Pentatricopeptide Motifs in the N-Terminal Extension Domain of Yeast Mitochondrial RNA Polymerase Rpo41p Are Not Essential for Its Function
J. Kruszewski1 and P. Golik1,2*
1University of Warsaw, Institute of Genetics and Biotechnology, Faculty of Biology, Pawinskiego 5A, 02-106 Warsaw, Poland; E-mail: pgolik@igib.uw.edu.pl2Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Pawinskiego 5A, 02-106 Warsaw, Poland
* To whom correspondence should be addressed.
Received March 30, 2016; Revision received May 10, 2016
The core mitochondrial RNA polymerase is a single-subunit enzyme that in yeast Saccharomyces cerevisiae is encoded by the nuclear RPO41 gene. It is an evolutionary descendant of the bacteriophage RNA polymerases, but it includes an additional unconserved N-terminal extension (NTE) domain that is unique to the organellar enzymes. This domain mediates interactions between the polymerase and accessory regulatory factors, such as yeast Sls1p and Nam1p. Previous studies demonstrated that deletion of the entire NTE domain results only in a temperature-dependent respiratory deficiency. Several sequences related to the pentatricopeptide (PPR) motifs were identified in silico in Rpo41p, three of which are located in the NTE domain. PPR repeat proteins are a large family of organellar RNA-binding factors, mostly involved in posttranscriptional gene expression mechanisms. To study their function, we analyzed the phenotype of strains bearing Rpo41p variants where each of these motifs was deleted. We found that deletion of any of the three PPR motifs in the NTE domain does not affect respiratory growth at normal temperature, and it results in a moderate decrease in mtDNA stability. Steady-state levels of COX1 and COX2 mRNAs are also moderately affected. Only the deletion of the second motif results in a partial respiratory deficiency, manifested only at elevated temperature. Our results thus indicate that the PPR motifs do not play an essential role in the function of the NTE domain of the mitochondrial RNA polymerase.
KEY WORDS: mitochondria, RNA polymerase, pentatricopeptide motifs, yeastDOI: 10.1134/S0006297916100084
Abbreviations: mtRNAP, mitochondrial RNA polymerase; NTE, N-terminal extension (domain); ORF, open reading frame; POLRMT, human mitochondrial DNA-directed RNA polymerase; PPR, pentatricopeptide (motif).
Mitochondria of Eukaryotes are derived from an alpha-proteobacterial
endosymbiont [1, 2], but in the
course of evolution their genomes were drastically reduced and acquired
many unique characteristics that distinguish them from their
eubacterial relatives. With the exception of the primitive mitochondria
of jakobid protists that still encode a multisubunit eubacterial-type
RNA polymerase in the organellar genome [3, 4], transcription and replication of the mitochondrial
DNA is assured by specific single-subunit enzymes related to the
proteins of the T-odd bacteriophages and encoded in the nuclear genome
[5].
The first typical mitochondrial RNA polymerase (mtRNAP) described was that in the yeast Saccharomyces cerevisiae [6]. It is a complex composed of a 153-kDa catalytic subunit encoded by the RPO41 gene and a single accessory 40-kDa transcription factor encoded by the MTF1 gene [7-12]. Whereas Rpo41p can recognize and initiate transcription from pre-melted promoter sequences in vitro [13], Mtf1p is required for transcription initiation from duplex DNA [14, 15]. The Rpo41p–Mtf1p complex recognizes the promoter sequence by a mechanism reliant on induced fit and DNA bending [14]. The interaction between Rpo41p and Mtf1p depends on multiple regions of both proteins [16] and is maintained during initiation but not the elongation phase of transcription [17].
Transcription in S. cerevisiae mitochondria is initiated from a simple nonanucleotide promoter [18-20] giving rise to 11 polycistronic primary transcripts [21, 22]. Mitochondrial transcription by mtRNAP is coupled to respiratory activity by a simple ATP sensing kinetic mechanism [23], while RNA degradation and translational regulation provide regulatory mechanisms necessary to fine tune the expression of particular genes [21, 24-26]. Additionally, mtRNAP primes mitochondrial DNA replication [27] and is thus essential for maintaining the functional ([rho+]) mtDNA.
Structural studies of the human mitochondrial RNA polymerase (POLRMT), which is orthologous to Rpo41p, revealed that it consists of three distinct domains: the C-terminal catalytic domain (CTD) that exhibits the typical polymerase right-hand fold, the N-terminal domain (NTD) resembling the promoter-binding domain of the T7 RNA polymerase, and an N-terminal extension (NTE) that includes a domain containing PPR (pentatricopeptide) motifs [28-30]. The catalytic domain is highly conserved between the phage polymerase and the mitochondrial RNA polymerases from diverse eukaryotes. The N-terminal domain is more divergent, but a structural similarity among orthologous mtRNAP sequences and the phage protein is still apparent [29]. In the human protein, this region has been implicated in transcription initiation through interactions with the promoter and the two transcription factors [28]. In the yeast Rpo41 protein, this region also appears to be involved in promoter recognition and in interactions with the sole transcription factor (Mtf1p) [31, 32].
The first ~400 amino acids (a.a.) at the amino terminus of mtRNAP form the NTE that has no counterpart in the bacteriophage RNA polymerase [33] and is generally not conserved between orthologous sequences, although weak similarity in this region can be detected in ascomycetous fungi [34]. In the human mtRNAP, this extension is joined to the NTD by α-helical domain containing two PPR motifs [29]. Whereas no significant sequence similarity can be found between the NTE of human POLRMT and yeast Rpo41p, application of an algorithm fine-tuned for detection of yeast pentatricopeptide proteins indicated that up to five putative PPR motifs can be found in the N-terminal part of Rpo41p [35].
Proteins containing the PPR motifs are a large family of RNA-binding factors that can be found in all eukaryotes [36-42]. They are particularly abundant in plants, where they constitute the most numerous paralogous families containing hundreds of sequences per genome [38, 42]. The vast majority of known PPR proteins are involved in various posttranscriptional mechanisms of organellar gene expression (reviewed in [36, 37, 39, 40]). They have been extensively studied in both model yeast systems – S. cerevisiae [39] and S. pombe [43], where they play significant roles in maintaining and regulating the expression of mitochondrial genes.
