Binding of DNA with Abf2p Increases Efficiency of DNA Uptake by Isolated Mitochondria
E. O. Samoilova, I. A. Krasheninnikov, E. N. Vinogradova, P. A. Kamenski*, and S. A. Levitskii
Lomonosov Moscow State University, Faculty of Biology, 119991 Moscow, Russia; E-mail: kamenski_pa@mail.bio.msu.ru* To whom correspondence should be addressed.
Received February 16, 2016; Revision received March 23, 2016
Mutations in mitochondrial DNA often lead to severe hereditary diseases that are virtually resistant to symptomatic treatment. During the recent decades, many efforts were made to develop gene therapy approaches for treatment of such diseases using nucleic acid delivery into the organelles. The possibility of DNA import into mitochondria has been shown, but this process has low efficiency. In the present work, we demonstrate that the efficiency of DNA import can be significantly increased by preforming its complex with a mitochondria-targeted protein nonspecifically binding with DNA. As a model protein, we used the yeast protein Abf2p. In addition, we measured the length of the DNA site for binding this protein and the dissociation constant of the corresponding DNA–protein complex. Our data can serve as a basis for development of novel, highly efficient approaches for suppressing mutations in the mitochondrial genome.
KEY WORDS: mitochondria, nucleic acid import, Abf2p, binding site, dissociation constant, mitochondrial diseases, gene therapyDOI: 10.1134/S0006297916070087
Mitochondria are very important organelles of eukaryotic cells, catalyzing the biosynthesis of ATP via oxidative phosphorylation. Although the majority of mitochondrial proteins are encoded in the nuclear genome, synthesized in the cytoplasm, and then delivered into the organelles, mitochondria possess their own apparatus for storage and expression of genetic information encoded in mitochondrial DNA (mtDNA) and have their own transcription and translation systems. The mitochondrial genome is usually a circular DNA molecule containing genes of ribosomal RNAs of mitochondria, of the majority of tRNAs, and of some proteins of the oxidative phosphorylation system. The number of mtDNA copies in a mitochondrion varies from one to ten in different organisms [1], and these few copies of DNA together with some proteins are organized in special structures, nucleoids, which are usually associated with the mitochondrial inner membrane [2].
It is known that mtDNA is significantly more prone to mutagenesis than nuclear DNA because it is subjected to action of increased concentrations of reactive oxygen species produced in the reactions of oxidative phosphorylation [3]. The arising and accumulation of mutations in the mitochondrial genome lead to development of many diseases, such as muscle dystrophies and neurodegenerative diseases. Moreover, the accumulation of mutations in mtDNA is also thought to be associated with aging and carcinogenesis [4].
Some gene therapy approaches for treatment of such diseases are now under development. The majority of these approaches suggest modification of nuclear DNA by inserting into it genes whose expression products would be delivered into mitochondria through the apparatus of protein import [4]. Direct modification of the mitochondrial genome seems difficult because of complications associated with direct delivery of genetic material into mitochondria. In fact, methods for transformation of mitochondria have been developed only for the yeast Saccharomyces cerevisiae and for the unicellular alga Chlamydomonas reinhardtii, but these methods are based on a ballistic transformation, which is a technically difficult and expensive procedure [5].
Nevertheless, rather recently it was shown that isolated mitochondria have a natural competence for internalization of exogenous DNA [6]. This ability was shown for mitochondria of mammals, plants, and yeasts; it was also demonstrated that such DNA could be transcribed and then the resulting mRNA may be used for protein synthesis [7]. The uptake of DNA by mitochondria depended on the potential difference on membranes of the organelles and was inhibited by high concentrations of KCl and NaCl [7]. According to the available data, the transfer of DNA across the outer mitochondrial membrane is realized under the influence of the voltage-dependent anion channel (VDAC), or porin, and this process does not depend on the protein import apparatus. DNA was transferred across the inner mitochondrial membrane in plants with involvement of subunit CuBPp of respiratory chain complex I, whereas in yeast it seemed to occur with involvement of protein Cox13p, which is a component of complex III. Certainly, it seems very attractive to use the existing natural mechanism of mitochondria transformation for correcting mitochondrial mutations through expression of exogenous DNA in these organelles in order to change the heteroplasmy level in various cells.
