Glutathione Reductase Gene Expression Depends on Chloroplast Signals in Arabidopsis thaliana
E. Yu. Garnik*, V. I. Belkov, V. I. Tarasenko, M. A. Korzun, and Yu. M. Konstantinov
Siberian Institute of Plant Physiology and Biochemistry, Siberian Branch of the Russian Academy of Sciences, 664033 Irkutsk, Russia; fax: +7 (3952) 51-0754; E-mail: elga74@yandex.ru* To whom correspondence should be addressed.
Received August 7, 2015; Revision received November 5, 2015
Glutathione reductase (EC 1.6.4.2) is one of the main antioxidant enzymes of the plant cell. In Arabidopsis thaliana, glutathione reductase is encoded by two genes: the gr1 gene encodes the cytosolic-peroxisomal form, and the gr2 gene encodes the chloroplast-mitochondrial form. Little is known about the regulation of expression of plant glutathione reductase genes. In the present work, we have demonstrated that gr2 (but not gr1) gene expression in Arabidopsis leaves changes depending on changes in redox state of the photosynthetic electron transport chain. Expression of both the gr1 and gr2 genes was induced by reactive oxygen species. In heterotrophic suspension cell culture of Arabidopsis, expression of both studied genes did not depend on H2O2 level or on changes in the redox state of the mitochondrial electron transport chain. Our data indicate that chloroplasts are involved in the regulation of the glutathione reductase gene expression in Arabidopsis.
KEY WORDS: reactive oxygen species, hydrogen peroxide, glutathione reductase, redox-regulation, chloroplast-nuclear regulation of gene expression, Arabidopsis thalianaDOI: 10.1134/S0006297916040064
Glutathione reductase (EC 1.6.4.2) is a component of the antioxidant defense system of cells in both pro- and eukaryotes [1]. In plants, glutathione reductase functions (and seems to control the limiting reaction) as a component of the so-called ascorbate-glutathione cycle, which is responsible for maintaining the pool of reduced forms of two most important low molecular weight antioxidants of the plant cell, ascorbic acid and reduced glutathione [2]. Holoenzymes of plant glutathione reductases are FAD-containing NADPH-binding mono-, homodi-, homotri-, or homotetramers [3]. Their protein components in higher plants are encoded by small multigenic families: two genes of glutathione reductase were identified in Nicotiana tabacum [4] and Pisum sativum [5], and three genes – in Oryza sativa and Populus trichocarpa [6]. In Arabidopsis thaliana, glutathione reductases are encoded by two nuclear genes: gr1 (AT3G24170) and gr2 (AT3G54660). The gr1 gene product functions in the cytosol and peroxisomes [7], and the gr2 gene product acts in chloroplasts and mitochondria [1].
Notwithstanding great interest in glutathione reductase as a key enzyme responsible for plant resistance against unfavorable environmental factors, there are very few data on regulation of expression of the genes encoding this enzyme. Data on induction of the genes in various plant species under abiotic stress conditions [1, 8, 9] do not reveal anything about the molecular mechanisms of this induction. Involvement of signals related to abscisic acid was supposed [10], but the promoter regions of the glutathione reductase gene seem to contain no regulatory sequences that are traditionally associated with regulation depending on abscisic acid [11]. It was supposed that genes encoding chloroplast-mitochondrial forms of plant glutathione reductases should be constantly expressed at a high level to efficiently detoxify large amounts of reactive oxygen species (ROS), which are constantly produced in these organelles as byproducts of respiration and/or photosynthesis [1]. In fact, it was shown that various abiotic stresses caused pronounced induction of the gr1 gene, whereas the gr2 gene was induced weakly or was not at all [12, 13]. Under stress conditions, the amount of EST (expressed sequence tag) in barley Hordeum vulgare significantly increased in the case of genes encoding cytosolic but not chloroplast isoforms of glutathione reductases [1]. However, mechanisms responsible for such rigidity of expression of chloroplast-mitochondrial glutathione reductases of plants are unknown.
Thus, the regulation of expression of genes encoding glutathione reductases of plants is still unclear. In the present work, we studied a possible role of reactive oxygen species and of the redox state of the electron transport chains of chloroplasts and mitochondria in the induction of genes encoding the cytosolic-peroxisomal (gr1) and plastid-mitochondrial (gr2) forms of glutathione reductase in Arabidopsis. For the first time, we show both genes to be inducible under certain conditions, but to be regulated independently, and their regulation seems to be mediated through chloroplasts.
