Determination of Regions Involved in Amyloid Fibril Formation for Aβ(1-40) Peptide
A. K. Surin1,2, E. I. Grigorashvili1, M. Yu. Suvorina1, O. M. Selivanova1, and O. V. Galzitskaya1*
1Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; E-mail: ogalzit@vega.protres.ru2State Research Center for Applied Microbiology and Biotechnology, 142279 Obolensk, Moscow Region, Russia; E-mail: alan@vega.protres.ru
* To whom correspondence should be addressed.
Received April 7, 2016; Revision received April 18, 2016
The studies of amyloid structures and the process of their formation are important problems of biophysics. One of the aspects of such studies is to determine the amyloidogenic regions of a protein chain that form the core of an amyloid fibril. We have theoretically predicted the amyloidogenic regions of the Aβ(1-40) peptide capable of forming an amyloid structure. These regions are from 16 to 21 and from 32 to 36 amino acid residues. In this work, we have attempted to identify these sites experimentally by the method of tandem mass spectrometry. As a result, we show that regions of the Aβ(1-40) peptide from 16 to 22 and from 28 to 40 amino acid residues are resistant to proteases, i.e. they are included in the core of amyloid fibrils. Our results correlate with the results of the theoretical prediction.
KEY WORDS: Aβ peptide, amyloid fibril, amyloidogenic regions, mass spectrometry, Alzheimer’s diseaseDOI: 10.1134/S0006297916070130
Abbreviations: a.a., amino acid; APP, β-amyloid precursor protein; DMSO, dimethylsulfoxide; EM, electron microscopy; HPLC-MS, high performance liquid chromatography/mass spectrometry; NMR, nuclear magnetic resonance.
The problem of protein folding is a principal in current structural
biology. It is important to explain how protein from an unfolded
polypeptide chain acquires quickly and correctly the spatial structure
providing the performance of its specific function. It is now clear
that practically any protein can form amyloids, and small specific
regions, called aggregating or amyloidogenic, are responsible for the
formation of amyloid fibrils [1]. The
characteristic peculiarity of these regions makes it possible to
predict such regions from the amino acid sequence [1-6].
Today we know what proteins are associated with the development of certain diseases: Tau protein and Aβ peptide are responsible for the development of Alzheimer’s disease, α-synuclein – for Parkinson’s disease, huntingtin – for Huntington’s disease, prions – for Creutzfeldt–Jakob disease, amylin – for type II diabetes, etc. [7]. Thus, all neurodegenerative diseases, whose pathogenesis includes aggregation of proteins with consequent formation of protein depositions, are classified as proteinopathies. In addition, proteinopathies are subdivided according to the type of the key protein that causes the proteinopathy.
Studies of the mechanism of amyloidogenesis of Aβ peptide will reveal factors promoting pathogenesis of Alzheimer’s disease. Different methods described in the literature, both experimental and theoretical, are used for this aim [8-10]. The accumulated data show high polymorphism of amyloid structures depending on the conditions of experiments and the amino acid sequence of the peptide itself [11]. Therefore, it is challenging to clarify how the mechanism of amyloid formation depends on the amino acid sequence of a certain amyloidogenic region.
During the years of studies of Aβ peptide and processes of its aggregation, different researchers have obtained the following voluminous information. Structures of different forms of Aβ peptide were determined using nuclear magnetic resonance spectroscopy (NMR) (Fig. 1) [12, 13] as well as structures of amyloid fibrils (Fig. 2) [14, 15]. The process of Aβ peptide formation in the cell was studied and described [16]. Different oligomers of Aβ peptide (containing from 2 to 24 monomers) were discovered and studied, and it was demonstrated that these oligomers have a toxic effect on neurons [17-20]. It was clarified that Aβ peptide in the normal state is present in organisms and performs physiological functions [21, 22]. Mechanisms of amyloid formation of Aβ peptide were proposed: a nucleation mechanism [23] and a mechanism without specific states [24]. Data were obtained demonstrating that divalent metal ions bind actively with amyloid fibrils, affecting the process of amyloidogenesis: in particular, zinc accelerates the formation of amyloid fibrils [25], while copper forms coordination bonds with histidine residues [26]; Aβ peptide can be degraded by neprilysin (an insulin-degrading enzyme) and endothelin-transforming enzyme [27]. Worthy of special mention is a study where the use of a combination of the hydrogen–deuterium exchange by mass spectrometry demonstrated that the middle region (20-35) in the amino acid sequence of the peptide is prime for the process of aggregation, and then follows the C-terminal region (36-42), and finally the hydrophilic N-terminal region (1-19) for Aβ(1-42). At 37°C and under agitation, Aβ(1-42) aggregates faster than at 25°C. On the contrary, in the presence of Cu2+, the kinetics slows due to the formation of a complex at the C-terminus stabilizing the soluble Aβ(1-42) form [28].
