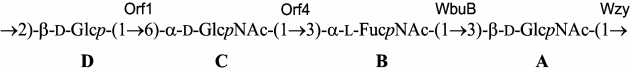Structure and Biosynthesis Gene Cluster of the O-Antigen of Escherichia coli O12
S. N. Senchenkova1, Yuanyuan Zhang2, A. V. Perepelov1, Xi Guo2, A. S. Shashkov1, Bin Liu2, and Yu. A. Knirel1*
1Zelinsky Institute of Organic Chemistry, Russian Academy of Sciences, 119991 Moscow, Russia; fax: +7 (499) 135-5328; E-mail: yknirel@gmail.com2Nankai University, TEDA Institute of Biological Sciences and Biotechnology, 300457 Tianjin, P. R. China; E-mail: striker198126@aliyun.com
* To whom correspondence should be addressed.
Received October 5, 2015; Revision received November 21, 2015
Two polysaccharides were isolated from Escherichia coli O12, the major being identified as the O12-antigen and the minor as the K5-antigen. The polysaccharides were studied by sugar analysis, Smith degradation, and one- and two-dimensional 1H and 13C NMR spectroscopy. As a result, the following structure of the O12-polysaccharide was elucidated, which, to our knowledge, has not been hitherto found in bacterial carbohydrates: →2)-β-d-Glcp-(1→6)-α-d-GlcpNAc-(1→3)-α-l-FucpNAc-(1→3)-β-d-GlcpNAc-(1→. The →4)-β-d-GlcpA-(1→4)-α-d-GlcpNAc-(1→ structure established for the K5-polysaccharide (heparosan) is previously known. Functions of genes in the O-antigen biosynthesis gene cluster of E. coli O12 were assigned by comparison with sequences in the available databases and found to be consistent with the O12-polysaccharide structure.
KEY WORDS: Escherichia coli, O-polysaccharide structure, O-antigen gene cluster, glycosyltransferaseDOI: 10.1134/S0006297916040106
Abbreviations: ABC, ATP-binding cassette; COSY, correlation spectroscopy; FucNAc, 2-acetamido-2-deoxyfucose; GlcA, glucuronic acid; GlcNAc, 2-acetamido-2-deoxyglucose; HSQC, heteronuclear single-quantum coherence; ROESY, rotating-frame nuclear Overhauser effect spectroscopy; TOCSY, total correlation spectroscopy.
Escherichia coli bacteria are the predominant species in the
human intestinal microflora. They include both pathogenic and commensal
strains and are one of the most common causes of diarrheal diseases [1]. Escherichia coli isolates are classified
into serotypes by a combination of their O-, H-, and sometimes
K-antigens, which together define the serological specificity of the
bacteria. The O-antigen represents a polysaccharide chain of the
lipopolysaccharide (O-polysaccharide) and the K-antigen is a capsular
polysaccharide. Antigen diversity helps survival of various bacterial
clones in their specific niche. The surface antigens also are an
important virulence factor, and their loss makes many pathogens serum
sensitive or otherwise seriously impaired in virulence. Specific genes
involved in their biosynthesis are generally localized in clusters on
the chromosome [2, 3], whose
variations define the antigenic diversity of bacteria to a large
extend. Currently, more than 180 E. coli O-serogroups are
recognized [4].
This work was performed in the framework of a project devoted to elucidation of the structural and genetic diversity of E. coli O-antigens. Data on such a large number of various clones of one species are expected to shed light on the evolution history of the O-antigen diversification of enteric bacteria, to enable assignment of functions of genes involved in the O-antigen biosynthesis, including hundreds of glycosyltransferases, and improvement of classification scheme of E. coli isolates necessary for diagnostics of infectious diseases, epidemiological monitoring, and elaboration of vaccines against pathogenic strains. By now, O-antigen structures have been elucidated for most of the O-serogroups (http://nevyn.organ.su.se/ECODAB/), but some remain unknown. Here we report data on the O-antigen of E. coli O12, which belonged to one of the O-serogroups that had not been investigated yet. The O12-polysaccharide structure was established, and the gene cluster for its biosynthesis was characterized. In addition to the O-polysaccharide, a minor polysaccharide was isolated from the bacterium and identified as the K5-antigen.