However, the natural process has low efficiency: even at high concentrations of both mitochondria and DNA, no more than 0.1% of the nucleic acid added penetrated into the isolated organelles [8]. Based on the modern model of nonspecific DNA delivery into mitochondria, we supposed that the efficiency of this process could be increased by creating elevated concentrations of DNA near the surface of the mitochondria. This could be achieved using a DNA-binding protein containing a mitochondrial targeting signal (MTS). To test this idea, we decided to use the major protein of the yeast S. cerevisiae mitochondrial nucleoid, Abf2p. This protein, capable of non-specific DNA binding, consists of two domains (HMG1 and HMG2, High-Mobility Groups 1 and 2) and by now has been studied more carefully than all other proteins of the yeast mitochondrial nucleoid. Yeasts lacking Abf2p are capable of supporting their mtDNA during growth on medium containing glycerol as a non-fermentable source of carbon. However, when such cells were cultivated on media with fermentable carbon sources, e.g. with glucose, they gradually lost mtDNA [9]. It has also been shown that deletion of the ABF2 gene is associated with significant lowering of recombination acts in mitochondria [10, 11]. Moreover, Abf2p provided the stabilization of intermediates of Holliday structure formation, which also confirmed an important role of this protein in mitochondrial recombination [12]. It has been shown by high-performance atomic-force microscopy that interactions between Abf2p and mtDNA are weaker than the interactions between nuclear DNA and histones [13]. Nevertheless, these interactions are sufficiently strong to compact mtDNA due to a large number of sequence-nonspecific interactions.
During this work, we demonstrated that Abf2p binds with double-stranded DNA as a monomer and occupies a 13-bp region in DNA. We also show that DNA binding with Abf2p significantly increases the efficiency of its import into isolated yeast mitochondria.
MATERIALS AND METHODS
Escherichia coli and S. cerevisiae strains. The E. coli strains Top-10 OneShot® (Invitrogen, USA) and B834(DE3) (Novagen, USA) were used, as well the S. cerevisiae strain W303.
Preparation of recombinant proteins Abf2p and TrxA–Abf2p. The ABF2 gene was amplified with genomic DNA of S. cerevisiae using the primers ABF2_MTS_F (5′- cgatcatatgcaccaccaccaccaccacaaggcttccaagagaacgc-3′) and ABF2R (5′- gactctcgaggttgagagggtagcgagc-3′), and the resulting fragment was hydrolyzed by endonucleases NdeI and XhoI (Thermo Scientific, USA) and cloned within expression vector pET30a (Novagen) treated with the same enzymes. The correspondence of the cloned sequence to the ABF2 was confirmed by sequencing according to Sanger (the sequencing was performed in the Federal Medical Biological Agency of Russia). The resulting vector pET30a_ABF2 was used for transforming the E. coli strain B834(DE3) cells. The grown transformants were inoculated into 200 ml of the 2× YT medium containing kanamycin (50 µg/ml) (Sigma, USA). The bacterium was cultured at 37°C in a thermoshaker at 200 rpm until reaching OD ~ 0.7, then the temperature was decreased to 30°C, an expression inducer (isopropyl-β-D-thiogalactopyranoside, IPTG) was added to the concentration of 0.25 mM, and the cultivation was continued for 4 h. Then the cells were precipitated by centrifugation at 3000g for 10 min, washed with 50 ml of phosphate-saline buffer (10 mM Na2HPO4, 1.76 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4), and resuspended in 5 ml of the starting chromatographic buffer (25 mM sodium phosphate buffer, pH 7.4, 0.5 M NaCl, 25 mM imidazole). The cells were broken by ultrasonication (six 10-s pulses with 15-s intervals); the lysate was clarified by centrifugation at 30,000g for 30 min. The recombinant protein was purified by metal-affinity chromatography on a column with nickel-Sepharose (HiTrap; GE Healthcare, USA) using an AKTA Purifier chromatographic system (GE Healthcare) according to the manufacturer’s instructions. The fraction containing the recombinant protein was immediately transferred into a buffer for storage (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, l.5 mM EDTA) using a Sephadex G-25-containing 5-ml HiTrap Desalting column.
The vector for expression of the TrxA–Abf2p fusion protein gene was prepared by cloning the restriction fragment NdeI–NdeI from the plasmid pET32a (Novagen) within vector pET30a_ABF2 pretreated with the same enzymes. The recombinant protein was isolated and purified according to the above-described method.
