35S Promoter Methylation in Kanamycin-Resistant Kalanchoe (Kalanchoe pinnata L.) Plants Expressing the Antimicrobial Peptide Cecropin P1 Transgene
T. V. Shevchuk*, N. S. Zakharchenko, S. V. Tarlachkov, O. V. Furs, O. V. Dyachenko, and Y. I. Buryanov
Branch of Shemyakin–Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; fax: +7 (4967) 330-527; E-mail: shevchuk@bibch.ru* To whom correspondence should be addressed.
Received May 31, 2016
Transgenic kalanchoe plants (Kalanchoe pinnata L.) expressing the antimicrobial peptide cecropin P1 gene (cecP1) under the control of the 35S cauliflower mosaic virus 35S RNA promoter and the selective neomycin phosphotransferase II (nptII) gene under the control of the nopaline synthase gene promoter were studied. The 35S promoter methylation and the cecropin P1 biosynthesis levels were compared in plants growing on media with and without kanamycin. The low level of active 35S promoter methylation further decreases upon cultivation on kanamycin-containing medium, while cecropin P1 synthesis increases.
KEY WORDS: Kalanchoe pinnata, transgenic plants, antimicrobial peptide cecropin P1, DNA methylation, 35S cauliflower mosaic virus 35S RNA promoter, kanamycin, neomycin phosphotransferase IIDOI: 10.1134/S0006297916090054
Abbreviations: AMP, antimicrobial peptides; cecP1, cecropin P1 gene; Km, kanamycin; nptII, neomycin phosphotransferase II gene; 35S promoter, 35S cauliflower mosaic virus RNA promoter.
The adaptive response of plants, similar to all eukaryotic organisms, is
formed by the mechanisms of epigenetic regulation of chromatin
structure forming phenotypes (epigenomes) maximally adapted to
environmental conditions. These mechanisms involve reversible
epibiochemical modifications of various structural components of
chromatin, which comprises DNA, histones, other nuclear proteins and
noncoding RNAs [1]. Postreplicative DNA methylation
is an epibiochemical modification of chromatin. DNA methylation
performs different functions in a cell, including involvement in plant
adaptation to abiotic and biotic stresses [2-5]. The enhancement of plant resistance to pests and
phytopathogens is an important problem of plant bioengineering. In this
current area of research, attention is drawn to the possibility of
using the transgenes of antimicrobial peptides (AMP). It has been shown
that the expression of AMP transgenes not only increases plant
resistance to bacterial and fungal phytopathogens, but also enhances
the expression of some native plant genes and plant resistance to
various stresses [6-8]. It
seems that AMP transgenes can be integrated into the system of stress
resistance in plants and activate it. Gene expression can be controlled
by the level of gene promoter methylation [9].
Antibiotic resistance marker genes including the neomycin
phosphotransferase II gene (nptII) are widely used in the
selection of transgenic plants. The selectable marker gene nptII
provides plant resistance to kanamycin and supposedly can also be
integrated into the system of stress resistance of plants. The question
arises whether AMP biosynthesis can be enhanced by using the
nptII gene during the growth of transgenic plants on a
kanamycin-containing medium.
In the present work, we tested this assumption by analyzing the content of cecropin P1 and the methylation of 35S cauliflower mosaic virus RNA (35S) promoter controlling the cecropin P1 (cecP1) gene expression in nptII-cecP1 plants of Kalanchoe pinnata L. growing on kanamycin-containing medium.
MATERIALS AND METHODS
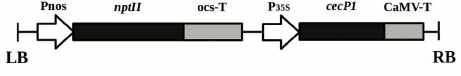
Plant material. The research objects were the previously obtained transgenic plants of the Kalanchoe pinnata L. line with the selective neomycin phosphotransferase II (nptII) gene under the nopaline synthase promoter and the antimicrobial peptide cecP1 gene under the 35S cauliflower mosaic virus RNA promoter [10] (Fig. 1). The plants were grown in vitro for a year, with grafting once in three weeks. The plants were cultivated on MS medium [11] containing 7 g/liter agar, 30 g/liter sucrose, and a standard set of salts (pH 5.8), without kanamycin (Km–) and with kanamycin (Km+, 25 mg/liter), at 22-24°C with a 16-h day and illumination of 100 µE·m–2 ·s–1.