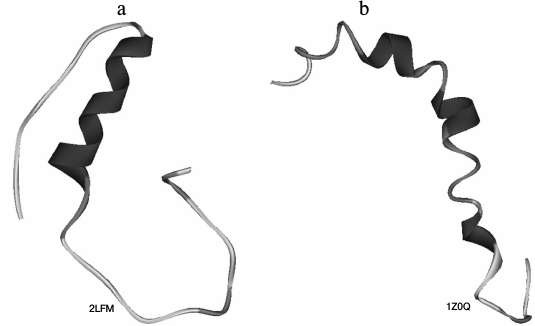
Fig. 1. Three-dimensional structures of Aβ peptide obtained by NMR spectroscopy for (a) Aβ(1-40) [12] and (b) Aβ(1-42) [13]. PDB codes are given on the figure.
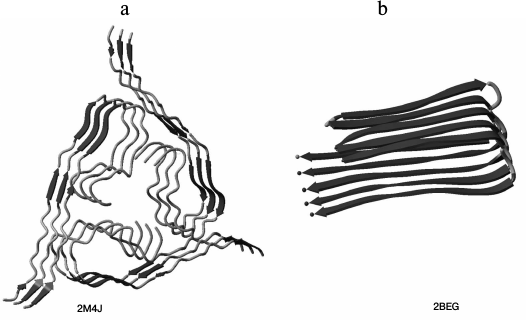
Fig. 2. Three-dimensional structures of amyloid fibril sections (a) for Aβ(1-40) peptide (the PDB code for the protein is 2M4J [14]; three layers of the protofibril are shown) and (b) for Aβ(1-42) peptide (the PDB code for the protein is 2BEG [15]; five layers of the protofibril are shown). NMR model No. 1 is presented for both structures.
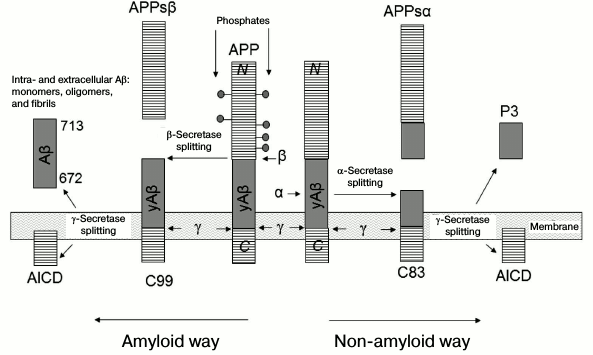
The source of Aβ peptide is β-amyloid precursor protein (APP). This is a transmembrane glycoprotein whose N-terminal fragment is outside the cell, while the C-terminal fragment is inside the cell. APP is a protein that undergoes controlled intramembrane proteolysis. This process for APP can occur in two ways: non-amyloid and amyloid. In the first case, α-secretase imbedded in the membrane cleaves the N-terminal fragment of APPsα that contains a part of the Aβ peptide sequence, and the remaining fragment C83 is processed by γ-secretase, the results of which are peptide p3 and the intracellular domain of APP (AICD).
When the process is amyloidogenic, β-secretase competing with α-secretase cleaves by approximately 16 a.a. less from the N-terminus (fragment APPsβ), leaving the sequence of Aβ peptide undamaged [29]. The remaining fragment C99 is also processed by γ40- and γ42-secretases, forming peptides Aβ(1-40) and Aβ(1-42) and AICD (Fig. 3) [29].
