Peptide Aβ(16-25) Forms Nanofilms in the Process of Its Aggregation
O. M. Selivanova1, E. Yu. Gorbunova2, L. G. Mustaeva2, E. I. Grigorashvili1, M. Yu. Suvorina1, A. K. Surin1,3, and O. V. Galzitskaya1*
1Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; E-mail: ogalzit@vega.protres.ru2Shemyakin–Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia
3State Research Center for Applied Microbiology and Biotechnology, 142279 Obolensk, Moscow Region, Russia
* To whom correspondence should be addressed.
Received April 4, 2016; Revision received April 8, 2016
A method for the synthesis and high purification of fragments of Aβ(1-42) peptide has been elaborated. We have synthesized the amyloidogenic fragment Aβ(16-25) predicted by us and studied the process of its aggregation by electron microscopy and X-ray analysis. Electron microscopy images show that the peptide forms a film, which is not characteristic of amyloid fibrils. At the same time, according to the X-ray diffraction data, its preparations display the presence of two main reflections (4.6-4.8 and 8-12 Å) characteristic of cross-β structure of amyloid fibrils. Thus, the fragment Aβ(16-25) that we predicted is a promising object not only for studying the process of polymerization of the peptides/proteins, but also for using it as a nanomaterial to study a number of biological processes.
KEY WORDS: amyloid fibril, protofibril, oligomer, Aβ peptide, Alzheimer’s disease, electron microscopyDOI: 10.1134/S0006297916070129
A great number of proteins and peptides have been determined whose polymerization into fibril formations causes the development of different diseases (amyloidosis). However, not all fibril formations are toxic for organisms. There are numerous functional amyloids. Such amyloids have been discovered in many representatives of the animal and plant worlds as well as in yeasts and bacteria. For some peptide and protein hormones, a standard functional state is their long preservation in secretory granules of the endocrine system (glucagon, somatostatin, etc.) [1-3]. Many microorganisms contain amyloid proteins on their surfaces [4-9]. They help microorganisms not only interact with inert surfaces (plastic, glass) [10], but also are necessary for adhesion on surfaces of host cells [11]. This increases the risk of development of some diseases, for example, tuberculosis [12]. Much attention has been paid lately to studies of biofilms formed by amyloid proteins of microorganisms, for example, E. coli protein curlin [6]. Inasmuch as biofilms serve to facilitate the attachment of bacterial cells to surfaces of host cells, thus promoting penetration of infection in the organism, conditions are sought which prevent this process. Recently attention has been focused on the possibility of using biofilms as nanobiomaterial. As a result, a method is proposed for using biofilms to enhance retroviral transduction or direct visualization of viral particles on the surface of nanofilms [13]. This is just the beginning of the application of nanofilms. However, different variations in the design of nanofilms will allow changing their properties, which is undoubtedly a promising field of studies.
Bioinformatics methods have made it possible to reveal that proteins contain so-called amyloidogenic regions whose presence leads to the formation of amyloid fibrils. Examination of the process of fibril formation on such fragments using mutation analysis suggests different mechanisms of formation of amyloid structures. A simplified scheme of fibril formation involves three states: a change in the monomer state, formation of an oligomer structure, and formation of mature fibrils [14]. The most puzzling moment in the scheme is the transformation of the oligomer into a mature fibril. Oligomer forms that can frequently be adsorbed on the cell wall as ring structures and play the role of a pore are the most toxic for organisms. Damage of the cell wall in such an unusual way can result in cell death. Therefore, some authors believe that amyloid formation is a defense mechanism, which allows the organism to decrease the number of toxic oligomer structures through their aggregation as fibril formations.
