Dioxygenases of Chlorobiphenyl-Degrading Species Rhodococcus wratislaviensis G10 and Chlorophenol-Degrading Species Rhodococcus opacus 1CP Induced in Benzoate-Grown Cells and Genes Potentially Involved in These Processes
I. P. Solyanikova1*, O. V. Borzova1,2, E. V. Emelyanova1, E. S. Shumkova1,3, N. V. Prisyazhnaya1, E. G. Plotnikova4, and L. A. Golovleva1,2
1Skryabin Institute of Biochemistry and Physiology of Microorganisms, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; E-mail: innas@ibpm.pushchino.ru2Pushchino State Natural Science Institute, 142290 Pushchino, Moscow Region, Russia
3Bach Institute of Biochemistry, Research Center of Biotechnology, Russian Academy of Sciences, 119071 Moscow, Russia
4Institute of Ecology and Genetics of Microorganisms, Ural Branch, Russian Academy of Sciences, 614081 Perm, Russia
* To whom correspondence should be addressed.
Received May 30, 2016; Revision received June 9, 2016
Dioxygenases induced during benzoate degradation by the actinobacterium Rhodococcus wratislaviensis G10 strain degrading haloaromatic compounds were studied. Rhodococcus wratislaviensis G10 completely degraded 2 g/liter benzoate during 30 h and 10 g/liter during 200 h. Washed cells grown on benzoate retained respiration activity for more than 90 days, and a high activity of benzoate dioxygenase was recorded for 10 days. Compared to the enzyme activities with benzoate, the activity of benzoate dioxygenases was 10-30% with 13 of 35 substituted benzoate analogs. Two dioxygenases capable of cleaving the aromatic ring were isolated and characterized: protocatechuate 3,4-dioxygenase and catechol 1,2-dioxygenase. Catechol inhibited the activity of protocatechuate 3,4-dioxygenase. Protocatechuate did not affect the activity of catechol 1,2-dioxygenase. A high degree of identity was shown by MALDI-TOF mass spectrometry for protein peaks of the R. wratislaviensis G10 and Rhodococcus opacus 1CP cells grown on benzoate or LB. DNA from the R. wratislaviensis G10 strain was specifically amplified using specific primers to variable regions of genes coding α- and β-subunits of protocatechuate 3,4-dioxygenase and to two genes of the R. opacus 1CP coding catechol 1,2-dioxygenase. The products were 99% identical with the corresponding regions of the R. opacus 1CP genes. This high identity (99%) between the genes coding degradation of aromatic compounds in the R. wratislaviensis G10 and R. opacus 1CP strains isolated from sites of remote location (1400 km) and at different time (20-year difference) indicates a common origin of biodegradation genes of these strains and a wide distribution of these genes among rhodococci.
KEY WORDS: Rhodococcus wratislaviensis G10, Rhodococcus opacus 1CP, benzoate, catechol 1,2-dioxygenase, protocatechuate 3,4-dioxygenase, biodegradation genesDOI: 10.1134/S000629791609008X
Abbreviations: Amax, maximal activity at saturation with substrate; BDO, benzoate 1,2-dioxygenase; Cat, catechol; Cat-1,2-DO, catechol 1,2-dioxygenase; Cat-2,3-DO, catechol 2,3-dioxygenase; 4-CCat, 4-chlorocatechol; 3,5- and 4,5-DCCat, 3,5- and 4,5-dichlorocatechols; Ki, inhibition constant; Km, Michaelis constant; MALDI-TOF, matrix-assisted laser desorption/ionization-time of flight mass spectrometry; 3MCat and 4MCat, 3- and 4-methylcatechol; OD, optical density; PCA, protocatechuate; PCA-3,4-DO, protocatechuate 3,4-dioxygenase; pHBA, para-hydroxybenzoate.
Bacteria are capable of degrading aromatic compounds due to possession
of high specificity enzymes. To completely utilize growth substrates,
bacteria need to have initial attack enzymes with a relatively wide
substrate specificity that allow the cell to realize its primary attack
on a substrate, and a set of enzymes catalyzing the conversion of
resulting products into the tricarboxylic acid cycle intermediates.
Studies on bacterial degradation of natural and anthropogenic compounds
have shown that bacteria have a great variety of enzymes capable of
degrading these compounds via different pathways [1-3].
Benzoic acid is widely distributed in the environment. It can be produced by living organisms themselves and be a result of human activities leading to accumulation of this compound. The wide use of benzoate as a preservative explains increased interest in the behavior and fate of this compound in the environment. An enzyme initiating benzoate degradation by microorganisms under aerobic conditions, benzoate 1,2-dioxygenase (BDO; EC 1.14.12.10) [4], is a bicomponent system consisting of reductase and oxidase components; the latter is presented in α- and β-type subunits [5]. The α-subunit is believed to be responsible for the substrate specificity of BDO [6, 7]. Aerobic degradation of benzoate occurs via multiple pathways, with production as key intermediates of catechol (Cat) (mainly in bacteria), protocatechuate (PCA) (mainly in fungi), hydroquinone, gentisate (2,5-dihydroxybenzoate), and CoA-derivatives [4, 8-10]. Cleavage of the catechol ring in position 1,2 is catalyzed by catechol 1,2-dioxygenase (Cat-1,2-DO; EC 1.13.11.1) and by catechol 2,3-dioxygenase (Cat-2,3-DO; EC 1.13.11.2) in position 2,3. Both variants of the reaction are widely distributed in bacteria. Our earlier studies revealed that 11 strains of four genera of actinobacteria degraded benzoate by cleaving the produced catechol in the ortho-position [11]. Using specific primers, these strains were tested for the presence of the gene encoding the BDO α-subunit. Sequencing the fragments obtained from specific amplification showed that these regions were highly homologous. A dendrogram was obtained in which fragments of BDO from two strains, Rhodococcus opacus 1CP and Rhodococcus wratislaviensis G10, were clustered together. The similarity with the corresponding regions of BDO from the strain Rhodococcus jostii RHA1 that belongs to the most completely studied strains degrading wide spectrum pollutant was, respectively, 88 and 97% in the R. opacus 1CP and R. wratislaviensis G10 strains, which indicated that these genes should have a common origin [11, 12]. Cultures of R. opacus 1CP and R. wratislaviensis G10 intact cells were used for preliminary assessment of the substrate specificity of BDOs, and BDO from R. wratislaviensis G10 was shown to have a slightly wider specificity than the enzyme from R. opacus 1CP [11].
During benzoate degradation by actinobacteria, there is another poorly studied feature: the type of dioxygenase cleaving the aromatic ring of catechol produced during the transformation of benzoate. As mentioned above, the aerobic degradation of benzoate by bacteria in the great majority of cases is associated with production of catechol. However, in some works there are data indicating that in addition to catechol-specific dioxygenases, the cell-free extract contains protocatechuate 3,4-dioxygenase (PCA-3,4-DO; EC 1.13.11.3), but its role during this process is unclear. The PCA-3,4-DO activity in the cell-free extract varies significantly, from zero in Streptomyces spp. to 25-28 nmol/min per mg in Amycolatopsis spp. [13]. Activities of the two dioxygenases, Cat-1,2-DO and PCA-3,4-DO, were detected in the cell-free extract of R. opacus 1CP and R. wratislaviensis G10 cultures grown on benzoate [11]. The specific activity of PCA-3,4-DO in the G10 strain cell-free extract was comparable with the Cat-1,2-DO activity. Both strains are very interesting because they degrade aromatic pollutants with high activity, and thus can be used in biotechnology for removal of resistant compounds from polluted soils.