Typical PPR proteins contain multiple tandem repeats of a 35-a.a. motif forming two parallel α-helices, resulting in a solenoid superhelical structure of the entire protein [37, 39, 42]. Most are composed solely of the PPR repeats, while some (found mostly in plants) contain additional C-terminal domains conferring enzymatic activities [37, 40]. The mitochondrial RNA polymerase is thus not a typical PPR protein, as it contains only 2-5 PPR motifs that constitute only a minor portion of the entire sequence.
The role of the unconserved N-terminal extension of the mitochondrial RNA polymerases, including the PPR motifs found therein, is not entirely clear. In the human POLRMT protein, the PPR domain interacts with the promoter recognition loop of the NTD domain and is essential for transcription initiation [29]. It also interacts with the upstream N-terminal extension that binds the transcription factor TFAM [28]. In yeast, deletions of the N-terminal extension of Rpo41p result in a respiratory deficiency connected with a loss of mitochondrial DNA stability that is, however, not caused by a major transcriptional defect [33]. Remarkably, while a complete or near complete (472 or 373 a.a., respectively) NTE deletion resulted in an unconditional defect, deleting as many as 185 a.a. gave only a conditional (temperature sensitive) phenotype [33]. Subsequent studies suggested that this domain couples transcription to RNA processing and translation through interactions with the Nam1 and Sls1 proteins [34, 44, 45]. Deletion of the first 270 a.a. of the Rpo41p NTE domain does not affect the catalytic activity of the polymerase in vitro, in fact enhancing the productive/abortive ratio of RNA synthesis, whereas a 380-a.a. deletion decreases initiation from duplex, but not pre-melted promoters [46].
These results suggest that the function of the N-terminal extension domain of the yeast mitochondrial RNA polymerase is separate from the polymerase activity, but it can be related to the coordination of transcription with the subsequent steps of gene expression. However, available data do not indicate whether the putative PPR motifs found in this domain are involved in its function. We therefore decided to investigate their role by constructing a series of small deletions involving only the PPR motif sequences and assessing the activity of the resultant Rpo41 variants in vivo.
MATERIALS AND METHODS
Media and genetic techniques. Complete (YP) media supplemented with 2% glucose (YPD), 2% glycerol (YPG), or 2% glycerol and 0.1% glucose (YPDG), as well as synthetic complete (SC) media, were prepared as described in our recent work [47]. Standard yeast genetic methods were as described previously [48, 49]. Yeasts were transformed using the LiAc/PEG/ssDNA protocol [50].
Plasmids and construction of deletants. Plasmids pJJ1148 and pJJ1149 [51] contained the RPO41 gene cloned in the YCplac33 (ARS-CEN, URA3) and YCplac111 (ARS-CEN, LEU2) vectors, respectively.
Plasmid pJJ1149 (YCplac111:RPO41) was used as the starting point for deletion mutagenesis. The PCR-mediated Site-directed, Ligase-Independent Mutagenesis (SLIM) protocol [52] was used do delete segments encoding respective PPR motifs. With this method, two pairs of primers, termed RT, FT and RS, FS, are used for each deletion (primers are named according to the description in [52]).
To delete the first PPR motif (PPR1, a.a. from 47 to 82), nucleotides 139 to 446 of the RPO41 ORF (open reading frame) were deleted using primers dPPR1FT (5′-ATCTGAAGACCCCCTTGTATGGAAAAATCCTTCAGAATTAGAG), dPPR1RT (5′-TACAAGGGGGTCTTCAGATATTAATATTGTACTTGTTGAGTCCG), dPPR1FS (5′-TGGAAAAATCCTTCAGAATTAGAG), and dPPR1RS (ATTAATATTGTACTTGTTGAGTCCG), to yield vector YCplac111:RPO41ΔPPR1.
To delete the second PPR motif (PPR2, a.a. from 112 to 146), nucleotides 334 to 438 of the RPO41 ORF were deleted using primers dPPR2FT (5′-GACAAGAACTAGAGACGTTTTTATCGAGGATTATAACATGTAC), dPPR2RT (5′-AACGTCTCTAGTTCTTGTCATCGAGGTTACTGCTGAGGCATG), dPPR2FS (5′-TTTATCGAGGATTATAACATGTAC), and dPPR2RS (5′-ATCGAGGTTACTGCTGAGGCATG), to yield vector YCplac111:RPO41ΔPPR2.
To delete the third PPR motif (PPR3, a.a. from 291 to 325), nucleotides 871 to 975 of the RPO41 ORF were deleted using primers dPPR3FT (5′-CAAAGTGGAAGCTGAGAACTCAAAAGACGCTGAAAAGCTTATAG), dPPR3RT (5′-GTTCTCAGCTTCCACTTTGTTCAAACCCTCCTCATTTTC), dPPR3FS (5′-TCAAAAGACGCTGAAAAGCTTATAG), and dPPR3RS (5′-TTCAAACCCTCCTCATTTTC), to yield vector YCplac111:RPO41ΔPPR3.
Numbering starts with the first amino acid (M1) and the first nucleotide (A1) of the protein and the ORF, respectively.
All the constructs were verified by Sanger sequencing performed in the Laboratory of DNA Sequencing and Oligonucleotide Synthesis, Institute of Biochemistry and Biophysics, Polish Academy of Science.
Yeast strains. The rpo41::KanMX4 cassette amplified from the DNA of the EUROSCARF Y15657 strain (MATα; ura3Δ0; leu2Δ0; his3Δ1; lys2Δ0; YFL036w::kanMX4) with primers YFL036w_A (5′-TCTCCGTGAAGATATTGGTAAAAAG) and YFL036w_D (5′-ACAAAATTGTGCACTTGTCATTAGA) was used to transform strain W303-1B [53] (MATα, ade2-1, leu2-3, 112 ura3-,1 trp1-1, his3-11, 15 can1-100 [rho+]) to obtain a strain carrying a deletion of the entire RPO41 ORF. This strain was subsequently crossed to two isonuclear W303 MATα derivative strains CW04 [53] (MATα, ade2-1, leu2-3, 112 ura3-,1 trp1-1, his3-11, 15 can1-100 [rho+ 13 introns]) and CW252 [54] (MATα, ade2-1, leu2-3, 112 ura3-,1 trp1-1, his3-11, 15 can1-100 [rho+ intronless]), yielding heterozygous diploids HZRPO41 and HZRPO41int, respectively. The heterozygous diploids were transformed with pJJ1148 (YCplac33:RPO41), and the transformants sporulated. Haploid MATα spores carrying the rpo41::KanMX4 deletion and the wild type RPO41 gene on pJJ1148 were selected, yielding strains DRPO41 (MATα, ade2-1, leu2-3, 112 ura3-,1 trp1-1, his3-11, 15 can1-100, rpo41::KanMX4 [YCplac33:RPO41] [rho+ 13 introns]) and DRPO41int (MATα, ade2-1, leu2-3, 112 ura3-,1 trp1-1, his3-11, 15 can1-100, rpo41::KanMX4 [YCplac33:RPO41] [rho+ intronless]).