One approach in studying amyloid formation of Aβ peptide is comparative investigation of properties of its amyloidogenic fragments and different mutant forms. Such studies provided data showing that Aβ peptide fragments of different lengths have differences in characteristic parameters of amyloid formation (rate, lag period, fibril morphology) and suggested what regions of the peptide are responsible for aggregation and what regions are responsible for the morphology of the formed fibrils. It has been demonstrated for fragments of an extracellular region of Aβ peptide that depending on the presence of residues of histidine and glutamic and aspartic acids, the morphology of the formed fibrils changes, and a fragment containing the minimal number of such residues forms fibrils in a pH range not typical of Aβ peptide (<3). Besides, fragments containing no so-called hydrophobic cluster (residues 17-21 in the Aβ peptide sequence) also differ in their properties [15]. Further evidence for the importance of the charged amino acid residues in the process of amyloid formation is that when they are substituted for uncharged residues, the morphology of the fibrils formed by the mutant form of the peptide also changes [16].
MATERIALS AND METHODS
Chemical synthesis of fragment Aβ(16-25). Solid-phase chemical synthesis was used to obtain preparative amounts of the Aβ(16-25) peptide fragment. Aβ(16-25) peptide fragments are chemical compounds of peptide nature that contain hydrophobic “complex sequences” consisting of fragments with a chain of four and more hydrophobic residues [17]. The chemical synthesis and purification of such peptides are a complicated task due to low solubility in both water and organic solvents. During the experiments, the effect of different conditions of condensation and chromatography regimes of product purification on the chemical yield was studied. The main accountable indices were the high content of the endproduct in reaction mass samples and the absence of side products found earlier.
The synthesis was performed using the Fmoc-methodology [18], i.e. the 9-fluorenylmethoxycabonyl group determining the corresponding protective groups for side functions of amino acids was used as temporary protection of the α-amino group. The docking of the first amino acid on the polystyrene polymer with a p-benzyloxybenzyl hydroxyl linker was done using the carbodiimide method in the presence of 0.1 equivalent of 4-dimethylaminopyridine (DMAP) [19]. The level of the docking was determined by a quantitative picric acid test [20]. Capping of unreacted hydroxyl residues on the polymer was performed using benzoic anhydride in pyrimidine. Condensation of amino acids was conducted by activation of TBTU (O-(benzotriazol-1-yl)- N,N,N′,N′-tetramethyluronium tetrafluoroborate) [21] in N-methylpyrrolidone to inhibit aggregation. Repeated condensation was conducted using the carbodiimide method. The completeness of docking was controlled using the ninhydrin test [20]. Peptides were cleaved from the polymer support using trifluoroacetic acid (TFA) in an argon flow with addition of different scavengers such as TIS (triisopropyl silane) and EDT (ethanedithiol-1,2) [22]. After preliminary purification with repeated precipitation, raw reaction mixtures were analyzed using high performance liquid chromatography. The peptides were purified and analyzed on a Waters high-pressure chromatograph with a Waters 2487 detector (Waters, USA). Preparative purification was done on a Reprosil-Pur C18-AQ 10 µm (250 × 20 mm; Dr. Maisch GmbH, Germany) column in an acetonitrile gradient. Analytical chromatography was performed on a Luna 5u C18(2) 100 Å (250 × 4.6 mm; Phenomenex, USA) column. Detection was conducted at the wavelength of 226 nm. The purified peptide was tested using a mass spectrometer (Thermo Fisher Scientific, Germany). The estimated peptide mass coincided with the calculated one.
Mass spectrometry. Mass-spectrometric analysis was performed using an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). Ionization by nanoelectrospray with direct introduction of the sample was used as a method for ionization of the sample. The analyzer was an orbital trap with resolution of 240,000. The voltage supplied to the capillary varied from 0.8 to 1.5 kV. The mass spectrum was recorded in the range from 500 to 2000 m/z.
Sample preparation. The Aβ(16-25) fragment sample was first dissolved in a minimal volume of 100% dimethylsulfoxide (DMSO) (with final concentration of 5%), and then it was reduced to the required concentration (0.25-0.50 mg/ml) using 50 mM Tris-HCl, pH 7.5.