MATERIALS AND METHODS
Bacterial strain and growth. Escherichia coli O12 type strain (laboratory stock number G1280) was obtained from the Institute of Medical and Veterinary Science, Adelaide, Australia (IMVS). The bacterium was grown to late log phase in 8 liters of Luria–Bertani broth using a 10-liter fermenter (BIOSTAT C-10; B. Braun Biotech International, Germany) under constant aeration at 37°C and pH 7.0. Bacterial cells were washed and dried as described [5].
Isolation of the lipopolysaccharide and polysaccharides. The lipopolysaccharide was isolated in a yield 8.5% from bacterial cells by the phenol–water method [6], and to remove phenol, the crude extract was dialyzed without separation of the layers and freed from nucleic acids and proteins by treatment with 50% aqueous CCl3CO2H to pH 2.0 at 4°C. The supernatant was dialyzed and lyophilized.
Mild acid degradation of the lipopolysaccharide (150 mg) was performed with 2% aqueous HOAc at 100°C until precipitation of lipid (80 min). The precipitate was removed by centrifugation (13,000g, 20 min), and a mixture of polysaccharides (40 mg) was isolated by gel-permeation chromatography on a column (56 × 2.6 cm) of Sephadex G-50 Superfine (Amersham Biosciences, Sweden) in 0.05 M pyridinium acetate buffer, pH 4.5, monitored using a differential refractometer (Knauer, Germany).
Chemical analyses. A sample of the polysaccharides (0.5 mg) was hydrolyzed with 2 M CF3CO2H (120°C, 2 h). Monosaccharides were identified by GLC of the alditol acetates on a Maestro (Agilent 7820) chromatograph (Interlab, Russia) equipped with an HP-5 column (0.32 mm × 30 m) and a temperature program of 160°C (1 min) to 290°C at rate 7°C/min. The absolute configurations of the monosaccharides were determined by GLC of the acetylated (S)-2-octyl glycosides as described [7].
Smith degradation. A sample of the polysaccharides (20 mg) was oxidized with 0.1 M NaIO4 (2 ml) in the dark for 48 h at 20°C, reduced with an excess of NaBH4, and desalted by dialysis. The products were hydrolyzed with 2% aqueous HOAc for 2 h at 100°C and fractionated by gel-permeation chromatography on a column (85 × 2.5 cm) of TSK HW-40 (S) in aqueous 1% HOAc to give oligosaccharide 1 (5 mg) and glycoside 2 (3.5 mg).
NMR spectroscopy. Samples were deuterium-exchanged by freeze-drying from 99.9% D2O and then examined as solution in 99.95% D2O. NMR spectra were recorded on an Avance II 600 MHz spectrometer (Bruker, Germany) at 40°C for the polysaccharides or 25°C for compounds 1 and 2 using sodium 3-trimethylsilylpropanoate-2,2,3,3-d4 (δH 0, δC —1.6) as internal reference for calibration. Two-dimensional NMR spectra were obtained using standard Bruker software, and the Bruker TopSpin 2.1 program was used to acquire and process the NMR data. Mixing times of 100 and 150 ms were used in TOCSY and ROESY experiments, respectively.
RESULTS AND DISCUSSION
Elucidation of polysaccharide structures. Lipopolysaccharide was obtained from E. coli O12 cells by phenol–water extraction and degraded with mild acid to give a mixture of polysaccharides isolated by Sephadex G-50 gel-permeation chromatography. Sugar analysis using GLC of the alditol acetates after full acid hydrolysis of the polysaccharides revealed Glc, GlcNAc, and FucNAc. GLC analysis of the acetylated (S)-2-octyl glycosides demonstrated the d-configuration of Glc and GlcNAc and the l-configuration of FucNAc.
The 1H and 13C NMR spectra showed two series of signals, belonging to major and minor polysaccharides consisting of tetrasaccharide and disaccharide repeating units, respectively. In the major series, there were signals for four anomeric atoms, one CH3-C group (C6 of FucNAc), three HOCH2-C groups (C6 of hexoses), and three nitrogen-bearing carbons (C2 of amino sugars). In the minor series, there were signals for two anomeric atoms, one HOCH2-C group, and one nitrogen-bearing carbon. The 1H and 13C NMR chemical shifts are given in the table.
