Interaction between Saccharomyces cerevisiae Mitochondrial DNA-Binding Protein Abf2p and Cce1p Resolvase
E. O. Samoilova, I. A. Krasheninnikov, and S. A. Levitskii*
Lomonosov Moscow State University, Faculty of Biology, 119991 Moscow, Russia; E-mail: krolick@yandex.ru* To whom correspondence should be addressed.
Received June 27, 2016; Revision received July 19, 2016
Mitochondrial DNA is susceptible to the action of reactive oxygen species generated by the reactions of oxidative phosphorylation. Homologous recombination is one of the mechanisms providing integrity of the mitochondrial genome. Some proteins that take part in this process in budding yeast mitochondria have been identified. These include Abf2p, the major protein of the mt-nucleoid that specifically binds cruciform DNA, and Cce1p – Holliday junction resolvase. Here we show that Abf2p does not significantly affect either binding of Cce1p to branched DNA or rate and specificity of Holliday junction resolution. These data suggest the existence of an alternative homologous recombination pathway in yeast mitochondria.
KEY WORDS: mitochondrion, homologous recombination, Cce1p, Abf2pDOI: 10.1134/S0006297916100096
Mitochondria supply energy to eukaryotic cells by synthesizing ATP during oxidative phosphorylation processes. Most of the mitochondrial proteins are encoded in the nucleus and are imported into mitochondria after synthesis in the cytoplasm. However, mitochondria possess their own genome encoding several proteins of the electron transport chain, mitochondrial tRNAs, and rRNAs. The mitochondrial genome of the budding yeast Saccharomyces cerevisiae is a circular DNA that is organized into a nucleoprotein complex (the so-called nucleoid). The nucleoid contains, according to various data, one to several DNA molecules [1] and several dozens of different proteins [2] depending on the cultivation conditions.
Due to the increased vulnerability of mitochondrial DNA (mtDNA) [3] compared to nuclear DNA, caused by the high concentration of reactive oxygen species in mitochondria, the system of mitochondrial genome repair plays a pivotal role in providing normal functioning of mitochondria. Homologous recombination is one of the most important ways of repairing double-strand breaks in both nuclear and mitochondrial DNAs. Its mechanisms are believed to be largely similar in nuclei and mitochondria [4]. Despite extensive study of this process in various organisms, the precise mechanisms of homologous recombination in mitochondria remain incompletely explored [4].
The major mitochondrial nucleoid protein Abf2p is one of the participants in the homologous recombination process [5]. This protein belongs to a HMG (High Mobility Group) family of proteins. Proteins of this family perform numerous functions. Namely, they participate in DNA compaction, act as transcription factors, and contribute to DNA repair and recombination; they also take part in cell differentiation [6]. By diversity of their functions, HMG proteins of eukaryotes resemble bacterial histone-like DNA-binding HU proteins. Their functional resemblance is further confirmed by experiments on complementarity of the corresponding genes [7].
Abf2p protein, which is encoded by the nuclear genome, contains two HMG domains. According to some data, only one domain, located at the N-terminal part of the protein, plays a key role in DNA binding [8]. Deletion of the ABF2 gene decreases stability of mtDNA of yeast cultivated in media supplemented with fermentable carbon sources, while the mitochondrial genome is completely eliminated during prolonged growth in such media [9]. It was also shown that ABF2 deletion causes an increase in occurrence of homologous recombination events without affecting occurrence of point mutations [10]. Apparently, Abf2p acts at the late stages of homologous recombination, namely, at Holliday junction resolution, as this protein specifically binds to branched DNA structures [11].