Analysis of changes in electrophoretic mobility (EMSA). The binding of recombinant proteins Abf2p and TrxA–Abf2p was studied using the gel retention assay. As substrates for the binding, synthetic duplexes were used with lengths of 20, 24, 28, and 40 bp, which were prepared by complementation of the corresponding pairs of oligonucleotides X20 (5′- AGTCTAGACTGCAGTTGAGT-3′) and X20_C (5′-ACTCAACTGCAGTCTAGACT-3′), X24 (5′- AGTCTAGACTGCAGTTGAGTCCTT-3′) and X24_C (5′- AAGGACTCAACTGCAGTCTAGACT-3′), X28 (5′- AGTCTAGACTGCAGTTGAGTCCTTGCTA-3′) and X28_C (5′- TAGCAAGGACTCAACTGCAGTCTAGACT-3′), X40 (5′- AGTCTAGACTGCAGTTGAGTCCTTGCTAGGACGGATCCCT-3′) and X40_C (5′- AGGGATCCGTCCTAGCAAGGACTCAACTGCAGTCTAGACT-3′) [14]. In experiments for determination of oligomerization degree and dissociation constant, oligonucleotide X40 labeled with 6-FAM (6-carboxyfluorescein) was used. Oligonucleotides were mixed at the concentration of 1-3 µM in buffer for annealing (25 mM Tris-HCl, pH 8.0, 200 mM NaCl, 2 mM EDTA), heated to 95°C, and then cooled slowly.
Binding of DNA with the protein was performed in buffer containing 25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, BSA (0.1 mg/ml), and 7% glycerol. To DNA at the concentration of 10-100 nM, different amounts of recombinant proteins were added, and the mixtures were incubated at room temperature and then separated on 6% nondenaturing polyacrylamide gel (29 : 1, 18 × 20 × 0.075 cm, 400 V, 2 h). Then the gel was either stained with ethidium bromide and photographed in UV, or (using FAM-labeled DNA) scanned in the blue fluorescence regimen with a Storm865 scanner (GE Healthcare, USA). The resulting images were processed using ImageJ software (NIH, USA). Dissociation constants were calculated as described published [15].
Isolation of mitochondria. Active mitochondria were isolated from S. cerevisiae according to a described protocol [16]. Briefly, a colony of yeast strain W303 was inoculated into 10 ml of liquid medium YPGal and cultured for 16-18 h in a 50-ml tube at 30°C and 200 rpm. The resulting culture was transferred into 1 liter of the YPGal medium in a 5-liter Erlenmeyer flask and cultured at 30°C and 200 rpm for 16-18 h until OD600 reached 1.5-2.0. The cells were precipitated by centrifugation at 3000g for 10 min, washed in 1/10 of water volume, and then resuspended in SCE buffer (0.6 M sorbitol, 0.3 M mannitol, 20 mM K2HPO4, 20 mM citric acid, 1 mM EDTA) at the ratio of 1 ml of the buffer per gram wet cell mass. The resulting suspension was combined with the same volume of glass spheres (0.45-0.6 mm; Sigma, USA), and the cells were destroyed with a FastPrep disintegrator (MP, USA) twice for 20 s with an interruption of 3 min for cooling on ice. The resulting lysate was centrifuged 8-10 times at 3000g for 10 min at 4°C until the complete disappearance of precipitate. The fraction enriched with mitochondria was precipitated at 15,000g for 15 min at 4°C. The precipitate was resuspended in FB buffer (10 mM Tricine-KOH, 0.1 mM EDTA, 50 mM NaCl, pH 7.5) with 65% sucrose, transferred into a centrifugal tube, deposited above with layers of 55, 43, and 15% sucrose in buffer FB, and then centrifuged at 40,000g for 1 h. The mitochondria occurring at the interface between the 55 and 43% sucrose layers were gathered, diluted three times with FB buffer containing 15% sucrose, precipitated at 18,000g for 5 min, and then resuspended in SCE buffer and diluted to total mitochondrial protein concentration of 1-2 mg/ml.
Import of DNA into mitochondria. As a molecule to be imported, we used the γ[32P]ATP-labeled linear fragment with length of 520 bp that was prepared as a result of PCR amplification. Before realizing the import, 150 fmol DNA was incubated for 15 min at room temperature with different concentrations of the recombinant protein Abf2p in 10 µl of buffer (25 mM Tris-HCl, 50 mM NaCl, 1 mM EDTA). Then the mixture was combined with 90 µl of suspension of freshly isolated mitochondria and incubated for 30 min at 25°C. The reaction mixtures were then supplemented with MgCl2 to the concentration of 10 mM and 5 units of DNase I (Thermo Scientific) and incubated for 15 min at 37°C. Then the mitochondria were precipitated as described above, washed twice with five volumes of SCE buffer, and resuspended in 25 mM Tris-HCl, 250 mM NaCl, 2 mM EDTA, 0.1% SDS; DNA was extracted with phenol–chloroform mixture. The isolated DNA was separated in 0.9% nondenaturating agarose gel and transferred onto a nitrocellulose filter. The filter was exposed for 8-12 h onto a reusable photoluminescent screen (Phosphor Storage Screen; GE Healthcare), and then the screen was visualized with a Storm 865 scanner (GE Healthcare).