Fig. 1. Scheme of transgenic structure incorporated into the genome of Kalanchoe pinnata plants. LB and RB, left and right boundaries of T-DNA; Pnos, nopaline synthase gene promoter; nptII, neomycin phosphotransferase II gene; ocs-T, octopine synthase gene polyadenylation signal; P35S, 35S cauliflower mosaic virus RNA promoter; cecP1, cecropin P1-encoding gene; CaMV-T, cauliflower mosaic virus polyadenylation signal.
Western blot analysis. Cecropin P1 was assayed in cell-free extracts of the transgenic plants. This method is completely described in [7]. Electrophoresis was performed in the Tricine-SDS-PAGE system; proteins were transferred onto a PVDF nylon membrane (Amersham Pharmacia Biotech, Great Britain) [12]. Enzyme immunoassay of cecropin P1 was carried out with rabbit polyclonal antibodies against this synthetic peptide and horseradish peroxidase-conjugated anti-rabbit immunoglobulins. Synthetic cecropin P1 was obtained by solid phase synthesis [13]. The membrane was developed using the ECL chemiluminescence system (Pierce, USA). The content of cecropin P1 was determined by densitometry.
Analysis of antimicrobial activity of plant extracts. The content of cecropin P1 in Km– plant extracts was determined by agar diffusion assay by the absence of growth of E. coli D22 test culture, with synthetic cecropin as a standard [10]. The Km+ plants were not analyzed due to potential presence of still active kanamycin in their extracts. The measurements were made in three biological and three analytical replications and are presented as the arithmetic mean of the values obtained and their standard errors (±SE).
Bisulfite treatment of genomic DNA. The DNA from the leaf mixture of three plants (analyzed for the presence of cecropin P1) was isolated using a Genomic DNA Purification Kit (ThermoScientific, Lithuania) (OD260/OD280 1.93, fragments of more than 10 kb). The genomic DNA of the kalanchoe plants was bisulfate-treated using an EpiTect reagent kit (Qiagen, USA). The DNA obtained after bisulfite conversion was amplified using the following pair of primers: 5′-gagggttattgagattttttaataaaggg-3′ (direct) and 5′-ttttaaacaaccaaaaacccataaatccc-3′ (reverse). The reaction mixture (20 µl) contained 1× sulfate buffer (75 mM Tris-HCl, pH 8.8, 20 mM (NH4)2SO4, 0.01% Tween 20), 1.5 mM MgCl2, 100 mM dNTP, 50 pmol of each primer, and 1 U of Taq polymerase (ThermoScientific). The program was as follows: 95°C – 30 s; 35 cycles: 95°C – 30 s, 58°C – 30 s, 72°C – 50 s; then 72°C – 3 min in a TC-24/H(B) amplifier (BIOER TECHNOLOGY, China). The PCR product corresponding to the 414 bp 35S promoter fragment was purified using a Zymoclean™ Gel DNA Recovery Kit (Zymo Research, USA).
Construction of recombinant vector and analysis of clones. The resultant PCR product was incorporated by ligation into the TA vector (Evrogen, Russia), which was then used to transform E. coli XL-1 cells (Stratagene, USA) with a Transform Aid Bacterial Transformation Kit (ThermoScientific). The DNAs from separate bacterial colonies were analyzed for the presence of the target insert by PCR with the primers used for amplification of the products of bisulfite conversion. The amplification conditions were as follows: 95°C – 4 min; 25 cycles: 95°C – 30 s, 58°C – 30 s, 72°C – 50 s; then 72°C – 3 min. The cells of selected colonies were grown in a liquid LB medium [14]; the plasmid DNA was isolated from these cells with a Gene JET Plasmid Miniprep Kit (ThermoScientific), and the target insert orientation was determined by PCR with the standard primer T7 and the reverse conversion primer. The amplification conditions were as follows: 95°C – 2 min; 25 cycles: 95°C – 30 s, 58°C – 30 s, 72°C – 50 s; then 72°C –3 min.