Holliday junction resolution in S. cerevisiae mitochondria is performed by Cce1p resolvase [12], which introduces consecutive breaks into symmetrically situated angles of the junction [13]. It is worth mentioning that, similarly to other resolvases, Cce1p is selective for both certain DNA structure and its sequence. Therefore, it hydrolyzes phosphodiester bonds after CT nucleotide pair [14]. It was demonstrated earlier that double deletion of the ABF2 and CCE1 genes encoding mitochondrial resolvase causes significant accumulation of branched DNAs in mitochondria, as compared to a single deletion of CCE1 [5]. These facts indicate that Abf2p and Cce1p may somehow interact with each other. Upon such interaction, Abf2p may stabilize Holliday junctions by affecting the selectivity or rate of their resolution by Cce1p. To test this suggestion, we dissected the influence of Abf2p on Cce1p binding to DNA and the efficiency of in vitro hydrolysis of Holliday junctions by this resolvase.
MATERIALS AND METHODS
Escherichia coli and S. cerevisiae strains. In the present work, E. coli strains Top-10 OneShot® (Invitrogen, USA) and B834(DE3) (Novagen, USA) and S. cerevisiae strain W303 were used.
Gene cloning and purification of recombinant proteins Abf2p and Cce1p. Recombinant Abf2p was prepared as we described previously [15]. The CCE1 gene was amplified with primers CCE1F (5′-tcgacatatgtcgacagcacag) and CCE1R (5′-tcgactcgagttagtcattgttgtaagtgttc) using S. cerevisiae genome DNA as a template. The resulting amplicon was treated with endonucleases NdeI and XhoI (Thermo Scientific, USA) and cloned in expression vector pET30a (Novagen) treated with the same enzymes. The sequence of the cloned CCE1 was confirmed by Sanger sequencing (sequencing was performed at the Federal Research and Clinical Center of Physical-Chemical Medicine, Federal Medical Biological Agency of Russia). The resulting plasmid pET30a_Cce1p was used for transformation of E. coli strain B834(DE3). The transformed colonies were inoculated into 200 ml of 2× YT supplemented with 50 µg/ml kanamycin (Sigma, USA). The bacteria were cultivated at 37°C and 200 rpm until OD reached ~0.7. Then, the temperature was decreased to 30°C, and isopropyl β-D-thiogalactopyranoside was added to the concentration of 0.25 mM, and cultivation was continued for 4 h. The cells were collected by centrifugation at 3000g for 10 min, washed with 50 ml of PBS (10 mM Na2HPO4, 1.76 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4), and suspended in 5 ml of starting buffer for chromatography (25 mM sodium phosphate buffer, pH 7.4, 0.5 M NaCl, 25 mM imidazole). The cells were disrupted by ultrasonic treatment (six pulses 10 s each with 15 s pauses). The lysates were clarified by centrifugation at 30,000g for 30 min. The recombinant protein was purified with immobilized metal ion affinity chromatography on a Ni-Sepharose HisTrap column (GE Healthcare, USA) using an ÄKTAPurifier chromatography system (GE Healthcare) according to the manufacturer’s manual. Buffer in the fraction containing the recombinant protein was immediately changed to the storage buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1.5 mM EDTA) using a 5 ml HiTrap desalting column packed with Sephadex G-25.
Electrophoretic mobility shift assay (EMSA). To study binding of recombinant proteins Abf2p and Cce1p, a gel retardation assay was used. As substrates, DNA structures corresponding to the Holliday junction variants analogous to the ones described in [14] obtained by annealing the synthetic oligonucleotides (sequences are given in the table) were used. The oligonucleotides were mixed at 1-3 µM concentrations in the assembly buffer (25 mM Tris-HCl, pH 8.0, 200 mM NaCl, 2 mM EDTA). The samples were heated to 95°C and cooled slowly.
Sequences of oligonucleotides used for assembly of the analyzed
structures

* Potential sites for cleavage with Cce1p are indicated with
“/”.