RESULTS
Although Abf2p is one of the best-studied proteins of the yeast mitochondrial nucleoid, some characteristics of its binding with DNA are still poorly investigated. Thus, the length of the DNA binding site of the protein is unknown, and it is still under discussion, whether Abf2p is a monomer or dimer when being bound to DNA. The dissociation constant of the Abf2p–DNA complex also has not been determined reliably. Because our idea on the possibility of using Abf2p for increasing the efficiency of DNA penetration into mitochondria needed knowledge of sufficiently precise values of these parameters, at the beginning of the work we determined them as described below.
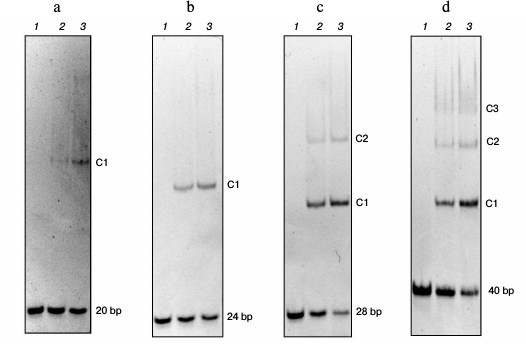
Length of DNA binding site of Abf2p is 13 nucleotide pairs. The length of the Abf2p-binding site on mtDNA was determined by analyzing changes in the electrophoretic mobility of double-stranded DNA fragments with different lengths as described in [17] (Fig. 1). Thus, analysis of the interaction of the recombinant Abf2p with a 20-bp fragment of DNA revealed on the gel one “retained” band that corresponded to generation of one DNA–protein complex. Consequently, the protein-binding site on mtDNA was longer than 10 bp. Abf2p interacted similarly with a 24-bp fragment with production of one complex, which meant that the binding site length was more than 12 bp. With a 28-bp fragment, Abf2p produced two easily detectable retention bands that corresponded to production of two complexes – with one and with two molecules of the protein. Consequently, the Abf2p-binding site on mtDNA was no longer than 14 bp. Finally, the interaction of the protein with a 40-bp fragment of DNA resulted in three retention bands on the gel, which suggested a possibility of three molecules of the protein binding to this fragment. Thus, the length of the protein-binding site on mtDNA could not be more than 13 bp. Therefore, we concluded that the length of the Abf2p site for binding double-stranded DNA was 13 bp.
Fig. 1. Determination of the length of the Abf2p-binding site on mtDNA. Linear fragments (50 nM) of double-stranded DNA with different lengths were incubated with recombinant Abf2p at concentrations of 150 and 300 nM in 25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 0.1 mg/ml BSA, 7% glycerol and then were separated in 6% polyacrylamide gel. a-d) 20-, 24-, 28-, and 40-bp DNA fragments, respectively: 1) without Abf2p; 2) with 100 nM Abf2p; 3) with 250 nM Abf2p. C1-C3, complexes of DNA with one, two or three molecules of the protein.
Abf2p interacts with DNA as a monomer. The oligomerization degree of proteins is often determined by gel filtration. However, many proteins with asymmetric fold show gel-filtration elution profiles that are not specific for their weights. In particular, such data were obtained for TFAM, which is an ortholog of Abf2p in human mitochondria [18]. It seems also to refer to Abf2p, because the gel filtration of free Abf2p and of Abf2p in an equimolar mixture with a 20-bp fragment of double-stranded DNA resulted in calculated values of molecular weights of 31.5 and 42.8 kDa, respectively (data not presented). These values correspond neither to the monomer (19.816 kDa) nor to the dimer (39.632 kDa) of the protein, nor to complexes of DNA with the monomer or dimer.