The selected plasmid DNA preparations were sequenced. The clones with direct orientation of the insert were sequenced using the standard primer SP6; the clones with reverse orientation of the insert were sequenced using the standard primer T7. The level of the 35S promoter methylation presented as the arithmetic mean and standard deviations (±SE) was determined from the data obtained for nine individual DNA clones of the Km+ and Km– plants under study.
RESULTS AND DISCUSSION
The transformed Kalanchoe pinnata L. plants expressing the cecP1 gene and the selective neomycin phosphotransferase II (nptII) gene were grown in vitro on the MS medium with and without kanamycin.
The amounts of cecropin P1 in the plants grown with and without kanamycin were compared by Western blot analysis. The content of P1 in plant extracts was determined after preliminary calibration using synthetic cecropin P1. The plants were shown to contain the desired peptide with molecular weight of about 3.4 kDa, corresponding to the mature form of cecropin P1 (Fig. 2). The higher intensity of positive cecropin signals was found in the plants growing on kanamycin-containing medium. The calculation by densitometry showed that the amount of cecropin P1 in Km+ and Km– plant extracts to be 0.052 ± 0.004 and 0.018 ± 0.005% of total soluble protein, respectively. The agar diffusion assay of the content of cecropin P1 in Km– plant extracts by the absence of microbial growth [10] yielded a similar value: 0.022 ± 0.004%. This method was not used to determine the content of cecropin P1 in Km+ plants due to potential presence of some portion of still active (not phosphorylated) kanamycin in their tissues.
Fig. 2. Western blot analysis of cecropin P1 from kalanchoe plant extracts (50 µg of total soluble protein). Lanes: K1) synthetic cecropin P1 (30 ng); K2) synthetic cecropin P1 (10 ng); 1) nontransformed plant; 2-4) Km+ plants; 5-7) Km– plants.
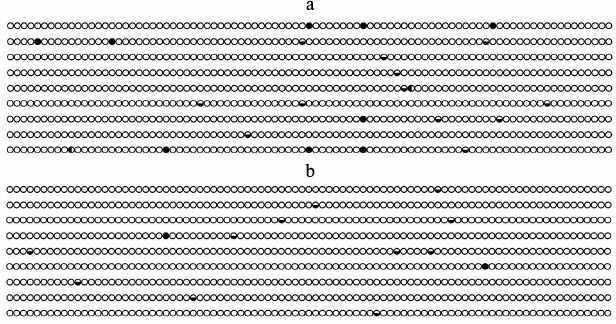
In the present work, bisulfite conversion was used to study the methylation of the 35S promoter controlling the synthetic cecP1 gene expression. The sequenced region of the promoter was a 333-bp fragment (upstream of adenine of the start codon). This sequence is in agreement with the published data on the structure of the 35S promoter [15]. This fragment has 87 potential sites of methylation in different nucleotide contexts. Among them, 13 are CG sites, 5 are CHG sites, and the rest are asymmetric CHH contexts (H = A, T, or C). A schematic representation of cytosine methylation at these sites is shown in Fig. 3.
Fig. 3. Schematic positioning of 5-methylcytosine among all cytidyl nucleotides in the 35S promoter controlling the cecP1 gene expression. a) DNA of Km– plants; b) DNA of Km+ plants; ○ – nonmethylated cytosine; ◒ – CH; ● – CG; ◐ – CHG (H = A, T, or C).
