Free Initiation Factors eIF4A and eIF4B Are Dispensable for Translation Initiation on Uncapped mRNAs
P. A. Sakharov and S. Ch. Agalarov*
Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; E-mail: sultan@vega.protres.ru* To whom correspondence should be addressed.
Received July 14, 2016; Revision received August 26, 2016
The formation of ribosomal 48S initiation complexes at the start AUG codon of uncapped mRNA leader sequences was studied using the methodology of primer extension inhibition (toe-printing). The experiments were performed in the system composed of purified individual components required for translation initiation. The formation of ribosomal 48S initiation complexes at the initiation codon was tested depending on the presence of the initiation factors eIF4F, eIF4A, and eIF4B. Several mRNAs containing short leader sequences lacking the extended secondary structure were studied. It was found that 48S ribosomal complexes at mRNAs with such leaders were not formed in the absence of eIF4F. In contrast, the removal of either eIF4A or eIF4B from the experimental system was found to be dispensable for the formation of the 48S complex.
KEY WORDS: translation initiation, 48S ribosomal initiation complex, initiation factors, eIF4F, eIF4A, eIF4B, toe-printingDOI: 10.1134/S0006297916100175
The process of translation initiation in eukaryotes goes through three phases. The first phase is the interaction and binding of the preinitiation 43S ribosomal complex, which is the 40S ribosomal subunit and several associated initiation factors, with the 5′-terminal cap structure of the mRNA leader sequence with the formation of the 48S ribosomal complex. The second phase is the movement (sliding) of the 48S ribosomal complex downstream along the 5′-untranslated region (5′-UTR) of the mRNA chain. The functional role of the movement is the scanning of the sequence until reaching and recognition of the initiation AUG codon [1-3]. As the scanning process strictly requires the unidirectional movement along the mRNA chain, energy expenses become necessary; accordingly, in the case of the capped mRNAs the moving of the ribosomal complex along the mRNA chain and the corresponding scanning are known to be ATP-dependent [4]. The unidirectional scanning of the eukaryotic mRNA leader sequence has been proved experimentally [5]. The third phase starts immediately upon the recognition of the initiation AUG codon that induces the stop of the 48S ribosomal complex at the codon.
A general model for translation initiation in eukaryotes has been proposed that describes different modes of ribosome binding to mRNAs [6]. The cap-dependent initiation starts with the recognition of the cap structure at the 5′-end of mRNA by the eIF4E subunit of the large heterotrimeric protein eIF4F (eIF4G·eIF4E·eIF4A) associated with the preinitiation 43S ribosomal particle via eIF3. The cap-dependent binding of the 43S preinitiation complex to mRNA is mediated by several initiation factors, eIF4F, eIF4A, and eIF4B, and requires energy derived from ATP hydrolysis [7]. For naturally capped rabbit β-globin mRNA, it was demonstrated that eIF4F [8] and free eIF4A [9] are needed for effective formation of the 48S initiation complexes at the initiation AUG codon, whereas the necessity for eIF4B was questionable [10]. In the present work, we have shown that the capped β-globin mRNA strictly requires eIF4B for scanning.
Besides, it is known that some naturally capped 5′-UTRs can provide a successful translation initiation when used as uncapped constructs. Such a case has been demonstrated for the poly(A) leader sequence of pox virus mRNAs [9], as well as the omega leader of TMV RNA [11] and the 5′-UTR of mRNA encoding for obelin (a light-emitting protein of the hydroid polyp Obelia longissima) [12]. Also, it was shown that the initiation of translation of mRNAs with uncapped leader sequences such as the synthetic (CAA)n leader [8], the poly(A) leader [9], and the omega leader [13] does not require the presence of eIF4F. An alternative cap- and energy-independent mechanism of internal translation initiation on the omega leader was described [13]. The question arises whether the eIF4F/ATP-independent scanning mechanism can work in eukaryotic systems only in special cases mentioned above, or it also can be realized with all short uncapped leader sequences.