Fig. 3. Scheme of controlled intramembrane proteolysis of amyloid precursor proteins (APP) [29]. Residues 672-713 in APP correspond to Aβ(1-42).
Mechanisms of amyloid formation of Aβ peptide are studied intensively. Based on the experimental data, mechanisms with and without special states have been described. However, a generally accepted hypothesis is nucleation-dependent polymerization, suggesting that the formation of fibrils requires a structured nucleus whose generation is followed by cooperative inclusion of new molecules of Aβ peptide when the concentration of monomers in solution becomes critical. According to this hypothesis, nucleation is a limiting stage. This is corroborated by the presence of a lag-period in observations of the kinetics of amyloid formation of Aβ peptide [30]. A simplified scheme of the process shows that the formation of fibrils for Aβ peptide has three main stages: a changed state of the monomer, formation of an oligomer structure, and formation of mature fibrils [31].
Determination of regions responsible for aggregation of the peptide and formation of amyloid fibrils is one of the primary aims in studying the formation of such structures. Knowledge of these regions will facilitate the choice of compounds interacting with a given region and thus preventing its aggregation. Therefore, in this study our task was to find amyloidogenic regions in amyloid fibrils formed by Aβ(1-40) peptide and to compare the data with the theoretical prediction.
MATERIALS AND METHODS
Obtaining amyloid fibrils from Aβ(1-40) peptide. The Aβ(1-40) peptide preparation (Sigma Aldrich, USA) was dissolved to the concentration of 20 mg/ml in 100% dimethylsulfoxide (DMSO) and then diluted to the concentration of 1 mg/ml in 50 mM Tris-HCl (pH 7.5). The peptide solution was incubated for 24 h at 37°C.
Electron microscopy and X-ray analysis of fibrils. The presence of amyloid fibrils in solution was monitored by electron microscopy and X-ray analysis. The preparations of Aβ(1-40) peptide were dissolved in 5% DMSO, 50 mM Tris-HCl, pH 7.5, and incubated at 37°C, and the concentration was adjusted to 0.2 mg/ml. Aliquots of the preparation were collected for EM analysis at selected times. Negative staining was used to prepare samples for this analysis. A copper grid (400 Mesh) covered by a formvar film (0.2% formvar solution in chloroform) was placed on a sample drop (~10 µl). After adsorption (5 min), the grids with the preparation were stained with 1% (w/v) aqueous solution of uranyl acetate (1.5-2.0 min). The samples were analyzed using a JEM-1200EX microscope (JEOL, Japan) at accelerating voltage of 80 kV. Images were recorded at 40,000× magnification on Kodak films (SO-163) for EM studies.
For the X-ray analysis, the samples of Aβ(1-40) peptide were incubated under the same conditions for 7 days. After that, the preparation was concentrated to 20 mg/ml at room temperature using an Eppendorf 5301 vacuum concentrator (Eppendorf, Germany). The resulting drops were placed between the tips of glass rods (~1 mm in diameter) coated with paraffin. Thus, the fibrils in the preparation were oriented along the axis of the rods. X-Ray images were obtained using a Microstar emitter with HELIOX optics equipped with a Platinum 135 CCD detector (X8 Proteum System; Bruker AXS, Germany). The Cu Kα emission was used (λ = 1.54 Å). To position the preparation at the correct angle to the X-ray beam, a four-circle kappa-goniometer was used.
Sedimentation and proteolysis of amyloid fibrils from Aβ(1-40) peptide. After incubation, the preparation was centrifuged for 20 min at 10,000 rpm and then washed twice with 100 mM NH4HCO3 (pH 7.5). The amyloid fibrils precipitated in this way were dissolved in 100 mM NH4HCO3 (pH 7.5) to the concentration of 1 mg/ml and were incubated for 3 h with a mixture of proteases (trypsin, chymotrypsin, proteinase K at Aβ(1-40) peptide/protease ratio of 1 : 25). To raise the activity of proteinase K, CaCl2 was added to the mixture to adjust the concentration to 5 mM. After termination of incubation with the mixture of proteases, the solution was centrifuged for 20 min at 10,000 rpm, washed twice with 100 mM NH4HCO3 (pH 7.5), and then washed with 0.1% trifluoroacetic acid (TFA). Then the sample was dried in a vacuum concentrator.