Electron microscopy. The Aβ(16-25) peptide samples in 5% DMSO, 50 mM Tris-HCl, pH 7.5, at concentration 0.25-0.50 mg/ml were incubated at 37°C. At fixed time intervals, aliquots were taken for the electron microscopy (EM) analysis. The samples for this analysis were negatively stained. A copper grid (400 Mesh) covered with a formvar film (0.2% formvar solution in chloroform) was placed on a preparation drop (~10 µl). After adsorption, the girds with the sample were stained with 1% (w/v) aqueous solution of uranyl acetate. The excess of the sample and uranyl acetate were removed with filter paper. The samples were analyzed on a JEM-1200EX (JEOL, Japan) microscope at accelerating voltage of 80 kV. The recording was made at 40,000× magnification onto Kodak film (SO-163) for EM studies.
X-Ray analysis. The Aβ(16-25) peptide was incubated in 5% DMSO, 50 mM Tris-HCl, pH 7.5, concentration 0.5 mg/ml at 37°C during 48 h. After that, its concentration was brought to 5 mg/ml using centrifugation at 12,000 rpm for 30 min. The supernatant was removed, and 5-7 µl of the pellet of mature fibrils was placed in the clearance (~1.0-1.5 mm) between the tips of two glass rods (of ~1 mm in diameter) covered with wax [23]. The preparation was dried for several hours. X-Ray analysis of the preparations was conducted using a Microstar X-ray emitter with HELIOX optics equipped with a Platinum 135 CCD detector (X8 Proteum System; Bruker AXS, Germany). Cu Kα radiation (λ = 1.54 Å) was used (the sample was oriented perpendicular to the X-ray beam).
RESULTS AND DISCUSSION
Studies of amyloid formation by the Aβ(16-25) fragment. Amyloids are fibril formations with specific properties described by different methods. First of all, the presence of a fibril structure is revealed by electron microscopy methods including EM, cryoEM, and atomic force microscopy (AFM). These methods make it possible to observe fibrils elongated to several micrometers and on average 10 nm in diameter with characteristic polymorphous morphology in the form of helices of different periods, ribbons, and bundles. Polymorphism is a specific feature of amyloid structures and is revealed for many proteins, not only under changes in the conditions of preparing the samples but also under the same conditions. Another important characteristic of amyloids is the presence of a cross-β-structure, which is determined by X-ray analysis. The fluorescence analysis can reveal (though not in all cases) characteristic kinetics of binding thioflavin T or S. Besides these basic methods, observations of birefringence upon interaction of preparations with fluorescent Congo red are also used. In addition, circular dichroism spectroscopy is used for observing the increase in β-structure upon formation of amyloids.
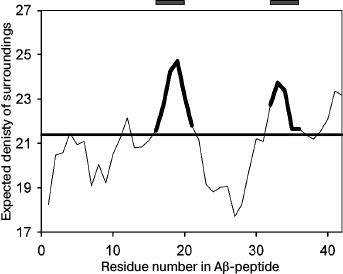
Though under different conditions different proteins can form amyloid structures, the presence of charged and hydrophobic amino acids in the protein sequence is of significance for amyloid formation. In the Aβ(1-40) peptide, sites from the 16th to the 21st amino acid and from the 32nd to the 36th amino acid were predicted to be amyloidogenic by the FoldAmyloid program (http://bioinfo.protres.ru/fold-amyloid/) [24] (Fig. 1). The PASTA2 program (http://protein.bio.unipd.it/pasta2) [25] predicts only one amyloidogenic site (30-40 a.a.), while the WALTZ program (http://waltz.switchlab.org/) [26] predicts another site (16-21 a.a.). In this work, we analyzed the first predicted amyloidogenic peptide (16-25, KLVFFAEDVG) to study the process of its aggregation.