The DRPO41 and DRPO41int strains were transformed with vectors YCplac111:RPO41ΔPPR1, YCplac111:RPO41ΔPPR2, YCplac111:RPO41ΔPPR3. The pJJ1148 plasmid, carrying the wild type RPO41 gene, was then eliminated by counter selection on 5-FOA plates [55], leaving the deletion variant as the only mitochondrial RNA polymerase in the cells. The strains thus obtained were dPPR1int (MATα, ade2-1, leu2-3, 112 ura3-,1 trp1-1, his3-11, 15 can1-100, rpo41::KanMX4 [YCplac111:RPO41ΔPPR1] [rho+ intronless]), dPPR2int (MATα, ade2-1, leu2-3, 112 ura3-,1 trp1-1, his3-11, 15 can1-100, rpo41::KanMX4 [YCplac111:RPO41ΔPPR2] [rho+ intronless]), dPPR3int (MATα, ade2-1, leu2-3, 112 ura3-,1 trp1-1, his3-11, 15 can1-100, rpo41::KanMX4 [YCplac111:RPO41ΔPPR3] [rho+ intronless]), dPPR1 (MATα, ade2-1, leu2-3, 112 ura3-,1 trp1-1, his3-11, 15 can1-100, rpo41::KanMX4 [YCplac111:RPO41ΔPPR1] [rho+ 13 introns]), dPPR2 (MATα, ade2-1, leu2-3, 112 ura3-,1 trp1-1, his3-11, 15 can1-100, rpo41::KanMX4 [YCplac111:RPO41ΔPPR2] [rho+ 13 introns]), and dPPR3 (MATα, ade2-1, leu2-3, 112 ura3-,1 trp1-1, his3-11, 15 can1-100, rpo41::KanMX4 [YCplac111:RPO41ΔPPR3] [rho+ 13 introns]).
D273-10B/51 [56] (MATα ade5, [rho0]) was used as a tester strain in mtDNA stability assays.
Respiratory growth assay. Portions of 10 µl from 10–1, 10–2, 10–3, and 10–4 dilutions of each strain (OD600 = 1) were spotted on YPD and YPG plates and incubated at 30°C (normal temperature) or 37°C (elevated temperature) for three days.
Assays of mtDNA stability. Stability of the mitochondrial genome was assessed by assaying the frequency of petite colony generation using the tetrazolium overlay method [57] as described previously [47]. Confirmation that the petite phenotype was due to mtDNA defects was obtained by crossing to the D273-10B/51 tester strain.
Mitochondrial RNA preparation and Northern blots. RNA was isolated from mitochondrial preparations enriched by differential centrifugation as described previously [58]. A Total RNA Mini kit (A&A Biotechnology, Poland) was used in the RNA purification step. RNA was electrophoresed in 1% agarose-formaldehyde gel and blotted onto nylon membrane (Nytran N; GE Healthcare, England) as described previously [59]. Methylene blue staining of blots was used to normalize loading. Oligonucleotide and dsDNA probes were labeled with 32P as described previously [60]. Results were visualized using a Typhoon FLA 9000 laser imaging system (GE Healthcare).
The following probes were used: for COX1, plasmid pYGT21 containing exon sequences [61]; for COX2, plasmid pJM2 [62]; for COB, oligonucleotide 5′-CATGGCATGCTCCAATTTGTCCTAATAATACGAAATTGAAA; for ATP6/8, plasmid pBZ6 (B. Zapisek) – ATP6 ORF cloned into pGEM®-T Easy (Promega, USA); for ATP9, PCR product (325 bp) amplified using primers Atp9F (5′-TATGCAATTAGTATTAGCAGC) and Atp9R (5′-GAATGTTATTAATTTAATCAAATGAG).
RESULTS
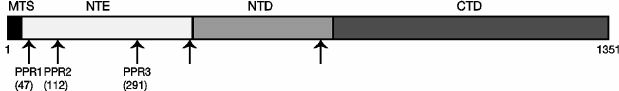
Construction of deletions of PPR motifs in the N-terminal extension domain of Rpo41p. Unlike in human POLRMT [29], PPR motifs in the yeast Rpo41 sequence are not readily detected by profile matching tools. However, using yeast-specific profiles and orthologous alignments from different yeast genomes [35], five divergent PPR motifs can be identified in S. cerevisiae Rpo41p (Fig. 1). Three of them, named PPR1, PPR2, and PPR3 here, are located within the unconserved N-terminal extension domain. The fourth motif lies at the boundary of the N-terminal extension and the conserved N-terminal domain, and the fifth is close to the beginning of the C-terminal conserved catalytic domain. As the N-terminal extension is functionally independent from the bacteriophage-like RNA polymerase activity confined to the conserved regions of the protein, we decided to investigate the function of the three PPR motifs located therein by generating deletions encompassing each of these motifs.
Fig. 1. Schematic structure of the S. cerevisiae Rpo41p mitochondrial RNA polymerase. MTS, mitochondrial targeting sequence; NTE, N-terminal extension; NTD, N-terminal domain; CTD, C-terminal (catalytic) domain. Arrows indicate the positions of pentatricopeptide (PPR) motifs predicted in silico [35]. Numbers indicate amino acid positions for the three named PPR motifs that were studied here; the position of the first amino acid is shown.
To obtain variants bearing deletions of the first three PPR motifs, sequences encoding 35 a.a. (beginning at the first amino acid of each motif, as shown in Fig. 1) were deleted from the RPO41 ORF in pJJ1149 (see “Materials and Methods” for details of plasmid and strain construction) using the PCR-mediated Site-directed Ligase-Independent Mutagenesis (SLIM) protocol [52], yielding the YCplac111:RPO41dPPR1, YCplac111:RPO41dPPR2, and YCplac111:RPO41dPPR3 vectors.