MATERIALS AND METHODS
Plant material and experimental conditions. Seeds of Arabidopsis thaliana (L.) Heynh. ecotype Columbia (further Col-0) were obtained from the Arabidopsis Biological Resource Center (The Ohio State University, USA). The seeds were sterilized for 8 min in solution containing 70% ethanol and 0.1% Triton X-100, washed thrice in sterile water, and placed on a solid medium in Petri dishes along the dish diameter. The nutritional medium contained mineral salts MS [14] (half of the quantity) and Phytagel (Sigma-Aldrich, USA) – 0.8%. After stratification at 4°C for three days, the dishes were installed vertically and seedlings were grown for 14 days at 23°C under illumination of 100 µmol·m—2·s–1 for 16 h daily. To study the influence of 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), all dishes before treatment were kept in the dark for 18 h. Treatment with 20 µM DCMU (Sigma-Aldrich) and 10 mM H2O2 was realized by spraying leaves of intact plants with the corresponding solutions, and RNA was isolated 2 h (H2O2) or 4 h (H2O2, DCMU) after the spraying. Plants treated with deionized water containing corresponding concentrations of solvents were used as controls; all solutions contained 0.05% Triton X-100. A suspension cell culture Col-0 was grown in the dark at 26°C on MS medium supplemented with thiamine (1 mg/liter), pyridoxine (0.5 mg/liter), nicotinic acid (0.5 mg/liter), inositol (100 mg/liter), 2,4-dichlorophenoxyacetic acid (0.3 mg/liter), and sucrose (30 g/liter); subcultivation was performed once per 10 days. The suspension culture was used for experiments 7 days after the transfer of the cells into fresh medium. The cells were treated with antimycin A (10 µM; Sigma-Aldrich) for 4 h, and with hydrogen peroxide (10 mM) for 2 and 4 h. Pretreatment with N-acetylcysteine (2 mM; Sigma-Aldrich) was started 30 min before the addition of antimycin A. When the experiment was terminated, the cells were filtered from the cultural medium with a vacuum filter, washed in 3-5 volumes of deionized water, and RNA was immediately isolated. All experiments were conducted in at least three biological repeats.
Determination of ROS content. The hydrogen peroxide content was determined in the suspension culture cells from dichlorofluorescein (DCF) fluorescence as described [15]. Upon treatment with redox-agents, the culture was supplemented with H2DCF-diacetate (Fluka, USA) to the concentration of 5 µM and incubated for 40 min in the dark at 26°C with agitation. Then 1 ml of the culture medium separated from the cells and cellular fragments was diluted four-fold with deionized water, and fluorescence of the resulting solutions was measured (at exciting light wavelength 480 nm and emitted light wavelength 524 nm) with a Shimadzu RF 5301 PC spectrofluorometer (Japan). The result is presented as the ratio of fluorescence and the wet weight of the cells. For comparative evaluation, the results are expressed as percentage of the hydrogen peroxide level under control conditions. In leaves of 14-day-old plants, the superoxide radical content was determined qualitatively as described in work [16]. Before the experiment, the plants were kept in the dark for 18 h. The cut leaves were infiltrated with nitroblue tetrazolium solution (Fermentas, Lithuania) (1 mg/ml in 10 mM KH2PO4, pH 7.8) or with the same solution supplemented with DCMU (20 µM); the samples were kept in the dark or at the light (100 µmol·m–2·s–1) for 1 h and then washed free of chlorophyll in 70% ethanol at 80°C and photographed.
Extraction of RNA. RNA was extracted from the plant material with TRI-Reagent (Sigma-Aldrich) according to the producer’s protocol. The material was homogenized with TRI-Reagent in a TissueLyser II homogenizer (Qiagen, USA) for 2 min at 30 oscillations per second. Proteins were denatured with 1-bromo-3-chloropropane (Sigma-Aldrich). Nucleic acids were precipitated overnight with 2.5 volumes of 96% ethanol in the cold. Then the samples were centrifuged at 14,000g and 4°C for 10 min, the nucleic acid precipitate was dried at room temperature, dissolved in 25-40 µl of deionized sterile water, and used for synthesis of the first chain of cDNA. The quantity and quality of the isolated RNA was monitored by electrophoresis in 1% agarose under nondenaturing conditions.
Real-time reverse transcriptase PCR. The first chain of cDNA was synthesized using RNA pretreated with DNase I (Fermentas, Lithuania) as a template. The synthesis was realized using oligo(dT)15 primer and reverse transcriptase RevertAid H Minus M-MuLV (Fermentas) according to the producer’s protocol. The real-time reverse transcriptase PCR was conducted using the SYBR Select Master Mix mixture of reagents (Applied Biosystems, USA) and the C1000 Thermal Cycler CFX96 Real-Time System (Bio-Rad, USA). The reaction mixture volume was 10 µl. PCR was performed as follows: heating to 50°C for 2 min (as recommended by the producer), one denaturation cycle (95°C, 3 min), 36 amplification cycles (95°C for 20 s, 60°C for 30 s, 72°C for 30 s). The gene yls8 was used as a reference. The following primers were used: for the yls8 gene (AT5G08290) R – TGTCCTTGAGAGCCCAGTTGAT, L – GAGGTGCTTGCGTCTGTTGCT, for the gr1 gene (AT3G24170) R – GCAAAATACAGCACAGGCGACAT, L – GTAGGAGATGCCACAAACCGAA, for the gr2 gene (AT3G54660) R – AGGTGGCTGGGAGAAAACGG, L – TCTGGGCTGTTGGGGATGTT, for the gdh2 gene (AT5G07440) R – CTCCTCCTGCGTTTGCGTAGA, L – CGCTCTTGGTGGTGTCCTGAA.
Glucose content was determined enzymatically with a Glucose (HK) Assay Kit (Sigma-Aldrich). A weighed sample of Arabidopsis green tissue (300 mg) was homogenized in liquid nitrogen, supplemented with two weights of deionized water, and incubated on a water bath for 30 min at 80°C. Then the sample was centrifuged for 10 min at 15,000g. The supernatant was used for determination of glucose content according to the producer’s protocol, the optical density of solutions being determined with a SmartSpec Plus spectrophotometer (Bio-Rad). Glucose concentration in the supernatant (mg/ml) was determined as (ΔA × TV × F × 180.2)/(6.22 × d × SV × 1000), where ΔA is the difference between the optical density of the samples and the reference solution; TV is the total volume of the samples, ml; F is the dilution factor; 180.2 is the molar weight of glucose, g/mol; SV is the supernatant volume in the samples, ml; 6.22 is the molar absorption coefficient of NADH at 340 nm; d is the optical pathlength (1 cm); 1000 is the coefficient of µg calculation into mg. The result is presented in mg glucose per g wet weight of Arabidopsis tissues.