To answer the question, we studied several leader sequences of different nature, a common feature of which was the ability to initiate cap-independent translation. The investigation of the 48S complex formation at the initiation codon for these leaders revealed strong dependence of the complex formation on the presence of eIF4F. At the same time, the level of the complex formation was largely independent of the presence of free eIF4A and eIF4B. Thus, in the present work we describe a new mode of eukaryotic initiation that differs from both conventional cap-dependent and cap/eIF4F-independent [8, 9, 13] mechanisms.
MATERIALS AND METHODS
Plasmids. Plasmid pTZ10ΩLuc containing the firefly luciferase-coding region with omega sequence or its modified forms was described in [14]. To obtain RNA containing the leader sequence of mRNA encoding for obelin, the plasmid construct pObeLucTMV also containing the luciferase-coding region was used [15]. The construct encoding the fusion (CAA)4-5′-UTRglobin leader sequence [16] was kindly provided by E. Z. Alkalaeva.
Components of system for 48S initiation complex formation. Natural (eIF2, eIF3, eIF4F) and recombinant (eIF1, eIF1A, eIF4A, eIF4B) translation initiation factors, ribosomal subunits, and native mRNA of rabbit β-globin were obtained as described in [9].
In vitro transcription. Transcription was performed as described in [13, 14]. The incubation mixture contained 80 mM Tris-OAc, pH 7.5, 10 mM KCl, 10 mM DTT, 2 mM spermidine, 0.01% Triton X-100, 4 mM ATP, GTP, UTP and CTP each, 22 mM Mg(OAc)2, 1 U/µl of RNase inhibitor, 12 U/µl of T7 RNA polymerase and 100 ng/µl DNA template. The reaction was performed at 37°C for 2 h. The purity of polyribonucleotides obtained was verified by electrophoresis in 6% polyacrylamide gel with 7% urea.
Toe-printing assay. Formation of the ribosomal initiation complexes, the primer extension inhibition assay (toe-printing), and the analysis of the products of the primer extension reaction were performed as described in [13, 17]. The ribosomal 48S initiation complexes were assembled from individual purified components of the translation apparatus, namely 40S ribosomal subunits, mRNA, Met-tRNAi, and initiation factors eIF1, eIF2, eIF3, eIF4A, eIF4B, and eIF4F, as well as ATP and GMPPNP (the latter was added to block translation after reaching the initiation codon); the mixture was incubated at 37°C for 15 min. The primer extension reaction was performed using DNA primer with fluorescent label. The cDNAs formed in the primer extension reaction were analyzed by capillary gel electrophoresis. The collected data were processed with GeneMarker 1.5 software (SoftGenetics, USA). Fluorescence intensities corresponding to each cDNA peak were measured to determine the amount of reverse transcription products.
RESULTS
We have studied the 48S complex formation at the initiation codon using the technique of primer extension inhibition, or toe-printing [18], with modifications [9, 19].
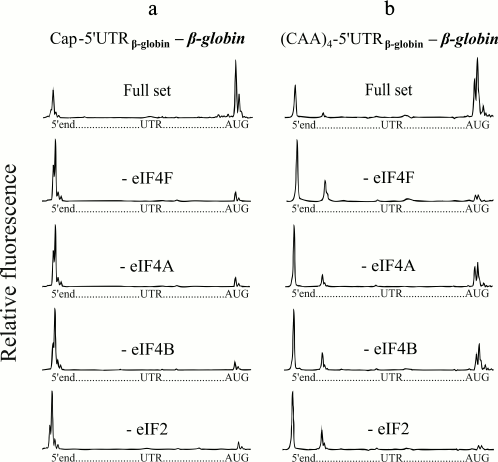
The native capped β-globin mRNA usually serves as a typical eukaryotic mRNA for in vitro studies of translation (5′-UTR of the β-globin mRNA is shown in Fig. 1). Figure 2a (with the exception of the second lowest plot) shows the results of the control tests in our experiments. The top electrophoregram (Fig. 2a) demonstrates the formation of 48S initiation complexes on the mRNA with capped β-globin mRNA in the presence of a full set of initiation factors. Omission of eIF4F or free eIF4A from the incubation mixture led to virtual abolishment of the formation of the initiation 48S complex (Fig. 2a). These results are compatible with previously published ones [8, 9]. The result of the omission of eIF4B in the case of β-globin mRNA is presented in the second lowest electrophoregram (Fig. 2a). It is seen that the omission of eIF4B also led to lack of the 48S complexes that reached the AUG codon. Thus, here we demonstrate the absolute necessity of eIF4B for scanning of the natural capped β-globin mRNA.