As deletion of RPO41 results in complete and irreversible loss of mtDNA [63], heterozygous diploid strains were used as a starting point. To test the possible involvement of the PPR motifs in Rpo41 in intronic RNA processing, strains with two variants of the mitochondrial genome were used – one containing 13 introns (CW04), and one devoid of all introns (CW252). Wild type RPO41 was introduced into the heterozygous strains by transformation with the pJJ1148 vector. The resulting transformants were subsequently sporulated, and haploid strains DRPO41 and DRPO41int carrying the deletion of the RPO41 in the genomic DNA, with the pJJ1148 plasmid providing the functional wild type allele, and a [rho+] mitochondrial genome containing 13 introns, or devoid of all introns, respectively, were selected.
DRPO41 and DRPO41int strains were subsequently transformed with the ARS-CEN, LEU2 vectors encoding the PPR deletion variants of the Rpo41 protein (YCplac111:RPO41dPPR1, YCplac111:RPO41dPPR2, and YCplac111:RPO41dPPR3). The pJJ1148 vector encoding the wild type Rpo41p was then removed by counter selection on 5-FOA plates [55], resulting in strains where the deletion variants were the only Rpo41 proteins in the cell. These strains were named dPPR1int, dPPR2int, and dPPR3int (strains with the intronless mtDNA) and dPPR1, dPPR2, and dPPR3 (strains with intron-containing mtDNA).
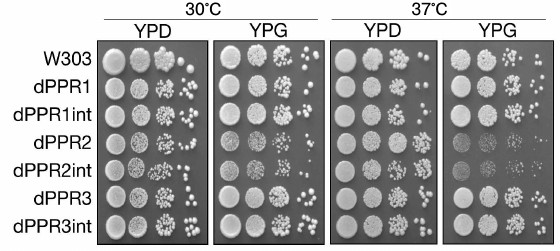
Effect of PPR motif deletions on respiratory growth of the yeast. Respiratory capacity of the strains carrying the deletion variants of the Rpo41p RNA polymerase was verified by testing their growth on a non-fermentable carbon source (glycerol) at normal (30°C) and elevated (37°C) temperature (Fig. 2). Deletions of the first and third PPR motifs had no observable effect on respiratory growth of the relevant strains at either temperature. Only in the dPPR2 and dPPR2int strains, bearing a deletion of the second PPR motif, a moderate respiratory defect could be observed. This strain still grows on glycerol, but the resultant colonies are visibly smaller (an effect observable also on glucose), suggesting a hypomorphic petite phenotype, similar to the defect observed in deletants missing the first 90 a.a. of Rpo41p [34]. This phenotype was more apparent at the elevated temperature, but even then, it did not amount to a complete loss of respiratory growth.
Fig. 2. Respiratory growth of S. cerevisiae strains bearing the wild type (W303) or deletant variants of RPO1. Strains dPPR1, dPPR2, and dPPR3 bear deletions of the first, second, and third PPR motif, respectively. Suffix “int” indicates strains with the intronless mitochondrial DNA. Fermentative growth was tested on YPD (YP with 2% glucose) plates at the normal (30°C) and elevated (37°C) temperature. Respiratory growth was tested on YPG (YP with 2% glycerol) plates at the normal (30°C) and elevated (37°C) temperature. Serial dilutions of appropriate cultures were spotted on plates as described in “Materials and Methods”. Only the dPPR2 and dPPR2int strains show slower respiratory growth, particularly at 37°C.
As no difference between the intron-containing and intronless mtDNA could be observed in any of the deletants, subsequent experiments were performed using only the intron-containing strains.
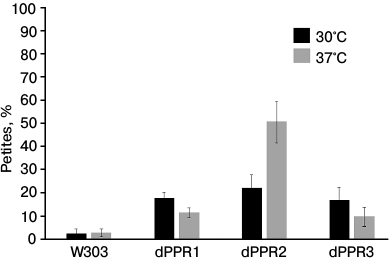
Effect of PPR motif deletions on the stability of the mitochondrial genome. Deletions of large segments of the N-terminal extension result in a loss of mitochondrial genome stability [33] manifesting itself in a complete loss [rho0] or extensive deletions [rho−] of mtDNA. These so-called cytoplasmic petite colonies arise also in wild type yeast cultures, albeit at a low rate of about 1-5%. We measured the petite frequencies in dPPR1, dPPR2, and dPPR3 mutants (Fig. 3) using the tetrazolium test [57] and by crossing to a [rho0] tester strain. The dPPR1int and dPPR3int deletants show a moderate but significant (p < 0.01) increase in petite generation (17-18%) that is apparent at both 30 and 37°C. Interestingly, the effect is slightly more pronounced at 30°C. The dPPR2 deletant also shows a similar increase in petite generation at 30°C (~22%), but a much stronger (up to ~50%) effect at 37°C, which is consistent with a visible respiratory growth impairment at this temperature (Fig. 2). In the wild type strain there is no statistically significant difference in mtDNA stability in cells grown at the normal and elevated temperature (2.6 and 2.9%).
Fig. 3. Mitochondrial genome stability in S. cerevisiae strains bearing the wild type (W303) or deletant variants of RPO1. Frequencies of petite mutants in strains carrying the wild type RPO41 or alleles expressing variants with PPR motif deletions grown at the normal (30°C) and elevated (37°C) temperature were measured as described in “Materials and Methods”. All three deletants show significantly (p < 0.01) increased frequencies of petite cells relative to the wild type. Differences between dPPR cultures grown at 30 and 37°C are also significant (p < 0.01), while the elevated temperature does not significantly affect mtDNA stability of the strain carrying the wild type RPO41 gene.