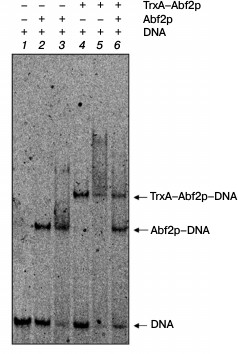
To elucidate the oligomerization degree of Abf2p, we used an approach similar to that used in work [18]. We obtained the recombinant fused protein consisting of thioredoxin A of E. coli and Abf2p (TrxA–Abf2p) and analyzed changes in the electrophoretic mobility of DNA on its interaction with Abf2p, TrxA–Abf2p, and with their equimolar mixture (Fig. 2). The fused protein TrxA–Abf2p had the same ability of binding DNA as Abf2p, but the retention band in the gel was located significantly higher. If Abf2p had interacted with DNA as a dimer, the binding of the oligonucleotide duplex with the mixture of two recombinant proteins would have resulted in three retention bands, the lower one corresponding to the complex of Abf2p dimer with DNA, the upper one corresponding to the complex of the TrxA–Abf2p dimer with DNA, and the intermediate one indicating the presence of the complex of the Abf2p plus TrxA–Abf2p plus DNA. In the electrophoregram, one can distinctly see that no intermediate retention band was produced. Thus, Abf2p interacted with linear DNA as a monomer.
Fig. 2. Abf2p interacts with DNA in a monomeric form. The 40-bp DNA fragment (10 nM) was incubated with the recombinant proteins Abf2p and TrxA–Abf2p separately or together and then separated in 6% polyacrylamide gel. Lanes: 1) DNA without proteins; 2) DNA with 50 nM Abf2p; 3) DNA with 100 nM Abf2p; 4) DNA with 50 nM TrxA–Abf2p; 5) DNA with 100 nM TrxA–Abf2p; 6) DNA with 75 nM Abf2p and 75 nM TrxA–Abf2p. The DNA complexes with Abf2p and TrxA–Abf2p are different in electrophoretic mobility; the absence of the intermediate band on binding with the protein mixture indicates the absence of oligomerization.
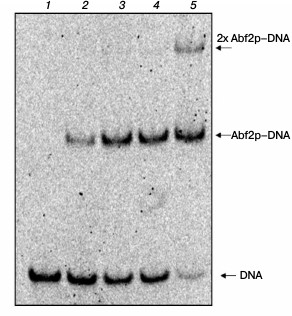
Determination of dissociation constant of Abf2p complex with linear DNA. The dissociation constant of the complex of double-stranded linear DNA with recombinant Abf2p was also determined by analyzing changes in the electrophoretic mobility as described in “Materials and Methods” (Fig. 3). The calculated Kd of the complex of 40-bp DNA fragment with the protein was ~40 nM that, considering the binding site length found by us gives the Kd value of ~120 nM for one site. Binding cooperativity was not established, the cooperativity coefficient being determined as described in work [19].
Fig. 3. Abf2p binding with double-stranded DNA. A 40-bp DNA fragment was incubated with increasing concentrations of Abf2p (25, 50, 100, 150 nM) and then the mixtures were separated in 6% polyacrylamide gel. Lanes: 1) DNA without proteins; 2) DNA with 25 nM Abf2p; 3) DNA with 50 nM Abf2p; 4) DNA with 100 nM Abf2p; 5) DNA with 150 nM Abf2p. The calculated apparent dissociation constant is ~40 nM.
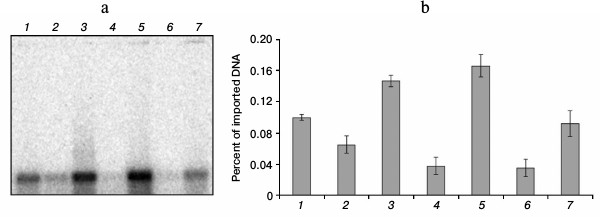
Influence of binding with Abf2p on DNA import into isolated yeast mitochondria. To find out whether the binding of DNA with the protein Abf2p influences its ability to be imported into mitochondria, we determined the efficiency of the import of a 520-bp fragment of double-stranded DNA in complexes with different amounts of the recombinant protein into isolated mitochondria of S. cerevisiae. We used our data on the binding site length and on the dissociation constant of the complex to choose optimal concentrations of the protein and its amount relatively to the number of binding sites on the fragment used. Because the binding site length was 13 bp, the fragment we used contained 40 such sites. Respectively, for the site/protein ratio of 1 : 1, we used 40 molecules of the protein per each DNA molecule, and when the working concentration of DNA in the reaction mixture for the import was 15 nM, the protein concentration at such ratio was 600 nM. For the control, equivalent amounts of buffer with the protein but without DNA were added into the parallel reactions. Results of the experiment are presented in Fig. 4.