1H and 13C NMR chemical shifts (δ, ppm)
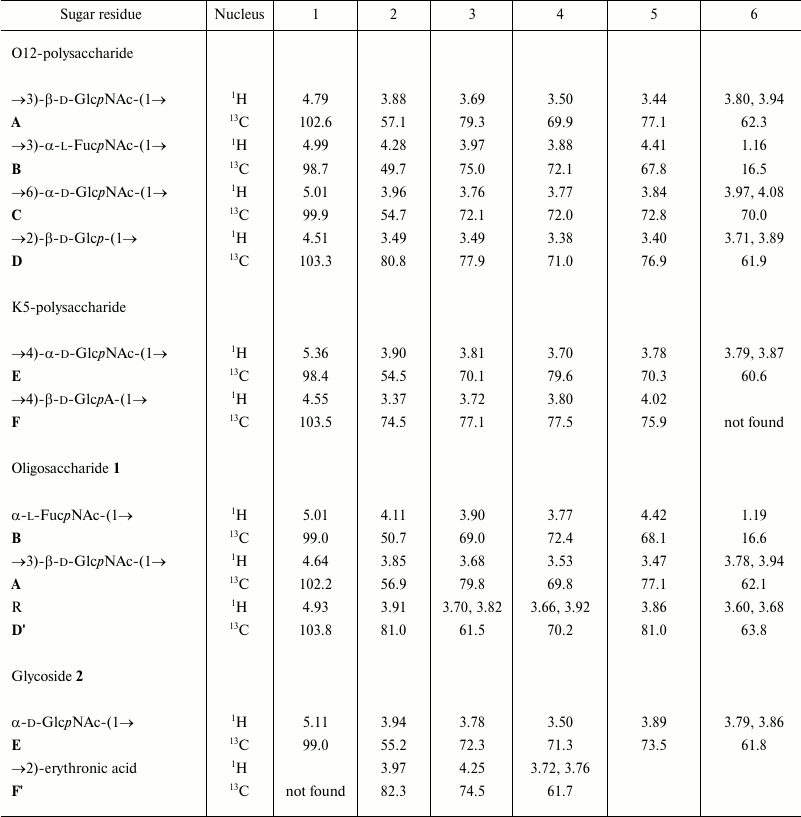
Note: Chemical shifts for the N-acetyl groups are for the K5- and
O12-polysaccharides: δH 1.97-2.04, δC
23.2-23.6 (Me) and 174.9-175.2 (CO); for oligosaccharide 1:
δH 1.98 and 2.04, δC 23.4, 23.5 (both
Me), 175.4 and 175.7 (both CO); for glycoside 2:
δH 2.04, δC 23.2 (Me) and 175.7
(CO).
The 1H and 13C NMR spectra of the polysaccharides were assigned using 2D 1H,1H COSY, TOCSY, and 1H,13C HSQC experiments, and spin systems for the major monosaccharides (β-GlcpNAc A, α-FucpNAc B, α-GlcpNAc C, β-Glcp D) and minor monosaccharides (α-GlcpNAc E, β-GlcpA F) were recognized by characteristic chemical shifts [8, 9] and 3JH,H coupling constants. Significant downfield displacements of the signals for C2 of unit D, C3 of units A and B, and C6 of unit C in the major series as well as C4 of units E and F in the minor series, as compared with their positions in the spectra of the corresponding non-substituted monosaccharides [8, 9], were due to glycosylation and enabled determination of the positions of substitution of sugar residues in the repeating units.
A two-dimensional ROESY spectrum of the polysaccharides showed correlations between the anomeric protons and protons at the linkage carbons, which were assigned as follows: in the major series, for D H1/C H6, C H1/B H3, B H1/A H3, A H1/D H2; in the minor series, for F H1/E H4, E H1/F H4. These data are in agreement with positions of substitution and define the sequences of the monosaccharides in the repeating units.
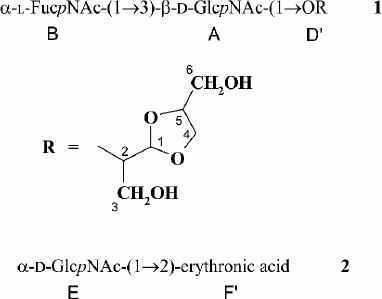
The polysaccharide structures were confirmed by Smith degradation, which resulted in destruction of the Glc and α-GlcNAc residues in the major polymer and the GlcA residue in the minor polymer. As a result, oligosaccharide 1 and glycoside 2 were obtained from the major and minor polysaccharides, respectively, and were separated by gel chromatography. Their structures shown in Fig. 1 were established by one- and two-dimensional NMR spectroscopy as described above and were consistent with the structures of the initial polysaccharides.
Fig. 1. Structures of Smith degradation products from the O-polysaccharide (1) and K-polysaccharide (2) of E. coli O12.