Fig. 1. Leader sequences studied. (CAA)n→polyU, the omega sequence in which CAA repeats of the central part are completely replaced by uridylic residues. 3A→3C, the omega sequence, in which 3C are substituted for 3A. The substitutions are shown in underlined bold.
Fig. 2. Formation of ribosomal 48S initiation complexes at initiation AUG codon of native capped β-globin mRNA (a) and recombinant uncapped mRNA with the fusion (CAA)4-5′-UTRglobin leader sequence (b). The top electrophoregram in each panel shows the result of incubation of mRNA with ribosomal particles and the full set of initiation factors. The subsequent electrophoregrams demonstrate the results of the omission of eIF4F, eIF4A, or eIF4B from the complex formation mixture. The lowest electrophoregrams show the results of a negative control (omission of eIF2 from the system). On each electrophoregram, the integral fluorescence of the left-hand major peaks reflects the amount of the full-length product when mRNA was read out by reversed transcription up to the 5′-end without stop. The integral fluorescence of the right-hand major peaks, here described as a “trident”, corresponds to the product of the reversed transcription stopped by the initiation 48S ribosomal complex formed at the initiation AUG codon.
Unlike native capped β-globin mRNA, the 48S complex formation on synthetic uncapped β-globin mRNA transcripts is extremely inefficient [8]. Addition of four or six CAA triplets to the 5′-end of uncapped β-globin mRNAs greatly enhanced the 48S complex formation up to the level of the 48S complex formation on native capped β-globin mRNA [8]. That is, this fusion (CAA)n-5′-UTRglobin leader sequence became cap-independent. Figure 2b demonstrates that the (CAA)4-5′-UTRglobin leader provides efficient formation of initiating 48S complexes at initiation AUG codons in the presence of a full set of the initiation factors (top electrophoregram), which is in agreement with the previous data [8]. It can be seen that like in the case of the native capped β-globin mRNA, omission of the ATP-dependent factor eIF4F abolished the formation of the initiation 48S complex at mRNA with the (CAA)4-5′-UTRglobin leader (Fig. 2b, second electrophoregram from the top). However, in the absence of either eIF4A or eIF4B, the (CAA)4-5′-UTRglobin leader still provides a substantial level of the 48S complex formation (Fig. 2b, the second and third electrophoregrams from the bottom). The results as a negative control are also presented: as it is well known, in the absence of eIF2, 48S complexes are not formed (Fig. 2, lowest electrophoregrams).
