REVIEW: DNA Import into Mitochondria
Yu. M. Konstantinov1,2*, A. Dietrich3, F. Weber-Lotfi3, N. Ibrahim3, E. S. Klimenko1, V. I. Tarasenko1, T. A. Bolotova1, and M. V. Koulintchenko1,3
1Siberian Institute of Plant Physiology and Biochemistry, Siberian Branch of the Russian Academy of Sciences, 664033 Irkutsk, Russia; fax: +7 (3952) 510-754; E-mail: yukon@sifibr.irk.ru2Irkutsk State University, 664003 Irkutsk, Russia; fax: +7 (3952) 241-855; E-mail: isuplantphysiology@mail.ru
3Université Louis Pasteur, Institut de Biologie Moléculaire des Plantes du CNRS, 67084 Strasbourg, France; fax: +33 (0) 367155300; E-mail: andre.dietrich@ibmp-cnrs.unistra.fr
* To whom correspondence should be addressed.
Received May 30, 2016; Revision received June 24, 2016
In recent decades, it has become evident that the condition for normal functioning of mitochondria in higher eukaryotes is the presence of membrane transport systems of macromolecules (proteins and nucleic acids). Natural competence of the mitochondria in plants, animals, and yeasts to actively uptake DNA may be directly related to horizontal gene transfer into these organelles occurring at much higher rate compared to the nuclear and chloroplast genomes. However, in contrast with import of proteins and tRNAs, little is known about the biological role and molecular mechanism underlying import of DNA into eukaryotic mitochondria. In this review, we discuss current state of investigations in this area, particularly specificity of DNA import into mitochondria and its features in plants, animals, and yeasts; a tentative mechanism of DNA import across the mitochondrial outer and inner membranes; experimental data evidencing several existing, but not yet fully understood mechanisms of DNA transfer into mitochondria. Currently available data regarding transport of informational macromolecules (DNA, RNA, and proteins) into the mitochondria do not rule out that the mechanism of protein and tRNA import as well as tRNA and DNA import into the mitochondria may partially overlap.
KEY WORDS: mitochondria, DNA, import, mitochondrial permeability transition pore, porin, adenine nucleotide translocator, plants, yeasts, mammalsDOI: 10.1134/S0006297916100035
Abbreviations: ANT, adenine nucleotide translocator; HGT, horizontal gene transfer; MPT, mitochondrial permeability transition; MPTP, mitochondrial permeability transition pore; mtDNA, mitochondrial DNA; OXPHOS, oxidative phosphorylation; PBR/TSPO, peripheral benzodiazepine receptor/translocator protein; plDNA, plasmid-like DNA; VDAC, voltage-dependent anion channel or mitochondrial porin.
In all eukaryotes, mitochondria possess their own genetic system
preserved to some extent from an ancestral endosymbiont. Systematic
studies of mitochondrial nucleotide sequences suggest that ancestors of
green plants had a compact mitochondrial genome [1]. Despite this, higher plants, particularly
angiosperms vs. animals and fungi, have a remarkably larger
mitochondrial genome (average 300-800 kb vs. average 16-100 kb,
respectively). Based on the data on nucleotide sequence of the
mitochondrial genome, it was found that in mtDNA only 11-18% represent
genes encoding proteins or structural RNAs, and more than 5% of mtDNA
sequence is of chloroplast, nuclear, or viral origin. Most
intriguingly, neither function nor origin has been determined so far
for more than a half of the total mitochondrial sequences [2-4]. Apart from that,
mitochondria in many plant species are characterized by another trait
that distinguishes them from the mitochondria of mammals and many other
eukaryotes. It was found that in addition to the main high molecular
weight mtDNA, the former possess one or several types of circular
and/or linear plasmids having features of an autonomous replicon in the
form of DNA or RNA ranging from 0.7 up to 20 kb [5-10]. Most of the currently
examined plant species were found to possess mitochondrial plasmid-like
DNA (plDNA) in the form of a species-specific set of circular and
linear DNAs with uncertain functions, which replicate independently of
the main genomic DNA [6, 8, 11]. The majority of mitochondrial plasmids has no
homology with the main mitochondrial genome and do not seem to be
necessary for its functioning. However, compared to the main genome,
their abundance in mitochondria may be quite high, depending on
developmental stage: for instance, in maize (Zea mays L.) a 6 :
1 stoichiometric ratio can be reached for linear plasmid S1 and S2 to
the main mitochondrial chromosome. In some way, these data resemble the
quantitative ratio for chromosomal and plasmid DNA in bacteria. Linear
mitochondrial plDNA may carry expressed sequences encoding proteins or
tRNA [11, 12]. The origin of
mitochondrial plasmids is unknown. It is assumed that double-stranded
plasmids might be introduced into the cells of higher plants via a
symbiotic or pathogenic route. In support of this hypothesis, it is
reasoned that double-stranded linear plasmids are associated through
their 5′-end with protein, thus resembling a structure of some
viral nucleic acids [13].
According to the features of mitochondrial genome organization in plants, it seems quite probable that plant mitochondria exhibit a marked ability to take up and integrate foreign sequences into their genome. In particular, in maize, after excluding genes, introns, open reading frames, and plastid-derived DNA, approximately 75% of the mitochondrial genome is composed of DNA with unknown origin and function [14]. A horizontal gene transfer (HGT) as the most common process in prokaryotes implying transmission of genetic information between species via a non-sexual route may play some role in shaping the structure of the mitochondrial genome in plants. Using comparative genomics, horizontal gene transfer was found to play an important role not only in evolution of prokaryote genomes, but also in higher eukaryotes as well. By analyzing HGT rate, it was demonstrated that among the three kinds of DNA-containing compartments (nuclei, mitochondria, chloroplasts) in plants, horizontal DNA transfer occurs most extensively into the mitochondria [15]. Most probably, the mechanism for HGT between plants can be explained by parasitism [16], non-standard-pollination, flowering, bacterial or viral transfer, fungal pathogens, or symbionts [17, 18].
In plant mitochondria, the presence of integrated retrotransposons of nuclear origin, as well as sequences derived from viral and plasmid RNA, suggests that inter-organelle transfer probably occurred via RNA intermediates [3, 19]. This assumption is consistent with the data on naturally existing tRNA import into mitochondria [20]. In many species, the mitochondrial genome does not code for all transfer RNAs (tRNAs) required for their functioning; therefore, tRNAs encoded in the nuclear genome are imported into the mitochondria from the cytosol. This biological process originally discovered in the 1960s in Tetrahymena pyriformis [21] is still being explored in an ever-expanding range of eukaryotes [22-24]. Depending on the species, a single or even entire set of tRNAs necessary for mitochondrial translation and expression of mtDNA may be imported into the mitochondria. Moreover, in mammalian cells nucleus-encoded 5S rRNAs [25] as well as RNA components of RNase R/RNase MRP [26] are imported into the mitochondria, although a role for such RNAs inside the organelles remains unclear. Recent studies on mammals demonstrated that nucleus-encoded specific microRNAs (miRNAs) are found inside the mitochondria, presumably participating in special regulatory mechanisms [27]. By making use of a naturally existing mechanism for transport of lysine-tRNA by yeast mitochondria, it was demonstrated that import of modified tRNALys into mitochondria from humans with MERRF syndrome exhibiting deficient mitochondrial translation might partially rescue impaired mitochondrial functions [28]. tRNA expressed in the cytosol is imported into the mitochondria through various mechanisms differing among species, which include components of protein translocation complexes (for yeasts and mammals [29]) and nucleotide translocation channels (for plants [30]).
Nonetheless, although import of larger RNA molecules has been recently shown [31], such a mechanism cannot explain all cases when mitochondria take up genetic information as well as emergence of DNA-plasmids within mitochondria. Based on the assumption that mitochondria may be naturally competent to uptake foreign DNA similarly to bacteria, this phenomenon has been investigated since the end of the 1980s. The first evidence of a naturally existing mechanism for DNA translocation into plant mitochondria was obtained in a mitochondrial system of DNA synthesis in organello using a series of pBR bacterial vectors as import substrates [32-34] (Table 1). It was found that bacterial DNA was able not only to enter the matrix, but could also be utilized by the mitochondrial genetic system as a template for DNA synthesis. During further studies, it was demonstrated that mitochondria isolated not only from various plant species (potato, maize, cauliflower, tobacco suspension culture) [35-38], but from mammals (rat, human cultured cells) [39] and yeast Saccharomyces cerevisiae [40] were also able to import double-stranded linear DNA molecules via an active translocation pathway. It is important to note that exogenous DNA imported into isolated mitochondria of various origin was functionally active and involved in genetic processes inside the organelles (Table 2). For instance, it was found out that the foreign genetic material, particularly the GFP gene within the transfection vector based on maize plasmid replicon, controlled by mitochondrial regulatory sequences after being imported into the organelles, could be expressed as well as serve as a template for DNA synthesis [35]. Hence, the presence of regulatory mitochondrial sequences is crucial for synthesis of both RNA and DNA [39]. In addition, the ability of genetic constructs translocated into mitochondria to recombine with resident mtDNA is another key element: while transfecting plant mitochondria, it was found that if a foreign gene (GFP) was flanked with 500 bp-sequences homologous to resident mitochondrial DNA, its integration occurred into the organellar genome via homologous recombination [38]. Hence, the recombination process does not require precise homology between an inserted sequence and mtDNA. In addition, it was also found that imported DNA might serve as a substrate for the mitochondrial repair system in both plants and mammals [41, 42].