Data were processed statistically and figures were drawn on the quantitative determination of ROS using the Microsoft Excel program. The significance of differences was assessed using Student’s t-test at probability level p < 0.05. For the real-time reverse transcriptase PCR, the data were processed statistically and figures were drawn using CFX ManagerTM Software v.1.6 program (Bio-Rad). In all figures, bars represent standard deviations.
RESULTS
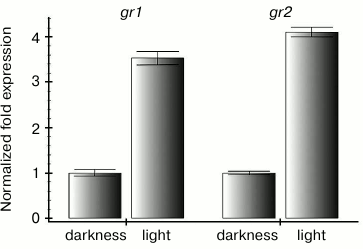
Maintaining a high level of reduced glutathione seemed to be especially important for green tissues of plants at light, because ROS are actively produced in photosynthesizing chloroplasts [17]. Therefore, it was reasonable to expect an increase in expression of genes encoding enzymes of the ascorbate-glutathione cycle when plants were transferred from darkness into light. In fact, we found that contents of transcripts of both genes encoding glutathione reductase of Arabidopsis increased 3-4-fold 4 h after the plants were transferred from darkness to the moderate light (Fig. 1).
Fig. 1. Differences in levels of gr1 and gr2 gene transcripts in Arabidopsis leaves under conditions of darkness and moderate light. Darkness: the plants were kept in the dark for 22 h; light: the plants were kept in the dark for 18 h and then under conditions of illumination (100 µmol·m–2·s–1) for 4 h.
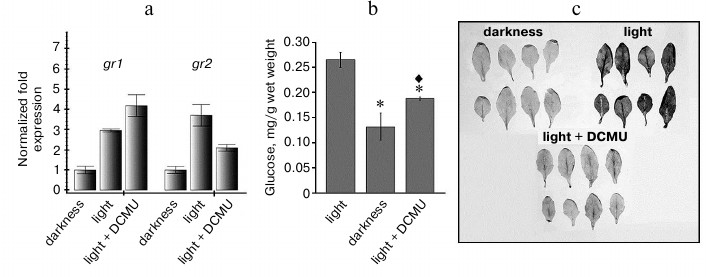
Such increase in expression could be caused by various events, in particular, by an increase in ROS generation and by changes in the redox state of the chloroplast electron transport chain components. To study the possible involvement of photosynthetic apparatus components in the regulation of expression of the studied genes, we applied the inhibitor of photosynthesis 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), which is routinely used in such experiments. DCMU binds with the QB-site of photosystem II (PS II) and thus inhibits electron transfer from PS II to plastoquinone of the thylakoid membrane, thus passing the plastoquinone thylakoid pool into the oxidized state. Hence, under conditions of moderate light, DCMU fixed the plastoquinone pool in the oxidized state [18].
We found a correlation between the level of the gr2 gene transcripts and the redox state of the photosynthetic chain part downstream PS II: the oxidized state of this part corresponded to lower level of the expression of this gene, whereas the reduced state correlated with its higher level (Fig. 2a). Successive increase in the level of gr1 gene transcripts was observed on the following changes in conditions: control in the dark – control at light – DCMU (Fig. 2a), i.e. the gr1 gene expression level did not correlate with the redox state of this region of the chain. Obviously, under conditions of our experiments, expressions of the gr1 and gr2 genes were regulated independently. It should be noted that the level of the gr2 gene transcripts in the presence of DCMU did not decrease to the level characteristic for darkness. The inhibitory effect of DCMU on photosynthesis was confirmed by a decrease in the glucose level in Arabidopsis green tissues in the presence of the inhibitor (Fig. 2b).
Inhibition of electron transport chains nearly inevitably leads to changes in ROS level in the corresponding organelles and then in the whole cell. However, in chloroplasts under light conditions not only various regions of the electron transport chain can be sources of ROS, but also components of the stroma [19], and this makes it difficult to accurately predict how the total cellular ROS level will change under particular conditions of illumination in the presence of inhibitors. To elucidate whether the changes in expression of the studied genes were associated with the level of ROS, and in particular, of superoxide radical, we determined the superoxide radical content during the treatment of Arabidopsis leaves with DCMU. We found that under our conditions, just the redox state of the chain components located downstream the PS II determined the superoxide radical content in the cells: when this region was oxidized (in darkness and during treatment with DCMU), the superoxide radical content was low, but it was high at the reduced state of this region during illumination (Fig. 2c).
Fig. 2. Expression of gr1 and gr2 genes and level of glucose and of ROS in Arabidopsis leaves in the presence of DCMU. a) Levels of gr1 and gr2 transcripts. Darkness: plants were kept in dark for 22 h; light – plants were kept at light (100 µmol·m–2·s–1) for 4 h; light + DCMU – plants were kept at light in the presence of 20 µM DCMU for 4 h. b) Glucose level in leaves. Light – plants were kept at light (100 µmol·m–2·s–1) for 4 h; darkness – plants were kept in dark for 22 h; light + DCMU – plants were kept at light in the presence of 20 µM DCMU for 4 h. Signs * and ♦ indicate significant difference at probability level p < 0.05: * this variant is different from control at light; ♦ this variant is different from control in the dark. c) ROS level in leaves. Darkness – leaves were infiltrated with nitroblue tetrazolium solution and kept in the dark for 1 h; light – leaves were kept at light (100 µmol·m–2·s–1) for 1 h; light + DCMU – leaves were infiltrated with nitroblue tetrazolium solution + 20 µM DCMU and kept at light (100 µmol·m–2·s–1) for 1 h.