The DNA was bound to the proteins in buffer that contained 25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 0.1 mg/ml BSA, and 7% glycerol. Variable amounts of the recombinant proteins were added to the DNA in the 10-100 nM concentration range and incubated at a room temperature. Then, the mixtures were fractionated in 6% non-denaturing polyacrylamide gel (29 : 1; 90 mM Tris-borate, 2 mM EDTA, 6 × 8 × 0.075 cm, 100 V, 1 h). After the run, the gel was imaged in a blue-fluorescence mode using a Storm865 scanner (GE Healthcare). The images were processed using ImageJ software (NIH, USA). During the study of specificity of interaction between Abf2p and the Holliday junction, a 20 bp dC:dI duplex was used as a competitor double-strand DNA at various ratios. Dissociation constants were calculated as described in [16].
Holliday junction cleavage by Cce1p protein. Hydrolytic activity of the recombinant Cce1p was measured as follows: first, oligonucleotides R2 (for fixed structure) and B (for mobile structure) (table) were labeled with [γ32P]ATP using T4 bacteriophage polynucleotide kinase (Thermo Scientific) according to the manufacturer’s manual. Then, junctions were assembled as described above. Hydrolysis was carried out in cleavage buffer that contained 20 mM Tris-HCl, pH 8.0, 30 mM NaCl, 10 mM MgCl2, 2 mM dithiothreitol (DTT), 1 mg/ml BSA. DNA structure (10 pmol) and Cce1p (final concentration of 50-200 nM) and (in some reactions) Abf2p (final concentration 50-200 nM) were added to the reaction mixture. Cleavage was performed at 37°C for 5, 10, 15, and 30 min. The reaction was stopped by adding EDTA to 10 mM concentration. Proteins in the reaction mixture were removed with proteinase K (20 µg/ml at 56°C for 20 min). Then, the DNAs were denatured by formamide and separated in denaturing 15% polyacrylamide gel (29 : 1; 90 mM Tris-borate, 8 M urea, 2 mM EDTA, 6 × 8 × 0.075 cm, 100 V, 1 h). After the run, the gel was fixed in 10% acetic acid, dried, and exposed for 2-4 h to a Storage Phosphor Screen (GE Healthcare). Then, the screen was scanned with the Storm865 scanner, and the images were processed using ImageJ software.
RESULTS AND DISCUSSION
Although there is a great collection of data confirming the participation of Abf2p in homologous recombination of mitochondrial DNA [5, 10, 17], the mechanism of its action remains obscure. We proposed that characterizing Abf2p interaction with yeast Cce1p mitochondrial resolvase might shed light on details of the participation of the former in homologous recombination.
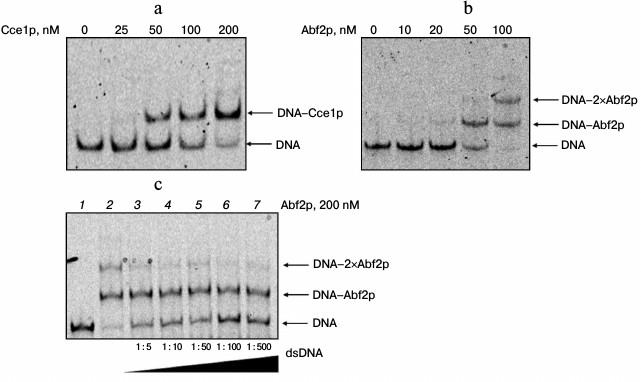
Interaction of Abf2p and Cce1p with branched DNA structures. The interaction of recombinant proteins Abf2p and Cce1p with branched DNA structures was characterized by monitoring changes of mobility of fixed Holliday junctions in a gel (Fig. 1). According to our data, both proteins bind to a Holliday junction in a similar manner, forming a clear DNA–protein complex even in the high-salt conditions used. In this case, Cce1p is more prone to forming 1 : 1 protein–DNA complex, while ABf2p is able to form higher order complexes on increasing its concentration in the reaction mixture. Apparently, this difference can be associated with Cce1p selectivity for the bound strand sequence. Alternatively, it could be explained by the fact that hydrolysis of symmetrical strands in the Holliday junction proceeds successively [13], and it is possible that binding of Cce1p to the symmetrical strand is determined by the cleavage of the strand that was initially bound to Cce1p.