Table 1. Characteristics of DNA substrates
used during examination of DNA translocation into plant
mitochondria
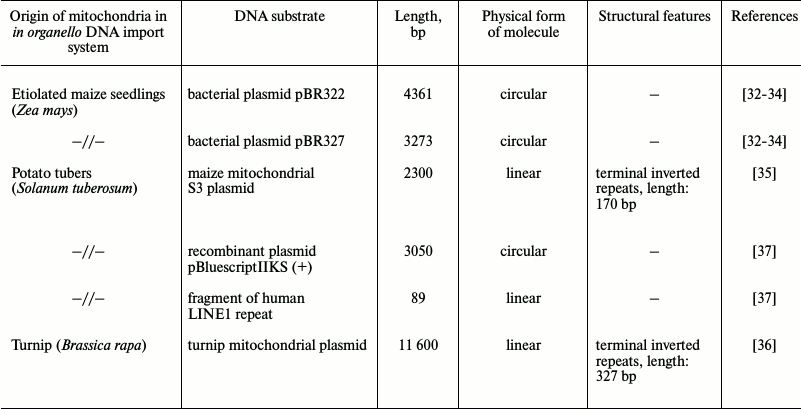
Table 2. Import of DNA into mitochondria of
various origin*
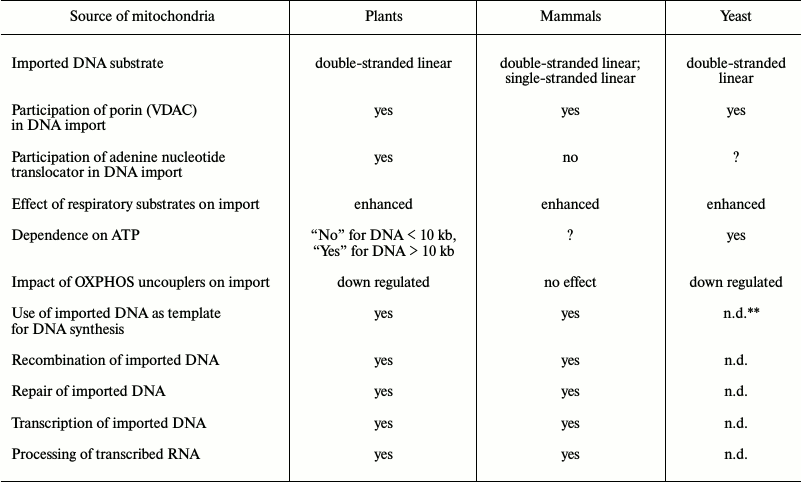
* The data were adapted from [35-42, 55].
** n.d., not determined.
SPECIFICITY OF DNA IMPORT INTO MITOCHONDRIA
Original studies devoted to investigation of the mechanism of DNA translocation across the mitochondrial membrane [32-37] were performed using a highly limited set of DNA substrates with size less than 10 kb, mainly focusing on a series of pBR bacterial plasmids and 2.3 kb linear mitochondrial plasmid derived from maize mitochondria (Table 1). Such substrates were shown to be imported into isolated mitochondria from both plants and mammals without exhibiting any specificity. Additionally, it was found that for plant mitochondria: (i) import of DNA does not depend on its nucleotide sequence; (ii) efficacy of import declines while the size of the imported DNA molecule is increasing; (iii) circular vs. linear DNA molecules are not imported very efficiently; (iv) single-stranded DNA is not imported into mitochondria [35]. Compared to DNA import into plant mitochondria, in mammals it was found to have one major difference: they can translocate both double- and single-stranded DNA molecules [39]. However, by analyzing the complete mitochondrial genome sequences from various species of higher plants, it was demonstrated that DNA fragments greatly varying in size and origin could undergo translocation with subsequent integration into the organellar genomes in vivo [43, 44].
The presence of additional plasmid-like DNA (plDNA) displaying properties of autonomous replicons (mitochondrial plasmids) in mitochondria from some plant species, whose origin and function are not fully clear, raises a question regarding the mechanism of their translocation across mitochondrial membranes as well as replication and expression. A linear 11,640-bp-long plasmid was found in mitochondria from rapeseed (Brassica napus L.) [45] that differed by containing six open reading frames, two of which code for DNA-polymerase (ORF5) and phage type RNA-polymerase (ORF6) expressed in this plant. Like a 2.3-kb-long mitochondrial plasmid from maize, this plasmid is characterized by having terminal inverted repeats (327 and 170 bp, respectively). It is known that inside mitochondria such repeats are involved in binding to proteins participating in replication and stabilization of plasmids. Using a linear 11.6-kb-long plasmid from B. napus as a substrate for DNA import into mitochondria from plants and human cell culture, it was found that import of DNA into plant mitochondria exhibits specificity with respect to longer substrates [36]. It was found that the efficacy of import for such DNA into plant mitochondria depends on the presence of certain motifs in its sequence, namely 5′- and 3′-terminal inverted repeats [36]. Hence, import of DNA into mammalian mitochondria still did not depend on either its sequence or its size.
Undoubtedly, import of nuclear encoded proteins and nucleic acids into mitochondria is among the most important mitochondrial processes. Understanding of mechanisms underlying these processes represents a difficult task, as it challenges current knowledge regarding properties of biological membranes. Solving this issue might result in emergence of novel ways to manipulate the mitochondrial genetic system. Nonetheless, at present there is no realistic understanding of which channel(s) and driving force(s) confer translocation of nucleic acids into mitochondria, especially of those with size more than 11 kb [36]. Over the two last decades, mechanisms underlying uptake of specific RNAs by mitochondria have been extensively studied, but the data also do not allow to make definitive conclusions and are still being debated [25, 27, 46].
MECHANISM OF DNA IMPORT ACROSS THE MITOCHONDRIAL OUTER
MEMBRANE
Mitochondrial functions depend on specific trafficking of metabolites and macromolecules executed by transport mechanisms across two mitochondrial membranes. Apparently, translocation across the outer membrane proceeds through a limited number of pores/channels, with a dominant role of mitochondrial porin or VDAC (Voltage-Dependent Anion Channel) [47, 48] upon transport of common metabolites as well as TOM40 during protein transport [49, 50]. Crossing the energized inner mitochondrial membrane impermeable to hydrogen ions by macromolecules is a more complexly organized process. Whereas proteins intended to be translocated into the matrix are transported through the TIM23 complex [49, 50], metabolite exchange occurs with participation of a large number of specialized MCF (Mitochondrial Carrier Family) transporters [51, 52]. Proteins of the MCF family largely have similar structural organization and contain six transmembrane domains. However, each of these proteins has its own preferable substrate(s). According to our observations, while considering translocation of nucleic acids into mitochondria a key role in transport of DNA across the mitochondrial outer membrane in plants [35], mammals [39], and the yeast Saccharomyces cerevisiae [40] is played by VDAC (Table 2).
Previously, S. cerevisiae was proposed as an informative genetic model for examining protein components involved in uptake of DNA by mitochondria [40]. By combining relevant biochemical data, such strategy revealed that a β-barrel structure of membrane protein VDAC (mitochondrial porin) may play a central role in translocation of DNA across the outer mitochondrial membrane [40]. In addition, it is known that VDAC is also required for import of tRNA into the organelles in plants [24], but not in trypanosomatids [53].
It is believed that VDAC is able to form 3-4 nm pores in the outer membrane, which match in size to the minimum diameter of double-stranded DNA helix [54]. In plant mitochondria, adenine nucleotide translocator (ANT) and ATPase β-subunit precursor located at the external side of the mitochondrial outer membrane may act as partners for VDAC during DNA import. This possibility is suggested from the data of proteomics analysis [55] based on masking cyanine-tagged proteins of the outer membrane in the presence of DNA as well as the available data on location of this protein [56]. After being translocated across the outer membrane into the intermembrane space of mitochondria, DNA is presumably recruited by Cu-binding protein (CuBP), a complex I subunit. The latter was shown to participate in the process both by examining profile of mitochondrial membrane-tagged proteins in the presence and absence of DNA, as well as in experiments assessing import of DNA into mitochondria isolated from an A. thaliana mutant containing an inactivated gene encoding CuBP [55].