HPLC-MS analysis of regions of amyloid fibrils protected from proteolysis. The sample dried in a vacuum concentrator was dissolved in 5 µl of 100% formic acid, which decomposed the structures, and then the sample volume was adjusted to the volume for the reaction of hydrolysis using solution of 20 mM ammonium acetate (C2H3O2NH4). The HPLC mass spectrometry analysis was performed using an Easy nLC 1000 nano-flow liquid chromatograph (Thermo Scientific, USA). Separation was done on a combined column of a concentrating Acclaim PepMap 100 (75 µm × 2 cm pre-column with the C18 3 µm 100 Å phase) and a separating Acclaim PepMap RSLC column (50 µm × 15 cm with the C18 2 µm 100 Å phase) (Thermo Scientific). The volume of the introduced sample varied from 2 to 10 µl. After initial loading at 5% solution B (organic phase), a gradient of solution B from 5 to 80% during 80 min was used at flow rate 250 nl/min. Solution A consisted of 0.1% of formic acid (FA) in water, whereas solution B was composed of 80% acetonitrile (ACN) and 0.1% FA in water. The liquid chromatography (LC) column outlet was online coupled with the electrospray (nESI) source of the mass spectrometer. The mixture was analyzed using an OrbiTrap Elite ETD high-performance mass spectrometer (Thermo Scientific). The difference of potentials in the emitter and the conical entrance was 2 kV, and the temperature of the heated capillary was 200°C. Ion fragmentation was performed by the methods of collision activated dissociation (CAD) and electron transfer dissociation (ETD). The mass of ions was recorded at a resolution of 240,000, while that of ion fragments was recorded at a resolution of 60,000. The fragmentation spectra were processed using the PEAKS Studio 7.5 software (Bioinformatics Solution Inc., Canada). Peptides whose signal intensity over the ion flux exceeded 106 were considered significant.
RESULTS AND DISCUSSION
Preparation of amyloid fibrils formed by synthetic Aβ(1-40) peptide. Aβ peptide monomers are soluble amphiphilic molecules. The mass of a monomer with length of 42 a.a. is 4513.1 Da. Its amino acid sequence is: DAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVVIA. The Aβ(1-40) peptide is distinguished by the absence of Ile and Ala residues at its C-terminus, and its molecular mass is 4329.2 Da (monoisotropic mass).
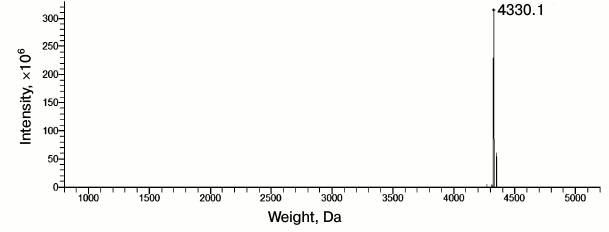
To carry out this study, we purchased the Aβ(1-40) preparation from Sigma-Aldrich (USA). Its purity was tested by mass spectrometry (Fig. 4) and found to correspond to that indicated by the manufacturer (≥90%, HPLC).
Fig. 4. Normalized mass spectrum of synthetic Aβ(1-40) peptide (Sigma-Aldrich). Index “4330.1” is the value of the most abundant mass of Aβ(1-40).