Fig. 1. Interaction of proteins Abf2p and Cce1p with a Holliday junction. Assembled fixed Holliday junctions (20 nM) were incubated with recombinant proteins Abf2p and Cce1p at various concentrations in 25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 0.1 mg/ml BSA, and 7% glycerol. They were then separated in 6% polyacrylamide gel. a) Interaction of branched DNA with Cce1p; b) interaction of branched DNA with Abf2p. Protein concentrations are presented above the electrophoregrams; c) competitive binding of Abf2p with branched DNA. Abf2p at 200 nM concentration and dsDNA (dC:dI, 20 bp) at increasing concentrations were added to an fixed Holliday junction. Molar ratios of Holliday junction to dC:dI are indicated at the bottom of the figure.
Further increase in Abf2p concentration in the reaction mixture led to appearance of bands that correspond to higher order complexes. Nevertheless, during the study of competition binding of different DNA forms (branched and linear), it was shown that these complexes disappear on increasing the double-strand DNA concentration in the reaction mixture (data not shown). Hence, the density of the band that corresponds to the complex of two Abf2p molecules per Holliday junction decreases (Fig. 1c). At the same time, even 500-fold excess of competitor DNA does not decrease intensity of the band that corresponds to the complex of one protein molecule with one DNA molecule. Similar data for Cce1p were reported [14].
We calculated apparent dissociation constants for complexes of the recombinant proteins with an immobile Holliday junction by analyzing the images. The Kd for the complex of Abf2p with such junction was 27 ± 4 nM, and the Kd for the junction–Cce1p complex containing protein monomer was 74 ± 7 nM. Taking into account that, in contrast to Abf2p, Cce1p binds to DNA as a dimer [14], the dissociation constant of the complex is approximately 37 nM. Similar dissociation constant values were obtained for complexes of the recombinant proteins with a mobile Holliday junction (data not shown).
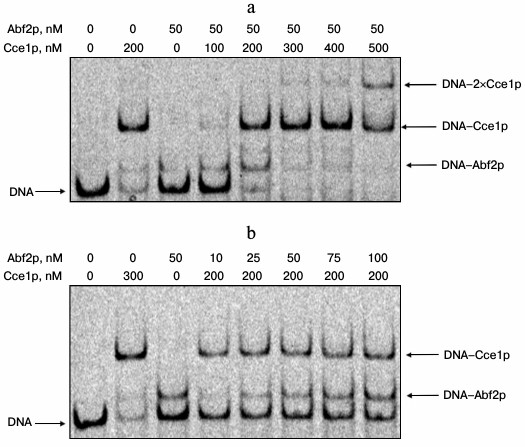
Mutual influence of Cce1p and Abf2p on their binding to a branched DNA. To determine whether one of the recombinant proteins affects the interaction of the other one with a Holliday junction, we analyzed changes in complex mobility first at constant Abf2p concentration and increasing Cce1p concentrations (Fig. 2a), and then at constant Cce1p concentration and increasing concentrations of Abf2p (Fig. 2b).
Fig. 2. Mutual influence of Cce1p and Abf2p on their binding to a branched DNA. Assembled fixed Holliday junctions (20 nM) were incubated with recombinant proteins Cce1p and Abf2p at various concentrations in 25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 0.1 mg/ml BSA, 7% glycerol; then, they were separated in 6% polyacrylamide gel. a) Interaction with branched DNA at constant Abf2p levels and increasing concentrations of Cce1p; b) interaction with branched DNA at constant Cce1p levels and increasing concentrations of Abf2p. Protein concentrations are presented above the electrophoregrams.
