Application of Created Restriction Site PCR-RFLP to Identify POT1 Gene Polymorphism
Tuanwei Wang1#, Sihua Wang1,2#, Xiaoran Duan1#, Xiaolei Feng1#, Pengpeng Wang1, Wu Yao1, Yongjun Wu3, Feifei Feng3, Songcheng Yu4, Yiming Wu1, and Wei Wang1*
1Zhengzhou University, College of Public Health, Department of Occupational Health and Occupational Disease, 450001 Zhengzhou, China; fax: +86-371-6778-1466; E-mail: ww375@126.com2Henan Institute of Occupational Health, Department of Occupational Health, 450052 Zhengzhou, China; E-mail: 13526723488@163.com
3Zhengzhou University, College of Public Health, Department of Hygiene Toxicology, 450001 Zhengzhou, China; E-mail: wuyongjun@zzu.edu.cn
4Zhengzhou University, College of Public Health, Department of Sanitary Chemistry, 450001 Zhengzhou, China; E-mail: scyu@zzu.edu.cn
# These authors contributed equally to this work.
* To whom correspondence should be addressed.
Received December 17, 2015; Revision received February 22, 2016
Protection of telomeres protein 1 (POT1) plays pivotal roles in protection of chromosome ends and regulation of telomere length with other telomere binding proteins; its genetic polymorphisms are associated with many diseases. In this study, we explored a novel PCR-RFLP method for typing the single nucleotide polymorphism (SNP) rs1034794 of the human POT1 gene. A new restriction enzyme site was introduced into a POT1 gene amplification product by created restriction site PCR (CRS-PCR). One primer was designed based on changed sequence; after PCR amplification, a new restriction enzyme site for AluI was introduced into the PCR products. One hundred and seventy eight samples from Han Chinese individuals were tested to evaluate this new method. The 3′-end of the forward primer was next to the polymorphic site, and the third base from the 3′-end was the mismatched base A. The final PCR product contained the AGCT sequence (AluI recognition site) when the ancestral POT1 alleles were amplified. The data obtained with the new method perfectly matched those obtained with the sequencing method. Thus, CRS-PCR is a new low-cost and high-efficiency alternative for rs1034794 typing.
KEY WORDS: created restriction site, single nucleotide polymorphism, protection of telomeres 1DOI: 10.1134/S0006297916060092
Abbreviations: CRS-PCR, created restriction site PCR; PCR-RFLP, PCR restriction fragment length polymorphism; POT1, protection of telomeres protein 1; SNP, single nucleotide polymorphism.
Telomere-related proteins play an important role in maintaining the
integrity of telomeric structure through protecting chromosome ends [1]. Protection of telomeres protein 1 (POT1) has been
shown to be involved in telomere elongation and to bind telomerase [2]. These proteins are able to bind the G-rich domains
in the telomeric repeat sequences to protect chromosome ends [3-5]. Recently, some studies
regarding POT1 gene polymorphism have been published.
Interestingly, certain types of polymorphism have been linked to
diseases [6, 7].
PCR-RFLP (PCR restriction fragment length polymorphism) is a commonly used method for detecting polymorphisms. This method is simple, fast, and effective. Many restriction enzymes are commercially available and inexpensive. However, some enzymes are very expensive, which limits their application for large-scale sample screening. For instance, such is the restriction enzyme HpyCH4V that could be used for POT1 gene rs1034794 typing. To circumvent this problem, we introduced a new restriction site into POT1 amplification products by use of a mismatched primer and examined the possibility of the application of this method for a large-scale screening in the Han Chinese population.
MATERIALS AND METHODS
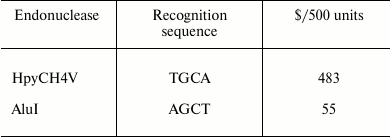
Gene information. The POT1 gene and rs1034794 single nucleotide polymorphism (SNP) data were obtained from the NCBI website (http://www.ncbi.nlm.nih.gov). The sequence analysis shows that the sequence TGCA in polymorphism site rs1034794 can be cleaved by expensive restriction enzyme HpyCH4V. To find a lower cost PCR-RFLP method, a new AluI enzyme site was created to detect the POT1 genotypes.