Fig. 4. Influence of DNA binding with Abf2p on its import into isolated S. cerevisiae mitochondria. Import of the 520-bp fragment pretreated with the recombinant Abf2p at different protein/binding site ratios. a) Radioautograph of the import reaction result (see “Materials and Methods”); b) diagram of quantitative assessment of imported DNA at different protein/binding site ratios (the data of three independent experiments with standard deviations are presented): 1) fragment introduced (520 bp, 0.1% of imported amount); 2, 4, 6) import of fragment without Abf2p; 3) fragment with Abf2p treated at ratio one protein molecule per three binding sites; 5) fragment with Abf2p treated at ratio one protein molecule per four binding sites; 7) fragment with Abf2p treated at ratio one protein molecule per 10 binding sites.
As is obvious from our data, binding with Abf2p (containing mitochondrial localization signal) increased the efficiency of DNA import into isolated mitochondria, and the maximal increase in imported DNA amount (1.6 times as compared to the control) was achieved at the ratio of one protein molecule per four binding sites on DNA (150 nM protein at 15 nM DNA). Further increase in the protein amount was associated with a decrease in the import efficiency that seemed to result from a decrease in the total charge of nucleic acid and, as a consequence, a decrease in its affinity for the mitochondrial membrane. Moreover, at high concentrations of the protein, high degree of binding could prevent DNA transport through the outer mitochondrial membrane via VDAC.
DISCUSSION
Mutations in the mitochondrial genome cause many diseases whose treatment is nowadays difficult or impossible. At present, mitochondrial mutations are corrected mainly by compensation of functions of the damaged mitochondrial genes through expression of their analogs in the nucleus and subsequent import of such expression products into the mitochondria [4]. Direct correction of mtDNA mutations is difficult because there is no reliable system of the genetic material delivery into mitochondria. Only one effective approach has been developed for modification of the mitochondrial genome – a biolistic transformation of mitochondria of two organisms: S. cerevisiae and C. reinhardtii [5]. It is known that during evolution mitochondria were derived from a prokaryotic precursor [20] and retained some prokaryotic biological features. In particular, one such feature is so-called “natural competence”, i.e. the ability of isolated mitochondria to internalize exogenous DNA. This ability has been demonstrated for mitochondria of yeast, plants, and mammals, but this process has low efficiency [8]. According to some data, the efficiency of transformation of prokaryotic cells and of transfecting eukaryotes can be increased using preliminary binding of the transforming nucleic acid with nonspecifically DNA-binding proteins [21]. We supposed that the binding of DNA with a protein carrying a mitochondrial localization signal would be able to increase the efficiency of DNA import into mitochondria due to creation of its locally elevated concentration near the external mitochondrial membrane. As such protein, we used the yeast mitochondrial DNA-binding protein Abf2p with its mitochondrial localization signal. We chose this protein because as a model organism we used S. cerevisiae, which is a standard model for studies on mitochondria.
Abf2p is the major protein of the yeast mitochondrial nucleoid and participates in both the packing of mtDNA and its replication and recombination. Although data on this protein were rather abundant, the exact character of its interaction with DNA remained unknown. To obtain complete and reliable data on the Abf2p interaction with DNA, we determined the dissociation constant of their complex and evaluated the length of the DNA region necessary for binding the protein. We found that the length of the Abf2p binding site is 13 bp. It is known that the length of binding site of the human mitochondrial HMG-protein TFAM is 30 bp, which is longer than the average length of the binding sites of HMG-proteins for which the length of 8-12 bp is more characteristic [18]. This seems to be associated with TFAM dimerization in the presence of double-stranded DNA, which was shown earlier [18]. The Abf2p binding site seems to better correspond to those of its bacterial functional analogs represented by HU proteins. For the majority of the latter, the binding site length is 10-12 bp [22], and the least of the known sites of bacterial functional analogs belongs to the HU protein of Acholeplasma laidlawii (9 bp) [17]. With this in mind, we supposed that Abf2p could bind with DNA as a monomer. The question of oligomerization degree of DNA-binding proteins of the HMG2 group, which includes Abf2p, is still under discussion. Thus, the Abf2p ortholog TFAM from human mitochondria was shown to interact with DNA as a dimer [18] and also as a monomer [23]. Our data indicate that Abf2p binds with linear double-stranded DNA as a monomer. A slightly longer binding site than in the HU proteins could be mediated by different affinities of HMG domains of this protein for DNA [24]: the first domain located nearer to the N-terminal part of the molecule had higher DNA-binding activity than the C-terminal domain. It is possible that the C-terminal HMG domain is responsible for some auxiliary functions and creates steric obstacles for binding another molecule of the protein in close vicinity of the first domain.