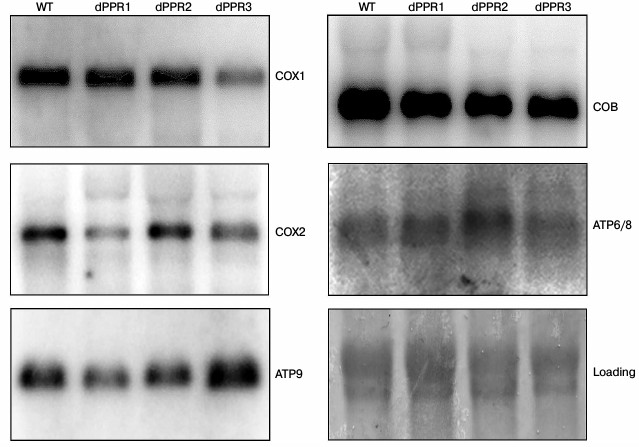
Effect of PPR motif deletions on steady-state levels of mitochondrial transcripts. Mutations in the N-terminal extension domain of Rpo41p can affect steady-state levels of specific mitochondrial RNAs [34, 45]. To determine whether deletions of PPR motifs could also specifically influence mitochondrial transcripts, we performed a series of Northern blots in deletant strains. Mitochondrial RNA from the wild type and the dPPR1, dPPR2, and dPPR3 strains grown at 30°C was hybridized to probes recognizing some of the mitochondrially encoded mRNAs: COX1 and COX2 encoding subunits of the cytochrome oxidase complex (Complex IV), COB encoding the apocytochrome b (Complex III), and ATP9 and ATP6-8 encoding subunits of the ATP synthase (Complex V).
The results (Fig. 4) show a moderate decrease in the levels of mature mRNAs encoding subunits of Complex IV, with each deletant following a different pattern. Deletion of the first motif (dPPR1) results in a marked decrease (to about 40% of wild type) of COX2 mRNA, with the remaining mRNAs mostly unaffected. Deletion of the second motif (dPPR2) does not show a clear effect on any mRNA. Deletion of the third motif (dPPR3) results in a visible decrease (to about 44% of wild type level) of COX1. The mRNAs encoding apocytochrome b (COB) and the subunits of the ATP synthase (ATP6/8 and ATP9) were not significantly affected in any of the deletants. Such a moderate change in the mature mRNA levels is likely to have little effect on respiratory growth, as yeast mutants with as little as 10% of wild type mature mitochondrial transcript were found to be respiratory competent [25, 64]. The results are therefore consistent with the respiratory function assays that show mostly unaffected respiratory growth at normal temperature (30°C).
Fig. 4. Northern blot analysis of mitochondrial mRNAs in S. cerevisiae strains bearing the wild type (W303) or deletant variants of RPO1. Mitochondrial RNA preparations from each strain, grown at 30°C, were separated in 1% agarose-formaldehyde gel, blotted onto nylon membrane, and hybridized to appropriate probes as described in “Materials and Methods”. With the exception of COX1 in dPPR3 and COX2 in dPPR1, all the mRNA levels are either unchanged or slightly increased relative to wild type. Blots were stained with methylene blue to normalize the amount of mitochondrial RNA preparations in each lane using rRNA bands (“loading”).
DISCUSSION
Of the five divergent PPR motifs that can be found in the sequence of the yeast mitochondrial RNA polymerase, three are located in the N-terminal extension (NTE) domain. Unlike the downstream N-terminal domain and the C-terminal domain, this region of the protein shows no homology to the T7 bacteriophage RNA polymerase, and it is not conserved in evolution, with some regions of similarity shared only with other fungi [34]. Other organellar RNA polymerases, like the human POLRMT [29], also have an N-terminal extension, but no sequence conservation can be detected. This domain is not essential for the main catalytic activity of the protein, and even large deletions therein result only in a temperature sensitive respiratory deficiency phenotype [33].
The results obtained in this study are consistent with these previous findings. Deletion of any of the three PPR motifs located in the NTE domain does not affect respiratory growth at the normal temperature, and only deletion of the second motif (a.a. from 112 to 146) gives a partial respiratory deficiency at the elevated temperature. All three PPR motif deletions result in a decreased mitochondrial DNA stability, but not in a complete loss of functional mtDNA. By way of comparison, deletion of up to 212 a.a. in the NTE domain results in a complete respiratory deficiency at the elevated temperature with concomitant mtDNA destabilization [34]. The phenotype observed for the PPR motif deletions in this study is therefore a milder, partial form of the deficiency observed for the NTE deletion. In fact, a single amino acid substitution (R129D) has a more pronounced effect on the function of the protein compared to the PPR motif deletions [34]. It can thus be concluded that these motifs do not play a particularly significant role in the functioning of this domain.
The observed phenotypes, together with previously reported data obtained using large deletions in the N-terminal extension in vivo [33, 44, 45] and in vitro [46], also support the conclusion that this region of the protein is not involved in the main enzymatic activity. Deletions of PPR motifs studied here, as well as other mutations and deletions in the NTE region [44, 45], affect the steady state levels of some mitochondrial mRNAs (mainly COX1 or COX2), while other transcripts remain unaffected. This suggests that the function of this region concerns RNA stability and translation rather than general transcription, and it is likely regulatory.
These observations are consistent with a model suggesting that the N-terminal extension of the mitochondrial RNA polymerase is involved in coupling transcription with RNA processing and translation. This function is probably achieved through interactions with two known accessory proteins: Nam1 and Sls1 [34, 44, 45]. Sls1p interacts with the N-terminal part of Rpo41p, anchoring the RNA polymerase and the nascent transcript to the inner membrane complexes that include translational activators [44]. Nam1p is a putative RNA chaperone [65, 66] that interacts with Sls1p, NTE domain of Rpo41p, and the nascent RNA [44]. The PPR motifs appear to participate in these interactions as their deletion results in a decrease of mtDNA stability, changes in mitochondrial mRNA levels, and (in the case of the second motif), partial temperature-sensitive phenotype. However, they are not essential, as the change in phenotype is relatively mild compared to the larger N-terminal extension deletions.
In typical PPR proteins, the pentatricopeptide motifs form RNA binding surfaces and usually act as intermediates in interactions between organellar transcripts and other proteins [37-42]. However, typical RNA-binding PPR proteins are composed of multiple adjacent repeats, whereas in Rpo41p the motifs are few and dispersed. It is currently not clear whether they participate in RNA binding, or even whether the entire N-terminal extension interacts directly with RNA. Crystallographic studies of the human mitochondrial RNA polymerase revealed that the two PPR motifs, found at the boundary of the N-terminal extension and the conserved N-terminal domain, do not interact with RNA, but they participate in intramolecular interactions between different domains of the protein [29]. In yeast Rpo41p, the fourth and fifth motif, also located near domain boundaries (see Fig. 1), could arguably play a similar role, although without crystallographic data it remains only a speculation. It is also conceivable that the three PPR motifs in the NTE domain that were the subject of the present study also participate in interactions with accessory proteins, consistent with the postulated function of the entire domain. The PPR motif shares evolutionary and structural relationship with other α-α repeat sequences, like TPR (tetratricopeptide), SEL1, and HAT motifs forming solenoid folds [42, 67, 68], many of which participate in protein–protein interactions.
