Disulfide bridges and blockage of Shaker B K(+)-channels
by another butantoxin peptide purified from the Argentinean scorpion
Tityus trivittatus, Toxicon, 41, 173-179.
195.Chagot, B., Pimentel, C., Dai, L., Pil, J.,
Tytgat, J., Nakajima, T., Corzo, G., Darbon, H., and Ferrat, G. (2005)
An unusual fold for potassium channel blockers: NMR structure of three
toxins from the scorpion Opisthacanthus madagascariensis,
Biochem. J., 388, 263-271.
196.Zhao, R., Dai, H., Qiu, S., Li, T., He, Y., Ma,
Y., Chen, Z., Wu, Y., Li, W., and Cao, Z. (2011) SdPI, the first
functionally characterized Kunitz-type trypsin inhibitor from scorpion
venom, PLoS One, 6, e27548.
197.Gao, B., Harvey, P. J., Craik, D. J., Ronjat,
M., De Waard, M., and Zhu, S. (2013) Functional evolution of scorpion
venom peptides with an inhibitor cystine knot fold, Biosci.
Rep., 33.
198.Zhijian, C., Yun, X., Chao, D., Shunyi, Z.,
Shijin, Y., Yingliang, W., and Wenxin, L. (2006) Cloning and
characterization of a novel calcium channel toxin-like gene BmCa1 from
Chinese scorpion Mesobuthus martensii Karsch, Peptides,
27, 1235-12340.
199.Varga, Z., Gurrola-Briones, G., Papp, F.,
Rodriguez de la Vega, R. C., Pedraza-Alva, G., Tajhya, R. B., Gaspar,
R., Cardenas, L., Rosenstein, Y., Beeton, C., Possani, L. D., and
Panyi, G. (2012) Vm24, a natural immunosuppressive peptide, potently
and selectively blocks Kv1.3 potassium channels of human T-cells,
Mol. Pharmacol., 82, 372-382.
200.Dudina, E. E., Korolkova, Y. V., Bocharova, N.
E., Koshelev, S. G., Egorov, T. A., Huys, I., Tytgat, J., and Grishin,
E. V. (2001) OsK2, a new selective inhibitor of Kv1.2 potassium
channels purified from the venom of the scorpion Orthochirus
scrobiculosus, Biochem. Biophys. Res. Commun., 286,
841-847.
201.Swartz, K. J. (2004) Towards a structural view
of gating in potassium channels, Nat. Rev. Neurosci., 5,
905-916.
202.Catterall, W. A., Cestele, S., Yarov-Yarovoy,
V., Yu, F. H., Konoki, K., and Scheuer, T. (2007) Voltage-gated ion
channels and gating modifier toxins, Toxicon, 49,
124-141.
203.Jimenez-Vargas, J. M., Restano-Cassulini, R.,
and Possani, L. D. (2012) Toxin modulators and blockers of hERG K(+)
channels, Toxicon, 60, 492-501.
204.Tenenholz, T. C., Klenk, K. C., Matteson, D.
R., Blaustein, M. P., and Weber, D. J. (2000) Structural determinants
of scorpion toxin affinity: the charybdotoxin (alpha-KTX) family of
K(+)-channel blocking peptides, Rev. Physiol. Biochem.
Pharmacol., 140, 1351-1385.
205.Jiang, Y., Lee, A., Chen, J., Cadene, M.,
Chait, B. T., and MacKinnon, R. (2002) Crystal structure and mechanism
of a calcium-gated potassium channel, Nature, 417,
515-522.
206.MacKinnon, R., Cohen, S. L., Kuo, A., Lee, A.,
and Chait, B. T. (1998) Structural conservation in prokaryotic and
eukaryotic potassium channels, Science, 280, 106-109.
207.Legros, C., Pollmann, V., Knaus, H. G.,
Farrell, A. M., Darbon, H., Bougis, P. E., Martin-Eauclaire, M. F., and
Pongs, O. (2000) Generating a high affinity scorpion toxin receptor in
KcsA-Kv1.3 chimeric potassium channels, J. Biol. Chem.,
275, 16918-16924.