Fig. 1. Profile of the expected number of contacts for the Aβ(1-42) peptide obtained using the FoldAmyloid program [24]. The predicted amyloidogenic regions are marked bold in the profile and shown by gray rectangles above the profile.
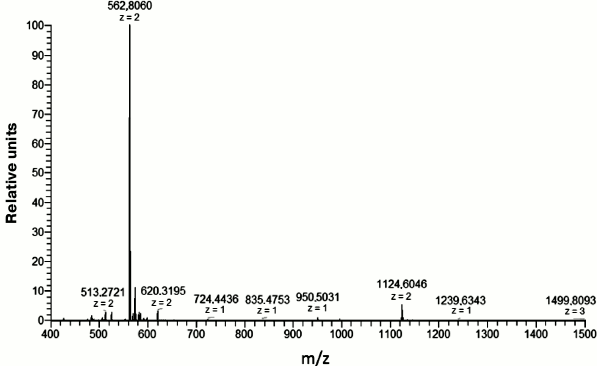
This fragment was chemically synthesized in sufficient amounts (see “Materials and Methods”). Its purity was checked using mass spectrometry (Fig. 2).
Fig. 2. Mass spectrum of the Aβ(16-25) fragment. The peptide mass is 1123.456 Da calculated for the [M + 2H+]2+ ion with m/z = 562.8060.
Electron microscopy. The data of EM analysis of the Aβ(16-25) preparation at concentration 0.25 mg/ml make it possible to observe straightaway (0 h incubation) the generation of small thin fibrils with length up to 300 nm and diameter of about 6-7 nm in the field. These fibrils can interact with each other forming planar ribbon-like fibrils (table). After 8 h incubation, structures appear in the form of wide ribbons up to 1 µm in length and width of about 50 nm. After 24 h incubation, the length of these planar ribbon- or film-like formations remains about 1 µm, whereas their width grows to 80 nm. Upon incubation of the Aβ(16-25) preparation at concentration of 0.5 mg/ml, it is possible to observe the generation of elongated particles up to 300-500 nm in length and up to 30 nm in width that look like ribbons. By 8 h incubation, the width of elongated polymer formations reaches 60-70 nm, and by 24 h incubation, it grows to 150 nm. The length of the particles does not change significantly upon incubation from 8 to 24 h, i.e. polymers grow predominantly perpendicular to their main axis.
Kinetics of amyloid formation of Aβ(16-25) peptide fragment using
electron microscopy
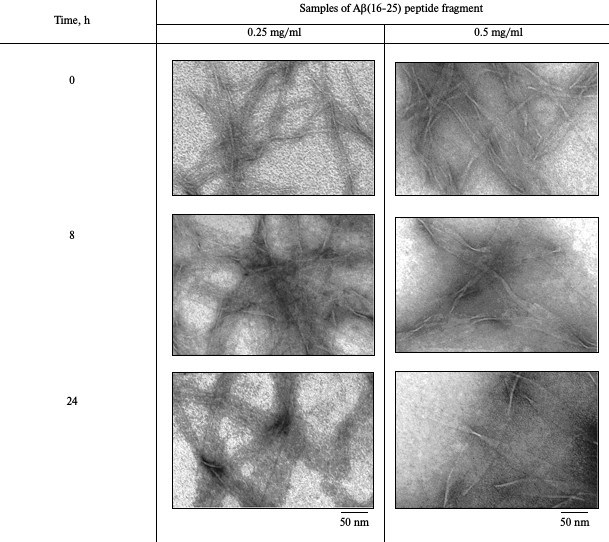
Note: Buffer – 50 mM Tris-HCl, pH 7.5; incubation at
37°C (5% DMSO).