The preparations of DNA clones obtained from the Km+ and Km– plants were sequenced (nine of each). The level of the 35S promoter methylation was low in both cases, indicating its active state. The 35S promoter of the cecP1 gene contained 2.87 ± 0.52% of methylated cytosine residues in plants grown on the medium without kanamycin and 1.49 ± 0.17% of methylated cytosines in plants grown on the medium with kanamycin. Thus, the level of the 35S promoter methylation in the plants growth in the presence of kanamycin was lower compared to the plants grown without kanamycin.
The methylation of the 35S promoter of separate clones is variable, reflecting the specificity of its methylation in individual plant cells. In the 35S promoter of plants cultivated in the presence of kanamycin, the methylation of CG and CHG sites decreases to a greater extent compared to the modification of asymmetric CHH sites: from 9 to 2 for CG sites, from 2 to 0 for CHG sites, and from 14 to 11 for CH sites. In the asymmetric CHH site overlapping the consensus sequence of the CAAT promoter, the cytosine residue remained methylated in the plants grown both with and without kanamycin. At the same time, marked methylation of CG sites (cytosine positions 45 and 53) is observed in the 35S promoter of the plants grown without kanamycin. Probably, the site-specific methylation of the 35S promoter determines the level of its activity. As known, the adaptation of Mesembryanthemum crystallinum plants to environmental stresses is associated with the CHG-type of DNA methylation [4]. However, no site-specific peculiarities of the 35S promoter methylation were revealed in the transgenic plants of Kalanchoe pinnata. In the inactive 35S promoter of Nicotiana attenuate plants, the level of methylation in different site contexts is 5-9 times higher compared to that of the active promoter [16]. The studied Kalanchoe pinnata plants also demonstrate a low level of methylation of this promoter in the active state. The decreased level of the 35S promoter methylation in these plants during their growth on kanamycin is accompanied by increased level of cecropin P1 biosynthesis. Thus, the activity of the 35S promoter of transgenic plants depends on the degree of its demethylation. This work shows that the synthesis of targeted antimicrobial peptides can be regulated by specific conditions of cultivation of transgenic plants.
Previously, we have shown that the expression of the cecropin P1 gene in plants contributes to the enhancement of their resistance to stress factors, both biological and abiotic, and indicates the fitting of this heterologous gene into the integral system of stress resistance [7]. The nptII gene encoding the kanamycin-inactivating neomycin phosphotransferase determines the growth of transgenic plants on the selective medium with kanamycin and belongs to antibiotic resistance genes. The results suggest that the nptII gene is a component of the integral plant system of stress resistance. The selective nptII gene widely used in plant genetic engineering, by analogy with the cecP1 gene, can induce protection from various stress factors. Apparently, in plants possessing two foreign “protective” transgenes, the stress factor specific for one transgene is able to induce enhanced coordinated expression of the other “protective” transgene. Hence, the question arises as to the nature of the signal regulating the synthesis of cecropin P1 and the molecular mechanisms of its transduction with the involvement of the nptII gene in this process.
The molecular-epigenetic interaction between the cecP1 and nptII transgenes and their expression in plants will be further investigated to understand the mechanisms of stress resistance of plants.
Acknowledgements
This work was supported by the Ministry of Education and Science of the Russian Federation (Agreement No. 14.607.21.0016, the unique identifier for applied scientific research (project) RFMEFI60714X0016; generation and biochemical analysis of transgenic kalanchoe plants) and by the Russian Foundation for Basic Research (project No. 16-04-00623; genome sequencing).
REFERENCES
1.Buryanov, Y. I. (2015) Adaptive epibiochemistry and
epigenetics, Biochemistry (Moscow), 80, 1145-1156.
2.Kovarik, A., Koukalova, B., Bezdek, M., and Opatrn,
Z. (1997) Hypermethylation of tobacco heterochromatic loci in response
to osmotic stress, Theor. Appl. Genet., 95, 301-306.