Despite functional differences between the PPR motifs in the mitochondrial RNA polymerase and typical PPR proteins, they share a similar broad evolutionary role. The mitochondrial RNA polymerase is a descendant of the bacteriophage RNAP [5], which is a standalone enzyme that does not depend on any additional factors. To integrate the phage enzyme into the more complex organellar genetic system, interactions with accessory regulatory proteins had to evolve. The N-terminal extension of mtRNAP, which has no homology to the bacteriophage polymerase, arose to allow such interactions and coordinate the transcriptional activity with organellar RNA processing and translation. Similarly, nuclear-encoded PPR proteins evolved in the Eukaryotic host to provide activities necessary to maintain and regulate organellar gene expression, replacing prokaryotic mechanisms that were lost in the course of the evolution of the endosymbiont. Both the NTE domain of the mitochondrial RNA polymerase and the PPR proteins in general are thus examples of evolutionary adaptations in domesticating the endosymbiont, allowing integrating it with the host system. Such adaptations often involve helical repeat sequences as their modularity imparts great evolutionary plasticity. Indeed, the N-terminal extension sequences of mitochondrial RNA polymerases, as well as the entire PPR protein family, show rapid divergent evolution that adapts them to the quickly changing evolutionary dynamics of the nucleo/organellar interaction.
Acknowledgements
We thank Bartosz Zapisek for the gift of plasmid pBZ6. Plasmids pJJ1148 and pJJ1149 were a gift from Dr. Judith Jaehning.
This work was supported by the EU Operational Program Innovative Economy via the Foundation for Polish Science grant TEAM/2010-6/6.
REFERENCES
1.Gray, M. W., Burger, G., and Lang, B. F. (1999)
Mitochondrial evolution, Science, 283, 1476-1481.
2.Lang, B. F., Gray, M. W., and Burger, G. (1999)
Mitochondrial genome evolution and the origin of eukaryotes, Annu.
Rev. Genet., 33, 351-397.
3.Burger, G., Gray, M. W., Forget, L., and Lang, B.
F. (2013) Strikingly bacteria-like and gene-rich mitochondrial genomes
throughout jakobid protists, Genome Biol. Evol., 5,
418-438.
4.Lang, B. F., Burger, G., O’Kelly, C. J.,
Cedergren, R., Golding, G. B., Lemieux, C., Sankoff, D., Turmel, M.,
and Gray, M. W. (1997) An ancestral mitochondrial DNA resembling a
eubacterial genome in miniature, Nature, 387,
493-497.
5.Shutt, T. E., and Gray, M. W. (2006) Bacteriophage
origins of mitochondrial replication and transcription proteins,
Trends Genet., 22, 90-95.
6.Masters, B. S., Stohl, L. L., and Clayton, D. A.
(1987) Yeast mitochondrial RNA polymerase is homologous to those
encoded by bacteriophages T3 and T7, Cell, 51, 89-99.
7.Jang, S. H., and Jaehning, J. A. (1991) The yeast
mitochondrial RNA polymerase specificity factor, MTF1, is similar to
bacterial sigma factors, J. Biol. Chem., 266,
22671-22677.
8.Schubot, F. D., Chen, C. J., Rose, J. P., Dailey,
T. A., Dailey, H. A., and Wang, B. C. (2001) Crystal structure of the
transcription factor sc-mtTFB offers insights into mitochondrial
transcription, Protein Sci., 10, 1980-1988.
9.Matsunaga, M., Jang, S.-H., and Jaehning, J. A.
(2004) Expression and purification of wild type and mutant forms of the
yeast mitochondrial core RNA polymerase, Rpo41, Protein Express.
Purif., 35, 126-130.
10.Levens, D., Lustig, A., and Rabinowitz, M. (1981)
Purification of mitochondrial RNA polymerase from Saccharomyces
cerevisiae, J. Biol. Chem., 256, 1474-1481.
11.Tracy, R. L., and Stern, D. B. (1995)
Mitochondrial transcription initiation: promoter structures and RNA
polymerases, Curr. Genet., 28, 205-216.
12.Karlok, M. A., Jang, S.-H., and Jaehning, J. A.
(2002) Mutations in the yeast mitochondrial RNA polymerase specificity
factor, Mtf1, verify an essential role in promoter utilization, J.
Biol. Chem., 277, 28143-28149.
13.Matsunaga, M., and Jaehning, J. A. (2004)
Intrinsic promoter recognition by a “core” RNA polymerase,
J. Biol. Chem., 279, 44239-44242.
14.Tang, G.-Q., Deshpande, A. P., and Patel, S. S.
(2011) Transcription factor-dependent DNA bending governs promoter
recognition by the mitochondrial RNA polymerase, J. Biol. Chem.,
286, 38805-38813.
15.Paratkar, S., and Patel, S. S. (2010)
Mitochondrial transcription factor Mtf1 traps the unwound non-template
strand to facilitate open complex formation, J. Biol. Chem.,
285, 3949-3956.
16.Cliften, P., Park, J. Y., Davis, B. P., Jang,
S.-H., and Jaehning, J. A. (1997) Identification of three regions
essential for interaction between a sigma-like factor and core RNA
polymerase, Genes Dev., 11, 2897-2909.
17.Mangus, D. A., Jang, S. H., and Jaehning, J. A.
(1994) Release of the yeast mitochondrial RNA polymerase specificity
factor from transcription complexes, J. Biol. Chem., 269,
26568-26574.
18.Osinga, K. A., and Tabak, H. F. (1982) Initiation
of transcription of genes for mitochondrial ribosomal RNA in yeast:
comparison of the nucleotide sequence around the 5′-ends of both
genes reveals a homologous stretch of 17 nucleotides, Nucleic Acids
Res., 10, 3617-3626.
19.Christianson, T. W., and Rabinowitz, M. (1983)
Identification of multiple transcriptional initiation sites on the
yeast mitochondrial genome by in vitro capping with
guanylyltransferase, J. Biol. Chem., 258,
14025-14033.