The purpose of the present work was to determine specific features of dioxygenases induced during cultivation of the bacterium R. wratislaviensis G10 on benzoate, to comparatively characterize enzymes of this strain and of those of R. opacus 1CP (which degrades a wide spectrum of aromatic toxicants), and to analyze the presence in these cultures of similar genes potentially involved in benzoate degradation.
MATERIALS AND METHODS
Microorganisms. The study was performed on the gram-positive non-sporogenous bacterium R. wratislaviensis G10 isolated from soil polluted with haloaromatic compounds (territory of the “Galogen” enterprise, Perm) [14], and on R. opacus 1CP isolated from an enrichment culture on 2,4-dichlorophenol and capable of degrading monochlorophenols, some dichlorophenols, benzoate, and some substituted benzoates [15].
Assessment of benzoate utilization ability. The strains were cultured on sodium benzoate using a liquid mineral medium of the following composition (g/liter): Na2HPO4 – 0.73; KH2PO4 – 0.35; MgSO4·7H2O – 0.1; NaHCO3 – 0.25; MnSO4 – 0.002; NH4NO3 – 0.75; FeSO4·7H2O – 0.02. The biomass grown on the slant agar (five tubes) was washed off with the mineral medium and cultured in 750-ml Erlenmeyer flasks containing 100 ml of the medium and benzoate in concentrations of 0.25, 0.5, 0.75, 1, 2, 4, 8, and 10 g/liter (29°C, 220 rpm). Experiments on the enzyme induction were performed on the cells, which under laboratory conditions were maintained on a rich medium to exclude traces of dioxygenase activity on the growth on aromatic substrates.
DNA was isolated using a DiatomTM DNA Prep reagent kit (Biokom, Russia) according to recommendations of the producer, with slight modifications.
Primers were chosen for amplification of genes encoding protocatechuate 3,4-dioxygenase and catechol 1,2-dioxygenase using the PrimerQuest Tool program (http://eu.idtdna.com/Primerquest/Home/Index). Thermodynamic parameters and tertiary structures of oligonucleotides were predicted using OligoAnalyzer 3.1 (eu.idtdna.com/calc/analyzer). The primer specificity was tested in primer-blast (http://www.ncbi.nlm.nih.gov/tools/primer-blast).
Primers for amplification of genes encoding PCA-3,4-DO α- and β-subunits were chosen by nucleotide sequences of the R. opacus 1CP genes pcaG and pcaH (AF003947.1) (Table 1), primers for screening Cat-1,2-DO (catA) genes were chosen by nucleotide sequences of the R. opacus 1CP genes encoding catechol 1,2-dioxygenases (GenBank data). Four pairs of primers were chosen specific for isofunctional genes encoding Cat-1,2-DO (Table 1).
Table 1. Oligonucleotides used in this
work

Genes pcaG and pcaH were amplified using a Bio-Rad T100 apparatus (Bio-Rad, USA) under the following conditions: 95°C – 5 min, then eight cycles decreasing annealing temperature by 2°C every second cycle: 95°C – 30 s, 62°C > 60°C > 58°C > 56°C – 30 s, 72°C – 40 s; then 20 cycles: 95°C – 30 s, 54°C – 30 s, 72°C – 40 s; the terminal building: 72°C – 5 min. The catA genes were amplified similarly, decreasing annealing temperature by 2°C every second cycle (from 58 to 52°C) – eight cycles. The next 22 cycles were performed at 52°C. Elongation was realized at 72°C for 50 s. The reaction products were separated by electrophoresis in 1% agarose gel at 10 V/cm, stained with ethidium bromide solution (5 µg/ml), and photographed in UV using the Gel DocTM XR gel-documentation system (Bio-Rad).
The PCR product was sequenced at the GENOM Shared Facilities Centre, Institute of Molecular Biology, Russian Academy of Sciences (Moscow) using a Dynamic Terminator Cycle Sequencing Ready Reaction Kit (Amersham, Great Britain) on an automatic ABI 3100 Avant Genetic Analyser sequencer (Applied Biosystems, USA) according to the producer’s recommendations.
Analysis of nucleotide sequences of functional genes. Homological sequences were searched for in the GenBank/DDBJ/EMBL database (http://www.ncbi.nlm.nih.gov). The nucleotide sequences were analyzed using the BLAST algorithm (http://www.ncbi.nlm.nih.gov).
Deposition of nucleotide sequences. Nucleotide sequences of fragments of the genes catA2, catA, pcaG, and pcaH were deposited into GenBank under accession numbers KX084387, KX084388, KX084389, and KX084390, respectively.
MALDI-TOF mass spectrometry analysis. Intact cells were used for the analysis. Cultures were grown on an agarized mineral medium containing benzoate (250 mg/liter) and on the rich LB medium. A thin layer of the cells was placed with a sterile spatula onto a steel target plate, mixed with 0.6 µl of 70% formic acid, and dried. Then 0.6 µl of the matrix (α-cyano-4-hydroxycinnamic acid (HCCA)) in 50% aqueous solution of acetonitrile containing 2.5% trifluoroacetic acid) was deposited, and the plate was air dried at room temperature. Spectra were recorded in linear mode with delayed ion extraction with an Autoflex Speed device (Bruker Daltonics, Germany) at delay time of 350 ns and accelerating voltage of 20 kV. The spectra were recorded in the positive ion mode, and masses were recorded in the range of 2-20 kDa. The device was calibrated externally using the Bruker Bacterial Test Standard protein mixture (Bruker Daltonics) at spectra resolution of ±2 Da (200 ppm). The resulting spectra of each preparation of the strains were obtained by summarizing the spectra recorded in 10-15 points of analyzed preparations under 500 laser pulses. The mass spectra were processed using the Flex Analysis 3.0 program (Bruker Daltonics).
Determination of respiration and BDO activity by polarography. To determine the cell respiration and BDO activity of the R. wratislaviensis G10 strain, the cells freshly grown on benzoate (0.2-6 g/liter) were separated by centrifugation (16,000g, 15 min, 4°C), washed twice with Tris-HCl buffer (pH 7.6), resuspended in similar buffer, and analyzed immediately. The measurements were performed in 50 mM Tris-HCl buffer (pH 7.6) saturated with air oxygen at room temperature in an open stirrer-equipped 5-ml cuvette.
To determine the BDO activity, after recording of the basal cell respiration, substrate solution was introduced into the cuvette, and the change in cellular oxygen consumption was measured using a Clark oxygen electrode. The oxygen electrode was used in combination with an Ingold 531 O2 Amplifier system (Instrumentation Laboratory (MI), Switzerland-USA). The signal was recorded using an XY Recorder-4103 two-coordinate recorder (Laboratorium Pristroje, Czech Republic). This signal characterized the rate of BDO enzymatic reaction with substrates. The rate was expressed in pA/s (1 pA/s ~ 0.153 µg O2/liter per s). The cells remaining after the analysis were frozen and stored at –20°C until use.