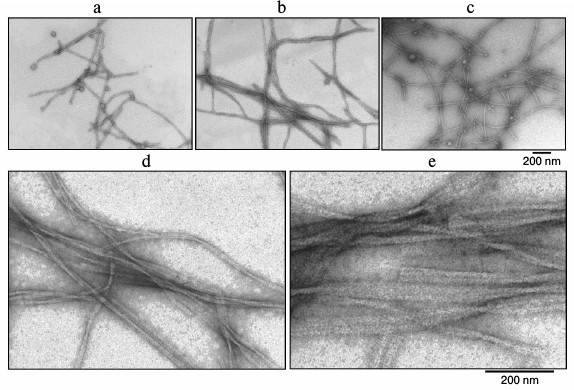
According to the EM data, the studied preparations contain fibril structures. Electron microscopy was also used to analyze the formation and morphological properties of the fibrils. Figure 5 shows amyloid fibrils formed by Aβ(1-40) at different times of incubation. As seen, even at the zero point (0 h incubation) there are fibrils (Fig. 5a) with even surface. The fibrils are not branched, and their diameter is about 8 nm. As the time of incubation increases, the number of fibrils grows (Fig. 5, b and c). At higher magnification, it is seen that fibrils form ribbons (Fig. 5, d and e). Such morphology of fibrils formed by Aβ(1-40) peptide was described earlier [32].
Fig. 5. EM images of fibrils of synthetic Aβ(1-40). The preparation dissolved in 5% DMSO, 50 mM Tris-HCl, pH 7.5, was incubated at 37°C (0.2 mg/ml): a) 0 h; b, c) 390 and 540 min, respectively; d, e) 390 and 540 min incubation, respectively (the images are shown at high magnification).
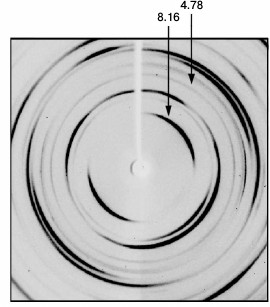
The presence of cross-β structure, described for amyloid fibrils, in the composition of fibrils was determined using X-ray analysis (by the presence of reflections at 4.8 and 8-10 Å characteristic of this structure in X-ray images). The data of X-ray analysis corroborate that the above fibrils contain cross-β structure (Fig. 6) as indicated by reflections at 8.16 and 4.78 Å.
Fig. 6. Data of diffraction analysis of the preparation of synthetic Aβ(1-40) peptide (Sigma-Aldrich). Arrows indicate reflections characteristic of amyloid structures.
Prediction of amyloidogenic regions of Aβ(1-40) by different methods. The FoldAmyloid method (http://bioinfo.protres.ru/fold-amyloid/) [2-4] developed specially for this purpose was used to predict amyloidogenic regions of the Aβ(1-40) peptide. We calculated regions in the sequence that from the theoretical point of view can be present in the core of amyloid structures formed by this peptide. These are regions from amino acid residues from 16 to 21 and from 32 to 36. Predictions made with other programs were as follows. Program PASTA2 (http://protein.bio.unipd.it/pasta2) [6] predicts only one region (30-40 a.a.), and WALTZ (http://waltz.switchlab.org/) [5] also predicts only one region (16-21 a.a.).
Determination of amyloidogenic regions in Aβ(1-40) using limited proteolysis. As known, the core of an amyloid fibril is resistant to the action of proteases. Therefore, it is possible to suggest that regions of the polypeptide chain that are involved in the core of the amyloid structure will be protected from the action of proteases, while the other parts of the polypeptide chain will be cleaved by proteases. Fibrils formed by this peptide were treated with a mixture of proteases consisting of trypsin, chymotrypsin, and proteinase K. When proteolysis was terminated, the mixture was centrifuged. The cleaved peptides (not included in the core of the amyloid fibril) remained in the supernatant, whereas amyloid structures were precipitated. The precipitate was washed, dried in a vacuum concentrator, and dissolved in formic acid. The sample was then layered on a reversed-phase column and eluted with an acetonitrile gradient. A mass spectrometer was used as the detector. The eluted peptides were ionized in the mass spectrometer, after which ions with the same mass were isolated and fragmented in an automatic mode using a specific algorithm. As a result, sets of masses of ions and their fragmentation spectra were obtained. After the analysis of the mass spectra and their fragments, the PEAKS Studio program was used to determine amino acid sequences of Aβ peptide fragments precipitated in the complex of amyloid fibrils.