20.Christianson, T. W., Edwards, J., Levens, D.,
Locker, J., and Rabinowitz, M. (1982) Transcriptional initiation and
processing of the small ribosomal RNA of yeast mitochondria, J.
Biol. Chem., 257, 6494-6500.
21.Turk, E. M., Das, V., Seibert, R. D., and
Andrulis, E. D. (2013) The mitochondrial RNA landscape of
Saccharomyces cerevisiae, PLoS One, 8,
e78105.
22.Foury, F., Roganti, T., Lecrenier, N., and
Purnelle, B. (1998) The complete sequence of the mitochondrial genome
of Saccharomyces cerevisiae, FEBS Lett.,
440, 325-331.
23.Amiott, E. A., and Jaehning, J. A. (2006)
Mitochondrial transcription is regulated via an ATP
“sensing” mechanism that couples RNA abundance to
respiration, Mol. Cell, 22, 329-338.
24.Herrmann, J. M., Woellhaf, M. W., and Bonnefoy,
N. (2013) Control of protein synthesis in yeast mitochondria: the
concept of translational activators, Biochim. Biophys. Acta,
1833, 286-294.
25.Rogowska, A. T., Puchta, O., Czarnecka, A. M.,
Kaniak, A., Stepien, P. P., and Golik, P. (2006) Balance between
transcription and RNA degradation is vital for Saccharomyces
cerevisiae mitochondria: reduced transcription rescues the
phenotype of deficient RNA degradation, Mol. Biol. Cell,
17, 1184-1193.
26.Krause, K., and Dieckmann, C. L. (2004) Analysis
of transcription asymmetries along the tRNAE-COB operon: evidence for
transcription attenuation and rapid RNA degradation between coding
sequences, Nucleic Acids Res., 32, 6276-6283.
27.Sanchez-Sandoval, E., Diaz-Quezada, C.,
Velazquez, G., Arroyo-Navarro, L. F., Almanza-Martinez, N.,
Trasvina-Arenas, C. H., and Brieba, L. G. (2015) Yeast mitochondrial
RNA polymerase primes mitochondrial DNA polymerase at origins of
replication and promoter sequences, Mitochondrion, 24,
22-31.
28.Morozov, Y. I., Agaronyan, K., Cheung, A. C. M.,
Anikin, M., Cramer, P., and Temiakov, D. (2014) A novel intermediate in
transcription initiation by human mitochondrial RNA polymerase,
Nucleic Acids Res., 42, 3884-3893.
29.Ringel, R., Sologub, M., Morozov, Y. I., Litonin,
D., Cramer, P., and Temiakov, D. (2011) Structure of human
mitochondrial RNA polymerase, Nature, 478, 269-273.
30.Schwinghammer, K., Cheung, A. C. M., Morozov, Y.
I., Agaronyan, K., Temiakov, D., and Cramer, P. (2013) Structure of
human mitochondrial RNA polymerase elongation complex, Nat. Struct.
Mol. Biol., 20, 1298-1303.
31.Nayak, D., Guo, Q., and Sousa, R. (2009) A
promoter recognition mechanism common to yeast mitochondrial and phage
t7 RNA polymerases, J. Biol. Chem., 284, 13641-13647.
32.Yang, X., Chang, H. R., and Yin, Y. W. (2015)
Yeast mitochondrial transcription factor Mtf1 determines the precision
of promoter-directed initiation of RNA polymerase Rpo41, PLoS
One, 10, e0136879.
33.Wang, Y., and Shadel, G. S. (1999) Stability of
the mitochondrial genome requires an amino-terminal domain of yeast
mitochondrial RNA polymerase, Proc. Natl. Acad. Sci. USA,
96, 8046-8051.
34.Rodeheffer, M. S., Boone, B. E., Bryan, A. C.,
and Shadel, G. S. (2001) Nam1p, a protein involved in RNA processing
and translation, is coupled to transcription through an interaction
with yeast mitochondrial RNA polymerase, J. Biol. Chem.,
276, 8616-8622.
35.Lipinski, K. A., Puchta, O., Surendranath, V.,
Kudla, M., and Golik, P. (2011) Revisiting the yeast PPR proteins
– application of an Iterative Hidden Markov Model algorithm
reveals new members of the rapidly evolving family, Mol. Biol.
Evol., 28, 2935-2948.
36.Lightowlers, R. N., and Chrzanowska-Lightowlers,
Z. M. (2013) Human pentatricopeptide proteins, RNA Biol.,
10, 1433-1438.
37.Filipovska, A., and Rackham, O. (2013)
Pentatricopeptide repeats, RNA Biol., 10, 1426-1432.
38.Giege, P. (2013) Pentatricopeptide repeat
proteins, RNA Biol., 10, 1417-1418.
39.Herbert, C. J., Golik, P., and Bonnefoy, N.
(2013) Yeast PPR proteins, watchdogs of mitochondrial gene expression,
RNA Biol., 10, 1477-1494.
40.Schmitz-Linneweber, C., and Sluyter, F. (2008)
Pentatricopeptide repeat proteins: a socket set for organelle gene
expression, Trends Plant Sci., 13, 663-670.
41.Delannoy, E., Stanley, W. A., Bond, C. S., and
Sluyter, F. (2007) Pentatricopeptide repeat (PPR) proteins as
sequence-specificity factors in post-transcriptional processes in
organelles, Biochem. Soc. Trans., 35, 1643-1647.
42.Small, I. D., and Peeters, N. (2000) The PPR
motif – a TPR-related motif prevalent in plant organellar
proteins, Trends Biochem. Sci., 25, 46-47.
43.Kuhl, I., Dujeancourt, L., Gaisne, M., Herbert,
C. J., and Bonnefoy, N. (2011) A genome wide study in fission yeast
reveals nine PPR proteins that regulate mitochondrial gene expression,
Nucleic Acids Res., 39, 8029-8041.
44.Rodeheffer, M. S., and Shadel, G. S. (2003)
Multiple interactions involving the amino-terminal domain of yeast
mtRNA polymerase determine the efficiency of mitochondrial protein
synthesis, J. Biol. Chem., 278, 18695-18701.