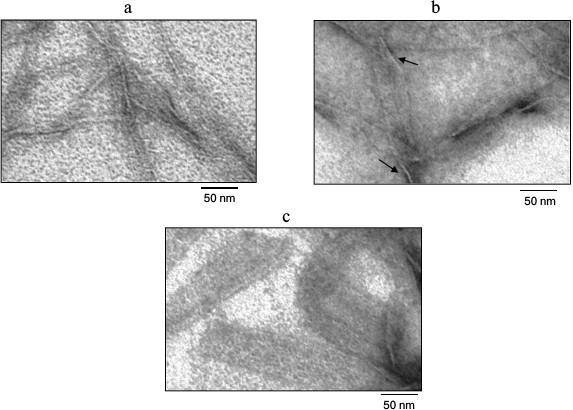
Figure 3 shows the Aβ(16-25) fragment preparation at concentration of 0.25 mg/ml under high magnification. As seen, polymerization of the peptide proceeds from small fibril formations with diameter of about 6 nm to rather elongated wide aggregates. These aggregates have small thickness that can be estimated approximately by the sites of polymer bends. Since such polymers are rather wide and thin (thickness of about 3 nm), it can be stated that the Aβ(16-25) fragment is polymerized in the form of thin and long films.
Fig. 3. Electron microscopy image of the Aβ(16-25) peptide sample under high magnification. The sample was prepared in 5% DMSO, 50 mM Tris-HCl, pH 7.5, at 37°C (0.25 mg/ml). a, b, c) 0, 8, and 24 h incubation, respectively. As seen in (b) and (c), the films are thin and are nearly mingled against the background. The film thickness in their bending sites is about 3 nm (arrows).
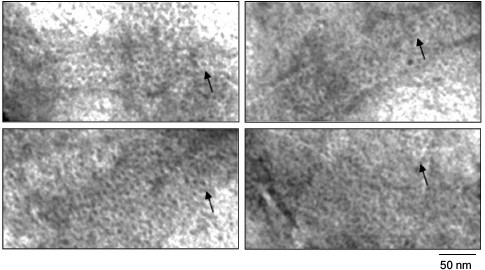
The most remarkable finding is that if polymer films are viewed under high magnification, it is seen that they are formed of ring structures with diameter of about 6 nm (Fig. 4) packed like a honeycomb.
Fig. 4. EM image of film fragments of the Aβ(16-25) peptide. The sample was prepared in 5% DMSO, 50 mM Tris-HCl, pH 7.5, 8 h incubation at 37°C (0.25 mg/ml). As seen, the films are formed of ring-like structures with diameter of about 6 nm (arrows).
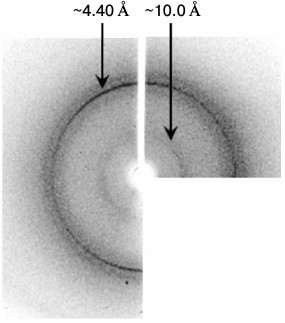
X-Ray analysis. Though the morphology of Aβ(16-25) polymers is uncommon, the data of X-ray analysis allow considering the presence of cross-β structure because the X-ray pattern has two main reflections (4.5-4.8 and 8-12 Å) (Fig. 5). Based on the X-ray diffraction patterns, we conclude that polymers of the Aβ(16-25) peptide are amyloids.
Fig. 5. Data of X-ray analysis of the Aβ(16-25) sample prepared in 50 mM Tris-HCl, pH 7.5, 48 h incubation at 37°C, 5 mg/ml.
An important feature of investigations of amyloid structures is the determination of conditions affecting the process of amyloidogenesis, such as temperature, pH, ionic strength, the presence of ligands, and others. It is possible that in spite of a great number of conflicting theoretical and experimental data on the mechanism of amyloid structure formation there should exist common key moments in this process inherent to all proteins. To exclude the effect of different context in proteins, it is convenient to use model peptides in which the major portion of their sequences is a priori an amyloidogenic region. Our experience studying different amyloidogenic proteins and peptides (insulin, Aβ(1-40) and Aβ(1-42) peptides, amyloidogenic fragments of protein Bgl2p, different fragments of Aβ(1-42) peptides) shows that the morphology of amyloid fibrils changes depending on the protein used. However, among the diversity of studied objects, for the first time we have observed polymers in the form of films using the Aβ(16-25) peptide predicted as an amyloidogenic fragment of integral Aβ(1-42) peptide. Thus, the Aβ(16-25) fragment that we predicted to be amyloidogenic is a promising object not only for establishing the process of polymerization of peptides/proteins with the formation of fibrils, but also for studying peptides as nanomaterial and biofilms challenging for the development of nanotechnologies.