To determine the respiration activity, the stirring was stopped after the basal cellular respiration was recorded and the rate of the oxygen concentration change was measured with the Clark oxygen electrode. This signal characterized the culture respiration rate. The respiration rate was expressed in µg O2/liter per s.
Experimental data on enzymatic kinetics were processed and Michaelis–Menten equation constants were determined using the SigmaPlot program (v. 12).
Activity determination in cell-free extracts and enzyme purification. The cells were cultivated in 750-ml flasks (28°C, 220 rpm) containing 200 ml of medium with benzoate (200-250 mg/liter) up to OD545 of 3.0-3.5. The biomass was collected and broken, and activities of the enzymes Cat-1,2-DO and PCA-3,4-DO were determined as described earlier [11, 12]. The activity unit was determined as the amount of the enzyme catalyzing conversion of 1 µmol substrate or production of 1 µmol product in 1 min.
The protein concentration was determined by a modified Bradford method [16] with BSA as a standard.
The measurements were repeated thrice in at least three independent series of experiments. The results present the averaged values. The data were processed using Student’s t-test test taking p < 0.05 as significant.
RESULTS
Growth of R. wratislaviensis G10 on benzoate and induction of dioxygenases. Cultivation of the R. wratislaviensis G10 strain revealed that benzoate (2 g/liter) was completely degraded in 30 h. The strain could degrade up to 10 g/liter of benzoate, but the lag-phase was sharply increased to 50 h at benzoate concentration of 4 g/liter and higher.
The cell growth on benzoate was accompanied by induction of benzoate 1,2-dioxygenase, catechol 1,2-dioxygenase, and protocatechuate 3,4-dioxygenase. Considering the oxygen dependence of these enzymes, dynamics of their induction were compared on growing and intact non-growing cells.
During cultivation of the R. wratislaviensis G10 and R. opacus 1CP strains on the rich LB medium and on the mineral medium with benzoate (300 mg/liter) and protocatechuate (0.25 mM), the optical density increased 3-5-fold (Table 2) in three days. No growth was observed during this time on the catechol-containing medium. Determination of the enzyme activities in the cell-free extracts showed that there was no dioxygenase activity or only its traces were detected during growth of strains on LB, and in the R. opacus 1CP strain the background values were slightly higher than those in the G10 strain. When both strains were grown on benzoate, the specific activities of Cat-1,2-DO and PCA-3,4-DO increased by two orders of magnitude. Cultivation in the presence of protocatechuate was accompanied by induction of both dioxygenases, but the trace level activity of Cat-1,2-DO was recorded that was typical for the cell growth on LB.
Table 2. Activities of dioxygenases
determined by polarography and spectrophotometry methods for the R.
wratislaviensis G10 and R. opacus 1CP strain cells cultured
on different substrates
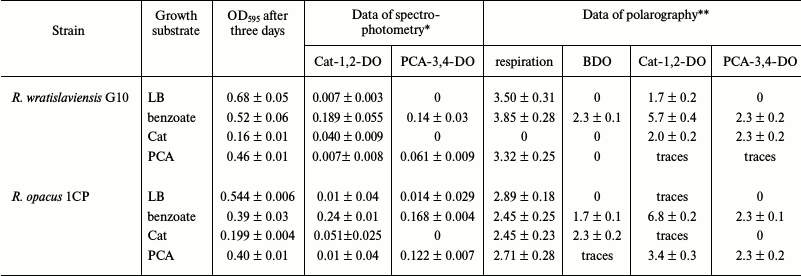
Notes: Mean results of at least three independent experiments are
presented.
* Specific activity, U/mg.
** Respiration, µg O2/liter per s; activity,
pA/s.
Table 2 presents results of the enzyme activity determination by polarography and spectrophotometry in the same preparations. Activities of BDO and PCA-3,4-DO were not detected by polarography in the cells of R. opacus 1CP and R. wratislaviensis G10 and of Cat-1,2-DO in the R. opacus 1CP cells grown on LB, which corresponded to the data of the spectrophotometric analysis. In the biomass of both strains grown on the mineral medium with benzoate, the activities of all three dioxygenases could be determined by polarography, whereas the BDO activity was not detected by spectrophotometry. Thus, the data presented show that the enzymatic pool of the cell can be assessed by either of the methods, notwithstanding limitations of each. For spectrophotometry, these limitations are associated with the possibility or impossibility of determining the enzyme activity after destruction of the native structure of complex proteins as in the case of BDO. The measurement accuracy of the polarographic method is determined by density of the cell suspension introduced into the reaction cuvette and by the physiological conditions of the cells.
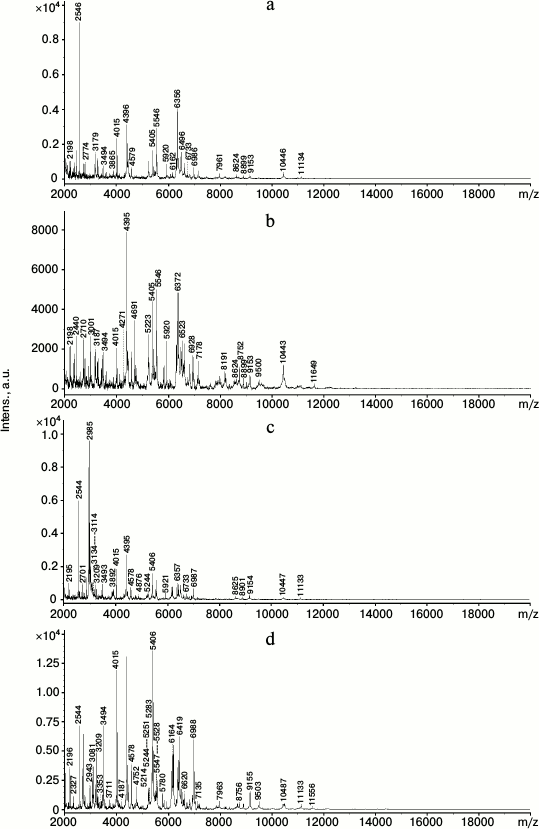
The strong similarity of the two strain cells was also confirmed by MALDI-TOF mass spectrometry analysis of the intact cells. Figure 1 shows that the cells of each strain grown on LB and on benzoate were different in their protein profiles in the limits of m/z values from 2 to 20,000 Da. Only 35 peaks with the same m/z value were found in the R. opacus 1CP strain cells grown on benzoate and on LB. The R. wratislaviensis G10 strain cells cultivated on benzoate and LB had still lower number of peaks with the same m/z value – only 27. On growth of R. opacus 1CP and R. wratislaviensis G10 on LB there were, respectively, 98 and 90 peaks, and 88 of them were the same in both cultures. When benzoate was used as a growth substrate, 94 peaks were recorded in the R. opacus 1CP and 87 peaks in the R. wratislaviensis G10 cells, and 77 of them were coincidental. In reality, the number of coincidences was higher, but some of the peaks gave very weak signals. Thus, according to the MALDI-TOF-mass spectrometry, the protein profiles of the 1CP and G10 strains grown under the same conditions and on the same substrate were only slightly different. The growth substrate influenced significantly and similarly the protein profile of the two strain cells, which suggests a high likeness of metabolic processes in the bacteria under comparison.