45.Bryan, A. C., Rodeheffer, M. S., Wearn, C. M.,
and Shadel, G. S. (2002) Sls1p is a membrane-bound regulator of
transcription-coupled processes involved in Saccharomyces
cerevisiae mitochondrial gene expression, Genetics,
160, 75-82.
46.Paratkar, S., Deshpande, A. P., Tang, G.-Q., and
Patel, S. S. (2011) The N-terminal domain of the yeast mitochondrial
RNA polymerase regulates multiple steps of transcription, J. Biol.
Chem., 286, 16109-16120.
47.Kaliszewska, M., Kruszewski, J., Kierdaszuk, B.,
Kostera-Pruszczyk, A., Nojszewska, M., Lusakowska, A., Vizueta, J.,
Sabat, D., Lutyk, D., Lower, M., Piekutowska-Abramczuk, D.,
Kaniak-Golik, A., Pronicka, E., Kaminska, A., Bartnik, E., Golik, P.,
and Tonska, K. (2015) Yeast model analysis of novel polymerase gamma
variants found in patients with autosomal recessive mitochondrial
disease, Hum. Genet., 134, 951-966.
48.Burke, D., Dawson, D., and Stearns, T. (2000)
Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course
Manual.
49.Dujardin, G., Pajot, P., Groudinsky, O., and
Slonimski, P. P. (1980) Long-range control circuits within mitochondria
and between nucleus and mitochondria. I. Methodology and phenomenology
of suppressors, Mol. Gen. Genet., 179, 469-482.
50.Gietz, R. D., and Woods, R. A. (2002)
Transformation of yeast by lithium acetate/single-stranded carrier
DNA/polyethylene glycol method, Methods Enzymol.,
350, 87-96.
51.Cliften, P., Jang, S.-H., and Jaehning, J. A.
(2000) Identifying a core RNA polymerase surface critical for
interactions with a sigma-like specificity factor, Mol. Cell.
Biol., 20, 7013-7023.
52.Chiu, J., March, P. E., Lee, R., and Tillett, D.
(2004) Site-directed, Ligase-Independent Mutagenesis (SLIM): a
single-tube methodology approaching 100% efficiency in 4 h,
Nucleic Acids Res., 32, e174.
53.Chiron, S., Suleau, A., and Bonnefoy, N. (2005)
Mitochondrial translation: elongation factor Tu is essential in fission
yeast and depends on an exchange factor conserved in humans but not in
budding yeast, Genetics, 169, 1891-1901.
54.Saint-Georges, Y., Bonnefoy, N., Di Rago, J. P.,
Chiron, S., and Dujardin, G. (2002) A pathogenic cytochrome b
mutation reveals new interactions between subunits of the mitochondrial
bc1 complex, J. Biol. Chem., 277, 49397-49402.
55.Sikorski, R. S., and Boeke, J. D. (1991) In
vitro mutagenesis and plasmid shuffling: from cloned gene to mutant
yeast, Methods Enzymol., 194, 302-318.
56.Groudinsky, O., Dujardin, G., and Slonimski, P.
P. (1981) Long-range control circuits within mitochondria and between
nucleus and mitochondria. II. Genetic and biochemical analyses of
suppressors which selectively alleviate the mitochondrial intron
mutations, Mol. Gen. Genet., 184, 493-503.
57.Ogur, M., St. John, R., and Nagai, S. (1957)
Tetrazolium overlay technique for population studies of respiration
deficiency in yeast, Science, 125, 928-929.
58.Malecki, M., Jedrzejczak, R., Puchta, O.,
Stepien, P. P., and Golik, P. (2008) In vivo and in vitro
approaches for studying the yeast mitochondrial RNA degradosome
complex, Methods Enzymol., 447, 463-488.
59.Tomecki, R., Dmochowska, A., Gewartowski, K.,
Dziembowski, A., and Stepien, P. P. (2004) Identification of a novel
human nuclear-encoded mitochondrial poly(A) polymerase, Nucleic
Acids Res., 32, 6001-6014.
60.Kolondra, A., Labedzka-Dmoch, K., Wenda, J. M.,
Drzewicka, K., and Golik, P. (2015) The transcriptome of Candida
albicans mitochondria and the evolution of organellar
transcription units in yeasts, BMC Genomics, 16, 1.
61.Szczepanek, T., and Lazowska, J. (1996)
Replacement of two non-adjacent amino acids in the S. cerevisiae
bi2 intron-encoded RNA maturase is sufficient to gain a
homing-endonuclease activity, EMBO J., 15, 3758-3767.
62.Mulero, J. J., and Fox, T. D. (1993) Alteration
of the Saccharomyces cerevisiae COX2 mRNA
5′-untranslated leader by mitochondrial gene replacement and
functional interaction with the translational activator protein PET111,
Mol. Biol. Cell, 4, 1327-1335.
63.Greenleaf, A. L., Kelly, J. L., and Lehman, I. R.
(1986) Yeast RPO41 gene product is required for transcription and
maintenance of the mitochondrial genome, Proc. Natl. Acad. Sci.
USA, 83, 3391-3394.
64.Schmidt, U., Maue, I., Lehmann, K., Belcher, S.
M., Stahl, U., and Perlman, P. S. (1998) Mutant alleles of the MRS2
gene of yeast nuclear DNA suppress mutations in the catalytic core of a
mitochondrial group II intron, J. Mol. Biol., 282,
525-541.
65.Manthey, G. M., Przybyla-Zawislak, B. D., and
McEwen, J. E. (1998) The Saccharomyces cerevisiae Pet309
protein is embedded in the mitochondrial inner membrane, Eur. J.
Biochem., 255, 156-161.
66.Wallis, M. G., Groudinsky, O., Slonimski, P. P.,
and Dujardin, G. (1994) The NAM1 protein (NAM1p), which is selectively
required for cox1, cytb and atp6 transcript
processing/stabilization, is located in the yeast mitochondrial matrix,
Eur. J. Biochem., 222, 27-32.
67.Main, E. R. G., Lowe, A. R., Mochrie, S. G. J.,
Jackson, S. E., and Regan, L. (2005) A recurring theme in protein
engineering: the design, stability and folding of repeat proteins,
Curr. Opin. Struct. Biol., 15, 464-471.
68.Kobe, B., and Kajava, A. V. (2000) When protein
folding is simplified to protein coiling: the continuum of solenoid
protein structures, Trends Biochem. Sci., 25,
509-515.