By using mutant strains om14 and qcr2 of S. cerevisiae, it was found that DNA translocation across the mitochondrial outer membrane seems to be accomplished by protein complex VDAC1/OM14/OM45, probably involving QCR2 (subunit 2 of the respiratory complex III) as an additional factor [55]. OM14 is an abundant cysteine-rich protein of the mitochondrial outer membrane, with its N-terminus being located in the cytosol and C-terminus protruding into the intermembrane space. The putative function of this protein is not yet determined, but it has no homologs in other species except budding yeast [56]. Recent interactome studies with yeasts demonstrated that VDAC1 and OM14 together with the additional protein OM45 build up a core unit of a transport channel in the outer membrane [57].
MECHANISM OF DNA IMPORT ACROSS THE MITOCHONDRIAL INNER
MEMBRANE
Despite intense ongoing research [23, 25, 58, 59], no clear understanding regarding translocation of nucleic acids across the energized mitochondrial inner membrane impermeable to hydrogen ions exists. Translocation of tRNALys (CUU) into yeast mitochondria [30] and 5S rRNA into the human organelles [59] is believed to be accomplished with a complex that normally imports protein precursors. An RNA import complex (RIC) was isolated from the inner membrane of mitochondria from Leishmania tropica [60], but its existence is still not generally accepted [25, 61-63]. The intermembrane space of mammalian mitochondria contains polynucleotide phosphorylase (PNPASE), which may function as an RNA receptor [26].
It is known that specific translocation of molecules across the mitochondrial inner membrane is performed with the help of numerous carrier proteins that constitute a mitochondrial carrier family (MCF) based on similarity in their amino acid sequence containing three repeats of about 100 amino acid residues in length [51, 52]. Such a classic MCF member as adenine nucleotide translocator in the mitochondrial inner membrane seems to be a direct partner to VDAC while a DNA-translocating pore in plant mitochondria is formed [35]. Apart from ANT, the evolutionarily conserved MCF family also includes phosphate transporter as well as transporters for pyruvate, α-ketoglutarate, malate-aspartate shuttle, transporters for glutamate and other amino acids, as well as other carrier proteins [51, 52]. Undoubtedly, ANT holds a special place among MCF proteins as it is a most abundant protein of mitochondrial inner membrane [62, 64, 65].
By using a set of highly selective inhibitors and ligands such as carboxyatractyloside, atractyloside, bongkrekic acid, and ADP, it was convincingly demonstrated that ANT takes part in translocating DNA across the mitochondrial inner membrane in plants [35]. However, the same inhibitors and ANT effectors had no impact on DNA translocation in mammalian [39] and yeast [40] mitochondria (Table 2), although ANT was detected in the DNA–protein complex separated by native electrophoresis during the analysis of mitochondrial samples isolated from S. cerevisiae [40]. Thus, in mammalian mitochondria, DNA translocation may occur in a way similar to that in yeasts but not in plants.
At the beginning of the new century, VDAC and ANT were considered as the main components in the mammalian mitochondrial permeability transition pore (MPTP) [66, 67], apparently being the only pore-forming structure in the mitochondrial inner membrane (which, however, does not exclude potential polymorphism in this unit even within the same species). It was believed that a similar pore involving VDAC and ANT might also exist in plant mitochondria [67]. In this connection, we assumed and then proved that plant MPTP (more precisely, its components) might participate in translocation of nucleic acids [35]. It is crucial that translocation of DNA into the organelles involving the above mentioned pore components was not accompanied with MPTP opening, and it was not shown to exhibit features typical of mitochondrial permeability transition (MPT), which responds to stressful factors and participates in apoptosis [68-70].
Previously, it was demonstrated that upon reconstructing mammalian or fungal ANT in proteoliposomes it could form large channels [62, 64]. Nonetheless, the ability of ANT to make a pore accommodating the size of a DNA molecule in a physiological conditions remains to be established, as it is not so evident based on the known structure of this protein [71]. Regarding the MPTP, it was found that in mice with inactivated ANT gene this pore may exist even in the absence of the major ANT1/ANT2 isoforms [68]. Thus, ANT should not be considered as the only protein of the inner membrane forming the MPTP. In recent years, the potential nature and structure of the MPTP complex in the inner membrane of mammalian mitochondria has been attracting significant attention due to its important role in developing mitochondrial impairments during ischemia/reperfusion in tissue and organs [69, 70, 72] as well as opening up perspectives for developing therapeutic cell technologies by targeting mitochondria with new-generation antioxidants [73]. In mammals [74], Leishmania [75], and yeasts [76], ANT is associated with ATP-synthase and phosphate transporter by forming a supercomplex called the ATP-synthasome. The idea of this supercomplex did not yet become a generally accepted model, but apparently, there is some agreement regarding formation of MPTP structure in the inner membrane. Depending on the proposed model, pore formation in mammalian mitochondria occurs by involving ATP-synthase dimers, c-subunit ring of ATP-synthase, or conformational changes and structural rearrangements within the supercomplex [68-70]. The ANT and phosphate transporter mainly contribute to controlling the pore, particularly via exchanging adenine nucleotides and Pi.
By using yeast mutants, an attempt to examine the impact of mitochondrial membrane dynamics on activity of DNA import into the organelles was made [55]. Among a number of proteins in S. cerevisiae identified to participate in fusion and fission of mitochondria, special attention was paid to membrane proteins FIS1 and MDM33 [77, 78]. FIS1 is anchored via its tail domain in the mitochondrial outer membrane, and it recruits fission factors from the cytosol [79]. MDM33 detected in the ~300 kDa complex was demonstrated to contain coiled-coil structures potentially suitable for protein–protein interactions [77]. This protein is also able to form oligomers [80]. Mutations in the MDM33 gene resulted in significantly decreased activity of DNA translocation across the outer membrane of mitochondria from S. cerevisiae, which points at the importance of presumptive intermembrane interaction for translocating nucleic acids [55]. In contrast, in case of mutated FIS1 gene, upregulated DNA transfer across the outer and inner membrane was observed compared to the wild type yeast mitochondria, which might be explained by impaired regulation of transport machinery for nucleic acids.
It is known that yeast mitochondria also contain a homolog of mammalian MPTP, generally known as Yeast Mitochondrial Unspecific Channel (YMUC) [67, 81]. The complete structure of YMUC has not been solved, but it is assumed that ANT and phosphate transporter are involved in its formation [67, 81]. Along with this, mitochondria from S. cerevisiae were found to contain ATP-synthasome supercomplexes [76]. Data suggesting that COX13 protein, a subunit of the III complex, might be involved in formation of YMUC in yeast mitochondria were obtained in [55].
Compared to plants, import of DNA into S. cerevisiae mitochondria was found to be activated by ATP [40]. However, in the case of a mutated FIS1 gene, no stimulatory effect of ATP was observed [55]. Perhaps, such stimulatory effect of ATP with regard to DNA mitochondrial import might be explained by phosphorylation of certain protein factors involved in this process [36].
Thus, the data of both earlier [35] and recent studies [55] clearly suggest the idea that active uptake of nucleic acids occurs by involving structural components of the MPTP rather than its opening. Indeed, translocation of DNA across mitochondrial inner membrane in plants is sensitive to highly specific inhibitors and ligands acting on both ANT as well as phosphate transporter, ATP-synthase, and the MPTP (adenine nucleotides at physiological concentration, atractyloside, carboxyatractyloside, bongkrekic acid, long-chain fatty acids, acyl-CoA, oligomycin, mersalyl, Mg2+, Ca2+, Pi). Moreover, it was found that addition of DNA results in mitochondrial swelling by increasing proton conductance of the inner membrane [55]. This effect may be explained by interaction of DNA molecules with the adenine nucleotide-binding site in the ANT with subsequent formation of a DNA-conducting channel, when its activity is accompanied by increased permeability of the mitochondrial inner membrane to hydrogen ions. It was shown that ANT is a checkpoint regulating permeability of the mitochondrial inner membrane to hydrogen ions [82]. Hence, any ANT ligands may serve as regulators (modulators) of this process [82].
The fact that the same inner membranes contain other MCF proteins [51, 52] suggests that MPTP might be polymorphic. Alternatively, it could be hypothesized that any MCF protein under certain circumstances may form a pore. Therefore, one should not consider that the most abundant carrier proteins of the inner membrane such as ANT and phosphate transporter exclusively participate in formation of the majority of mitochondrial pores. Overall, certain structural similarity among individual MCF members [51, 52], which might result in some overlapping of transport functions, might explain why during earlier studies DNA import was not completely suppressed after using either inhibitor/modulator or in case of mutations in transporter genes negatively affecting uptake of DNA by mitochondria [35-40].