Fig. 1. MALDI-TOF mass spectra of intact cells of the R. wratislaviensis G10 (a, c) and R. opacus 1CP (b, d) strains grown on LB (a, b) and on benzoate (c, d).
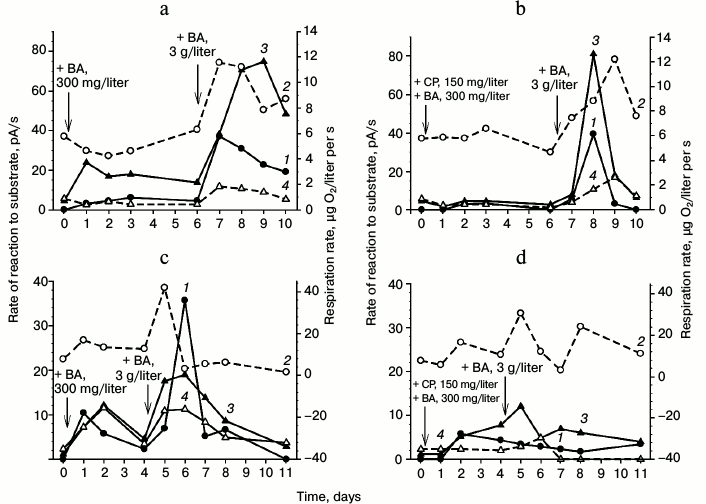
Induction of dioxygenases under non-growth conditions. Figure 2 shows results of induction of oxygen-dependent oxygenases in two variants of G10 culture: in cells freshly grown on LB and in cells grown on 6-10 g/liter benzoate with subsequent prolonged starvation for more than two months. Analysis by polarography of the enzymatic activity in both variants of the cells resuspended in buffer revealed virtually full absence of the dioxygenase activity. Not depending on the physiological conditions (freshly grown on LB or after the starvation for 2.5 months with aeration and 29°C), the cells manifested virtually the same respiration activity.
Fig. 2. Induction of dioxygenases by benzoate (BA) in the R. wratislaviensis G10 cells grown on the LB medium (a, b) and on benzoate (c, d) (starvation for 2.5 months after the growth on benzoate): a, c) induction in the absence of chloramphenicol (CP); b, d) induction in the presence of chloramphenicol. Curves: 1) rate of the reaction to benzoate; 2) respiration rate; 3) rate of the reaction to Cat; 4) rate of the reaction to PCA. The cells were resuspended in buffer. The moment of benzoate or chloramphenicol addition is indicated by the arrow. The substrate concentrations for testing the cell reactions were 3.47 mM of benzoate and 0.25 mM for Cat and PCA.
Analysis of the cell respiration activity revealed an interesting regularity observed upon benzoate addition to the cell suspension: the curve of cell respiration change repeated in the shape the curves of dioxygenase activity growth, but it was usually ahead of them by 0.5-1 day. The respiration activity increased in the presence of the substrate and then decreased to virtually the initial level.
The dioxygenase activity level also depended on the concentration of added benzoate. In all cases, the addition to the cells of 300 mg/liter benzoate led in less than 20 h to an increase in the reaction to benzoate, catechol, and protocatechuate. By the fourth day, the enzyme activities decreased, but their activities remained higher than those of the non-induced cells. The addition of 3 g/liter benzoate to such preadapted culture resulted in a sharp increase in the dioxygenase activities. We observed an induction of dioxygenase activities (inducible enzymes), which was confirmed by data obtained on the addition of chloramphenicol (an inhibitor of protein synthesis) to the cell suspension before the introduction of benzoate (Fig. 2, b and d). During the first day, chloramphenicol suppressed completely the BDO induction and virtually completely suppressed the induction of Cat-1,2-DO.
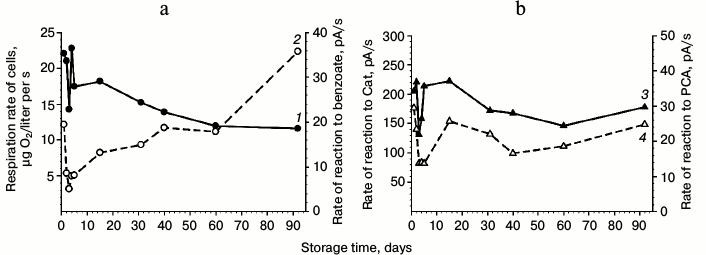
Benzoate 1,2-dioxygenase. The influence of storage duration of benzoate-grown cells on their respiration activity and on the activity of dioxygenases was studied. The results are presented in Fig. 3. Cells that had been washed and stored in buffer at 4°C retained viability for more than 90 days. More pronounced fluctuations in the respiration rates and in the cell reaction to benzoate, Cat, and PCA were observed during the first 5 days, and the changes in the cell reactions to these substrates occurred synchronously during the whole period under study. After the cell storage in buffer for 15 days, the cell reactions to benzoate, Cat, and PCA decreased. After storage for 35 days, the activities virtually stabilized. After storage for 90 days at 4°C, the BDO activity of the cells decreased twofold but remained reliably above zero (Fig. 3a). A slight reaction of the cells to PCA was also recorded, whereas the activity to Cat was high (Fig. 3b). However, during cell starvation (benzoate as the sole source of carbon and energy was exhausted) during 75 days under continuous aeration by stirring the culture liquid at 29°C, the BDO activity fell to zero, whereas the cell reaction to Cat and PCA under the same conditions could be identified as traces, i.e. they were virtually on the level of measurement error (Fig. 2c, curves 1, 3, 4).
Fig. 3. Influence of storage duration of benzoate-grown R. wratislaviensis G10 cells on their respiration activity (a) and on the activity of dioxygenases (b). Curves: 1) rate of reaction to benzoate; 2) respiration rate; 3) rate of reaction to Cat; 4) rate of reaction to PCA. The cells were suspended in buffer. The substrate concentrations for testing the cell reactions were as follows: benzoate – 3.47 mM; Cat and PCA – 0.25 mM.
The cell suspension was resistant on storage not only in buffer but also in tap water (artesian water in Pushchino, Moscow Region). Storage for 12 days at 4°C led to decrease in the cell respiration by less than twofold, from 17 to 13 µg O2/liter per s. The BDO activity during the same time decreased from 29 to 8 pA/s, but it remained on the level obtained after BDO induction by benzoate in cells grown on LB.
The influence of buffer pH on cell respiration and BDO activity was studied. The respiration of cells resuspended in buffer displayed virtually the same oxygen consumption and weakly depended on pH of the reaction buffer (50 mM Tris-HCl) in the range of 7.0-9.5 (the respiration changed from 20 to 23 µg O2/liter per s, respectively). The difference in the oxygen consumption of the cells resuspended in water at different pH values changed slightly stronger (17 µg O2/liter per s at pH 7.2 and 25 µg O2/liter per s at pH 9.0). The BDO activity in the cells stored in water was about a quarter lower than in the cells stored in the buffer (22-29 and 36-44 pA/s, respectively) within the pH range of the reaction buffer.