Based on the data obtained, it was concluded that the major polysaccharide from E. coli O12 has the structure shown in Fig. 2. Genetic analysis indicated that this polysaccharide represents the O-antigen of E. coli O12 (see below).
Fig. 2. Structure of the O-polysaccharide of E. coli O12 and assignment of glycosyltransferases to glycosidic linkages.
The minor polysaccharide from E. coli O12 has the structure of heparosan:
It is known as the capsular polysaccharide of E. coli (K5-antigen) [3, 10] and Pasteurella multocida type D [11]. Heparosan is a biosynthetic precursor of biologically important mammalian polysaccharides heparin and heparan sulfate [12].
Characterization of the O-antigen biosynthesis gene cluster. Most heteropolysaccharide O-antigens of E. coli are synthesized by the flippase Wzx/polymerase Wzy-dependent pathway, including the assembly of the O-unit on a phospholipid carrier on the inner side of the cytoplasmic membrane, its translocation through the membrane, and subsequent polymerization on the outer side of the membrane. Most genes involved in the O-antigen biosynthesis in E. coli and related bacteria are generally localized in a cluster on the chromosome between conserved galF and gnd genes [13].
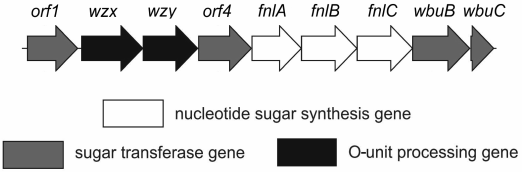
The O-antigen gene cluster of E. coli O12 (GenBank accession number AB811600) [2] includes nine genes (orfs1-9) having the same transcriptional direction from galF to gnd (Fig. 3). Proteins encoded by three genes (orfs5-7) share high level identity to FnlA, FnlB, and FnlC of E. coli O15, which have been identified as enzymes for the biosynthesis of UDP-l-FucNAc from UDP-d-GlcNAc [14]. Therefore, these genes are responsible for the synthesis of the nucleotide precursor of l-FucNAc in E. coli O12 too, and were named accordingly [2]. Genes for synthesis of the nucleotide precursors of common sugars, Glc and GlcNAc, are usually located outside the O-antigen gene cluster [15].
Fig. 3. Organization of the O-antigen gene cluster of E. coli O12.
From the remaining genes, two genes (orf2 and orf3) coded for transmembrane proteins Wzx (flippase) and Wzy (O-antigen polymerase), respectively, which are responsible for the O-antigen processing. The occurrence of wzx and wzy genes indicates that the O-polysaccharide is synthesized by translocation of the pre-assembled O-unit through the membrane followed by polymerization by the Wzx/Wzy-dependent pathway. Three genes (orfs1, 4, 8) encoded putative glycosyltransferases for the O-unit assembly, which is consistent with the tetrasaccharide O-unit.
The predicted protein Orf8 shares 96% identity and 98% similarity to glycosyltransferase WbuB of E. coli O4, which catalyzes formation of the α-l-FucpNAc-(1→3)-d-GlcNAc linkage [16]. This linkage is present in the O12-antigen too, and therefore, we propose that Orf8 has the same function and name it accordingly. When present in E. coli, the wbuB gene is accompanied by the wbuC gene at the 3′ end of the O-antigen gene cluster (Fig. 3) [2], whose function is unknown. Orf9 shares 97% identity and 98% similarity to WbuC of E. coli O4, and it is suggested that Orf9 be named WbuC.
Orf1 is 31% identical and 51% similar to predicted glycosyltransferase WfeQ of E. coli O164 [15] (GenPept accession number ACD37144.1). Orf4 shares 30% identity and 50% similarity to predicted glycosyltransferase WfbF of E. coli O123 [17] (GenPept accession number ABG81791.1). The polysaccharides of O164 [15, 18] and O123 [19] have the β-d-Glcp-(1→6)-α-d-Glcp and α-d-GlcpNAc-(1→3)-α-l-QuipNAc disaccharide fragments, which are similar to the β-d-Glcp-(1→6)-α-d-GlcpNAc and α-d-GlcpNAc-(1→3)-α-l-FucpNAc disaccharide fragments, respectively, of the O12-polysaccharide. Therefore, Orf1 is evidently responsible for the transfer of d-Glcp from UDP-d-Glcp to GlcpNAc and Orf4 for the transfer of d-GlcpNAc from UDP-d-GlcpNAc to l-FucpNAc (Fig. 2).