Our data demonstrate that Abf2p and Cce1p do not significantly affect the binding of each other with branched DNA. However, they compete for the substrate. Therefore, a band corresponding to the DNA–Abf2p complex loses density (Fig. 2a) at constant concentration of Abf2p during increase in concentration of Cce1p. At the same time, it should be noted that an additional band appears, which we did not observe in other experiments. We consider this band to be a complex of DNA with two Cce1p molecules. Perhaps Abf2p promotes formation of such complex. However, this issue is still to be addressed. It also should be mentioned that an increase in concentration of Abf2p at constant Cce1p levels facilitates formation of DNA–Abf2p complex without significant decrease in density of the Cce1p–DNA band (Fig. 2b).
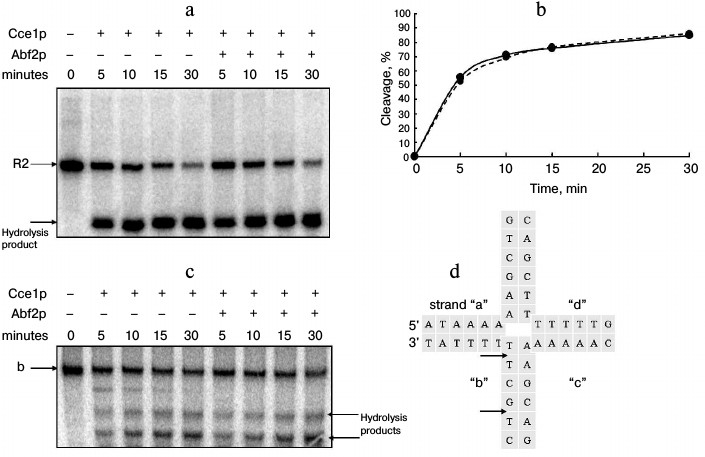
Effect of Abf2p on rate and selectivity of Holliday junction cleavage by Cce1p. The effect of Abf2p on the cleavage rate of one of the strands in an fixed Holliday junction with Cce1p resolvase was estimated by quantitative analysis of time-dependent accumulation of hydrolysis product at various concentrations and molar ratios of the recombinant proteins. Typical results are presented in Fig. 3. The data indicate that the presence of Abf2p does not affect the hydrolysis rate, as the cleavage product accumulation curves are actually the same in either the presence or absence of Abf2p.
Fig. 3. Effect of Abf2p on rate and selectivity of Holliday junction cleavage by Cce1p resolvase. a) Fixed Holliday junction (10 pmol) was incubated with 200 nM Cce1p in 20 mM Tris-HCl, pH 8.0, 30 mM NaCl, 10 mM MgCl2, 2 mM DTT, and 1 mg/ml BSA either in the absence or in the presence of 200 nM Abf2p at 37°C for 5, 10, 15, and 30 min. The reaction was stopped by adding EDTA to 10 mM; proteins were removed by proteinase K (20 µg/ml at 56°C for 20 min). Then, sample DNA was denatured with formamide and separated in denaturing 15% polyacrylamide gel. b) Time dependence of degree of strand “b” hydrolysis. Solid line – without Abf2p, dashed line – in the presence of 200 nM Abf2p. c) Mobile Holliday junction (10 pmol) was incubated with 200 nM Cce1p in 20 mM Tris-HCl, pH 8.0, 30 mM NaCl, 10 mM MgCl2, 2 mM DTT, and 1 mg/ml BSA either in the absence or in the presence of 200 nM Abf2p at 37°C for 5, 10, 15, and 30 min. The reaction was stopped by adding EDTA to 10 mM; proteins were removed by proteinase K (20 µg/ml at 56°C for 20 min). Then, the DNA was denatured with formamide and separated in denaturing 15% polyacrylamide gel. d) Structure of central part of the mobile junction. Potential hydrolysis sites in radioactively labeled strand are indicated with arrows.