The substrate specificity of BDO from R. wratislaviensis G10 was assessed using intact cells. The cell suspension activity with benzoate was 7.3 ± 0.5 µg O2/liter per s. The substrate specificity of BDO from benzoate-grown R. wratislaviensis G10 was assessed earlier using 15 substrate analogs [11]. In the present work, we broadened twofold, to 35, the spectrum of compounds for testing, including potential substrates: isomeric monochloro-, monomethyl-, and hydroxy-substituted benzoates, 2-methoxybenzoate, all isomeric monochloro- and monomethyl-salicylates, dihydroxy- and dichloro-benzoates, 2- and 4-methoxy- and 3,4-dimethoxycinnamic acids, phenol, and isomeric monochlorophenols. In addition to the earlier found activity with 3-chlorobenzoate, phenol, and 3- and 4-chlorophenols, the R. wratislaviensis G10 cells were shown to actively consume oxygen on addition of 2-methylbenzoate (10%), 3-methylbenzoate (21%) and 4-methylbenzoate (30%), 3-methylsalicylate (14%), 2-methoxybenzoate (10%), 2-methoxycinnamic (15%), and 4-methoxycinnamic (15%) acids (in parentheses changes in the O2 consumption are shown in percent from its value on benzoate addition). These findings show that BDO induced in benzoate-grown R. wratislaviensis G10 is characterized by narrow substrate specificity. In general, the reaction of benzoate-grown cells to the substrate analogs with different number and properties of the substituents was insignificant compared to the activity to benzoate.
Activities of two dioxygenases cleaving the aromatic ring, Cat-1,2-DO and PCA-3,4-DO, were found in both R. wratislaviensis G10 and R. opacus 1CP grown on benzoate. Depending on the benzoate concentration, the specific activity of Cat-1,2-DO from R. wratislaviensis G10 varied in the limits 0.07-0.23 U/mg, and for the enzyme from R. opacus 1CP these values were 0.22-0.25 U/mg. The activity of PCA-3,4-DO from R. wratislaviensis G10 was 0.12-0.43 U/mg, and the activity of PCA-3,4-DO from R. opacus 1CP was 0.09-0.44 U/mg.
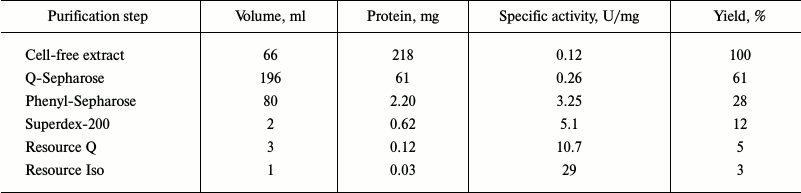
Both dioxygenases capable of cleaving the aromatic ring were isolated from R. wratislaviensis G10 cells. Table 3 presents a typical scheme of PCA-3,4-DO purification from cells grown at benzoate concentration of 200-250 mg/liter. The main characteristics of this enzyme were determined. The Km value of PCA-3,4-DO for protocatechuate was 25 ± 1 µM, the maximal activity was 26 ± 1 U/mg, the temperature optimum of its activity was 50°C, and the pH optimum was 9.25. Thermostability: {{anchor|GoBack}} the activity did not decrease for 2 h at 40°C, and at 50°C the activity decreased by 40% after 5 min and by 100% after 10 min. At 60°C, the enzyme was completely inactivated in 5 min.
Table 3. Results of purification of
PCA-3,4-DO from R. wratislaviensis G10 culture grown on
benzoate
The activity of PCA-3,4-DO from R. wratislaviensis G10 was slightly inhibited by catechol. The inhibition type was characterized as reversible competitive inhibition. The calculated value of Ki was 89 µM.
Typical results of purification of Cat-1,2-DO from the R. wratislaviensis G10 strain are presented in Table 4. Because the last stage of the purification (hydrophobic chromatography) was accompanied by a dramatic inactivation of the enzyme, the preparation was characterized after ion-exchange chromatography. The enzyme is a homodimer with subunit weight of 32.9 kDa. The temperature optimum was 40°C, the pH optimum was weakly pronounced – its value being 7.5, and at pH 5.5 the activity was only 27% lower and at pH 9.5 it was 10% lower. This enzyme was active with catechol and with 3- and 4-methylcatechol (3MCat, 4MCat). The activity with methylcatechols was higher than with catechol (Table 5). However, the Km values found from graphs were higher for substituted MCats than for Cat. The calculated values of the specificity constant showed that Cat was the preferable substrate for this enzyme. The activity with 4-chlorocatechol (4-CCat) was low, and there was no activity with dichlorocatechols. PCA neither inhibited nor activated the reaction of Cat-1,2-DO from R. wratislaviensis G10 with catechol.
Table 4. Results of purification of
Cat-1,2-DO from R. wratislaviensis G10 culture grown on
benzoate
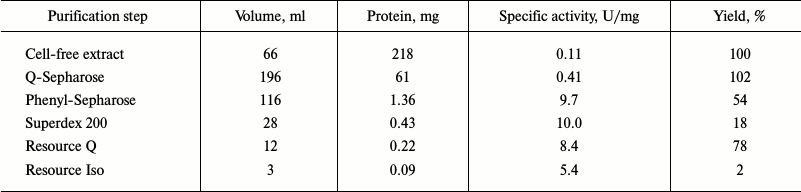
Table 5. Catalytic characteristics of
Cat-1,2-DO from R. wratislaviensis G10 strain
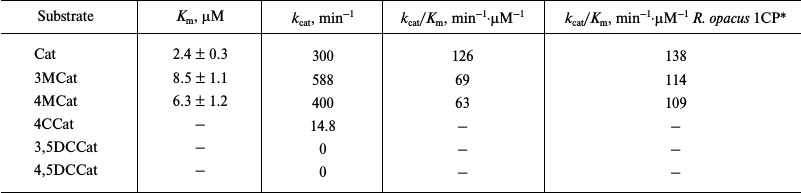
Notes: Values of catalytic constant (kcat) were
calculated based on the subunit weight of 32.9 kDa. 4CCat,
4-chlorocatechol; 3,5DCCat and 4,5DCCat, 3,5- and 4,5-dichlorocatechol,
respectively.
* For comparison, values of specificity constant are presented for
Cat-1,2-DO from R. opacus 1CP cells grown on benzoate [12].
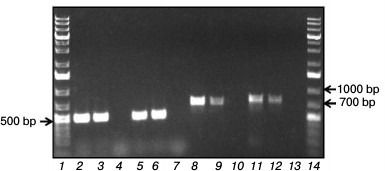
Search for and analysis of genes pcaG, pcaH, and catA in the R. wratislaviensis G10 genome. Primers chosen by nucleotide sequences of the R. opacus 1CP genes from the R. wratislaviensis G10 strain DNA-template were used to amplify fragments of genes pcaG and pcaH (coding α- and β-subunits of protocatechuate 3,4-dioxygenase) with length of ~500 bp (Fig. 4, lanes 5 and 2, respectively), as well as fragments of two of the four genes under study encoding isofunctional Cat-1,2-DO: catA2 (with primers cat4F/cat4R) and catA (with primers cat6F/cat6R) with length, respectively, of ~720 bp (lane 8) and 780 bp (lane 11).