There is a certain structural similarity between the F1-ATPase component of mammalian mitochondrial ATP-synthase and protein complexes providing DNA transport upon bacterial conjugation [83]. Nonetheless, bacterial molecular components involved in DNA translocation such as TrwB form a hexameric structure with vacant central channel, whereas an equivalent cavity in mitochondrial F1-ATPase is filled with coiled γ-subunit [83, 84]. Based on this, it might be assumed that plant ATP-synthase is also able to translocate DNA in a similar way; however, most probably this process would still require substantial structural rearrangements.
MODEL OF DNA TRANSLOCATION INTO MITOCHONDRIA
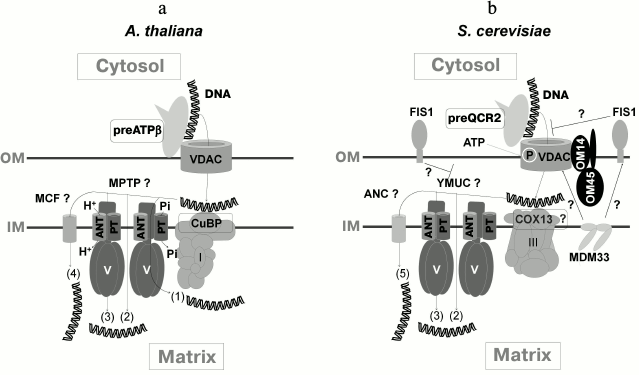
While creating a working model for DNA import into mitochondria according to currently available data [35, 37, 40, 55], it may be assumed that uptake of DNA is initiated by VDAC as well as by protein precursor (ATP-synthase β-subunit in plant mitochondria and complex III Qcr2p subunit in yeast) located on the external side of the outer membrane, normally residing in the inner membrane. Further, in the intermembrane space DNA is recruited by complex I CuBP subunit in plant mitochondria and perhaps by complex III COX13 subunit in yeast [55]. The channel-forming complex mediates subsequent DNA translocation across the inner membrane [35, 40, 55] (figure). Because tRNA transcripts stimulated DNA import into plant mitochondria, it may be that mechanisms responsible for import of tRNA and DNA into mitochondria partially overlap [55]. By considering various opportunities for formation of MPTP in different ways, the available data presume the existence of more than one mechanism for DNA import into mitochondria [37, 55] (figure). To confirm this, it may be noted that probably neither of the revealed DNA import factors plays a decisive role. Indeed, by using negatively influencing effector or in the absence of an expected import factor, activity of DNA import into mitochondria was only decreased, but not completely suppressed. Examination of DNA import into mitochondria from an A. thaliana line containing a mutation in the gene encoding Cu-binding protein (CuBP) as well as finding the effect of competitive inhibition of mitochondrial DNA import in the presence of tRNA also suggest the existence of various pathways of DNA import depending on its size [55]. In this respect, it is necessary to take into consideration additional and incompletely studied aspects regarding structure and functioning of MPTP. In particular, it was recently suggested that it would be appropriate to study a potential link between MPTP and complexes of mitochondrial protein import [81, 85]. Remarkably, functional protein import seems to be required for import of cytosolic tRNALys (CUU) into yeast mitochondria [30] and 5S rRNA into human organelles [86]. By all accounts, proteins and tRNA are translocated into plant mitochondria via different routes, but such components of the protein import machinery in the outer membrane as TOM20 and TOM40 were reported to be crucial for recruiting tRNA on the surface of the organelles [24].
Working model of DNA import into mitochondria. a) In plant mitochondria, DNA interacts with ATPase β-subunit precursor (preATPβ) and VDAC, then crosses the inner membrane through the channel made by VDAC, interacts with protein CuBP in the intermembrane space, undergoes translocation across the inner membrane via one of the potential mechanisms involved in formation of MPTP through (1) conformational changes and structural rearrangements within ATP-synthase supercomplex, (2) formation of ATP-synthase dimers, (3) rearrangement of c-subunit ring of ATP-synthase; (4) DNA translocation across the inner membrane seems to be carried out with participation of other members of the MCF family (mitochondrial carrier family) able to form pores. b) In yeast mitochondria, DNA may interact with QCR2 precursor (preQCR2) on the outer membrane, and in the intermembrane space, COX13 probably participates in translocation across yeast MPTP (YMUC). Upon MPTP formation, mechanism (1) is less feasible for yeasts; (5) adenine nucleotide carriers (ANC) probably serve as alternative pore-forming transporters. The presence of FIS1 is able to limit transport via VDAC and/or YMUC. ATP might stimulate DNA import via phosphorylation of proteins of the outer membrane. MDM33 may interact with components of the outer membrane contributing to DNA import. OM, outer membrane; IM, inner membrane; PT, phosphate transporter (modified from [55])
A question regarding a mechanism of sterol import into mitochondria remains beyond the scope of the current review. Sterols serve both as structural components in cell membranes and precursors for synthesis of steroid hormones inside mitochondria of the relevant specialized tissues [87, 88]. However, it is important to note that current mechanism of sterol import into mitochondria relies on close partner interactions between VDAC and ANT with peripheral benzodiazepine receptor (PBR) [89], which in recent years was called translocator protein (TSPO), as well as with five more proteins whose activity is directly related to functioning and/or regulation of the entire machinery of transmembrane sterol translocation into mitochondria [88]. Earlier, it was hypothesized that PBR/TSPO has an important role in DNA import into animal mitochondria [90]. Thus, one may think that depending on tissue and/or organ origin of mitochondria as well as membership in certain taxon, partnership between VDAC, ANT, and PBR/TSPO, evolutionarily established in mitochondria of higher organisms, is able to accomplish transmembrane translocation of various types of macromolecules by recruiting a set of additional import factors depending on type of the transported substrate. It is obvious that in case of DNA import into mitochondria of plants, animals, and yeast the set of necessary import factors differs [55], and its full composition for representatives of these groups of organisms remains to be established.
IS MITOCHONDRIAL DNA TRANSPORT INVOLVED IN INTRACELLULAR AND
INTERCELLULAR TRANSFER OF GENETIC MATERIAL?
It is necessary to consider that in contrast to import of proteins and tRNA, DNA import into mitochondria is probably a reversible process. The main purpose for releasing DNA from the mitochondria may be to provide a substrate for intercellular exchange with mitochondrial DNA as one of the conditions for horizontal gene transfer (HGT) into mitochondria. By now, an extended bulk of experimental data has accumulated supporting existence of horizontal gene transfer into mitochondria in eukaryotes (mainly plants) [15-19, 91-99]. However, whether imported and subsequently integrated into the mitochondrial genome foreign mtDNA is a result of controlled release (export) from mitochondria of donor host rather than programmed cell death remains unclear. Previously, in isolated potato (Solanum tuberosum) mitochondria it was demonstrated that imported foreign DNA of various length might be released (exported) [37]. If it is quite common in vivo, a controlled release of mtDNA from the organelles might take part in maintaining homoplasmy in the mitochondrial population by exchanging mtDNA between cells of various tissues and organs. The same process might also be enabled upon spreading new mtDNA variants within the organelle population, which therefore would represent a potential factor in evolution of the mitochondrial genome in both populations and species. After mtDNA exits from mitochondria into the cytosol, along with other nucleic acids it may be released by any means into the extracellular space [100, 101], thereby joining the pool of free circulating nucleic acids of extracellular fluids (blood, phloem sap, etc.) [100-103]. Assuming that circulating mtDNA like other circulating nucleic acids [103] is able to enter the cells, it could be expected that mtDNA may be imported into the mitochondria with subsequent integration into the mitochondrial genome via homologous recombination. The principle possibility of integration of imported foreign DNA flanked with sequences homologous to resident mtDNA into the organellar genome was earlier demonstrated in mitochondria isolated from higher plants [38].
In recent years, it has been found that in patients with acute myocardial infarction, tumor diseases, diabetes, major trauma, and some other pathological conditions the amount of free circulating mtDNA in the blood serum and plasma was significantly higher compared to a control group [101, 104-110]. During such conditions, free circulating mtDNA is able to act as an activator of inflammatory processes and factors of innate immunity [111-115]. It should be emphasized that neutrophils after releasing mtDNA into the circulation within so-called “extracellular traps” that kill bacteria remain vital [116]. So far, the accumulated data about varying amount of blood circulating mtDNA in a range of pathologies steadily extending over the last years suggest it as an important biomarker of pathologic conditions, not only being used for diagnostics, but also for monitoring efficacy of therapeutic procedures and making prognosis [117, 118]. One should not exclude that development of novel cell technologies allowing manipulation of genetic functions of mitochondria in vivo would result in an opportunity to regulate such vital systemic parameter in the future.