Thus, the changes in gr2 gene expression correlate not only with the redox state of the thylakoid membrane components, but also with the superoxide radical level in the leaves: higher expression corresponds to higher level of superoxide radical in the leaves (Fig. 2, a and c). The profile of gr1 gene expression does not correlate either with the redox state of the photosynthetic electron transport chain or with the superoxide radical level (Fig. 2, a and c). Thus, the level of gr1 transcripts in the variant “Light + DCMU” was fourfold higher than in the dark, whereas these variants were not significantly different in superoxide radical level. Thus, on the transition from darkness to light, gr1 gene expression was induced not by an increase in the superoxide radical level, but by some other factor(s).
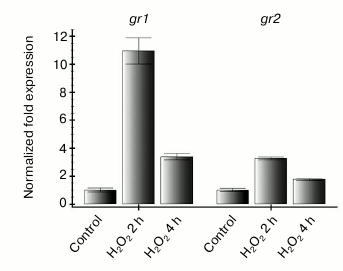
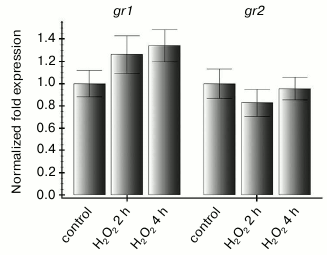
Therefore, it was interesting to learn how expression of the studied genes changes on increase in ROS level that is not associated with functioning of the photosynthetic electron transport chain. To determine this, leaves of intact Arabidopsis plants were treated with 10 mM hydrogen peroxide and then kept in the dark for 2 and 4 h. Such treatment resulted in an increase in the level of transcripts of both studied genes 2 h after beginning the treatment and in decrease to the initial level after 4 h (Fig. 3). Thus, we supposed that the dependence of gr2 gene expression on the redox state of the photosynthetic electron transport chain should be mediated through ROS. In our experiments with inhibitors of photosynthesis, the ROS level depended on the redox state of the thylakoid membrane components.
Fig. 3. Effect of exogenous hydrogen peroxide on gr1 and gr2 gene expression in Arabidopsis leaves. The experiment was performed in the dark. Control — leaves of intact plants were treated with sterile water; H2O2 — leaves were treated with 10 mM hydrogen peroxide and incubated for 2 and 4 h.
Expression of the gr1 gene was also induced after plant leaves were treated with hydrogen peroxide in the dark (Fig. 3), although at light we observed no association of the expression of this gene with ROS level (Fig. 2, a and c).
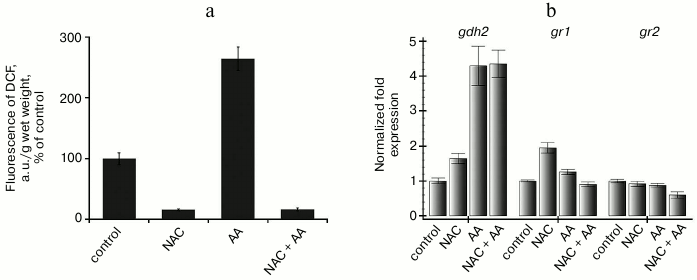
To prevent the possible influence of chloroplasts on changes in gr1 and gr2 gene expression on changes in ROS content, we performed some experiments with a heterotrophic suspension cell culture of Arabidopsis. The cells of this culture are lacked differentiated chloroplasts and incapable of autotrophic nutrition even after being transferred into light conditions. In our experiments, the culture was maintained in complete darkness, which prevented any influence not only of chloroplasts, but also even of photoreceptors. The ROS content and the redox state of the mitochondrial electron transport chain in the Arabidopsis suspension culture cells was modeled using an inhibitor of respiratory complex III, antimycin A, and a scavenger of ROS, N-acetylcysteine. Antimycin A inhibits electron transfer from the ubiquinone pool of the inner mitochondrial membrane to respiratory complex III, which leads to an over-reduction of the respiratory chain complex components located before the site of action of the inhibitor and to an increase in ROS generation [20]. In fact, the hydrogen peroxide content in the suspension culture after treatment with antimycin A was 2-3 times higher than in the control, and in the presence of N-acetylcysteine (combined with antimycin A and without it) was 5-6 times lower than the control (Fig. 4a). The level of gr1 gene transcripts increased slightly in the presence of N-acetylcysteine (without antimycin A) and was unchanged in the other cases. The level of the gr2 gene transcripts did not change in all variants of the experiment (Fig. 4b).
Fig. 4. ROS content and expression of the gdh2, gr1, and gr2 genes in cells of Arabidopsis suspension cell culture on inhibition of respiratory complex III. a) Assessment of ROS content by fluorescence of dichlorofluorescein; b) level of transcripts of gdh2, gr1, and gr2 genes. Control – the cell culture was supplemented with sterile water with corresponding concentration of ethanol; NAC – N-acetylcysteine (2 mM) for 4.5 h; AA – antimycin A (10 µM) for 4 h; (NAC+AA) – N-acetylcysteine (2 mM) for 30 min and then antimycin A (10 µM) for 4 h.