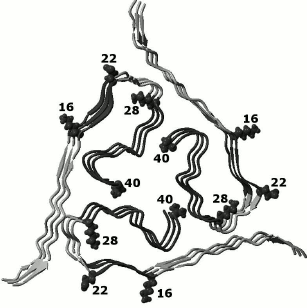
Figure 7 shows peptides accumulated after hydrolysis of Aβ(1-40). It is seen that peptides of the C-terminal region in the Aβ peptide are accumulated. In all C-terminal peptides, the region from amino acid residues 28 to 40 is resistant to the action of proteases. In addition, peptides are accumulated where the region from amino acid residues 16 to 22 also has increased resistance to the action of the proteases. In this case, control monomer (unassociated) structures of Aβ(1-40) are hydrolyzed completely into very short di-, tri-, and tetrapeptides. This indicates that in Aβ(1-40) the formation of the amyloid spine involves regions of amino acid residues 16-22 and 28-40. These data are compatible with those determining amyloid regions for this peptide that we predicted earlier (amino acid residues 16-21 and 32-36) using our program FoldAmyloid for finding amyloid forming regions in a protein chain. Besides, the sequences of amyloidogenic regions in peptide Aβ(1-40) obtained in our studies are supported by the results of solid-state NMR analysis [28] (Fig. 8).
Fig. 7. Sets of peptides accumulated after hydrolysis of Aβ(1-40) with a mixture of proteases (trypsin, chymotrypsin, proteinase K). The regions of Aβ(1-40) most protected from proteolysis in the amyloid structure are framed. Letters f, a, and o designate automatically determined modifications of amino acid regions (correspondingly formylation, acetylation, and oxidation). Amyloidogenic regions predicted theoretically by the FoldAmyloid program are underlined (http://bioinfo.protres.ru/fold-amyloid/) [4].
Fig. 8. Three-dimensional structure of a section of an amyloid fibril from the Aβ(1-40) peptide (the protein PDB code is 2M4J [14], three layers of a protofibril are shown) with regions (gray) included in the spine of the amyloid fibril according to our experimental data.
Thus, using mass-spectrometry we determined regions of the chain that are protected from the action of proteases in amyloid fibrils formed by the Aβ(1-40) peptide. We demonstrated that for the synthetic Aβ(1-40) peptide, amino acid regions 16-22 and 28-40 are protected from the action of proteases upon formation of the amyloid structure. These data are in agreement with the amyloidogenic regions (amino acid residues 16-21 and 32-36) in the sequence of the Aβ(1-40) peptide predicted theoretically.
We are grateful to Dr. A. D. Nikulin for his help in X-ray analysis of the peptide and Dr. A. V. Glyakina for her help in figures preparation.
This study was supported by the Russian Science Foundation (project No. 14-14-00536).
REFERENCES
1.Dovidchenko, N. V., and Galzitskaya, O. V. (2015)
Computational approaches to identification of aggregation sites and the
mechanism of amyloid growth, Adv. Exp. Med. Biol., 855,
213-239.
2.Galzitskaya, O. V., Garbuzynskiy, S. O., and
Lobanov, M. Y. (2006) Is it possible to predict amyloidogenic regions
from sequence alone? J. Bioinform. Comput. Biol., 4,
373-388.
3.Galzitskaya, O. V., Garbuzynskiy, S. O., and
Lobanov, M. Y. (2006) Prediction of amyloidogenic and disordered
regions in protein chains, PLoS Comput. Biol., 2,
e177.
4.Garbuzynskiy, S. O., Lobanov, M. Y., and
Galzitskaya, O. V. (2010) FoldAmyloid: a method of prediction of
amyloidogenic regions from protein sequence, Bioinformatics,
26, 326-332.
5.Maurer-Stroh, S., Debulpaep, M., Kuemmerer, N.,
Lopez de la Paz, M., Martins, I. C., Reumers, J., Morris, K. L.,
Copland, A., Serpell, L., Serrano, L., Schymkowitz, J. W. H., and
Rousseau, F. (2010) Exploring the sequence determinants of amyloid
structure using position-specific scoring matrices, Nat.
Methods, 7, 237-242.