We are grateful to Dr. A. D. Nikulin for his help in obtaining X-ray images.
This work was supported by the Russian Science Foundation (project No. 14-14-00536).
REFERENCES
1.Fowler, D. M., Koulov, A. V., Balch, W. E., and
Kelly, J. W. (2007) Functional amyloid – from bacteria to humans,
Trends Biochem. Sci., 32, 217-224.
2.Maji, S. K., Perrin, M. H., Sawaya, M. R.,
Jessberger, S., Vadodaria, K., Rissman, R. A., Singru, P. S., Nilsson,
K. P. R., Simon, R., Schubert, D., Eisenberg, D., Rivier, J.,
Sawchenko, P., Vale, W., and Riek, R. (2009) Functional amyloids as
natural storage of peptide hormones in pituitary secretory granules,
Science, 325, 328-332.
3.Shewmaker, F., McGlinchey, R. P., and Wickner, R.
B. (2011) Structural insights into functional and pathological amyloid,
J. Biol. Chem., 286, 16533-16540.
4.Barnhart, M. M., and Chapman, M. R. (2006) Curli
biogenesis and function, Annu. Rev. Microbiol., 60,
131-147.
5.Chapman, M. R., Robinson, L. S., Pinkner, J. S.,
Roth, R., Heuser, J., Hammar, M., Normark, S., and Hultgren, S. J.
(2002) Role of Escherichia coli curli operons in
directing amyloid fiber formation, Science, 295,
851-855.
6.Collinson, S. K., Doig, P. C., Doran, J. L.,
Clouthier, S., Trust, T. J., and Kay, W. W. (1993) Thin, aggregative
fimbriae mediate binding of Salmonella enteritidis to
fibronectin, J. Bacteriol., 175, 12-18.
7.Gibson, D. L., White, A. P., Rajotte, C. M., and
Kay, W. W. (2007) AgfC and AgfE facilitate extracellular thin
aggregative fimbriae synthesis in Salmonella enteritidis,
Microbiology, 153, 1131-1140.
8.Jonas, K., Tomenius, H., Kader, A., Normark, S.,
Romling, U., Belova, L. M., and Melefors, O. (2007) Roles of curli,
cellulose and BapA in Salmonella biofilm morphology studied by
atomic force microscopy, BMC Microbiol., 7, 70.
9.Lapidot, A., and Yaron, S. (2009) Transfer of
Salmonella enterica serovar Typhimurium from contaminated
irrigation water to parsley is dependent on curli and cellulose, the
biofilm matrix components, J. Food Prot., 72,
618-623.
10.Prigent-Combaret, C., Prensier, G., Le Thi, T.
T., Vidal, O., Lejeune, P., and Dorel, C. (2000) Developmental pathway
for biofilm formation in curli-producing Escherichia coli
strains: role of flagella, curli and colanic acid, Environ.
Microbiol., 2, 450-464.
11.Danne, C., and Dramsi, S. (2012) Pili of
gram-positive bacteria: roles in host colonization, Res.
Microbiol., 163, 645-658.
12.Alteri, C. J., Xicohtencatl-Cortes, J., Hess, S.,
Caballero-Olin, G., Giron, J. A., and Friedman, R. L. (2007)
Mycobacterium tuberculosis produces pili during human infection,
Proc. Natl. Acad. Sci. USA, 104, 5145-5150.