In recent decades, mainly owing to advances in genomics, it was possible to find massive transfer of quite long (ranging from 3 to 620 kb) fragments of mtDNA into the nucleus ending up with their insertion into various sites of chromosomes [15, 119, 120] and resulting in development of so-called “nuclear mitochondrial DNA segments” (NUMTs, sequences of mitochondrial origin located in the nuclear genome). While examining ten inbred maize strains, it became possible to find big interstrain differences in location of integration sites for mtDNA fragments in nuclear chromosomes, which might be explained by continuously ongoing processes such as transfer of mtDNA into the nucleus and loss of the earlier transferred NUMTs [119]. However, it is still unclear to what extent such horizontal DNA transfer is controlled, and if mitochondrial machinery of nucleic acid translocation is related to this in any way.
NATURAL COMPETENCE OF MITOCHONDRIA FOR DNA UPTAKE AND PROBLEM IN
CREATING TRANS-CHONDRIOMIC SPECIES
Overall, for at least two reasons, the issue of nucleic acid translocation into cellular organelles was and is still in focus for quite a large number of researchers. First, because various processes existing in nature are related to transfer of genetic information, e.g. transformation and transfection in bacteria, movement of viruses and transposable elements, inter-organellar exchange with nucleic sequences in many species, and, finally, poorly investigated interactions between cell genomes and the microbiome in humans and other mammals, it is evident that mechanisms allowing the nucleic acid translocation in such events should be investigated. On the other hand, it is quite attractive to create transgenic species via transformation of cell organelles by controlling spread of inserted foreign genes through their maternal inheritance. A similar system in plants has already been developed for chloroplasts by introducing genetic constructs via biolistic transformation, after which recipients were selected according to resistance of transformed plants to antibiotics spectinomycin or streptomycin, which selectivity is limited to biogenesis of chloroplasts [121]. In this case, nucleic acids enter chloroplasts either due to the mechanical disruption of intact membrane followed by restoration of its structure or import via some unknown transport mechanisms. However, for mitochondria no efficient system for genetic transformation has been developed so far due to much smaller size and lack of data, which might be used to create a genetic construct allowing selection of mitochondrial transformants. In contrast to chloroplasts, mitochondria are naturally competent to take up extramitochondrial DNA, which should be taken into consideration, and a currently missing appropriate system for selection of cells with transformed organelles represents the most serious obstacle for obtaining trans-mitochondrial species. Despite significant progress observed over recent years in developing methods for studying mitochondria such as isolation, biochemical and microscopic analysis, and genome sequencing, the stable in vivo transfection of mitochondria was only achieved for two unicellular organisms such as baker’s yeast Saccharomyces cerevisiae [122, 123] and the green alga Chlamydomonas reinhardtii [124]. In these experiments, genetic transformation of mitochondria was achieved using mutants with defective respiration as recipient strains.
Generally, by assessing the situation with investigations on DNA translocation into mitochondria, it is obvious that, soon or later, the consistent study on natural mechanism(s) underlying DNA import into mitochondria will result in knowledge that will allow to apply such phenomenon for genetic transformation of mitochondria of higher plants in vivo. No doubt, solving this issue will have a crucial importance for biology, medicine, and agricultural biotechnology.
Acknowledgements
This work was performed with financial support from the Russian Foundation for Basic Research (projects Nos. 15-04-05046, 15-54-16010).
REFERENCES
1.Turmel, M., Otis, C., and Lemieux, C. (2002) The
complete mitochondrial DNA sequence of Mesostigma viride
identifies this green alga as the earliest green plant divergence and
predicts a highly compact mitochondrial genome in the ancestor of all
green plants, Mol. Biol. Evol., 19, 24-38.
2.Kubo, T., Nishizawa, S., Sugawara, A., Itchoda, N.,
Estiati, A., and Mikami, T. (2000) The complete nucleotide sequence of
the mitochondrial genome of sugar beet (Beta vulgaris L.)
reveals a novel gene for tRNAcys (GCA), Nucleic Acids
Res., 28, 2571-2576.
3.Marienfeld, J., Unseld, M., and Brennicke, A.
(1999) The mitochondrial genome of Arabidopsis is composed of
both native and immigrant information, Trends Plant Sci.,
4, 495-502.
4.Unseld, M., Marienfeld, J. R., Brandt, P., and
Brennicke, A. (1997) The mitochondrial genome of Arabidopsis
thaliana contains 57 genes in 366,924 nucleotides, Nat.
Genet., 15, 57-61.
5.Brennicke, A., and Blanz, P. (1982) Circular
mitochondrial DNA species from Oenothera with unique sequence,
Mol. Gen. Genet., 187, 461-467.
6.Brown, G. G., and Zhang, M. (1995) Mitochondrial
plasmids: DNA and RNA, in The Molecular Biology of Plant
Mitochondria (Levings, C. S., III, and Vasil, I. K., eds.) Kluwer,
Netherlands, pp. 61-91.
7.Fukuhara, T., Moriyama, H., and Nitta, T. (1995)
The unusual structure of a novel RNA replicon in rice, J. Biol.
Chem., 270, 18147-18149.
8.Kanazawa, A., Tsutsumi, N., and Hirai, A. (1998)
Differential changes in copy number of rice mitochondrial plasmid-like
DNAs and main mitochondrial genomic DNAs that depend on temperature,
Curr. Genet., 33, 437-444.
9.Leaver, G., and Gray, M. (1982) Mitochondrial
genome organization and expression in higher plants, Annu. Rev.
Plant Physiol., 33, 390-395.
10.Lonsdale, D. M., and Grienenberger, J.-M. (1992)
The mitochondrial genome of plants, in Plant Gene Research. Cell
Organelles (Herrmann, R. G., ed.) Springer-Verlag, Austria, pp.
181-218.
11.Leon, P., O’Brien-Vedder, C., and Walbot,
V. (1992) Expression of ORF1 of the linear 2.3 kb plasmid of maize
mitochondria: product localization and similarities to the 130 kDa
protein encoded by the S2 episome, Curr. Genet., 22,
61-67.
12.Leon, P., Walbot, V., and Bedinger, P. (1989)
Molecular analysis of the linear 2.3 kb plasmid of maize mitochondria:
apparent capture of tRNA genes, Nucleic Acids Res., 17,
4089-4099.
13.Saumitou-Laprade, P., Pannebecker, G., Maggouta,
F., Jean, R., and Michaelis, G. (1989) A linear 10.4 kb plasmid in the
mitochondria of Beta maritima, Curr. Genet., 16,
181-186.
14.Clifton, S. W., Minx, P., Fauron, C. M.-R.,
Gibson, M., Allen, J. O., Sun, H., Thompson, M., Barbazuk, W. B.,
Kanuganti, S., Tayloe, C., Meyer, L., Wilson, R. K., and Newton, K. J.
(2004) Sequence and comparative analysis of the maize NB mitochondrial
genome, Plant Physiol., 136, 3486-3503.
15.Kleine, T., Maier, U. W., and Leister, D. (2009)
DNA transfer from organelles to the nucleus: the idiosyncratic genetics
of endosymbiosis, Annu. Rev. Plant Biol., 60,
115-138.
16.Davis, C. C., Anderson, W. R., and Wurdack, K. J.
(2005) Gene transfer from a parasitic flowering plant to a fern,
Proc. R Soc. B Biol. Sci., 272, 2237-2242.
17.Bergthorsson, U., Adams, K. L., Thomason, B., and
Palmer, J. D. (2003) Widespread horizontal transfer of mitochondrial
genes in flowering plants, Nature, 424, 197-201.
18.Won, H., and Renner, S. S. (2003) Horizontal gene
transfer from flowering plants to Gnetum, Proc. Natl. Acad.
Sci. USA, 100, 10824-10829.
19.Marienfeld, J. R., Unseld, M., Brandt, P., and
Brennicke, A. (1997) Viral nucleic acid sequence transfer between fungi
and plants, Trends Genet., 13, 260-261.
20.Marechal-Drouard, L., Weil, J.-H., and Dietrich,
A. (1993) Transfer RNAs and transfer RNA genes in plants, Annu. Rev.
Plant Physiol. Plant Mol. Biol., 44, 13-32.
21.Suyama, Y. (1967) The origins of mitochondrial
ribonucleic acids in Tetrahymena pyriformis,
Biochemistry, 6, 2829-2839.
22.Alfonzo, J. D., and Soll, D. (2009) Mitochondrial
tRNA import – the challenge to understand has just begun,
Biol. Chem., 390, 717-722.
23.Salinas, T., Duchene, A. M., Delage, L., Nilsson,
S., Glaser, E., Zaepfel, M., and Marechal-Drouard, L. (2006) The
voltage-dependent anion channel, a major component of the tRNA import
machinery in plant mitochondria, Proc. Natl. Acad. Sci. USA,
103, 18362-18367.
24.Schneider, A. (2011) Mitochondrial tRNA import
and its consequences for mitochondrial translation, Annu. Rev.
Biochem., 80, 1033-1053.
25.Smirnov, A., Entelis, N., Martin, R. P., and
Tarassov, I. (2011) Biological significance of 5S rRNA import into
human mitochondria: role of ribosomal protein MRP-L18, Genes
Dev., 25, 1289-1305.