Fig. 4. Screening of R. wratislaviensis G10 (G10) genes encoding protocatechuate 3,4-dioxygenase (pcaG, pcaH) and catechol 1,2-dioxygenases (catA). Rhodococcus opacus 1CP (1CP) was used as a positive control. Lanes: 1, 14) molecular weight marker 1 kb Plus DNA Ladder (Thermo Fisher Scientific, USA); pcaH: 2) G10; 3) 1CP; 4) negative control; pcaG: 5) G10; 6) 1CP; 7) negative control; catA2: 8) G10; 9) 1CP; 10) negative control; catA: 11) G10; 12) 1CP; 13) negative control.
The pcaG gene had 99 and 98% identity with genes of the α-subunit of protocatechuate 3,4-dioxygenases from R. opacus 1CP (AF003947.1) and R. opacus PD630 (CP003949.1), respectively. The pcaH gene had the 99% identity with the genes of β-subunit of protocatechuate 3,4-dioxygenases from R. opacus 1CP and R. opacus PD630.
The catA2 gene amplified on the R. wratislaviensis G10 DNA-template had 98% identity with the catA2 gene encoding Cat-1,2-DO (FM877593.1) of R. opacus 1CP, a degrader of chlorophenols, and 94% identity with the homologous gene (CP000431.1) of R. jostii RHA1, a degrader of chlorobiphenyls. Another gene of R. wratislaviensis G10, catA, encoding isofunctional Cat-1,2-DO had 99 and 96% identity with homologous genes of R. opacus 1CP (X99622.2) and R. jostii RHA1 (CP000431.1), respectively.
DISCUSSION
The ability of bacteria to degrade natural and anthropogenic compounds attracts great attention because using such strains is effective and economically advantageous for purification of the environment. However, it is also very important to understand spreading mechanisms of the genes mediating biodegradation in microorganisms isolated from different natural ecosystems. The strain R. wratislaviensis G10 isolated earlier from soil polluted with haloaromatic compounds is able to degrade some chlorobiphenyls [14]. Benzoate is a usual intermediate of biphenyl degradation by bacteria; therefore, the ability to degrade benzoate is quite expected. However, as discriminated from R. wratislaviensis G10, not all bacteria can degrade benzoate at concentrations up to 10 g/liter. Similar data were obtained for a hybrid organism resulting due to fusion of Pseudomonas putida and Bacillus subtilis protoplasts, which is considered by the authors [8] to be one of the most effective degraders of benzoate. However, the maximal specific rate of this hybrid growth was five times lower (31·10–3 h–1) than of the G10 strain. Growth parameters of the G10 culture calculated based on the growth curves indicated that the concentration of 2 g/liter was critical for the growth (Table 6). For comparison, for R. opacus 1CP this value was lower – 500 mg/liter [12]. Comparison of the calculated values revealed that at the optimal growth concentrations the G10 culture growth parameters were slightly better than those of 1CP. However, on increase in benzoate concentration these parameters in G10 became worse than in 1CP. The higher maximal specific growth rate, up to 0.66 h–1, was recorded for Pseudomonas putida culture at benzoate concentrations up to 400 mg/liter [17]. However, at benzoate concentration up to 200 mg/liter catechol produced from benzoate was degraded via the ortho-pathway, and on increase in the substrate concentration catechol degradation switched to the meta-cleavage pathway [17]. In our case, only intradiol dioxygenase activities were determined in the R. wratislaviensis G10 strain cells grown on 0.2-6 g/liter benzoate: the metabolism did not switch from the ortho-cleavage to meta-cleavage.
Table 6. Growth parameters of R.
wratislaviensis G10 and R. opacus 1CP on benzoate
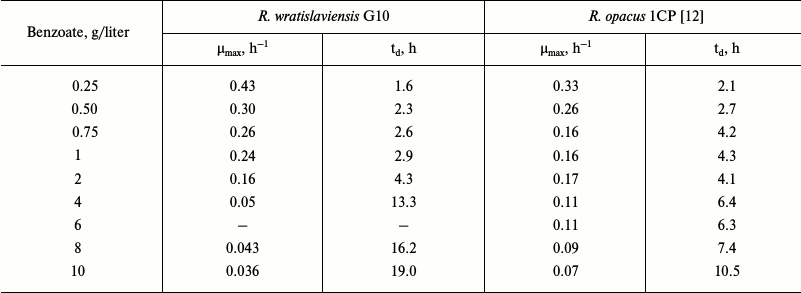
Notes: µmax, maximal specific growth rate;
td, doubling time.
In the present work, we found that in starving cells the activities of enzymes involved in degradation of the growth substrate were maintained for a long time. It is known that upon exhaustion of the substrate, synthesis of inducible enzymes is ended, and the existent protein molecules are degraded. However, there are no data on the rate of this process in strains that degrade toxicants. Our results revealed that benzoate-degrading enzymes retain their activities for a long time, in particular, under unfavorable conditions of cell suspension storage in water. These data partially explain the ability of cells to rapidly recommence growth on the recovery of favorable conditions. No doubt, the lower specificity of BDO from the G10 strain is its advantage compared with a similar enzyme from the R. opacus 1CP strain, which has extremely narrow substrate specificity. Because benzoate is an ingredient of some plants, it can be supposed that under natural conditions the culture can react to the presence of benzoate by induction of enzymes responsible for oxidative degradation. Although the activity of BDO from the G10 strain to substrates other than benzoate remains low under unfavorable conditions (in the absence of substrate specific for BDO), this can promote the culture to attack new substrates. In general, such tactics increases the survival probability of a given strain.
The comparative characterization of dioxygenases induced during R. wratislaviensis G10 and R. opacus 1CP growth on benzoate showed that despite the likeness of the enzymes, they have some differences. First of all, they are different in substrate specificity: Cat-1,2-DO from R. wratislaviensis G10 was more specific than the similar enzyme isolated from R. opacus 1CP cells. Table 7 presents the main features of the enzymes under comparison.
Table 7. Catalytic characteristics of
PCA-3,4-DO and Cat-1,2-DO from R. wratislaviensis G10 and R.
opacus 1CP
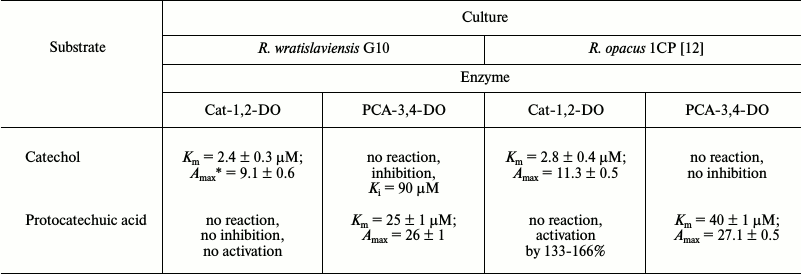
* Maximal activity at saturation with substrate (U/mg).