Effect of Abf2p on selectivity of cleavage of one of a mobile Holliday junction strands by Cce1p was analyzed electrophoretically by observing differences between hydrolysis products formed in the presence and in the absence of Abf2p (Fig. 3c). The labeled strand had two cleavage sites in the central part (Fig. 3d, indicated with arrows) of the junction. Due to the hydrolysis, one can see in the gel two corresponding products. It is worth mentioning that an additional product of higher size is formed in reactions without Abf2p. This product corresponds to hydrolysis at a potential cleavage site situated closer to the 3′-end of strand “b”. The ability of Cce1p to cleave DNA is largely dependent on the proximity of the cleavage site to the central part of the Holliday junction [14]. Thus, formation of such product in the absence of Abf2p may be explained by greater strand mobility in the junction; whereas the junction becomes more stable in the presence of Abf2p and a site that is so remote from the central part is not cleaved. However, the data do not demonstrate a significant influence of Abf2p on the selectivity of mobile Holliday junction cleavage by Cce1p resolvase.
Mitochondrial DNA is vulnerable to the action of aggressive compounds and reactive oxygen species, whose high concentration in matrix is determined by the functioning of the electron transport chain in the inner membrane. Due to the necessity of maintenance of mitochondrial genome integrity, the mechanisms for repair of mtDNA damages developed in the course of evolution [18]. Double-strand DNA break repair by homologous recombination is one such mechanism. It is now accepted that at least two independent recombination mechanisms exist in budding yeast mitochondria. The first is related to the replication of mtDNA and is mediated by Mhr1p protein. The second mechanism is associated with functioning of mitochondrial resolvase Cce1p. It is believed to be rather connected to mtDNA inheritance and repair [18]. Despite extensive studies of homologous recombination, the precise molecular mechanisms of the processes remain obscure.
The DNA-binding protein Abf2p is one of the participants of homologous recombination in yeast mitochondria [17]. Deletion of a gene encoding this protein causes fast accumulation of deletions in mtDNA and its further complete degradation [19] as well as increase in occurrence of recombination between repeated sequences [10]. Besides, upon double deletion of genes CCE1 and ABF2, accumulation of Holliday junctions, key intermediates of homologous recombination, proceedes at a higher rate compared to the case of a single deletion of either of these two genes [5]. On the basis of these facts, we assumed that Abf2p may somehow affect the ability of Cce1p to resolve Holliday junctions. To test this hypothesis, we obtained corresponding recombinant proteins and dissected their mutual influence upon binding to branched DNA and resolution of the Holliday junction.
Both Abf2p and Cce1p bind to the Holliday junction in a similar way forming complexes with dissociation constants of approximately 30 nM. It is worth mentioning an interesting difference: Abf2p binds DNA as a monomer [15], whereas Cce1p interacts with DNA as a dimer [14]. Besides that, we also noted an additional difference in binding of these proteins to branched DNA. Under in vitro conditions, Cce1p predominantly binds to DNA at 1 : 1 ratio, which may be explained either by successive nature of breaks that it introduces into central portions of symmetrical strands or by its selectivity for hydrolyzed sequence [13]. At the same time, Abf2p is able to bind the Holliday structure at 2 : 1 ratio when increasing concentration of the protein. However, it is important that the addition of competitor double-strand DNA, which is bound by Abf2p nonspecifically [11], formation of the complex of two protein molecules per DNA is substantially decreased. The latter suggests that formation of the second complex is less specific, which may be determined by alteration of the ternary structure of branched DNA caused by formation of the first complex. This pattern for binding branched DNA is typical for a number of HMG proteins, including the mitochondrial proteins of this family [11].
When both proteins are present in the reaction mixture, we did not observe any significant differences in their interaction with branched DNA. Nevertheless, it should be noted that in the presence of Abf2p, Cce1p becomes able to form a complex with DNA at 2 : 1 ratio, which may indicate some stabilization of the Holliday junction by Abf2p.