26.Wang, G., Chen, H. W., Oktay, Y., Zhang, J.,
Allen, E. L., Smith, G. M., Fan, K. C., Hong, J. S., French, S. W.,
McCaffery, J. M., Lightowlers, R. N., Morse, H. C., Koehler, C. M., and
Teitell, M. A. (2010) PNPASE regulates RNA import into mitochondria,
Cell, 142, 456-467.
27.Dietrich, A., Wallet, C., Iqbal, R. K.,
Gualberto, J. M., and Lotfi, F. (2015) Organellar non-coding RNAs:
emerging regulation mechanisms, Biochimie, 117,
48-62.
28.Kolesnikova, O. A., Entelis, N. S.,
Jacquin-Becker, C., Goltzene, F., Chrzanowska-Lightowlers, Z. M.,
Lightowlers, R. N., Martin, R. P., and Tarassov, I. (2004) Nuclear
DNA-encoded tRNAs targeted into mitochondria can rescue a mitochondrial
DNA mutation associated with the MERRF syndrome in cultured human
cells, Human Mol. Genet., 13, 2519-2534.
29.Tarassov, I., Entelis, N., and Martin, R. P.
(1995) An intact protein translocating machinery is required for
mitochondrial import of a yeast cytoplasmic tRNA, J. Mol. Biol.,
245, 315-323.
30.Delage, L., Dietrich, A., Cosset, A., and
Marechal-Drouard, L. (2003) In vitro import of a nuclearly
encoded tRNA into mitochondria of Solanum tuberosum, Mol.
Cell. Biol., 23, 4000-4012.
31.Wang, G., Shimada, E., Zhang, J., Hong, J. S.,
Smith, G. M., Teitell, M. A., and Koehler, C. M. (2012) Correcting
human mitochondrial mutations with targeted RNA import, Proc. Natl.
Acad. Sci. USA, 109, 4840-4845.
32.Konstantinov, Yu. M., Podsosonny, V. A., and
Lutsenko, G. N. (1988) Synthesis of bacterial vector plasmid pBR322 DNA
in isolated maize mitochondria, Dokl. AN SSSR, 298,
502-504.
33.Konstantinov, Yu. M., Podsosonny, V. A.,
Lutsenko, G. N., and Rivkin, M. I. (1989) Translocation of bacterial
vector plasmids into intact corn shoot mitochondria, Biochemistry
(Moscow), 54, 154-158.
34.Konstantinov, Y. M., Podsosonny, V. A., and
Lutsenko, G. N. (1990) Translocation of bacterial vector plasmids into
intact mitochondria of seedlings, Maize Genet. Cooper.
Newslett., 64, 67-68.
35.Koulintchenko, M., Konstantinov, Y., and
Dietrich, A. (2003) Plant mitochondria actively import DNA via the
permeability transition pore complex, EMBO J., 22,
1245-1254.
36.Ibrahim, N., Handa, H., Cosset, A.,
Koulintchenko, M., Konstantinov, Y., Lightowlers, R. N., Dietrich, A.,
and Weber-Lotfi, F. (2011) DNA delivery to mitochondria: sequence
specificity and energy enhancement, Pharm. Res., 28,
2871-2882.
37.Klimenko, E. S., Mileyko, V. A., Morozkin, E. S.,
Laktionov, P. P., and Konstantinov, Yu. M. (2011) Characteristics of
DNA export and import into potato (Solanum tuberosum)
mitochondria by using qualitative PCR, Biochemistry (Moscow) Suppl.
Ser. A Membr. Cell Biol., 28, 199-205.
38.Mileshina, D., Koulintchenko, M., Konstantinov,
Y., and Dietrich, A. (2011) Transfection of plant mitochondria and
in organello gene integration, Nucleic Acids Res.,
39, e115.
39.Koulintchenko, M., Temperley, R. J., Mason, P.
A., Dietrich, A., and Lightowlers, R. N. (2006) Natural competence of
mammalian mitochondria allows the molecular investigation of
mitochondrial gene expression, Human Mol. Genet., 15,
143-154.
40.Weber-Lotfi, F., Ibrahim, N., Boesch, P., Cosset,
A., Konstantinov, Y., Lightowlers, R. N., and Dietrich, A. (2009)
Developing a genetic approach to investigate the mechanism of
mitochondrial competence for DNA import, Biochim. Biophys. Acta,
1787, 320-327.
41.Boesch, P., Ibrahim, N., Paulus, F., Cosset, A.,
Tarasenko, V., and Dietrich, A. (2009) Plant mitochondria possess a
short-patch base excision DNA repair pathway, Nucleic Acids
Res., 37, 5690-5700.
42.Boesch, P., Ibrahim, N., Dietrich, A., and
Lightowlers, R. N. (2010) Membrane association of mitochondrial DNA
facilitates base excision repair in mammalian mitochondria, Nucleic
Acids Res., 38, 1478-1488.
43.Goremykin, V. V., Salamimi, F., Velasco, R., and
Viola, R. (2009) Mitochondrial DNA of Vitis vinifera and the
issue of rampant horizontal gene transfer, Mol. Biol. Evol.,
26, 99-110.
44.Alverson, A. J., Rice, D. W., Dickinson, S.,
Barry, K., and Palmer, J. D. (2011) Origins and recombination of the
bacterial-sized multichromosomal mitochondrial genome of cucumber,
Plant Cell, 23, 2499-2513.
45.Handa, H., Itani, K., and Sato, H. (2002)
Structural features and expression analysis of a linear mitochondrial
plasmid in rapeseed (Brassica napus L.), Mol. Genet.
Genom., 267, 797-805.
46.Bandiera, S., Mategot, R., Girard, M., Demongeot,
J., and Henrion-Caude, A. (2013) MitomiRs delineating the intracellular
localization of microRNAs at mitochondria, Free Radic. Biol.
Med., 64, 12-19.
47.Shoshan-Barmatz, V., De Pinto, V., Zweckstetter,
M., Raviv, Z., Keinan, N., and Arbel, N. (2010) VDAC, a
multi-functional mitochondrial protein regulating cell life and death,
Mol. Aspects Med., 31, 227-285.
48.Takahashi, Y., and Tateda, C. (2013) The
functions of voltage-dependent anion channels in plants,
Apoptosis, 18, 917-924.
49.Dudek, J., Rehling, P., and Van der Laan, M.
(2013) Mitochondrial protein import: common principles and
physiological networks, Biochim. Biophys. Acta, 1833,
274-285.
50.Murcha, M. W., Wang, Y., Narsai, R., and Whelan,
J. (2014) The plant mitochondrial protein import apparatus –
the differences make it interesting, Biochim. Biophys. Acta,
1840, 1233-1245.
51.Haferkamp, I., and Schmitz-Esser, S. (2012) The
plant mitochondrial carrier family: functional and evolutionary
aspects, Front. Plant Sci., 3, doi:
10.3389/fpls.2012.00002.
52.Palmieri, F., Pierri, C. L., De Grassi, A.,
Nunes-Nesi, A., and Fernie, A. R. (2011) Evolution, structure and
function of mitochondrial carriers: a review with new insights,
Plant J., 66, 161-181.
53.Pusnik, M., Charriere, F., Maser, P., Waller, R.
F., Dagley, M. J., Lithgow, T., and Schneider, A. (2009) The single
mitochondrial porin of Trypanosoma brucei is the main metabolite
transporter in the outer mitochondrial membrane, Mol. Biol.
Evol., 26, 671-680.
54.Mannella, C. A., Forte, M., and Colombini, M.
(1992) Toward the molecular structure of the mitochondrial channel,
VDAC, J. Bioenerg. Biomembr., 24, 7-19.
55.Weber-Lotfi, F., Koulintchenko, M. V., Ibrahim,
N., Hammann, P., Mileshina, D. V., Konstantinov, Y. M., and Dietrich,
A. (2015) Nucleic acid import into mitochondria: new insights into the
translocation pathways, Biochim. Biophys. Acta, 1853,
3165-3181.
56.Burri, L., Vascotto, K., Gentle, I. E., Chan, N.
C., Beilharz, T., Stapleton, D. I., Ramage, L., and Lithgow, T. (2006)
Integral membrane proteins in the mitochondrial outer membrane of
Saccharomyces cerevisiae, FEBS J., 273,
1507-1515.
57.Lauffer, S., Mabert, K., Czupalla, C., Pursche,
T., Hoflack, B., Rodel, G., and Krause-Buchholz, U. (2012)
Saccharomyces cerevisiae porin pore forms complexes with
mitochondrial outer membrane proteins Om14p and Om45p, J. Biol.
Chem., 287, 17447-17458.
58.Von Stedingk, E., Pavlov, P. F., Grinkevich, V.
A., and Glaser, E. (1999) The precursor of the F1beta subunit of the
ATP synthase is covalently modified upon binding to plant
mitochondrial, Plant Mol. Biol., 41, 505-515.