6.Walsh, I., Seno, F., Tosatto, S. C. E., and
Trovato, A. (2014) PASTA 2.0: an improved server for protein
aggregation prediction, Nucleic Acids Res., 42,
W301-307.
7.Aguzzi, A. (2009) Cell biology: beyond the prion
principle, Nature, 459, 924-925.
8.Cohen, S. I. A., Linse, S., Luheshi, L. M.,
Hellstrand, E., White, D. A., Rajah, L., Otzen, D. E., Vendruscolo, M.,
Dobson, C. M., and Knowles, T. P. J. (2013) Proliferation of
amyloid-β42 aggregates occurs through a secondary nucleation
mechanism, Proc. Natl. Acad. Sci. USA, 110,
9758-9763.
9.Dovidchenko, N. V., Finkelstein, A. V., and
Galzitskaya, O. V. (2014) How to determine the size of folding nuclei
of protofibrils from the concentration dependence of the rate and
lag-time of aggregation. I. Modeling the amyloid protofibril formation,
J. Phys. Chem. B, 118, 1189-1197.
10.Meisl, G., Yang, X., Hellstrand, E., Frohm, B.,
Kirkegaard, J. B., Cohen, S. I. A., Dobson, C. M., Linse, S., and
Knowles, T. P. J. (2014) Differences in nucleation behavior underlie
the contrasting aggregation kinetics of the Aβ40 and Aβ42
peptides, Proc. Natl. Acad. Sci. USA, 111, 9384-9389.
11.Suvorina, M. Y., Selivanova, O. M.,
Grigorashvili, E. I., Nikulin, A. D., Marchenkov, V. V., Surin, A. K.,
and Galzitskaya, O. V. (2015) Studies of polymorphism of
amyloid-β42 peptide from different suppliers, J.
Alzheimer’s Dis., 47, 583-593.
12.Sticht, H., Bayer, P., Willbold, D., Dames, S.,
Hilbich, C., Beyreuther, K., Frank, R. W., and Rosch, P. (1995)
Structure of amyloid A4-(1-40)-peptide of Alzheimer’s disease,
Eur. J. Biochem., 233, 293-298.
13.Tomaselli, S., Esposito, V., Vangone, P., Van
Nuland, N. A. J., Bonvin, A. M. J. J., Guerrini, R., Tancredi, T.,
Temussi, P. A., and Picone, D. (2006) The alpha-to-beta conformational
transition of Alzheimer’s Abeta-(1-42) peptide in aqueous media
is reversible: a step by step conformational analysis suggests the
location of beta conformation seeding, Chembiochem, 7,
257-267.
14.Lu, J.-X., Qiang, W., Yau, W.-M., Schwieters, C.
D., Meredith, S. C., and Tycko, R. (2013) Molecular structure of
β-amyloid fibrils in Alzheimer’s disease brain tissue,
Cell, 154, 1257-1268.
15.Luhrs, T., Ritter, C., Adrian, M., Riek-Loher,
D., Bohrmann, B., Dobeli, H., Schubert, D., and Riek, R. (2005) 3D
structure of Alzheimer’s amyloid-β(1-42) fibrils, Proc.
Natl. Acad. Sci. USA, 102, 17342-17347.
16.Kang, J., Lemaire, H. G., Unterbeck, A., Salbaum,
J. M., Masters, C. L., Grzeschik, K. H., Multhaup, G., Beyreuther, K.,
and Muller-Hill, B. (1987) The precursor of Alzheimer’s disease
amyloid A4 protein resembles a cell-surface receptor, Nature,
325, 733-736.
17.Selkoe, D. J. (2004) Cell biology of protein
misfolding: the examples of Alzheimer’s and Parkinson’s
diseases, Nat. Cell Biol., 6, 1054-1061.
18.Dahlgren, K. N., Manelli, A. M., Stine, W. B.,
Baker, L. K., Krafft, G. A., and LaDu, M. J. (2002) Oligomeric and
fibrillar species of amyloid-β peptides differentially affect
neuronal viability, J. Biol. Chem., 277, 32046-32053.