The next hypothesis to be tested was that Abf2p by means of stabilization of Holliday structure is able to increase accessibility of its central part for Cce1p and, therefore, to increase the hydrolysis rate of the corresponding strands. However, our data clearly showed that the rate of resolution of a Holliday junction is not changed in the presence of Abf2p. Apparently, marginal changes in the pattern of Cce1p binding to branched DNA in the presence of Abf2p do not significantly affect the accessibility of the hydrolysis site to resolvase.
We further checked whether Abf2p-mediated stabilization of the Holliday junction affects the specificity of Cce1p action. It is known that Cce1p hydrolyzes the DNA strand in the Holliday junction predominantly after a CT dinucleotide. Moreover, hydrolysis efficiency directly depends on the proximity of the site to the central part of the junction [14]. To dissect influence of Abf2p on selectivity of mitochondrial resolvase, we used a structure that simulated an intermediate of a branch migration process having no certain center of symmetry. Besides, there were several sites recognized by Cce1p in the studied strand. These sites were situated at different distances from the central part of the junction. It was found that Abf2p can somewhat affect Cce1p functioning by hampering hydrolysis at a site distant from the central part of the junction, whereas Abf2p did not significantly affect selectivity of Cce1p.
Thus, our data suggest that Abf2p significantly affected neither the pattern of Cce1p interaction with branched DNA, rate of enzyme functioning, nor selectivity of strand cleavage in a Holliday structure by this resolvase. However, as mentioned, the participation of Abf2p in homologous recombination of mtDNA is beyond doubt. Perhaps, this participation is restricted to the final stages of the process, as Abf2p specifically binds only branched structures that mimic Holliday junctions [11]. It should be mentioned here that the Cce1p-mediated recombination mechanism is not the only one in yeast mitochondria. Recombination proceeds (though less efficiently) in mitochondria even after deletion of the CCE1 gene; the same is true for a double deletion of the CCE1 and MHR1 genes [20]. Taking this into account as well as a significant influence of Abf2p on both quantity of recombination intermediates in mitochondria and occurrence of spontaneous deletions in mtDNA, we assume that Abf2p plays an important role in a yet unknown mechanism of homologous recombination in yeast mitochondria.
Acknowledgements
This work was carried out with financial support from the Ministry of Education and Science of the Russian Federation (FTP “Research and development in priority areas of development of the Russian scientific and technological complex for 2014-2020”, agreement 14.604.21.0113, identification number RFMEFI60414X0113).
REFERENCES
1.Miyakawa, I., Miyamoto, M., Kuroiwa, T., and Sando,
N. (2004) DNA content of individual mitochondrial nucleoids varies
depending on the culture conditions of the yeast Saccharomyces
cerevisiae, Cytologia (Tokyo), 69, 101-107.
2.Kucej, M., Kucejova, B., Subramanian, R., Chen, X.
J., and Butow, R. A. (2008) Mitochondrial nucleoids undergo remodeling
in response to metabolic cues, J. Cell Sci., 121,
1861-1868.
3.Brown, W. M., George, M., and Wilson, A. C. (1979)
Rapid evolution of animal mitochondrial DNA, Proc. Natl. Acad. Sci.
USA, 76, 1967-1971.
4.Chen, X. J. (2013) Mechanism of homologous
recombination and implications for aging-related deletions in
mitochondrial DNA, Microbiol. Mol. Biol. Rev., 77,
476-496.
5.MacAlpine, D. M., Perlman, P. S., and Butow, R. A.
(1998) The high mobility group protein Abf2p influences the level of
yeast mitochondrial DNA recombination intermediates in vivo,
Proc. Natl. Acad. Sci. USA, 95, 6739-6743.
6.Reeves, R. (2015) High mobility group (HMG)
proteins: modulators of chromatin structure and DNA repair in mammalian
cells, DNA Repair (Amst.), 36, 122-136.