As in many other O-serogroups [20], GlcNAc is likely the first sugar of the O-unit of E. coli O12. The wecA gene initiates the O-antigen biosynthesis by transfer of GlcpNAc 1-phosphate from UDP-d-GlcpNAc to undecaprenyl phosphate (UndP) to give UndPP-α-d-GlcpNAc. The wecA gene is located in the enterobacterial common antigen gene cluster [21] and is not duplicated in the O-antigen gene cluster.
The K5-polysaccharide (K5-antigen) belongs to group 2 capsules, which are commonly expressed in pathogenic extraintestinal E. coli and closely resemble the capsules of Neisseria meningitidis and Haemophilus influenzae [22]. The biosynthesis of the K5 polysaccharide in E. coli is well studied both biochemically and genetically [3, 22]. Briefly, it occurs by the ABC transporter-dependent pathway through the sequential addition of GlcA and GlcNAc residues to the nonreducing end of the growing polysaccharide chain catalyzed by two glycosyltransferases, KfiA and KfiC. The mature polymer is exported across the inner membrane by the ABC transporter. The proteins involved are encoded by kfiABCD genes in the kps locus located near the serA gene [23].
Therefore, two polysaccharides were found in bacteria E. coli O12. The major polysaccharide has a structure that, to our knowledge, has not been hitherto identified in bacterial carbohydrates. The genetic content of the O-antigen gene cluster of E. coli O12 is fully consistent with the established structure of this polysaccharide, and hence it represents the O12-antigen. Functions of genes and the corresponding proteins involved in biosynthesis of the O12-polysaccharides were determined. Particularly, each predicted glycosyltransferase was assigned to one of three glycosidic linkages that form the tetrasaccharide repeating unit. Our data extend the notion of the genetic basis of the structural diversity of the O-antigens of enteric bacteria and the database of carbohydrate biosynthesis enzymes with predicted functions. The minor polysaccharide was identified as the K5-antigen (heparosan), whose structure and genetics of biosynthesis have been elucidated earlier.
This work was supported by the Russian Science Foundation (project No. 14-14-01042).
Xi Guo was supported by the International Science and Technology Cooperation Program of China (2012DFG31680 and 2013DFR30640), the National Key Program for Infectious Diseases of China (2013ZX10004216-001-001 and 2013ZX10004221-003), the National Natural Science Foundation of China (NSFC) Program (31371259 and 81471904), and the Research Project of Chinese Ministry of Education (No. 113015A).
REFERENCES
1.Kaper, J. B., Nataro, J. P., and Mobley, H. L.
(2004) Pathogenic Escherichia coli, Nat. Rev. Microbiol.,
2, 123-140.
2.Iguchi, A., Iyoda, S., Kikuchi, T., Ogura, Y.,
Katsura, K., Ohnishi, M., Hayashi, T., and Thomson, N. R. (2014) A
complete view of the genetic diversity of the Escherichia coli
O-antigen biosynthesis gene cluster, DNA Res., 22,
105-107.
3.Whitfield, C. (2006) Biosynthesis and
assembly of capsular polysaccharides in Escherichia coli,
Annu. Rev. Biochem., 75, 39-68.
4.Joensen, K. G., Tetzschner, A. M., Iguchi, A.,
Aarestrup, F. M., and Scheutz, F. (2015) Rapid and easy in
silico serotyping of Escherichia coli isolates by use of
whole-genome sequencing data, J. Clin. Microbiol., 53,
2410-2426.
5.Robbins, P. W., and Uchida, T. (1962) Studies on
the chemical basis of the phage conversion of O-antigens in the E-group
Salmonellae, Biochemistry, 1, 323-335.
6.Westphal, O., and Jann, K. (1965) Bacterial
lipopolysaccharides. Extraction with phenol–water and further
applications of the procedure, Methods Carbohydr. Chem.,
5, 83-91.
7.Leontein, K., and Lonngren, J. (1993) Determination
of the absolute configuration of sugars by gas–liquid
chromatography of their acetylated 2-octylglycosides, Methods
Carbohydr. Chem., 9, 87-89.
8.Lipkind, G. M., Shashkov, A. S., Knirel, Y. A.,
Vinogradov, E. V., and Kochetkov, N. K. (1988) A computer-assisted
structural analysis of regular polysaccharides on the basis of
13C-n.m.r. data, Carbohydr. Res., 175,
59-75.