19.Garzon-Rodriguez, W., Sepulveda-Becerra, M.,
Milton, S., and Glabe, C. G. (1997) Soluble amyloid Aβ-(1-40)
exists as a stable dimer at low concentrations, J. Biol. Chem.,
272, 21037-21044.
20.Harper, J. D., Wong, S. S., Lieber, C. M., and
Lansbury, P. T. (1999) Assembly of Aβ amyloid protofibrils: an
in vitro model for a possible early event in Alzheimer’s
disease, Biochemistry, 38, 8972-8980.
21.Haass, C., Schlossmacher, M. G., Hung, A. Y.,
Vigo-Pelfrey, C., Mellon, A., Ostaszewski, B. L., Lieberburg, I., Koo,
E. H., Schenk, D., and Teplow, D. B. (1992) Amyloid β-peptide is
produced by cultured cells during normal metabolism, Nature,
359, 322-325.
22.Kamenetz, F., Tomita, T., Hsieh, H., Seabrook,
G., Borchelt, D., Iwatsubo, T., Sisodia, S., and Malinow, R. (2003) APP
processing and synaptic function, Neuron, 37,
925-937.
23.Jarrett, J. T., and Lansbury, P. T. (1993)
Seeding “one-dimensional crystallization” of amyloid: a
pathogenic mechanism in Alzheimer’s disease and scrapie?
Cell, 73, 1055-1058.
24.Carrotta, R., Manno, M., Bulone, D., Martorana,
V., and San Biagio, P. L. (2005) Protofibril formation of amyloid
β-protein at low pH via a non-cooperative elongation mechanism,
J. Biol. Chem., 280, 30001-30008.
25.Talmard, C., Leuma Yona, R., and Faller, P.
(2009) Mechanism of zinc(II)-promoted amyloid formation: zinc(II)
binding facilitates the transition from the partially α-helical
conformer to aggregates of amyloid β protein(1-28), J. Biol.
Inorg. Chem., 14, 449-455.
26.Ma, Q.-F., Hu, J., Wu, W.-H., Liu, H.-D., Du,
J.-T., Fu, Y., Wu, Y.-W., Lei, P., Zhao, Y.-F., and Li, Y.-M. (2006)
Characterization of copper binding to the peptide amyloid-β(1-16)
associated with Alzheimer’s disease, Biopolymers,
83, 20-31.
27.Turner, A. J., Fisk, L., and Nalivaeva, N. N.
(2004) Targeting amyloid-degrading enzymes as therapeutic strategies in
neurodegeneration, Ann. N. Y. Acad. Sci., 1035, 1-20.
28.Zhang, Y., Rempel, D. L., Zhang, J., Sharma, A.
K., Mirica, L. M., and Gross, M. L. (2013) Pulsed
hydrogen–deuterium exchange mass spectrometry probes
conformational changes in amyloid beta (Aβ) peptide aggregation,
Proc. Natl. Acad. Sci. USA, 110, 14604-14609.
29.Lichtenthaler, S. F., Haass, C., and Steiner, H.
(2011) Regulated intramembrane proteolysis – lessons from
amyloid precursor protein processing, J. Neurochem., 117,
779-796.
30.Finder, V. H., and Glockshuber, R. (2007)
Amyloid-β aggregation, Neurodegener. Dis., 4,
13-27.
31.Dovidchenko, N. V., Glyakina, A. V., Selivanova,
O. M., Grigorashvili, E. I., Suvorina, M. Y., Dzhus, U. F., Mikhailina,
A. O., Shiliaev, N. G., Marchenkov, V. V., Surin, A. K., and
Galzitskaya, O. V. (2016) One of the possible mechanisms of amyloid
fibrils formation based on the sizes of primary and secondary folding
nuclei of Aβ40 and Aβ42, J. Struct. Biol., 194,
404-414.
32.Kodali, R., Williams, A. D., Chemuru, S., and
Wetzel, R. (2010) Aβ(1-40) forms five distinct amyloid structures
whose β-sheet contents and fibril stabilities are correlated,
J. Mol. Biol., 401, 503-517.