7.Megraw, T. L., and Chae, C.-B. (1993) Functional
complementarity between the HMGl-like yeast mitochondrial histone HM
and the bacterial histone-like protein HU, J. Biol. Chem.,
268, 12758-12763.
8.Kurashenko, A. V., Samoilova, E. O., Baleva, M. V.,
Chicherin, I. V., Petrov, D. Yu., Kamenski, P. A., and Levitskii, S. A.
(2016) Two HMG domains of yeast mitochondrial protein Abf2p have
different affinity to DNA, Bull. Ross. State Med. Univ.,
1, 62-65.
9.Diffley, J. F., and Stillman, B. (1992) DNA binding
properties of an HMG1-related protein from yeast mitochondria, J.
Biol. Chem., 267, 3368-3374.
10.Sia, R. A., Carrol, S., Kalifa, L., Hochmuth, C.,
and Sia, E. A. (2009) Loss of the mitochondrial nucleoid protein,
Abf2p, destabilizes repetitive DNA in the yeast mitochondrial genome,
Genetics, 181, 331-334.
11.Bakkaiova, J., Marini, V., Willcox, S., Nosek,
J., Griffith, J. D., Krejci, L., and Tomaska, L. (2016) Yeast
mitochondrial HMG proteins: DNA-binding properties of the most
evolutionarily divergent component of mitochondrial nucleoids,
Biosci. Rep., 36, e00288.
12.West, S. C., Parsons, C. A., and Picksley, S. M.
(1987) Purification and properties of a nuclease from Saccharomyces
cerevisiae that cleaves DNA at cruciform junctions, J. Biol.
Chem., 262, 12752-12758.
13.Fogg, J. M., Schofield, M. J., Declais, A. C.,
and Lilley, D. M. (2000) Yeast resolving enzyme CCE1 makes sequential
cleavages in DNA junctions within the lifetime of the complex,
Biochemistry, 39, 4082-4089.
14.White, M. F., and Lilley, D. M. (1996) The
structure-selectivity and sequence-preference of the junction-resolving
enzyme CCE1 of Saccharomyces cerevisiae, J. Mol. Biol.,
257, 330-341.
15.Samoilova, E. O., Krasheninnikov, I. A.,
Vinogradova, E. N., Kamenski, P. A., and Levitskii, S. A. (2016) Abf2p
increases efficiency of DNA uptake by isolated mitochondria,
Biochemistry (Moscow), 81, 723-730.
16.Kamashev, D., and Rouviere-Yaniv, J. (2000) The
histone-like protein HU binds specifically to DNA recombination and
repair intermediates, EMBO J., 19, 6527-6535.
17.Zelenaya-Troitskaya, O., Newman, S. M., Okamoto,
K., Perlman, P. S., and Butow, R. A. (1998) Functions of the high
mobility group protein, Abf2p, in mitochondrial DNA segregation,
recombination and copy number in Saccharomyces cerevisiae,
Genetics, 148, 1763-1776.
18.Lipinski, K. A., Kaniak-Golik, A., and Golik, P.
(2010) Maintenance and expression of the S. cerevisiae
mitochondrial genome – from genetics to evolution and
systems biology, Biochim. Biophys. Acta, 1797,
1086-1098.
19.Newman, S. M., Zelenaya-Troitskaya, O., Perlman,
P. S., and Butow, R. A. (1996) Analysis of mitochondrial DNA nucleoids
in wild-type and a mutant strain of Saccharomyces cerevisiae
that lacks the mitochondrial HMG box protein Abf2p, Nucleic Acids
Res., 24, 386-393.
20.Mookerjee, S. A., and Sia, E. A. (2006)
Overlapping contributions of Msh1p and putative recombination proteins
Cce1p, Din7p, and Mhr1p in large-scale recombination and genome sorting
events in the mitochondrial genome of Saccharomyces
cerevisiae, Mutat. Res. Mol. Mech. Mutagen., 595,
91-106.