We showed earlier that expression of the gdh2 gene encoding the β-subunit of glutamate dehydrogenase of Arabidopsis is induced under conditions leading to reduction of the ubiquinone pool but does not depend on the ROS content in the cell [21]. In that series of experiments, the level of the gdh2 gene transcripts increased 4-6 times after treatment of the cells with antimycin A for 4 h, and the same effect was observed on addition of N-acetylcysteine (Fig. 4b). In the same experiments, levels of the gr1 and gr2 gene transcripts were not changed under the influence of antimycin A and seemed to have no dependence on either the redox sate of the mitochondrial electron transport chain or on the hydrogen peroxide content (Fig. 4, a and b). Treatment of the suspension culture cells with 10 mM hydrogen peroxide for 2 and 4 h did not have a pronounced influence on the expression character of the studied genes (Fig. 5). Thus, expression of both genes of Arabidopsis glutathione reductase in heterotrophic suspension culture cells did not depend on the hydrogen peroxide content in the cells.
Fig. 5. Effect of exogenous hydrogen peroxide on the gr1 and gr2 gene expression in suspension of Arabidopsis culture cells. Control – the culture was supplemented with sterile water; H2O2 – addition of 10 mM hydrogen peroxide and incubation for 2 and 4 h.
DISCUSSION
Both genes encoding glutathione reductases of Arabidopsis were induced by light when the plants were transferred from darkness into conditions of illumination (Fig. 1). As far as we know, this observation confirmed in databases of results of microarray experiments (e.g. www.genevestigator.com) was not reported earlier. It seems that such neglect can be explained by changes in expression of a huge number of plant genes (by some assessments up to 30% of the transcriptome [22]) on transition from darkness to light. This light-inducibility is due to various causes, including changes in the levels of metabolites (in particular, of sugars [23]), and changes in redox state of chloroplasts [24], mitochondria [25], and of the whole cell [26]. Some specific plastid-to-nucleus signaling cascades [27] and processes mediated through photoreceptors [28] can also contribute to light inducibility. It is interesting that the level of transcripts of both studied genes increased nearly equally when the plants were transferred from darkness into the light (Fig. 1). This contradicts the literature data on the lower sensitivity of the gr2 gene to changes in the environment [1, 12, 13]. However, these works described behavior of the glutathione reductase genes under stress conditions when the expression level at light and in the absence of the stress factor was taken as control. The change from complete darkness to conditions of moderate illumination (100 µmol·m–2·s–1 in our experiments) cannot be considered a stress factor. There are studies of metabolic and transcriptional responses of plants to transitions from light to darkness and from darkness to light [22, 23, 29], but the authors did not focus their attention on changes in expression of the genes that are traditionally thought to be associated with stress reactions of the cell. Thus, the absence of attention to reactions of the antioxidant defense genes on transitions from light to darkness and from darkness to light resulted in the concept that expression of genes encoding plastid-mitochondrial forms of plant glutathione reductases is poorly inducible or non-inducible [1]. Thus, it was still more interesting to elucidate what mechanisms are responsible for the transmission into the nucleus of information about the functional state of the chloroplast and finally led to induction of expression of the nuclear gr2 gene on the transition from darkness to light.
ROS [30] and changes in the redox state of the thylakoid membrane [24] are two main candidates for the role as a source of the primary signal triggering the induction of nuclear gene expression on changes in illumination. The first mechanism is usually thought to be associated with the regulation of expression of antioxidant and stress-inducible genes, whereas the second pathway in the majority of cases regulates expression of genes functionally associated with photosynthesis [31]. Glutathione reductase is an antioxidant and stress-inducible enzyme [3], but, on the other hand, the content of intrachloroplast reduced glutathione seems to be one of the most important metabolic factors influencing the efficiency of photosynthesis [32].
Our experiments showed for the first time that changes in expression of the gr2 gene encoding chloroplast-mitochondrial glutathione reductase of Arabidopsis were in good correlation with changes in the redox state of the photosynthetic electron transport chain components and in the content of reactive oxygen species during the treatment with the photosynthesis inhibitor DCMU (Fig. 2, a and c). Up to now, such regularities have been observed for genes encoding components of the electron transport chain of chloroplasts, the gene of the small subunit RUBISCO and genes of chlorophyll-binding proteins of the subfamily ELIP (early light inducible proteins), i.e. for genes whose products are immediately connected with the photosynthetic apparatus and photosynthesis [31], and also for the apx2 gene encoding cytosolic ascorbate peroxidase [30]. It should be noted that in all cases the redox state of the electron transport chain of chloroplasts was not the only factor determining the expression level of the corresponding genes. Thus, expression of the lhcb 1 gene, which is one of nuclear genes with expression depending on the redox state of plastoquinone [18], concurrently depends on signals mediated by sugars [33], abscisic acid [34], photoreceptors [35], and hydrogen peroxide [36]. Moreover, expression of this gene is subordinated to the internal circadian rhythm, independently of the light intensity [37].
Based on results of our experiments, we supposed that the gr2 gene expression at light should be controlled by the ROS level, which changed on change in the redox state of the thylakoid membrane components in chloroplasts. Such a mechanism was proposed for the apx2 gene encoding the cytosolic ascorbate peroxidase, which is a rare example of a gene whose product is neither a component of the photosynthetic apparatus nor an enzyme of the Calvin cycle. Nevertheless, expression of this gene changes in correspondence with changes in the redox state of the plastoquinone pool [30]. (Note that ascorbate peroxidase, as well as glutathione reductase, is a component of the ascorbate-glutathione cycle). However, it was shown in 2011 that the association between apx2 gene expression and the redox state of the plastoquinone pool was mediated through the level of 5′-phosphoadenosine 3′-phosphate, which, in turn, inhibited nuclear exoribonucleases acting as negative regulators of some stress-inducible genes [38]. Certainly, the mechanism of the light-dependent regulation of the gr2 gene expression unassociated with stress needs further studies. However, results of the present work show that the chloroplast-to-nucleus regulatory signals play an important role in this regulation.