208.Legros, C., Schulze, C., Garcia, M. L., Bougis,
P. E., Martin-Eauclaire, M.-F., and Pongs, O. (2002)
Engineering-specific pharmacological binding sites for peptidyl
inhibitors of potassium channels into KcsA, Biochemistry,
41, 15369-15375.
209.Goldstein, S. A., Pheasant, D. J., and Miller,
C. (1994) The charybdotoxin receptor of a Shaker K+ channel:
peptide and channel residues mediating molecular recognition,
Neuron, 12, 1377-1388.
210.Hidalgo, P., and MacKinnon, R. (1995) Revealing
the architecture of a K+ channel pore through mutant cycles
with a peptide inhibitor, Science, 268, 307-310.
211.Ranganathan, R., Lewis, J. H., and MacKinnon,
R. (1996) Spatial localization of the K+ channel selectivity
filter by mutant cycle-based structure analysis, Neuron,
16, 131-139.
212.Lecomte, C., Ferrat, G., Fajloun, Z., Van
Rietschoten, J., Rochat, H., Martin-Eauclaire, M. F., Darbon, H., and
Sabatier, J. M. (1999) Chemical synthesis and structure–activity
relationships of Ts kappa, a novel scorpion toxin acting on
apamin-sensitive SK channel, J. Pept. Res., 54,
369-376.
213.Shakkottai, V. G., Regaya, I., Wulff, H.,
Fajloun, Z., Tomita, H., Fathallah, M., Cahalan, M. D., Gargus, J. J.,
Sabatier, J. M., and Chandy, K. G. (2001) Design and characterization
of a highly selective peptide inhibitor of the small conductance
calcium-activated K+ channel, SkCa2, J. Biol. Chem.,
276, 43145-43151.
214.Pedarzani, P., D’Hoedt, D., Doorty, K.
B., Wadsworth, J. D. F., Joseph, J. S., Jeyaseelan, K., Kini, R. M.,
Gadre, S. V., Sapatnekar, S. M., Stocker, M., and Strong, P. N. (2002)
Tamapin, a venom peptide from the Indian red scorpion (Mesobuthus
tamulus) that targets small conductance Ca2+-activated
K+ channels and afterhyperpolarization currents in central
neurons, J. Biol. Chem., 277, 46101-46109.
215.Korolkova, Y. V., Bocharov, E. V., Angelo, K.,
Maslennikov, I. V., Grinenko, O. V., Lipkin, A. V., Nosyreva, E. D.,
Pluzhnikov, K. A., Olesen, S.-P., Arseniev, A. S., and Grishin, E. V.
(2002) New binding site on common molecular scaffold provides HERG
channel specificity of scorpion toxin BeKm-1, J. Biol. Chem.,
277, 43104-43109.
216.Pardo-Lopez, L., Zhang, M., Liu, J., Jiang, M.,
Possani, L. D., and Tseng, G.-N. (2002) Mapping the binding site of a
human ether-a-go-go-related gene-specific peptide toxin (ErgTx) to the
channel’s outer vestibule, J. Biol. Chem., 277,
16403-16411.
217.Dauplais, M., Lecoq, A., Song, J., Cotton, J.,
Jamin, N., Gilquin, B., Roumestand, C., Vita, C., De Medeiros, C. L.,
Rowan, E. G., Harvey, A. L., and Menez, A. (1997) On the convergent
evolution of animal toxins. Conservation of a diad of functional
residues in potassium channel-blocking toxins with unrelated
structures, J. Biol. Chem., 272, 4302-4309.
218.Gasparini, S., Danse, J. M., Lecoq, A.,
Pinkasfeld, S., Zinn-Justin, S., Young, L. C., De Medeiros, C. C.,
Rowan, E. G., Harvey, A. L., and Menez, A. (1998) Delineation of the
functional site of alpha-dendrotoxin. The functional topographies of
dendrotoxins are different but share a conserved core with those of
other Kv1 potassium channel-blocking toxins, J. Biol. Chem.,
273, 25393-25403.