Because Cat-1,2-DO and PCA-3,4-DO induced in the R. wratislaviensis G10 and R. opacus 1CP strains are rather similar in their physicochemical characteristics, their response to “alien” substrates PCA and Cat, respectively, was the most interesting. Dioxygenases of the strains were different in their reactions to nonspecific substrates. Data presented in Table 7 indicate that the concurrent presence of both dioxygenases in the 1CP strain is not a problem from the standpoint of the enzymatic catalysis efficiency. On the contrary, in R. wratislaviensis G10 catechol inhibits the PCA-3,4-DO reaction with PCA. Possibly, this protects the cells against branching of the metabolic flow at the stage of the aromatic ring opening and at the same time makes the culture less substrate-labile. Nevertheless, it should be taken into account that the activity of both dioxygenases significantly depends on concentration of the growth substrate.
The concurrent presence of Cat-1,2-DO and PCA-3,4-DO activities is known for a number of bacteria. Thus, in benzoate-grown Pseudomonas putida KT 2440 cells high activity of Cat-1,2-DO (1.26 U/mg) and traces of PCA-3,4-DO (0.6 U/mg) were detected [18]. On growth on para-hydroxybenzoate (pHBA) and vanillin, the PCA-3,4-DO activity increased 12-fold, and the authors concluded that this enzyme was not involved in the degradation of benzoate [18]. PCA-3,4-DO was found in Acinetobacter baumanii DU202 cells grown on pHBA [19], and in two of eight Amycolatopsis and Streptomyces strains grown on benzoate activities of Cat-1,2-DO and PCA-3,4-DO were detected; however, the PCA-3,4-DO activity was an order lower than that of Cat-1,2-DO and did not exceed 28 nmol/min per mg [13]. Both Cat-1,2-DO and PCA-3,4-DO were detected concurrently in the Rhodococcus sp. RHA1 strain grown on both benzoate and phthalate [20]. In this case, PCA-3,4-DO activity was found also in cells grown on pyruvate, which indicated a possible constitutive synthesis of the enzyme in this strain.
In addition to the profile likeness of dioxygenases induced in the benzoate-grown strains under comparison, another important feature was revealed concerning fundamentals of microbian metabolism: the strain R. wratislaviensis G10 was found to contain in its genome regions of genes coding α- and β-subunits of PCA-3,4-DO and two catechol 1,2-dioxygenases, which had 99% similarity with the corresponding regions of the R. opacus 1CP strain. The presence of high identity genes in the genome of strains isolated from distantly located regions suggests the distribution of genes mediating degradation of resistant toxicants. The possibility of horizontal spreading of genes on the level of both unicellular organisms and higher taxon representatives has been shown in many works [21-24]. It has been established also that closely related isolates of some species can contain different sets of genes because of horizontal passage or loss of genes. There are also many examples of the widely varied likeness of degradation enzymes in microorganisms of different taxonomic groups, which in general reflects the relation degree between these bacteria or a common tendency for adaptation of different microorganisms to the same influences of the environment. Thus, the likeness between genes coding amino oxidase (FlavAO) involved in putrescine degradation in Rhodococcus RHA1 with the corresponding genes of other actinobacteria varies from 52% in Actinoplanes sp. SE50/110 of the Micromonosporaceae family to 97% in Rhodococcus opacus B4/PD630 of the Nocardiaceae family that also includes Rhodococcus RHA1 [25]. The homology of genes coding catechol 1,2-dioxygenases in Burkholderia xenovorans LB400 (NZ_CP008760) with similar dioxygenases varies from 86% in Achromobacter xylosoxidans A8 (CP002287.1) to 99% in Cat-1,2-DO of Pseudomonas knackmussii B13 (HG322950.1) strains among virtually 200 sequences known from the database. The homology of the gene coding Cat-1,2-DO from R. opacus 1CP, which is present in the chromosomal cat-operon (GenBank X99622), with genes coding similar enzymes of other gram-positive bacteria varies from 73% in Mycobacterium rhodesiae NBB3 (the full genome, CP003169.1, with 74% overlapping), 75% in Pseudonocardia dioxanivorans CB1190 (the full genome, CP002593.1, with 70% overlapping) and in Gordonia sp. KTR9 (the full genome, CP002907.1, with 80% overlapping) to 99% in Rhodococcus opacus PD630 (the full genome, CP003949.1, with 100% overlapping) among 43 sequences present (http://blast.ncbi.nlm.nih.gov/).
The homology between catechol 1,2-dioxygenases of gram-positive and gram-negative bacteria are significantly lower. Thus, the isolated amino acid sequence of the gene encoding Cat-1,2-DO of the cat-operon of the R. opacus 1CP (GenBank X99622) chromosome has 32-38% identical positions with Cat-1,2-DO of the Pseudomonas knackmussii B13 strain (Sequence ID: ref|WP_043251440.1| and Sequence ID: emb|CDF83097.1|, respectively). The same can be said about many other enzymes mediating the biodegradation of pollutants. Based on the data presented, we conclude that the corresponding genes of gram-positive and gram-negative bacteria possessing a common ancestor in the group had an independent origin during convergent evolution. In the literature there is a prevalent opinion supporting the hypothesis that the ability of bacteria to degrade resistant pollutants is provided during horizontal evolution by transfer of biodegradation plasmids, including the transfer between cells of different taxonomic groups [26-28]. Our data have confirmed that genetic material is transferred between the Rhodococcus genus bacteria. This is confirmed by the finding in distantly located bacteria R. opacus 1CP and R. wratislaviensis G10 of high identity genes coding at least three enzymes involved in the degradation of benzoate: benzoate 1,2-dioxygenase, Cat-1,2-DO, and PCA-3,4-DO with identity above 95%. However, there is a specific feature common for these cultures and contradicting the statement about relative freedom of genetic material transfer. Here, we are dealing with detection in their genome of a gene coding chloromuconolactone dehalogenase, a unique enzyme found only in the degradation pathway of 3-chlorocatechol via 2-chloromuconate, 5-chloromuconolactone, and cis-dienelactone by rhodococci and absent in the genome of gram-negative bacteria [29]. However, among all rhodococci tested, this gene was detected only in R. opacus 1CP and in rhodococci of the R. wratislaviensis species. Thus, based on the earlier obtained data, we suppose that the biodegrading potential of bacteria of these two species should be very close. Moreover, repeated sequencing of R. opacus 1CP 16S rDNA allows us to identify it as belonging to the R. wratislaviensis species. Thus, the presence of gene clcF only in R. wratislaviensis bacteria and its absence in other species of rhodococci indicate that the distribution pathways of the biodegradation genes among different species of bacteria are restricted significantly stronger that previously believed.
Acknowledgements
This work was supported by the Russian Science Foundation (project No. 14-14-00368).
REFERENCES
1.Pieper, D. H. (2005) Aerobic degradation of
polychlorinated biphenyls, Appl. Microbiol. Biotechnol.,
67, 170-191.
2.Pieper, D. H., Gonzalez, B., Camara, B.,
Perez-Pantoja, D., and Reineke, W. (2010) Aerobic degradation of
chloroaromatics, in Handbook of Hydrocarbon and Lipid
Microbiology (Timmis, K. N., ed.) Springer-Verlag,
Berlin-Heidelberg, pp. 839-864.