9.Jansson, P.-E., Kenne, L., and Widmalm, G. (1989)
Computer-assisted structural analysis of polysaccharides with an
extended version of CASPER using 1H- and
13C-n.m.r. data, Carbohydr. Res., 188,
169-191.
10.Vann, W. F., Schmidt, M. A., Jann, B., and Jann,
K. (1981) The structure of the capsular polysaccharide (K5 antigen) of
urinary-tract-infective Escherichia coli 010:K5:H4, Eur. J.
Biochem., 116, 359-364.
11.DeAngelis, P. L., Gunay, N. S., Toida, T., Mao,
W. J., and Linhardt, R. J. (2002) Identification of the capsular
polysaccharides of type D and F Pasteurella multocida as
unmodified heparin and chondroitin, respectively, Carbohydr.
Res., 337, 1547-1552.
12.Sugahara, K., and Kitagawa, H. (2002) Heparin and
heparan sulfate biosynthesis, IUBMB Life, 54,
163-175.
13.Reeves, P. R., and Wang, L. (2002) Genomic
organization of LPS-specific loci, Curr. Top. Microbiol.
Immunol., 264, 109-135.
14.Kneidinger, B., O’Riordan, K., Li, J.,
Brisson, J. R., Lee, J. C., and Lam, J. S. (2003) Three highly
conserved proteins catalyze the conversion of
UDP-N-acetyl-d-glucosamine to precursors for the biosynthesis of
O antigen in Pseudomonas aeruginosa O11 and capsule in
Staphylococcus aureus type 5. Implications for the
UDP-N-acetyl-l-fucosamine biosynthetic pathway, J. Biol.
Chem., 78, 3615-3627.
15.Liu, B., Knirel, Y. A., Feng, L., Perepelov, A.
V., Senchenkova, S. N., Wang, Q., Reeves, P. R., and Wang, L. (2008)
Structure and genetics of Shigella O antigens, FEMS
Microbiol. Rev., 32, 627-653.
16.D’Souza, J. M., Samuel, G. N., and Reeves,
P. R. (2005) Evolutionary origins and sequence of the Escherichia
coli O4 O-antigen gene cluster, FEMS Microbiol. Lett.,
244, 27-32.
17.Beutin, L., Wang, Q., Naumann, D., Han, W.,
Krause, G., Leomil, L., Wang, L., and Feng, L. (2007) Relationship
between O-antigen subtypes, bacterial surface structures and O-antigen
gene clusters in Escherichia coli O123 strains carrying genes
for Shiga toxins and intimin, J. Med. Microbiol., 56,
177-184.
18.Linnerborg, M., Weintraub, A., and Widmalm, G.
(1999) Structural studies of the O-antigen polysaccharide from the
enteroinvasive Escherichia coli O164 cross-reacting with
Shigella dysenteriae type 3, Eur. J. Biochem.,
266, 460-466.
19.Perepelov, A. V., Liu, B., Shevelev, S. D.,
Senchenkova, S. N., Shashkov, A. S., Feng, L., Knirel, Y. A., and Wang,
L. (2010) Relatedness of the O-polysaccharide structures of
Escherichia coli O123 and Salmonella enterica O58, both
containing
4,6-dideoxy-4-{N-(S)-3-hydroxybutanoyl]-d-alanyl}amino-d-glucose;
revision of the E. coli O123 O-polysaccharide structure,
Carbohydr. Res., 345, 825-829.
20.Rojas-Macias, M. A., Stahle, J., Lutteke, T., and
Widmalm, G. (2015) Development of the ECODAB into a relational database
for Escherichia coli O-antigens and other bacterial
polysaccharides, Glycobiology, 25, 341-347.
21.Lehrer, J., Vigeant, K. A., Tatar, L. D., and
Valvano, M. A. (2007) Functional characterization and membrane topology
of Escherichia coli WecA, a glycosyltransferase initiating the
biosynthesis of enterobacterial common antigen and O-antigen
lipopolysaccharide, J. Bacteriol., 189, 2618-2628.
22.Whitfield, C., and Roberts, I. S. (1999)
Structure, assembly and regulation of expression of capsules in
Escherichia coli, Mol. Microbiol., 31,
1307-1319.
23.Rigg, G. P., Barrett, B., and Roberts, I. S.
(1998) The localization of KpsC, S and T, and KfiA, C and D proteins
involved in the biosynthesis of the Escherichia coli K5 capsular
polysaccharide: evidence for a membrane-bound complex,
Microbiology, 144, 2905-2914.