219.Banerjee, A., Lee, A., Campbell, E., and
Mackinnon, R. (2013) Structure of a pore-blocking toxin in complex with
a eukaryotic voltage-dependent K(+) channel, Elife, 2,
e00594.
220.Carlier, E., Avdonin, V., Geib, S., Fajloun,
Z., Kharrat, R., Rochat, H., Sabatier, J. M., Hoshi, T., and De Waard,
M. (2000) Effect of maurotoxin, a four disulfide-bridged toxin from the
chactoid scorpion Scorpio maurus, on Shaker K+
channels, J. Pept. Res., 55, 419-427.
221.Mouhat, S., Mosbah, A., Visan, V., Wulff, H.,
Delepierre, M., Darbon, H., Grissmer, S., De Waard, M., and Sabatier,
J.-M. (2004) The “functional” dyad of scorpion toxin Pi1 is
not itself a prerequisite for toxin binding to the voltage-gated Kv1.2
potassium channels, Biochem. J., 377, 25-36.
222.M’Barek, S., Mosbah, A., Sandoz, G.,
Fajloun, Z., Olamendi-Portugal, T., Rochat, H., Sampieri, F., Guijarro,
J. I., Mansuelle, P., Delepierre, M., De Waard, M., and Sabatier, J.-M.
(2003) Synthesis and characterization of Pi4, a scorpion toxin from
Pandinus imperator that acts on K+ channels, Eur.
J. Biochem., 270, 3583-3592.
223.Jouirou, B., Mosbah, A., Visan, V., Grissmer,
S., M’Barek, S., Fajloun, Z., Van Rietschoten, J., Devaux, C.,
Rochat, H., Lippens, G., El Ayeb, M., De Waard, M., Mabrouk, K., and
Sabatier, J.-M. (2004) Cobatoxin 1 from Centruroides noxius
scorpion venom: chemical synthesis, three-dimensional structure in
solution, pharmacology and docking on K+ channels,
Biochem. J., 377, 37-49.
224.Batista, C. V. F., Gomez-Lagunas, F., Rodriguez
de la Vega, R. C., Hajdu, P., Panyi, G., Gaspar, R., and Possani, L. D.
(2002) Two novel toxins from the Amazonian scorpion Tityus
cambridgei that block Kv1.3 and Shaker B K(+)-channels with
distinctly different affinities, Biochim. Biophys. Acta,
1601, 123-131.
225.Katoh, E., Nishio, H., Inui, T., Nishiuchi, Y.,
Kimura, T., Sakakibara, S., and Yamazaki, T. (2000) Structural basis
for the biological activity of dendrotoxin-I, a potent potassium
channel blocker, Biopolymers, 54, 44-57.
226.Lipkind, G. M., and Fozzard, H. A. (1997) A
model of scorpion toxin binding to voltage-gated K+
channels, J. Membr. Biol., 158, 187-196.
227.Gehlert, D. R., and Gackenheimer, S. L. (1993)
Comparison of the distribution of binding sites for the potassium
channel ligands [125I]apamin,
[125I]charybdotoxin, and [125I]iodoglyburide in
the rat brain, Neuroscience, 52, 191-205.
228.Shimony, E., Sun, T., Kolmakova-Partensky, L.,
and Miller, C. (1994) Engineering a uniquely reactive thiol into a
cysteine-rich peptide, Protein Eng., 7, 503-507.
229.Pardo, L. A., and Stuhmer, W. (2014) The roles
of K(+) channels in cancer, Nat. Rev. Cancer, 14,
39-48.
230.Lewis, R. J., and Garcia, M. L. (2003)
Therapeutic potential of venom peptides, Nat. Rev. Drug Discov.,
2, 790-802.
231.Vazquez, J., Feigenbaum, P., Katz, G., King, V.
F., Reuben, J. P., Roy-Contancin, L., Slaughter, R. S., Kaczorowski, G.
J., and Garcia, M. L. (1989) Characterization of high affinity binding
sites for charybdotoxin in sarcolemmal membranes from bovine aortic
smooth muscle. Evidence for a direct association with the high
conductance calcium-activated potassium channel, J. Biol. Chem.,
264, 20902-20909.