It seems that up to now there was only one attempt to study the possible involvement of retrograde plastid-to-nucleus signals in the induction of some genes encoding antioxidant enzymes of Arabidopsis (including the cytosolic glutathione reductase) under conditions of excess light [26]. It was concluded that neither signals mediated by abscisic acid nor ROS or the redox state of plastoquinone could play a significant role in providing the transcriptional response of the studied genes during at least 6 h after the transfer of leaf pieces under conditions of excess and extreme illumination. It was thought that the major role in the regulation of the transcriptional response under conditions of excess light should belong to the balance of major redox pairs of the cell and to lipid and other metabolic signals [26].
In our experiments, expression of the gr1 gene encoding the cytosolic glutathione reductase also did not correlate with either the redox state of the thylakoid membrane components or with ROS level on treatment of leaves with photosynthesis inhibitors (Fig. 2, a and c). Obviously, expression of the gr1 and gr2 genes was regulated by different mechanisms, which is in agreement with the literature data on the different accumulation of EST for these two genes under conditions of abiotic stress [1]. Although on treatment with photosynthesis inhibitors at light the gr1 gene expression did not correlate with the ROS level, on treatment of leaves with hydrogen peroxide in the dark the level of the gr1 gene transcripts increased even more strongly than that of gr2 (Fig. 3). Therefore, we suppose that the true signal for changing the expression of the gr1 gene (or expression of both studied genes) during treatment with hydrogen peroxide could be not the content itself of intracellular ROS but consequences of its sharp increase mediated by the chloroplasts of the cells. In fact, in heterotrophic culture expression of the gr1 and gr2 genes was not induced after the treatment with hydrogen peroxide (Fig. 5).
It is known that chloroplasts in the plant cell are involved not only in photosynthesis; such processes as synthesis and desaturation of fatty acids, nitrogen assimilation, biosynthesis of some amino acids, synthesis of pigments and tetrapyrrole compounds, isoprenoids, riboflavin and its derivatives, 3′-phosphoadenosine 5′-phosphate occur in chloroplasts [35]. Among the above-mentioned compounds, expression of nuclear genes is subjected to regulatory influence of intermediates of biosynthesis of tetrapyrroles and lipids, 3′-phosphoadenosine 5′-phosphate [27, 31]. The importance of completely differentiated chloroplasts for response of the plant cell to cold stress was clearly demonstrated by Dal Bosco and colleagues. Using barley mutants with defects in chloroplast biogenesis, they demonstrated that the induction of expression of some cold-inducible genes depended on the stage of plastid differentiation and was completely abolished in the chloroplast-deprived mutant albino [39].
It is likely that an increase in expression of glutathione reductase genes in green leaves of Arabidopsis in response to treatment with hydrogen peroxide reflects a complex reaction of the cell mediated through metabolic activity of the chloroplasts. Therefore, we consider very interesting the difference in the transcriptional responses of Arabidopsis cells with and without differentiated chloroplasts under conditions preventing functioning of the photosynthetic chain. We showed earlier that the cells of suspension cultures obtained from transgenic Arabidopsis plants with an increased and a decreased activity of the mitochondrial alternative oxidase have changed levels of ROS as compared to cultures obtained from wild-type plants, but this regularity was not observed for the plants themselves with increased and decreased activity of the alternative oxidase [40]. We have shown in the present work that the presence in the cells of differentiated chloroplasts has not only metabolic consequences, but also influences gene expression, including in the absence of electron transport in the photosynthetic chain. It is obvious that in our experiments just chloroplasts and not mitochondria acted as the major mediator modulating expression of the glutathione reductase genes. These data are also in agreement with a published hypothesis [26] that chloroplast-to-nucleus metabolic signals should be involved in the regulation of genes of plant antioxidant defense.
In this work, equipment of the Baikal Analytic Center of the Siberian Branch of the Russian Academy of Sciences was used, as well as equipment of the Center of Collective Use Phytotron of the Siberian Institute of Plant Physiology and Biochemistry, Siberian Branch of the Russian Academy of Sciences.
This work was supported by the Russian Foundation for Basic Research (project No. 14-44-04001).
REFERENCES
1.Tahmasebi, A., Aram, F., Ebrahimi, M.,
Mohammadi-Dehcheshmeh, M., and Ebrahimie, E. (2012) Genome-wide
analysis of cytosolic and chloroplastic isoforms of glutathione
reductase in plant cells, Plant Omics J., 5, 94-102.
2.Noctor, G., Queval, G., Mhamdi, A., Chaouch, S.,
and Foyer, C. H. (2011) Glutathione, The Arabidopsis Book, doi:
10.1199/tab.0142.
3.Chalapathi Rao, A. S. V., and Reddy, A. R. (2008)
Glutathione reductase: a putative redox regulatory system in plant
cells, in Sulfur Assimilation and Abiotic Stresses in Plants
(Khan, N. A., Singh, S., and Umar, S., eds.) Springer, The Netherlands,
pp. 111-147.