It should be noted that the dissociation constant of the Abf2p complex with double-stranded DNA determined by us by changes in the electrophoretic mobility is rather low as compared with constants for complexes of the HU proteins with DNA. This seems to reflect the necessity of protecting mitochondrial DNA against increased concentrations of reactive oxygen species in the mitochondrial matrix, which is likely to be realized by Abf2p tight interaction with DNA.
Finally, we have demonstrated that the binding of DNA by the recombinant Abf2p leads to an increase in the efficiency of DNA import into isolated mitochondria. To prove this, we based our study on the previously obtained data and on formation of the nucleoprotein complex and took into account the binding site/protein molar ratio but not the DNA/protein ratio. We demonstrated that the maximal increase in import efficiency was observed at the ratio of one protein molecule per four binding sites on DNA. However, on careful consideration of our experiments, a question arises whether the preliminary binding of the imported DNA with Abf2p can protect nucleic acid against enzymatic cleavage by nuclease and thus distort the data on the efficiency of DNA import into the mitochondria. However, the DNA/protein ratios that we used could not provide complete “screening” of the DNA molecule and could not prevent the nuclease degradation of a rather long region of nucleic acid. Moreover, in the case of distorting the data due to DNA protection by Abf2p, the efficiency would directly depend on the amount of protein introduced into the reaction mixture. On the contrary, our data indicated that the import efficiency decreased on increasing the protein amount relative to DNA (one molecule per three binding sites). This could be caused by a decrease in the total charge of the nucleoprotein complex and, as a consequence, a decrease in its affinity for the mitochondrial membrane.
According to the literature data, the efficiency of DNA import by yeast mitochondria does not depend on the length of the imported fragment [7]. Based on this, we suppose that our findings enlarge the current knowledge about the import of nucleic acids into mitochondria and will promote developing new systems for genetic material delivery into mitochondria, which will be useful for creating new approaches for both fundamental studies in molecular biology of mitochondria and applied research in this field.
This work was supported by the Ministry of Education and Science of the Russian Federation (Federal Targeted Program “Studies and Developments in Priority Lines of the Science and Technology Complex of Russia for 2014-2020”, agreement 14.604.21.0113, identifier RFMEFI60414X0113).
REFERENCES
1.Wiesner, R. J., Ruegg, J. C., and Morano, I. (1992)
Counting target molecules by exponential polymerase chain reaction:
copy number of mitochondrial DNA in rat tissues, Biochem. Biophys.
Res. Commun., 183, 553-559.
2.Legros, F., Malka, F., Frachon, P., Lombes, A., and
Rojo, M. (2004) Organization and dynamics of human mitochondrial DNA,
J. Cell Sci., 117, 2653-2662.
3.Chen, X. J. (2013) Mechanism of homologous
recombination and implications for aging-related deletions in
mitochondrial DNA, Microbiol. Mol. Biol. Rev., 77,
476-496.
4.Patrushev, M. V., Kamenski, P. A., and Mazunin, I.
O. (2014) Mutations in mitochondrial DNA and approaches for their
correction, Biochemistry (Moscow), 79, 1151-1160.
5.Bonnefoy, N., and Fox, T. D. (2007) Directed
alteration of Saccharomyces cerevisiae mitochondrial DNA by
biolistic transformation and homologous recombination, Methods Mol.
Biol., 372, 153-166.
6.Koulintchenko, M., Temperley, R. J., Mason, P. A.,
Dietrich, A., and Lightowlers, R. N. (2006) Natural competence of
mammalian mitochondria allows the molecular investigation of
mitochondrial gene expression, Hum. Mol. Genet., 15,
143-154.
7.Mileshina, D., Koulintchenko, M., Konstantinov, Y.,
and Dietrich, A. (2011) Transfection of plant mitochondria and in
organello gene integration, Nucleic Acids Res., 39,
e115.
8.Weber-Lotfi, F., Koulintchenko, M. V., Ibrahim, N.,
Hammann, P., Mileshina, D. V., Konstantinov, Y. M., and Dietrich, A.