232.Auguste, P., Hugues, M., Mourre, C., Moinier,
D., Tartar, A., and Lazdunski, M. (1992) Scyllatoxin, a blocker of
Ca(2+)-activated K+ channels: structure-function
relationships and brain localization of the binding sites,
Biochemistry, 31, 648-654.
233.Knaus, H. G., Schwarzer, C., Koch, R. O.,
Eberhart, A., Kaczorowski, G. J., Glossmann, H., Wunder, F., Pongs, O.,
Garcia, M. L., and Sperk, G. (1996) Distribution of high-conductance
Ca(2+)-activated K+ channels in rat brain: targeting to
axons and nerve terminals, J. Neurosci., 16, 955-963.
234.Knaus, H. G., Koch, R. O., Eberhart, A.,
Kaczorowski, G. J., Garcia, M. L., and Slaughter, R. S. (1995)
[125I]Margatoxin, an extraordinarily high affinity ligand
for voltage-gated potassium channels in mammalian brain,
Biochemistry, 34, 13627-13634.
235.Garcia-Calvo, M., Knaus, H. G., McManus, O. B.,
Giangiacomo, K. M., Kaczorowski, G. J., and Garcia, M. L. (1994)
Purification and reconstitution of the high-conductance,
calcium-activated potassium channel from tracheal smooth muscle, J.
Biol. Chem., 269, 676-682.
236.Knaus, H. G., Garcia-Calvo, M., Kaczorowski, G.
J., and Garcia, M. L. (1994) Subunit composition of the high
conductance calcium-activated potassium channel from smooth muscle, a
representative of the mSlo and slowpoke family of potassium channels,
J. Biol. Chem., 269, 3921-3924.
237.Bergeron, Z. L., and Bingham, J.-P. (2012)
Scorpion toxins specific for potassium (K+) channels: a
historical overview of peptide bioengineering, Toxins (Basel),
4, 1082-1119.
238.Jimenez-Gonzalez, C., McLaren, G. J., and Dale,
N. (2003) Development of Ca2+-channel and BK-channel
expression in embryos and larvae of Xenopus laevis, Eur. J.
Neurosci., 18, 2175-2187.
239.Pragl, B., Koschak, A., Trieb, M., Obermair,
G., Kaufmann, W. A., Gerster, U., Blanc, E., Hahn, C., Prinz, H.,
Schutz, G., Darbon, H., Gruber, H. J., and Knaus, H.-G. (2002)
Synthesis, characterization, and application of Cy-dye- and
Alexa-dye-labeled hongotoxin(1) analogues. The first high affinity
fluorescence probes for voltage-gated K+ channels,
Bioconj. Chem., 13, 416-425.
240.Bingham, J.-P., Bian, S., Tan, Z.-Y., Takacs,
Z., and Moczydlowski, E. (2006) Synthesis of a biotin derivative of
iberiotoxin: binding interactions with streptavidin and the BK
Ca2+-activated K+ channel expressed in a human
cell line, Bioconj. Chem., 17, 689-699.
241.Kudryashova, K. S., Nekrasova, O. V.,
Kuzmenkov, A. I., Vassilevski, A. A., Ignatova, A. A., Korolkova, Y.
V., Grishin, E. V., Kirpichnikov, M. P., and Feofanov, A. V. (2013)
Fluorescent system based on bacterial expression of hybrid KcsA
channels designed for Kv1.3 ligand screening and study, Anal.
Bioanal. Chem., 405, 2379-2389.
242.Clare, J. J. (2010) Targeting ion channels for
drug discovery, Discov. Med., 9, 253-260.
243.Overington, J. P., Al-Lazikani, B., and
Hopkins, A. L. (2006) How many drug targets are there? Nat. Rev.
Drug Discov., 5, 993-996.
244.Alexander, S. P., Benson, H. E., Faccenda, E.,
Pawson, A. J., Sharman, J. L., McGrath, J. C., Catterall, W. A.,
Spedding, M., Peters, J. A., Harmar, A. J., and CGTP Collaborators
(2013) The Concise Guide to PHARMACOLOGY 2013/14: overview, Br. J.