3.Du, L., Ma, L., Qi, F., Zheng, X., Jiang, C., Li,
A., Wan, X., Liu, S.-J., and Li, S. (2016) Characterization of a unique
pathway for 4-cresol catabolism initiated by phosphorylation in
Corynebacterium glutamicum, J. Biol. Chem., 291,
6583-6594.
4.Field, J. A., and Sierra-Alvarez, R. (2008)
Microbial transformation of chlorinated benzoates, Environ. Sci.
Biotechnol., 7, 191-210.
5.Neidle, E. L., Hartnett, C., Ornston, L. N.,
Bairoch, A., Rekik, M., and Harayama, S. (1991) Nucleotide sequences of
the Acinetobacter calcoaceticus benABC genes for benzoate
1,2-dioxygenase reveal evolutionary relationships among multicomponent
oxygenases, J. Bacteriol., 173, 5385-5395.
6.Parales, R. E., and Resnick, S. M. (2006) Aromatic
ring hydroxylating dioxygenases, Pseudomonas, 4,
287-340.
7.Kweon, O., Kim, S. J., Freeman, J. P., Song, J.,
Baek, S., and Cerniglia, C. E. (2010) Substrate specificity and
structural characteristics of the novel Rieske nonheme iron aromatic
ring-hydroxylating oxygenases NidAB and NidA3B3 from Mycobacterium
vanbaalenii PYR-1, mBio, 1, pii: e00135-10.
8.Li, M., Yi, P., Liu, Q., Pan, Y., and Qian, G.
(2013) Biodegradation of benzoate by protoplast fusant via intergeneric
protoplast fusion between Pseudomonas putida and Bacillus
subtilis, Int. Biodeterior. Biodegrad., 85,
577-582.
9.Zaar, A., Eisenreich, W., Bacher, A., and Fuchs, G.
(2001) A novel pathway of aerobic benzoate catabolism in the bacteria
Azoarcus evansii and Bacillus stearothermophilus, J.
Biol. Chem., 276, 24997-25004.
10.Rather, L. J., Knapp, B., Haehnel, W., and Fuchs,
G. (2010) Coenzyme A-dependent aerobic metabolism of benzoate via
epoxide formation, J. Biol. Chem., 285, 20615-20624.
11.Solyanikova, I. P., Emelyanova, E. V., Shumkova,
E. S., Egorova, D. O., Korsakova, E. S., Plotnikova, E. G., and
Golovleva, L. A. (2015) Peculiarities of the degradation of benzoate
and its chloro- and hydroxy-substituted analogs by actinobacteria,
Int. Biodeterior. Biodegrad., 100, 155-164.
12.Solyanikova, I. P., Emelyanova, E. V., Borzova,
O. V., and Golovleva, L. A. (2016) Benzoate degradation by
Rhodococcus opacus 1CP after a dormancy: characterization of
dioxygenases involved in the process, J. Environ. Sci. Health B,
5, 182-191.
13.Grund, E., Knorr, C., and Eichenlaub, R. (1990)
Catabolism of benzoate and monohydroxylated benzoates by
Amycolatopsis and Streptomyces spp., Appl. Environ.
Microbiol., 56, 1459-1464.
14.Plotnikova, E. G., Rybkina, D. O.,
Anan’ina, L. N., Yastrebova, O. V., and Demakov, V. A. (2006)
Characterization of microorganisms isolated from technogenic soils of
the Kama region, Russ. J. Ecol., 4, 233-240.
15.Gorlatov, S. N., Maltseva, O. V., Shevchenko, V.
I., and Golovleva, L. A. (1989) Degradation of chlorophenols by a
culture of Rhodococcus erythropolis, Mikrobiologiya,
58, 647-651.
16.Schlomann, M., Schmidt, E., and Knackmuss, H.-J.
(1990) Different types of dienelactone hydrolase in
4-fluorobenzoate-utilizing bacteria, J. Bacteriol., 172,
5112-5118.
17.Loh, K.-C., and Chua, S.-S. (2002) Ortho
pathway of benzoate degradation in Pseudomonas putida: induction
of meta pathway at high substrate concentrations, Enzyme
Microb. Technol., 30, 620-626.
18.Kim, Y. H., Cho, K., Yun, S.-H., Kim, J. Y.,
Kwon, K.-H., Yoo, J. S., and Kim, S. I. (2006) Analysis of aromatic
catabolic pathways in Pseudomonas putida KT 2440 using a
combined proteomic approach: 2-DE/MS and cleavable isotope-coded
affinity tag analysis, Proteomics, 6, 1301-1318.
19.Park, S. H., Kim, J. W., Yun, S. H., Leem, S. H.,
Kahng, H. Y., and Kim, S. I. (2006) Characterization of
β-ketoadipate pathway from multi-drug resistance bacterium,
Acinetobacter baumannii DU202 by proteomic approach, J.
Microbiol., 44, 632-640.
20.Patrauchan, M. A., Florizone, C., Dosanjh, M.,
Mohn, W. W., Davies, J., and Eltis, L. D. (2005) Catabolism of benzoate
and phthalate in Rhodococcus sp. strain RHA1: redundancies and
convergence, J. Bacteriol., 187, 4050-4063.
21.Crisp, A., Boschetti, C., Perry, M., Tunnacliffe,
A., and Micklem, G. (2015) Expression of multiple horizontally acquired
genes is a hallmark of both vertebrate and invertebrate genomes,
Genome Biol., 16, 50.
22.Polz, M. F., Alm, E. J., and Hanage, W. P. (2013)
Horizontal gene transfer and the evolution of bacterial and archaeal
population structure, Trends Genet., 29, 170-175.
23.Scholl, E. H., Thorne, J. L., McCarter, J. P.,
and Bird, D. M. (2003) Horizontally transferred genes in plant
parasitic nematodes: a high-throughput genomic approach, Genome
Biol., 4, R39.
24.Keeling, P. J., and Palmer, J. D. (2008)
Horizontal gene transfer in eukaryotic evolution, Nat. Rev.
Genet., 9, 605-618.
25.Foster, A., Barnes, N., Speight, R., and Keane,
M. A. (2013) Genomic organization, activity and distribution analysis
of the microbial putrescine oxidase degradation pathway, System.
Appl. Microbiol., 36, 457-466.
26.Dunning Hotopp, J. C. (2011) Horizontal gene
transfer between bacteria and animals, Trends Genet., 27,
157-163.
27.Syvanen, M. (2012) Evolutionary implications of
horizontal gene transfer, Annu. Rev. Genet., 46,
341-358.
28.Coleman, M. L., and Chisholm, S. W. (2010)
Ecosystem-specific selection pressures revealed through comparative
population genomics, Proc. Natl. Acad. Sci. USA, 107,
18635-18639.
29.Solyanikova, I. P., Plotnikova, E. G., Shumkova,
E. S., Robota, I. V., Prisyazhnaya, N. V., and Golovleva, L. A. (2014)
Chloromuconolactone dehalogenase ClcF of actinobacteria, J. Environ.
Sci. Health B, 49, 422-431.