(2015) Nucleic acid import into mitochondria: new insights into the
translocation pathways, Biochim. Biophys. Acta, 1853,
3165-3181.
9.Diffley, J. F., and Stillman, B. (1991) A close
relative of the nuclear, chromosomal high-mobility group protein HMG1
in yeast mitochondria, Proc. Natl. Acad. Sci. USA, 88,
7864-7868.
10.Okamoto, K., Perlman, P. S., and Butow, R. A.
(1998) The sorting of mitochondrial DNA and mitochondrial proteins in
zygotes: preferential transmission of mitochondrial DNA to the medial
bud, J. Cell Biol., 142, 613-623.
11.Zelenaya-Troitskaya, O., Newman, S. M., Okamoto,
K., Perlman, P. S., and Butow, R. A. (1998) Functions of the high
mobility group protein, Abf2p, in mitochondrial DNA segregation,
recombination and copy number in Saccharomyces
cerevisiae, Genetics, 148, 1763-1776.
12.MacAlpine, D. M., Perlman, P. S., and Butow, R.
A. (1998) The high mobility group protein Abf2p influences the level of
yeast mitochondrial DNA recombination intermediates in vivo,
Proc. Natl. Acad. Sci. USA, 95, 6739-6743.
13.Friddle, R. W., Klare, J. E., Martin, S. S.,
Corzett, M., Balhorn, R., Baldwin, E. P., Baskin, R. J., and Noy, A.
(2004) Mechanism of DNA compaction by yeast mitochondrial protein
Abf2p, Biophys. J., 86, 1632-1639.
14.Duckett, D. R., and Lilley, D. M. (1990) The
three-way DNA junction is a Y-shaped molecule in which there is no
helix-helix stacking, EMBO J., 9, 1659-1664.
15.Kamashev, D., Balandina, A., Mazur, A.,
Arrimondo, P., and Rouviere-Yaniv, J. (2008) HU binds and folds
single-stranded DNA, Nucleic Acids Res., 36,
1026-1036.
16.Kaufman, B. A., Newman, S. M., Hallberg, R. L.,
Slaughter, C. A., Perlman, P. S., and Butow, R. A. (2000) In organello
formaldehyde crosslinking of proteins to mtDNA: identification of
bifunctional proteins, Proc. Natl. Acad. Sci. USA, 97,
7772-7777.
17.Levitskii, S. A., Sycheva, A. M., Kharlampieva,
D. D., Oberto, J., Kamashev, D. E., Serebryakova, M. V., Moshkovskii,
S. A., Lazarev, V. N., and Govorun, V. M. (2011) Purification and
functional analysis of recombinant Acholeplasma laidlawii
histone-like HU protein, Biochimie, 93, 1102-1109.
18.Gangelhoff, T., Mungalachetty, T. S., Nix, J. C.,
and Churchill, M. E. (2009) Structural analysis and DNA binding of the
HMG domains of the human mitochondrial transcription factor A,
Nucleic Acids Res., 37, 3153-3164.
19.Huisman, O., Faelen, M., Girard, D., Jaffe, A.,
Toussaint, A., and Rouviere-Yaniv, J. (1989) Multiple defects in
Escherichia coli mutants lacking HU protein, J.
Bacteriol., 171, 3704-3712.
20.Poole, A. M., and Gribaldo, S. (2014) Eukaryotic
origins: how and when was the mitochondrion acquired? Cold Spring
Harb. Perspect. Biol., 6, a015990.
21.Luo, D., and Saltzman, W. M. (2000) Synthetic DNA
delivery systems, Nat. Biotech., 18, 33-37.
22.Guo, F., and Adhya, S. (2007) Spiral structure of
Escherichia coli HUalphabeta provides foundation for DNA
supercoiling, Proc. Natl. Acad. Sci. USA, 104,
4309-4314.
23.Ngo, H. B., Kaiser, J. T., and Chan, D. C. (2011)
The mitochondrial transcription and packaging factor Tfam imposes a
U-turn on mitochondrial DNA, Nat. Struct. Mol. Biol., 18,
1290-1296.
24.Kurashenko, A. V., Samoilova, E. O., Baleva, M.
V., Chicherin, I. V., Petrov, D. Y., Kamenski, P. A., and Levitskii, S.
A. (2016) Two hmg-domains of mitochondrial protein Abf2p are different
in the affinity for DNA, Bull. RSMU, 1, 68-72.