59.Salinas, T., Duchene, A. M., and
Marechal-Drouard, L. (2008) Recent advances in tRNA mitochondrial
import, Trends Biochem. Sci., 33, 320-329.
60.Adhya, S. (2008) Leishmania mitochondrial
tRNA importers, Int. J. Cell Biochem. Cell Biol., 40,
2681-2685.
61.Rubio, M. A., and Hopper, A. K. (2011) Transfer
RNA travels from the cytoplasm to organelles, Wiley Interdiscip.
Rev. RNA, 2, 802-817.
62.Schekman, R. (2010) Editorial expression of
concern: a bifunctional tRNA import receptor from Leishmania
mitochondria, Proc. Natl. Acad. Sci. USA, 107, 9476.
63.Niazi, A. K., Mileshina, D., Cosset, A., Val, R.,
Weber-Lotfi, F., and Dietrich, A. (2013) Targeting nucleic acids into
mitochondria: progress and prospects, Mitochondrion, 13,
548-558.
64.Brustovetsky, N., and Klingenberg, M. (1996)
Mitochondrial ADP/ATP carrier can be reversibly converted into a large
channel by Ca2+, Biochemistry, 35,
8483-8488.
65.Brustovetsky, N., Tropschug, M., Heimpel, S.,
Heidkamper, D., and Klingenberg, M. (2002) A large
Ca2+-dependent channel formed by recombinant ADP/ATP carrier
from Neurospora crassa resembles the mitochondrial permeability
transition pore, Biochemistry, 41, 11804-11811.
66.Zamzami, N., and Kroemer, G. (2001) The
mitochondrion in apoptosis: how Pandora’s box opens, Nat. Rev.
Mol. Cell Biol., 2, 67-71.
67.Vianello, A., Casolo, V., Petrussa, E., Peresson,
C., Patui, S., Bertolini, A., Passamonti, S., Braidot, E., and Zancani,
M. (2012) The mitochondrial permeability transition pore
(PTP) – an example of multiple molecular exaptation?
Biochim. Biophys. Acta, 1817, 2072-2086.
68.Kokoszka, J. E., Waymire, K. G., Levy, S. E.,
Sligh, J. E., Cai, J., Jones, D. P., MacGregor, G. R., and Wallace, D.
C. (2004) The ADP/ATP translocator is not essential for the
mitochondrial permeability transition pore, Nature, 427,
461-465.
69.Bernardi, P., and Di Lisa, F. (2015) The
mitochondrial permeability transition pore: molecular nature and role
as a target in cardioprotection, J. Mol. Cell. Cardiol.,
78, 100-106.
70.Halestrap, A. P., and Richardson, A. P. (2015)
The mitochondrial permeability transition: a current perspective on its
identity and role in ischemia/reperfusion injury, J. Mol. Cell.
Cardiol., 78, 129-141.
71.Pebay-Peyroula, E., Dahout-Gonzalez, C., Kahn,
R., Trezeguet, V., Lauquin, G. J., and Brandolin, G. (2003) Structure
of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside,
Nature, 426, 39-44.
72.Morciano, G., Giorgi, C., Bonora, M., Punzetti,
S., Pavasini, R., Wieckowski, M. R., Campo, G., and Pinton, P. (2015)
Molecular identity of the mitochondrial permeability transition pore
and its role in ischemia-reperfusion injury, J. Mol. Cell.
Cardiol., 78, 142-153.
73.Plotnikov, E. Y., Silachev, D. N., Jankauskas, S.
S., Rokitskaya, T. I., Chupyrkina, A. A., Pevzner, I. B., Zorova, L.
D., Isaev, N. K., Antonenko, Y. N., Skulachev, V. P., and Zorov, D. B.
(2012) Mild uncoupling of respiration and phosphorylation as a
mechanism providing nephro- and neuroprotective effects of penetrating
cations of the SkQ family, Biochemistry (Moscow), 77,
1029-1037.
74.Chen, C., Ko, Y., Delannoy, M., Ludtke, S. J.,
Chiu, W., and Pedersen, P. L. (2004) Mitochondrial ATP synthasome:
three-dimensional structure by electron microscopy of the ATP synthase
in complex formation with carriers for Pi and ADP/ATP, J.
Biol. Chem., 279, 31761-31768.
75.Detke, S., and Elsabrouty, R. (2008)
Identification of a mitochondrial ATP synthase–adenine nucleotide
translocator complex in Leishmania, Acta Trop.,
105, 16-20.
76.Clemencon, B. (2012) Yeast mitochondrial
interactosome model: metabolon membrane proteins complex involved in
the channeling of ADP/ATP, Int. J. Mol. Sci., 13,
1858-1885.
77.Messerschmitt, M., Jakobs, S., Vogel, F., Fritz,
S., Dimmer, K. S., Neupert, W., and Westermann, B. (2003) The inner
membrane protein Mdm33 controls mitochondrial morphology in yeast,
J. Cell Biol., 160, 553-564.
78.Westermann, B. (2008) Molecular machinery of
mitochondrial fusion and fission, J. Biol. Chem., 283,
13501-13505.
79.Van der Bliek, A. M., Shen, Q., and Kawajiri, S.
(2013) Mechanisms of mitochondrial fission and fusion, Cold Spring
Harb. Perspect. Biol., pii: a011072.
80.Zhang, Y., and Chan, D. C. (2007) Structural
basis for recruitment of mitochondrial fission complexes by Fis1,
Proc. Natl. Acad. Sci. USA, 104, 18526-18530.
81.Gutierrez-Aguilar, M., and Uribe-Carvajal, S.
(2015) The mitochondrial unselective channel in Saccharomyces
cerevisiae, Mitochondrion, 22, 85-90.
82.Panov, A., Filippova, S., and Lyakhovich, V.
(1980) Adenine nucleotide translocase as a site of regulation by ADP of
the rat liver mitochondria permeability to H+ and
K+ ions, Arch. Biochem. Biophys., 199,
420-426.
83.Cabezon, E., and De la Cruz, F. (2006) TrwB: an
F(1)-ATPase-like molecular motor involved in DNA transport during
bacterial conjugation, Res. Microbiol., 157, 299-305.
84.Tato, I., Matilla, I., Arechaga, I., Zunzunegui,
S., De la Cruz, F., and Cabezon, E. (2007) The ATPase activity of the
DNA transporter TrwB is modulated by protein TrwA: implications for a
common assembly mechanism of DNA translocating motors, J. Biol.
Chem., 282, 25569-25576.
85.Szabo, I., and Zoratti, M. (2014) Mitochondrial
channels: ion fluxes and more, Physiol. Rev., 94,
519-608.
86.Entelis, N. S., Kolesnikova, O. A., Dogan, S.,
Martin, R. P., and Tarassov, I. A. (2001) 5S rRNA and tRNA import into
human mitochondria. Comparison of in vitro requirements, J.
Biol. Chem., 276, 45642-45653.
87.Maxfield, F. R., and Wustner, D. (2002)
Intracellular cholesterol transport, J. Clin. Invest.,
110, 891-898.
88.Flis, V. V., and Daum, G. (2013) Lipid transport
between the endoplasmic reticulum and mitochondria, in The
Endoplasmic Reticulum. A Subject Collection from Cold Spring Harbor
Perspectives in Biology (Ferro-Novick, S., Rapoport, T. A., and
Schekman, R., eds.) Cold Spring Harbor Laboratory Press, Cold Spring
Harbor, New York, USA, pp. 109-130.
89.McEnery, M. W., Snowman, A. M., Trifiletti, R.
R., and Snyder, S. H. (1992) Isolation of the mitochondrial
benzodiazepine receptor: association with the voltage-dependent anion
channel and adenine nucleotide carrier, Proc. Natl. Acad. Sci.
USA, 89, 3170-3174.
90.Zorov, D. B. (1996) Mitochondrial transport of
nucleic acids. Involvement of the benzodiazepine receptor,
Biochemistry (Moscow), 61, 939-946.
91.Mower, J. P., Stefanovic, S., Hao, W., Gammow, J.
S., Jain, K., Ahmed, D., and Palmer, J. D. (2010) Horizontal
acquisition of multiple mitochondrial genes from a parasitic plant
followed by gene conversion with host mitochondrial gene, BMC
Biol., 8, 150.
92.Sanchez-Puerta, M. V., Cho, Y., Mower, J. P.,
Alverson, A. J., and Palmer, J. P. (2008) Frequent, phylogenetically
local horizontal transfer of the cox1 group intron in flowering plant
mitochondria, Mol. Biol. Evol., 25, 1762-1777.
93.Burger, G., Gray, M. W., and Lang, B. F. (2003)
Mitochondrial genomes: anything goes, Trends Genet., 19,
709-716.
94.Cho, Y., Qiu, Y. L., Kuhlman, P., and Palmer, J.