Primer design. The primers designed with Primer Premier 5.0 software are shown below. Forward: 5′-TTCAAACTTCAGGATCAAGC-3′; reverse: 5′-ATCATAGATGGAATAGACC-3′. In the forward primer sequence, the underlined capital A is the mismatch base, which replaces the natural base T. The strategies of base conversion are shown on the Fig. 1. The designed primers and corresponding amplification sequence are given below:
TTCAAACTTCAGGATCAAGC(5′-3′)
TTCAAACTTCAGGATCAAGCWactatcttaagaaaattgtcctgctttgttgtcattttaacctcttttggtgagtagtttagttcatttggacataaatctaaaatttatgtttgaactgcttgaatcttatagatcatcttcttgtttttgctcta
GGTCTATTCCATCTATGAT
CCAGATAAGGTAGATACTA(3′-5′).
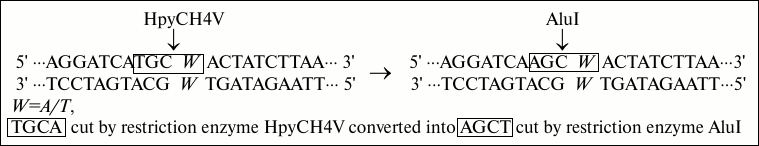
Fig. 1. Strategies of base conversion. In this base conversion strategy, W (A/T) was the polymorphic site, and the natural T base is replaced by mismatched A base. The recognition site TGCA cut by restriction enzyme HpyCH4V is converted into recognition site AGCT cut by restriction enzyme AluI.
In the sequence, the capital bases A, T, C, and G represent the same sequences as primers, and W indicates polymorphic locus A/T. Underlined is an artificially introduced A residue in the DNA sequence to produce a recognition site AGCT for restriction endonuclease AluI in the ancestral genotype allele POT1-AA (A – in the reverse POT1 gene encoding strand). The price of AluI is given in the table. As can be seen from the table, the price of AluI is nine times less than that of HpyCH4V.
Recognition sites and prices of respective endonucleases in NEB Co.
Genotyping of DNA. Genomic DNA was extracted from peripheral blood leukocytes using genomic DNA isolation kit (Bio Teke Corporation, China). PCR reactions were performed in a final volume of 15 µl containing 100 ng of genomic DNA, 0.3 µM of each primer (Sangon Biotech, China), 2× PCR mix (Lifefeng Biotech, China), and double-distilled water as follows: at 95°C for 3 min; at 95°C – 52°C – 72°C for 30 s each, 30 cycles; and then at 72°C for 5 min.
RFLP conditions. Enzyme digestion was carried out in a 20 µl final volume reaction mix consisting of 5 units (U) of AluI enzyme (New England Biolabs, USA), 2 µl 10× Buffer Tango, 7.5 µl double-distilled H2O, and 10 µl of PCR reaction mixture. The reaction was performed at 37°C overnight, and the digested products were electrophoresed on 3% agarose gel containing ethidium bromide (0.5 µg/ml) for 40 min. Agarose gels were observed under an UV transilluminator.
DNA sequencing. PCR products were sequenced using the reverse PCR primer provided by Sangon Biotech Company (China).
RESULTS
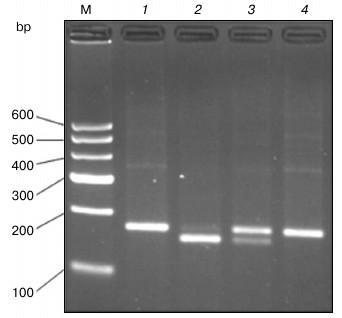
Enzyme digestion results. As can be seen in Fig. 2, the size of undigested PCR product is 177 bp as expected (lane 1). After digestion with AluI, the PCR product from homozygous alternative variant genotype (POT1-TT) allele could not be cut (lane 4). However, the homozygous ancestral genotype (POT1-AA) allele was cut into bands of 158 and 19 bp (lane 2), and the heterozygous genotype yielded three bands of 177, 158, and 19 bp (lane 3).
Fig. 2. PCR-RFLP for POT1 separated on a 3% agarose gel. The 177 bp POT1 product was digested with AluI, which does not cut the wild-type sequence but cleaves the T sequence into two smaller fragments of 158 and 19 bp, respectively. Lanes: 1) PCR product; 2) homozygous ancestral genotype (POT1-AA); 3) heterozygous genotype (POT1-AT); 4) homozygous alternative variant genotype (POT1-TT); M, molecular weight markers.