4.Creissen, G. P., Reynolds, H., Xue, Y., and
Mullineaux, P. M. (1995) Cloning and characterization of glutathione
reductase cDNAs and identification of two genes encoding the tobacco
enzyme, Plant J., 8, 167-175.
5.Stevens, R., Creissen, G. P., and Mullineaux, P. M.
(1997) Cloning and characterization of a cytosolic glutathione
reductase cDNA from pea (Pisum sativum L.) and its expression in
response to stress, Plant Mol. Biol., 35, 641-654.
6.Rouhier, N., Couturier, J., and Jacquot, J. (2006)
Genome-wide analysis of plant glutaredoxins systems, J. Exp.
Bot., 57, 1685-1696.
7.Romero-Puertas, M. C., Corpas, F. J., Sandalio, L.
M., Leterrier, M., Rodriguez-Serrano, M., Del Rio, L. A., and Palma, J.
M. (2006) Glutathione reductase from pea leaves: response to abiotic
stress and characterization of peroxisomal isozyme, New Phytol.,
170, 43-52.
8.Lee, H., Won, S.-H., Lee, B.-H., Park, H.-D.,
Chung, W.-I., and Jo, J. (2002) Genomic cloning and characterization of
glutathione reductase gene from Brassica campestris var.
Pekinensis, Mol. Cells, 13, 245-251.
9.Dghim, A. A., Mhamdi, A., Vaultier, M.-N.,
Hasenfratz-Sauder, M.-P., Le Thiec, D., Dizengremel, P., Noctor, G.,
and Jolivet, Y. (2013) Analysis of cytosolic isocitrate dehydrogenase
and glutathione reductase 1 in photoperiod-influenced responses to
ozone using Arabidopsis knockout mutants, Plant Cell
Environ., 36, 1981-1991.
10.Kaminaka, H., Morita, S., Nakajima, M., Masumura,
T., and Tanaka, K. (1998) Gene cloning and expression of cytosolic
glutathione reductase in rice (Oryza sativa L.), Plant Cell
Physiol., 39, 1269-1280.
11.Contour-Ansel, D., Torres-Franklin, M. L., Cruz,
D. E., Carvalho, M. H., D’Arcy-Lameta, A., and Zuily-Fodil, Y.
(2006) Glutathione reductase in leaves of cowpea: cloning of two cDNAs,
expression and enzymatic activity under progressive drought stress,
desiccation and abscisic acid treatment, Ann. Bot., 98,
1278-1287.
12.Trivedi, D. P., Gill, S. S., Yadav, S., and
Tuteja, N. (2013) Genome-wide analysis of glutathione reductase (GR)
genes from rice and Arabidopsis, Plant Signal. Behav.,
8, e23201.
13.Belin, C., Bashandy, T., Cela, J.,
Delorme-Hinoux, V., Riondet, C., and Reichheld, J. P. (2015) A
comprehensive study of thiol reduction gene expression under stress
conditions in Arabidopsis thaliana, Plant Cell Environ.,
38, 299-314.
14.Murashige, T., and Skoog, F. (1962) A revised
medium for rapid growth and bioassays with tobacco tissue cultures,
Physiol. Plant., 15, 473-497.
15.Meyer, E. H., Tomaz, T., Carroll, A. J.,
Estavillo, G., Delannoy, E., Tanz, S. K., Small, I. D., Pogson, B. J.,
and Millar, A. H. (2009) Remodeled respiration in ndufs4 with low
phosphorylation efficiency suppresses Arabidopsis germination and
growth and alters control of metabolism at night, Plant Physiol., 151,
603-619.
16.Maxwell, D. P., Wang, Y., and McIntosh, L. (1999)
The alternative oxidase lowers mitochondrial reactive oxygen production
in plant cells, Proc. Natl. Acad. Sci. USA, 96, 8271-8276.
17.Ivanov, B. N. (2014) Role of ascorbic acid in
photosynthesis, Biochemistry (Moscow), 79, 282-269.
18.Pfannschmidt, T., Shcutze, K., Brost, M., and
Oelmuller, R. (2001) A novel mechanism of nuclear photosynthesis gene
regulation by redox signals from the chloroplast during photosystem
stoichiometry adjustment, J. Biol. Chem., 276,
36125-36130.
19.Ivanov, B. N., Khorobrykh, S. A., Kozuleva, M.
A., and Borisova-Mubarakshina, M. M. (2013) The role of oxygen and its
reactive species in photosynthesis, in Photosynthesis: Questions to
Answer and What We Know Today (Allakhverdiev, S. I., Rubin, A. B.,
and Shuvalov, V. A., eds.) [in Russian], Institute of Computer-Aided
Studies, Izhevsk, pp. 243-298.
20.Zubo, Y. O., Potapova, T. V., Yamburenko, M. V.,
Tarasenko, V. I., Konstantinov, Yu. M., and Borner, T. (2014)
Inhibition of the electron transport strongly affects transcription and
transcript levels in Arabidopsis mitochondria,
Mitochondrion, 19, 222-230.
21.Tarasenko, V. I., Garnik, E. Yu., Shmakov, V. N.,
and Konstantinov, Yu. M. (2009) Induction of Arabidopsis gene
gdh2 expression during changes in redox state of the
mitochondrial respiratory chain, Biochemistry (Moscow),
74, 47-53.
22.Rasmusson, A. G., and Escobar, M. A. (2007) Light
and diurnal regulation of plant respiratory gene expression,
Physiol. Plant., 129, 57-67.