13.Dai, B., Li, D., Xi, W., Luo, F., Zhang, X., Zou,
M., Cao, M., Hu, J., Wang, W., Wei, G., Zhang, Y., and Liu, C. (2015)
Tunable assembly of amyloid-forming peptides into nanosheets as a
retrovirus carrier, Proc. Natl. Acad. Sci. USA, 112,
2996-3001.
14.Dovidchenko, N. V., Glyakina, A. V., Selivanova,
O. M., Grigorashvili, E. I., Suvorina, M. Y., Dzhus, U. F., Mikhailina,
A. O., Shiliaev, N. G., Marchenkov, V. V., Surin, A. K., and
Galzitskaya, O. V. (2016) One of the possible mechanisms of amyloid
fibrils formation based on the sizes of primary and secondary folding
nuclei of Aβ40 and Aβ42, J. Struct. Biol., 194,
404-414.
15.Fraser, P. E., Nguyen, J. T., Surewicz, W. K.,
and Kirschner, D. A. (1991) pH-dependent structural transitions of
Alzheimer amyloid peptides, Biophys. J., 60,
1190-1201.
16.Kirschner, D. A., Inouye, H., Duffy, L. K.,
Sinclair, A., Lind, M., and Selkoe, D. J. (1987) Synthetic peptide
homologous to beta protein from Alzheimer disease forms amyloid-like
fibrils in vitro, Proc. Natl. Acad. Sci. USA, 84,
6953-6957.
17.Bedford, J., Hyde, C., Johnson, T., Jun, W.,
Owen, D., Quibell, M., and Sheppard, R. C. (1992) Amino acid structure
and “difficult sequences” in solid phase peptide synthesis,
Int. J. Pept. Protein Res., 40, 300-307.
18.Fields, G. B., and Noble, R. L. (1990) Solid
phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino
acids, Int. J. Pept. Protein Res., 35, 161-214.
19.Beyermann, M., Henklein, P., Klose, A., Sohr, R.,
and Bienert, M. (1991) Effect of tertiary amine on the
carbodiimide-mediated peptide synthesis, Int. J. Pept. Protein
Res., 37, 252-256.
20.Stewart, J. M., and Young, J. D. (1984) Solid
Phase Peptide Synthesis, Pierce Chemical Co., Rockford, Illinois,
2nd Edn.
21.Reid, G. E., and Simpson, R. J. (1992) Automated
solid-phase peptide synthesis: use of
2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium tetrafluoroborate
for coupling of tert-butyloxycarbonyl amino acids, Anal.
Biochem., 200, 301-309.
22.Angell, Y. M., Alsina, J., Albericio, F., and
Barany, G. (2002) Practical protocols for stepwise solid-phase
synthesis of cysteine-containing peptides, J. Pept. Res.,
60, 292-299.
23.Serpell, L. C., Fraser, P. E., and Sunde, M.
(1999) X-ray fiber diffraction of amyloid fibrils, Methods
Enzymol., 309, 526-536.
24.Garbuzynskiy, S. O., Lobanov, M. Y., and
Galzitskaya, O. V. (2010) FoldAmyloid: a method of prediction of
amyloidogenic regions from protein sequence, Bioinformatics,
26, 326-332.
25.Walsh, I., Seno, F., Tosatto, S. C. E., and
Trovato, A. (2014) PASTA 2.0: an improved server for protein
aggregation prediction, Nucleic Acids Res., 42,
W301-307.
26.Maurer-Stroh, S., Debulpaep, M., Kuemmerer, N.,
Lopez de la Paz, M., Martins, I. C., Reumers, J., Morris, K. L.,
Copland, A., Serpell, L., Serrano, L., Schymkowitz, J. W. H., and
Rousseau, F. (2010) Exploring the sequence determinants of amyloid
structure using position-specific scoring matrices, Nat.
Methods, 7, 237-242.