3.Labra, M., Ghiani, A., Citterio, S., Sgorbati, S.,
Sala, F., Vannini, C., Ruffini-Castiglione, M., and Bracale, M. (2002)
Analysis of cytosine methylation pattern in response to water deficit
in pea root tips, Plant Biol., 4, 694-699.
4.Dyachenko, O. V., Zakharchenko, N. S., Shevchuk, T.
V., Bohnert, H. J., Cushman, J. C., and Buryanov, Y. I. (2006) Effect
of hypermethylation of CCWGG sequences in dna of
Mesembryanthemum crystallinum plants on their adaptation
to salt stress, Biochemistry (Moscow), 71, 461-465.
5.Boyko, A., Kathiria, P., Zemp, F. J., Yao, Y.,
Pogribny, I., and Kovalchuk, I. (2007) Transgenerational changes in the
genome stability and methylation in pathogen-infected plants
(virus-induced plant genome instability), Nucleic Acids Res.,
35, 1714-1725.
6.Campo, S., Manrique, S., Garcia-Martinez, J., and
San Segundo, S. (2008) Production of cecropin A in transgenic rice
plants has an impact on host gene expression, Plant Biotechnol.
J., 6, 585-608.
7.Zakharchenko, N. S., Buryanov, Ya. I., Lebedeva, A.
A., Pigoleva, S. V., Vetoshkina, D. V., Loktyushov, E. V., Chepurnova,
M. A., Kreslavsky, V. D., and Kosobryukhov, A. A. (2013) Physiological
peculiarities of transgenic rape plants expressing the gene of
antimicrobial peptide cecropin P1, Fiziol. Rast., 60,
424-433.
8.Goyal, R. K., Hancock, R. E. W., Mattoo, A. K., and
Misra, S. (2013) Expression of an engineered heterologous antimicrobial
peptide in potato alters plant development and mitigates normal abiotic
and biotic responses, PLoS One, 8, e77505.
9.Mette, M. F., Aufsatz, W., Van der Winden, J.,
Matzke, M. A., and Matzke, A. J. (2000) Transcriptional silencing and
promoter methylation triggered by double-stranded RNA, EMBO J.,
19, 194-201.
10.Zakharchenko, N. S., Rukavtsova, E. B., Shevchuk,
T. V., Furs, O. V., Pigoleva, S. V., Lebedeva, A. A., Chulina, I. A.,
Baydakova, L. K., and Buryanov, Ya. I. (2016) Generation and
characterization of Kalanchoe pinnata L. plants expressing the
gene of the antimicrobial peptide cecropin P1, Prikl. Biokhim.
Mikrobiol., 52, 1-8.
11.Murashige, T., and Skoog, F. (1962) A revised
medium for rapid growth and bioassays with tobacco cultures,
Physiol. Plant., 15, 473-497.
12.Schagger, H., and Von Jagow, G. (1987)
Tricine-sodium dodecyl sulfate-polyacrilamide gel electrophoresis for
the separation of proteins in the range from 1 to 100 kDa,
Anal. Biochem., 166, 368-379.
13.Rodionova, L. N., Zagranichny, V. E., Rodionov,
I. L., Lipkin, V. M., and Ivanov, V. T. (1997) Complete solid phase
synthesis of γ-subunit of bovine retinal cGMP phosphodiesterase
and some physicochemical properties of the synthesized protein,
Bioorg. Khim., 23, 933-948.
14.Sambrook, J., Fritsch, E. F., and Maniatis, T.
(1989) Molecular Cloning: A Laboratory Manual, Cold Spring
Harbor Laboratory Press, N. Y.
15.Topfer, R., Matzeit, V., Gronenborn, B., Schell,
J., and Steinbiss, H.-H. (1987) A set of plant expression vectors for
transcriptional and translational fusions, Nucleic Acids Res.,
15, 5890.
16.Weinhold, A., Kallenbach, M., and Baldwin, T.
(2013) Progressive 35S promoter methylation increases rapidly during
vegetative development in transgenic Nicotiana attenuate plants,
BMC Plant Biol., 13, 99-116.