23.Blasing, O. E., Gibon, Y., Gunter, M., Morcuende,
R., Osuna, D., Thimm, O., Usadel, B., Scheibe, W.-R., and Stitt, M.
(2005) Sugars and circadian regulation make major contributions to the
global regulation of diurnal gene expression in Arabidopsis, Plant Cell
Physiol., 17, 3257-3281.
24.Glasser, C., Haberer, G., Finkemeier, I.,
Pfannschmidt, T., Kleine, T., Leister, D., Dietz, K. J., Hausler, R.
E., Grimm, B., and Mayer, K. F. (2014) Meta-analysis of retrograde
signaling in Arabidopsis thaliana reveals a core module
of genes embedded in complex cellular signaling networks, Mol.
Plant, 7, 1167-1190.
25.Yoshida, K., Terashima, I., and Noguchi, K.
(2011) How and why does the mitochondrial respiratory chain respond to
light? Plant Signal. Behav., 6, 864-866.
26.Oelze, M. L., Vogel, M. O., Alsharafa, K.,
Kahmann, U., Viehhauser, A., Maurino, V. G., and Dietz, K. J. (2012)
Efficient acclimation of the chloroplast antioxidant defense of
Arabidopsis thaliana leaves in response to a 10- or 100-fold
light increment and the possible involvement of retrograde signals,
J. Exp. Bot., 63, 1297-1313.
27.Estavillo, G. M., Chan, K. X., Phua, S. Y., and
Pogson, B. J. (2013) Reconsidering the nature and mode of action of
metabolite retrograde signals from the chloroplast, Front. Plant Sci.,
doi: 10.3389/fpls.2012.00300.
28.Yurina, N. P., Mokerova, D. V., and Odintsova, M.
S. (2013) Light-induced stress proteins of phototrophs, Russ. J.
Plant Physiol., 60, 577-589.
29.Lee, C. P., Eubel, H., and Millar, A. H. (2010)
Diurnal changes in mitochondrial function reveal daily optimization of
light and dark respiratory metabolism in Arabidopsis, Mol. Cell.
Proteom., 9, 2125-2139.
30.Pogson, B. J., Woo, N. S., Forster, B., and
Small, I. D. (2008) Plastid signaling to the nucleus and beyond,
Trends Plant Sci., 13, 602-609.
31.Chi, W., Sun, X., and Zhang, L. (2013)
Intracellular signaling from plastid to nucleus, Annu. Rev. Plant
Biol., 64, 559-582.
32.Ogawa, K., Hatano-Iwasaki, A., Yanagida, M., and
Iwabuchi, M. (2004) Level of glutathione is regulated by ATP-dependent
ligation of glutamate and cysteine through photosynthesis in
Arabidopsis thaliana: mechanism of strong interaction of light
intensity with flowering, Plant Cell Physiol., 45, 1-8.
33.Cottage, A., Mott, E. K., Kempster, J. A., and
Gray, J. C. (2010) The Arabidopsis plastid-signaling mutant gun1
(genomes uncoupled1) shows altered sensitivity to sucrose and abscisic
acid and alterations in early seedling development, J. Exp. Bot., 61,
3773-3786.
34.Liu, R., Xu, Y.-H., Jiang, S.-C., Lu, K., Lu,
Y.-F., Feng, X.-J., Wu, Z., Liang, S., Yu, Y.-T., Wang, X.-F., and
Zhang, D.-P. (2013) Light-harvesting chlorophyll
a/b-binding proteins, positively involved in abscisic
acid signaling, require a transcription repressor, WRKY40, to balance
their function, J. Exp. Bot., 64, 5443-5456.
35.Waters, M. T., and Langdale, J. A. (2009) The
making of a chloroplast, EMBO J., 28, 2861-2873.
36.Borisova-Mubarakshina, M. M., Ivanov, B. N.,
Vetochkina, D. V., Lubimov, V. Y., Fedorchuk, T. P., Naydov, I. A.,
Kozuleva, M. A., Rudenko, N. N., Dall’Osto, L., Cazzaniga, S.,
and Bassi, R. (2015) Long-term acclimatory response to excess
excitation energy: evidence for a role of hydrogen peroxide in the
regulation of photosystem II antenna size, J. Exp. Bot., doi:
10.1093/jxb/erv410.
37.Xu, Y., and Johnson, C. H. (2001) A clock- and
light-regulated gene that links the circadian oscillator to LHCB
gene expression, Plant Cell, 13, 1411-1425.
38.Estavillo, G. M., Crisp, P. A., Pornsiriwong, W.,
Wirtz, M., Collinge, D., Carrie, C., Giraud, E., Whelan, J., David, P.,
Javot, H., Brearley, C., Hell, R., Marin, E., and Pogson, B. J. (2011)
Evidence for a SAL1-PAP chloroplast retrograde pathway that functions
in drought and high light signaling in Arabidopsis, Plant Cell, 23,
3992-4012.
39.Dal Bosco, C., Busconi, M., Govoni, C., Baldi,
P., Stanca, A. M., Crosatti, C., Bassi, R., and Cattivelli, L. (2003)
Cor gene expression in barley mutants affected in chloroplast
development and photosynthetic electron transport, Plant Physiol., 131,
793-802.
40.Tarasenko, V. I., Garnik, E. Yu., Shmakov, V. N.,
and Konstantinov, Yu. M. (2012) Modified alternative oxidase expression
results in different reactive oxygen species content in Arabidopsis
cell culture but not in whole plants, Biol. Plant., 56, 635-640.