D. (1998) Explosive invasion of plant mitochondria by a group I intron,
Proc. Natl. Acad. Sci. USA, 95, 14244-14249.
95.Bergthorsson, U., Richardson, A. O., Young, G.
J., Goertzen, L. R., and Palmer, J. D. (2004) Massive horizontal
transfer of mitochondrial genes from diverse land plant donors to the
basal angiosperm Amborella, Proc. Natl. Acad. Sci. USA,
101, 17747-17752.
96.Renner, S., and Bellot, S. (2012) Horizontal gene
transfer in eukaryotes: fungi-to-plant and plant-to-plant transfers of
organellar DNA, in Genomics of Chloroplast and Mitochondria
(Bock, R., and Knoop, V., eds.) Springer, Dordrecht, The Netherlands,
pp. 223-235.
97.Hao, W., Richardson, A. O., Zheng, Y., and
Palmer, J. D. (2010) Gorgeous mosaic of mitochondrial genes created by
horizontal transfer and gene conversion, Proc. Natl. Acad. Sci.
USA, 107, 21576-21581.
98.Bock, R. (2010) The give-and-take of DNA:
horizontal gene transfer in plants, Trends Plant Sci.,
15, 11-22.
99.Richardson, A. O., and Plamer, J. D. (2007)
Horizontal gene transfer in plants, J. Exp. Bot., 58,
1-9.
100.Bryzgunova, O. E., and Laktionov, P. P. (2015)
Development of circulating DNA pool in the blood: source, features of
composition and circulation, Biochemistry (Moscow) Suppl. Ser. B
Biomed. Chem., 61, 409-426.
101.Gahan, P. B. (2012) Biology of circulating
nucleic acids and possible roles in diagnosis and treatment in diabetes
and cancer, Infect. Disord. Drug Targets, 12,
360-370.
102.Gahan, P. B., and Swaminathan, R. (2008)
Circulating nucleic acids in plasma and serum, Ann. N. Y. Acad.
Sci., 1137, 1-6.
103.Gahan, P. (2013) Circulating nucleic acids:
possible inherited effects, Biol. J. Linn. Soc., 110,
931-948.
104.Wang, L., Xie, L., Zhang, Q., Cai, X., Tang,
Y., Wang, L., Hang, T., Liu, J., and Gong, J. (2015) Plasma nuclear and
mitochondrial DNA levels in acute myocardial infarction patients,
Coron. Artery Dis., 26, 296-300.
105.Bliksoen, M., Mariero, L. H., Ohm, I. K.,
Haugen, F., Yndestad, A., Solheim, S., Seljeflot, I., Ranheim, T.,
Andersen, G. O., Aukrust, P., Valen, G., and Vinge, L. E. (2012)
Increased circulating mitochondrial DNA after myocardial infarction,
Int. J. Cardiol., 158, 132-134.
106.Sudakov, N. P., Popkova, T. P., Novikova, M.
A., Katyshev, A. V., Nikiforov, S. B., Pushkarev, B. G., Goldberg, O.
A., Klimenkov, I. V., Lepekhova, S. A., Ezhikeeva, S. D., Ten, M. N.,
Osipov, V. G., and Konstantinov, Yu. M. (2012) The level of blood
plasma mitochondrial DNA upon acute myocardium damage in experiment,
Biopolym. Cell, 28, 321-324.
107.Chang, C. P., Chia, R. H., Wu, T. L., Tsao, K.
C., Sun, C. F., and Wu, J. T. (2003) Elevated cell-free serum DNA
detected in patients with myocardial infarction, Clin. Chim.
Acta, 327, 95-101.
108.Sudakov, N. P., Popkova, T. P., Novikova, M.
A., Katyshev, A. V., Goldberg, O. A., Nikiforov, S. B., Pushkarev, B.
G., Klimenkov, I. V., Lepekhova, S. A., Apartsin, K. A., Nevinsky, G.
A., and Konstantinov, Yu. M. (2015) Level of blood cell-free
circulating mitochondrial DNA as a novel biomarker of acute myocardial
ischemia, Biochemistry (Moscow), 80, 1387-1392.
109.Zhang, Q., Itagaki, K., and Hauser, C. J.
(2010) Mitochondrial DNA is released by shock and activates neutrophils
via p38 map kinase, Shock, 34, 55-59.
110.Itagaki, K., Kaczmarek, E., Lee, Y. T., Tang,
I. T., Isal, B., Adibnia, Y., Sandler, N., Grimm, M. J., Segal, B. Y.,
Otterbein, E., and Hauser, C. J. (2015) Mitochondrial DNA released by
trauma induces neutrophil extracellular traps, PLoS One,
10, e0120549.
111.Zhang, Q., Raoof, M., Chen, Y., Sumi, Y.,
Sursal, T., Junger, W., Brohi, K., Itagaki, K., and Hauser, C. J.
(2010) Circulating mitochondrial DAMPs cause inflammatory responses to
injury, Nature, 464, 104-107.
112.Sursal, T., Stearns-Kurosawa, D. J., Itagaki,
K., Oh, S. Y., Sun, S., Kurosawa, S., and Hauser, C. J. (2013) Plasma
bacterial and mitochondrial DNA distinguish bacterial sepsis from
sterile systemic inflammatory response syndrome and quantify
inflammatory tissue injury in nonhuman primates, Shock,
39, 55-62.
113.Krysko, D. V., Agostinis, P., Krysko, O., Garg,
A. D., Bachert, C., Lambrecht, B. N., and Vandenabeele, P. (2011)
Emerging role of damage-associated molecular patterns derived from
mitochondria in inflammation, Trends Immunol., 32,
157-164.
114.Nakahira, K., Haspel, J. A., Rathinam, V. A.,
Lee, S. J., Dolinay, T., Lam, H. C., Englert, J. A., Rabinovitch, M.,
Cernadas, M., Kim, H. P., Fitzgerald, K. A., Ryter, S. W., and Choi, A.
M. (2011) Autophagy proteins regulate innate immune responses by
inhibiting the release of mitochondrial DNA mediated by the NALP3
inflammasome, Nat. Immunol., 12, 222-230.
115.Collins, L. V., Hajizadeh, S., Holme, E.,
Jonsson, I. M., and Tarkowski, A. (2004) Endogenously oxidized
mitochondrial DNA induces in vivo and in vitro
inflammatory responses, J. Leucoc. Biol., 75,
995-1000.
116.Yousefi, S., Mihalache, C., Kozlowski, E.,
Schmid, I., and Simon, H. U. (2009) Viable neutrophils release
mitochondrial DNA to form neutrophil extracellular traps, Cell Death
Differ., 16, 1438-1444.
117.Ellinger, J., Muller, S. C., Wernert, N., Von
Ruecker, A., and Bastian, P. J. (2008) Mitochondrial DNA in serum of
patients with prostate cancer: a predictor of biochemical recurrence
after prostatectomy, BJU Int., 102, 628-632.
118.Arnalich, F., Codoceo, R., Lopez-Collazo, E.,
and Montiel, C. (2012) Circulating cell-free mitochondrial DNA: a
better early prognostic marker in patients with out-of-hospital cardiac
arrest, Resuscitation, 83, e162-163.
119.Lough, A. N., Roark, L. M., Kato, A., Ream, T.
S., Lamb, J. C., Birchler, J. A., and Newton, K. J. (2008)
Mitochondrial DNA transfer to the nucleus generates extensive insertion
site variation in maize, Genetics, 178, 47-55.
120.Stupar, R. M., Lilly, J. W., Town, C. D.,
Cheng, Z., Kaul, S., Buell, C. R., and Jiang, J. (2001) Complex mtDNA
constitutes an approximate 620-kb insertion on Arabidopsis
thaliana chromosome 2: implication of potential sequencing errors
caused by large-unit repeats, Proc. Natl. Acad. Sci. USA,
98, 5099-5103.
121.Bateman, J. M., and Purton, S. (2000) Tools for
chloroplast transformation in Chlamydomonas: expression vectors
and a new dominant selectable marker, Mol. Gen. Genet.,
263, 404-410.
122.Fox, T. D., Sanford, J. C., and McMullin, T. W.
(1988) Plasmids can stably transform yeast mitochondria lacking
endogenous mtDNA, Proc. Natl. Acad. Sci. USA, 85,
7288-7292.
123.Johnston, S. A., Anziano, P. Q., Shark, K.,
Sanford, J. C., and Butow, R. A. (1988) Mitochondrial transformation in
yeast by bombardment with microprojectiles, Science, 240,
1538-1541.
124.Remacle, C., Cardol, P., Coosemans, N., Gaisne,
M., and Bonnefoy, N. (2006) High-efficiency biolistic transformation of
Chlamydomonas mitochondria can be used to insert mutations in
complex I genes, Proc. Natl. Acad. Sci. USA, 103,
4771-4776.