Pharmacol., 170, 1449-1458.
245.Beeton, C., Wulff, H., Barbaria, J.,
Clot-Faybesse, O., Pennington, M., Bernard, D., Cahalan, M. D., Chandy,
K. G., and Beraud, E. (2001) Selective blockade of T-lymphocyte K(+)
channels ameliorates experimental autoimmune encephalomyelitis, a model
for multiple sclerosis, Proc. Natl. Acad. Sci. USA, 98,
13942-13947.
246.Beeton, C., Wulff, H., Standifer, N. E., Azam,
P., Mullen, K. M., Pennington, M. W., Kolski-Andreaco, A., Wei, E.,
Grino, A., Counts, D. R., Wang, P. H., LeeHealey, C. J., Andrews, B.,
Sankaranarayanan, A., Homerick, D., Roeck, W. W., Tehranzadeh, J.,
Stanhope, K. L., Zimin, P., Havel, P. J., Griffey, S., Knaus, H.-G.,
Nepom, G. T., Gutman, G. A., Calabresi, P. A., and Chandy, K. G. (2006)
Kv1.3 channels are a therapeutic target for T-cell-mediated autoimmune
diseases, Proc. Natl. Acad. Sci. USA, 103,
17414-17419.
247.Azam, P., Sankaranarayanan, A., Homerick, D.,
Griffey, S., and Wulff, H. (2007) Targeting effector memory T-cells
with the small molecule Kv1.3 blocker PAP-1 suppresses allergic contact
dermatitis, J. Invest. Dermatol., 127, 1419-1429.
248.Feske, S., Wulff, H., and Skolnik, E. Y. (2015)
Ion channels in innate and adaptive immunity, Annu. Rev.
Immunol., 33, 291-353.
249.DeCoursey, T. E., Chandy, K. G., Gupta, S., and
Cahalan, M. D. (1984) Voltage-gated K+ channels in human
T-lymphocytes: a role in mitogenesis? Nature, 307,
465-468.
250.Chandy, K. G., DeCoursey, T. E., Cahalan, M.
D., McLaughlin, C., and Gupta, S. (1984) Voltage-gated potassium
channels are required for human T-lymphocyte activation, J. Exp.
Med., 160, 369-385.
251.Lewis, R. S., and Cahalan, M. D. (1995)
Potassium and calcium channels in lymphocytes, Annu. Rev.
Immunol., 13, 623-653.
252.Feske, S., Skolnik, E. Y., and Prakriya, M.
(2012) Ion channels and transporters in lymphocyte function and
immunity, Nat. Rev. Immunol., 12, 532-547.
253.Wulff, H., Calabresi, P. A., Allie, R., Yun,
S., Pennington, M., Beeton, C., and Chandy, K. G. (2003) The
voltage-gated Kv1.3 K(+) channel in effector memory T-cells as new
target for MS, J. Clin. Invest., 111, 1703-1713.
254.Breland, A. E., and Currier, R. D. (1983)
Scorpion venom and multiple sclerosis, Lancet, 2,
1021.
255.Chi, V., Pennington, M. W., Norton, R. S.,
Tarcha, E. J., Londono, L. M., Sims-Fahey, B., Upadhyay, S. K., Lakey,
J. T., Iadonato, S., Wulff, H., Beeton, C., and Chandy, K. G. (2012)
Development of a sea anemone toxin as an immunomodulator for therapy of
autoimmune diseases, Toxicon, 59, 529-546.
256.Pennington, M. W., Beeton, C., Galea, C. A.,
Smith, B. J., Chi, V., Monaghan, K. P., Garcia, A., Rangaraju, S.,
Giuffrida, A., Plank, D., Crossley, G., Nugent, D., Khaytin, I.,
Lefievre, Y., Peshenko, I., Dixon, C., Chauhan, S., Orzel, A., Inoue,
T., Hu, X., Moore, R. V., Norton, R. S., and Chandy, K. G. (2009)
Engineering a stable and selective peptide blocker of the Kv1.3 channel
in T-lymphocytes, Mol. Pharmacol., 75, 762-773.