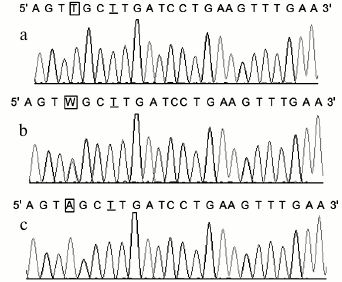
Consistency of the results. Polymorphisms of POT1 were typed in 178 Chinese Han individuals. The genotype frequencies were 116 for AA, 56 for AT, and 6 for TT. We also genotyped the 178 PCR products by Sanger sequencing, which is the gold standard for genotyping. The genotypes of the 178 samples obtained with the new method matched the gene sequencing data completely (the DNA sequencing results of the POT1 gene are shown in Fig. 3). Since we used the reverse primer to sequence the PCR products, the sequence from the sequencing map was the complementary sequence of PCR products. In Fig. 3, the underlined T was the mismatched base, which was consistent with mismatched A base in the forward primer; the T base in frame in Fig. 3a corresponds to POT1-AA genotype, and A base in frame in Fig. 3c corresponds to POT1-TT genotype. The W in frame in Fig. 3b surely corresponds to POT1-AT genotype. With the exception of the mismatched base, the sequences of the PCR products were consistent with those published in GenBank.
Fig. 3. Sequencing results of three rs1034794 SNP genotypes of POT1 gene. The T underlined is the mismatched base corresponding to the A base in the forward primer. The T base in-frame in Fig. 2a corresponds to POT1-AA genotype, and A base in-frame in Fig. 2c corresponds to POT1-TT genotype, respectively. The W in-frame in Fig. 2b corresponds to POT1-AT genotype.
DISCUSSION
PCR-RFLP method has been widely used since 1988 [8]. It proved to be a rapid and sensitive method for detection of gene polymorphisms [9]. However, this method has some limitations, for instance, some restriction enzymes, such as HpyCH4V or its isoschizomers, are very expensive. This circumstance has hindered application of the method, especially in developing countries. In this study, we introduced a new restriction enzyme site for AluI to type the POT1 polymorphism rs1034794. Restriction enzyme AluI is much less expensive than HpyCH4V; $428 would be saved when 500 U of enzyme is used to screen 100 samples. Besides, the new method proved quite reliable compared with sequencing.
In conclusion, here we report a simple and economical technique for typing of the POT1 gene polymorphism rs1034794, which allows for large-scale sample analyses.
We thank the National Natural Science Foundation of China for support (NSFC81001239).
REFERENCES
1.Nan, H., Qureshi, A. A., Prescott, J., De Vivo, I.,
and Han, J. (2011) Genetic variants in telomere-maintaining genes and
skin cancer risk, Hum. Genet., 129, 247-253.
2.Izgi, A., Gunal, A., Yalcin, S., and Gunduz, U.
(2014) Telomere 1 (POT1) gene expression and its association with
telomerase activity in colorectal tumor samples with different
pathological features, Biomed. Pharmacother., 68,
841-846.
3.Baumann, P., and Cech, T. R. (2001) Pot1, the
putative telomere end-binding protein in fission yeast and humans,
Science, 292, 1171-1175.
4.Baumann, P., Podell, E., and Cech, T. R. (2002)
Human Pot1 (protection of telomeres) protein: cytolocalization, gene
structure, and alternative splicing, Mol. Cell. Biol.,
22, 8079-8087.
5.Hockemeyer, D., Sfeir, A. J., Shay, J. W., Wright,
W. E., and De Lange, T. (2005) POT1 protects telomeres from a transient
DNA damage response and determines how human chromosomes end, EMBO
J., 24, 2667-2678.
6.Shen, J., Gammon, M. D., Terry, M. B., Bradshaw, P.
T., Wang, Q., Teitelbaum, S. L., Neugut, A. I., and Santella, R. M.
(2012) Genetic polymorphisms in telomere pathway genes, telomere
length, and breast cancer survival, Breast Cancer Res. Treat.,
134, 393-400.
7.Zee, R. Y., Ridker, P. M., and Chasman, D. I.
(2011) Genetic variants of 11 telomere-pathway gene loci and the risk
of incident type 2 diabetes mellitus: the Women’s Genome Health
Study, Atherosclerosis, 218, 144-146.
8.Deng, G. R. (1988) A sensitive non-radioactive
PCR-RFLP analysis for detecting point mutations at 12th codon of
oncogene c-Ha-ras in DNAs of gastric cancer, Nucleic Acids
Res., 16, 6231.
9.Fontecha, G. A., Garcia, K., Rueda, M. M.,
Sosa-Ochoa, W., Sanchez, A. L., and Leiva, B. (2015) A PCR-RFLP method
for the simultaneous differentiation of three entamoeba species,
Exp. Parasitol., 151-152, 80-83.