257.Beeton, C., Pennington, M. W., and Norton, R.
S. (2011) Analogs of the sea anemone potassium channel blocker ShK for
the treatment of autoimmune diseases, Inflamm. Allergy Drug
Targets, 10, 313-321.
258.Mouhat, S., Visan, V., Ananthakrishnan, S.,
Wulff, H., Andreotti, N., Grissmer, S., Darbon, H., De Waard, M., and
Sabatier, J.-M. (2005) K+ channel types targeted by
synthetic OSK1, a toxin from Orthochirus scrobiculosus scorpion
venom, Biochem. J., 385, 95-104.
259.Koo, G. C., Blake, J. T., Talento, A., Nguyen,
M., Lin, S., Sirotina, A., Shah, K., Mulvany, K., Hora, D., Cunningham,
P., Wunderler, D. L., McManus, O. B., Slaughter, R., Bugianesi, R.,
Felix, J., Garcia, M., Williamson, J., Kaczorowski, G., Sigal, N. H.,
Springer, M. S., and Feeney, W. (1997) Blockade of the voltage-gated
potassium channel Kv1.3 inhibits immune responses in vivo, J.
Immunol., 158, 5120-5128.
260.Leonard, R. J., Garcia, M. L., Slaughter, R.
S., and Reuben, J. P. (1992) Selective blockers of voltage-gated
K+ channels depolarize human T-lymphocytes: mechanism of the
antiproliferative effect of charybdotoxin, Proc. Natl. Acad. Sci.
USA, 89, 10094-10098.
261.Bradding, P., and Wulff, H. (2009) The
K+ channels K(Ca)3.1 and K(v)1.3 as novel targets for asthma
therapy, Br. J. Pharmacol., 157, 1330-1339.
262.Koshy, S., Huq, R., Tanner, M. R., Atik, M. A.,
Porter, P. C., Khan, F. S., Pennington, M. W., Hanania, N. A., Corry,
D. B., and Beeton, C. (2014) Blocking KV1.3 channels inhibits Th2
lymphocyte function and treats a rat model of asthma, J. Biol.
Chem., 289, 12623-1262.
263.Grgic, I., Wulff, H., Eichler, I., Flothmann,
C., Kohler, R., and Hoyer, J. (2009) Blockade of T-lymphocyte KCa3.1
and Kv1.3 channels as novel immunosuppression strategy to prevent
kidney allograft rejection, Transplant. Proc., 41,
2601-2606.
264.Schmitt, N., Grunnet, M., and Olesen, S.-P.
(2014) Cardiac potassium channel subtypes: new roles in repolarization
and arrhythmia, Physiol. Rev., 94, 609-653.
265.Kass, R. S. (2005) The channelopathies: novel
insights into molecular and genetic mechanisms of human disease, J.
Clin. Invest., 115, 1986-1989.
266.Robbins, C. A., and Tempel, B. L. (2012) Kv1.1
and Kv1.2: similar channels, different seizure models,
Epilepsia, 53, Suppl. 1, 134-141.
267.Maljevic, S., and Lerche, H. (2014) Potassium
channel genes and benign familial neonatal epilepsy, Prog. Brain
Res., 213, 17-53.
268.Guglielmi, L., Servettini, I., Caramia, M.,
Catacuzzeno, L., Franciolini, F., D’Adamo, M. C., and Pessia, M.
(2015) Update on the implication of potassium channels in autism: K(+)
channelautism spectrum disorder, Front. Cell. Neurosci.,
9, 34.
269.Huber, S. M. (2013) Oncochannels, Cell
Calcium, 53, 241-255.
270.Tsantoulas, C., and McMahon, S. B. (2014)
Opening paths to novel analgesics: the role of potassium channels in
chronic pain, Trends Neurosci., 37, 146-158.
271.Ocana, M., Cendan, C. M., Cobos, E. J.,
Entrena, J. M., and Baeyens, J. M. (2004) Potassium channels and pain:
present realities and future opportunities, Eur. J. Pharmacol.,
500, 203-219.