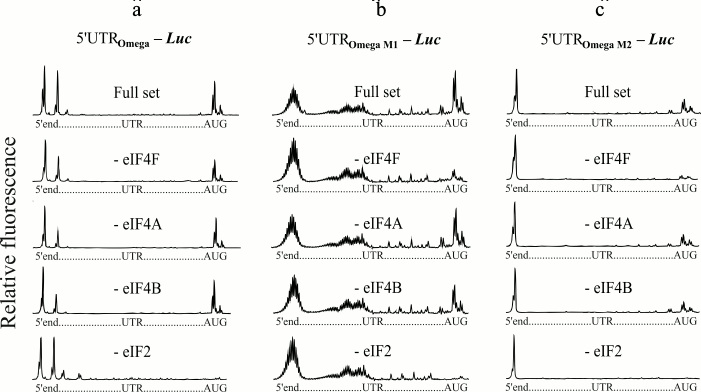
The 5′-untranslated sequence of TMV RNA (omega leader) is a well-known translational enhancer [20]. The uncapped omega sequence can support translation at the level comparable with that of the capped omega [11]. The omega RNA has been shown to have a unique compact and stable structure [21, 22]. Earlier, the polyribonucleotides containing modified omega sequences were synthesized, and it was demonstrated that changes made in the sequence led to strong destabilization of the omega RNA structure [14]. Despite this, these altered omega sequences retained their capability for effective cap-independent translation (unpublished results). In the present work, we studied initiation factor requirements for scanning of the modified forms of the omega RNA. Two forms were used (Fig. 1): the omega sequence in which CAA repeats of the central part are completely replaced by uridylic residues (M1 form), and the omega sequence in which 3C are substituted for 3A (M2 form) [14]. Efficient 48S complex formation on wild type omega RNA in the absence of eIF4F was previously shown [13] and is reproduced here as control tests (Fig. 3a). Additional experiments on omission of free eIF4A and eIF4B are also presented (Fig. 3a). The results of the omission of eIF4F in the case of the mRNA constructs with modified forms of the omega leader are shown in Fig. 3, b and c. The upper plots show the formation of 48S complexes on the modified forms in the presence of a full set of initiation factors. However, unlike wild type omega RNA, the omission of eIF4F led to lack of the formation of the initiation 48S complex on the modified forms (Fig. 3, b and c, the second plots from the top). At the same time, the omission of eIF4A or eIF4B does not exert significant influence on the amount of the complexes assembled at AUG (Fig. 3, b and c, plots second and third from the bottom).
Fig. 3. Formation of ribosomal 48S initiation complexes at initiation AUG codon of mRNA with wild type omega leader sequence (a) and modified omega leader sequences (b, c). The top electrophoregram in each panel shows the result of incubation of mRNA with ribosomal particles and the full set of initiation factors. The subsequent electrophoregrams demonstrate the results of the omission of eIF4F, eIF4A, or eIF4B from the complex formation mixture. The lowest electrophoregrams show the results of a negative control (omission of eIF2 from the system). It should be mentioned that the M1 modified form (b) displays length heterogeneity, as indicated by multiple stop points of the reversed transcription at the 5′-end of the M1 form. This stems from the fact that T7 polymerase exhibits a slippage effect when transcribing homopolymeric sequences (extended poly(U) track in our case).
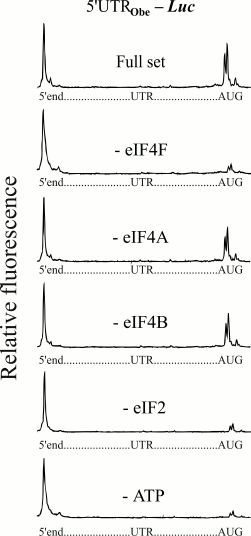
The next series of experiments were performed on the 5′-UTR of cellular mRNA of animal nature. The leader sequence of mRNA (Fig. 1) encoding for obelin (a light-emitting protein of hydroid polyp Obelia longissima) was used. This leader has been shown to provide a high level of translation when it is used as an uncapped version in mRNA constructs [12]. Figure 4 shows electrophoregrams demonstrating the formation of 48S initiation complexes on the mRNA with an uncapped obelin leader in the presence of a full set of initiation factors, and in the absence of eIF4F, eIF4A, or eIF4B. A similar pattern can be seen again that like in the cases of the (CAA)4-5′-UTRglobin and modified omega leaders, omission of the ATP-dependent factor eIF4F led to the loss of the 48S complex formation at mRNA with the obelin leader. In addition, correspondingly, the absence of free eIF4A or eIF4B did not influence the 48S complex formation. An additional experiment on removal of ATP from the system also did not reveal the formation of 48S ribosomal complexes (Fig. 4, lowest electrophoregram).
Fig. 4. Formation of ribosomal 48S initiation complexes at initiation AUG codon of mRNA with the leader sequence of mRNA (Fig. 1) encoding for obelin. An additional experiment on removal of ATP from the incubation mixture is shown here (lowest plot). For more details, see legend to Fig. 2.
DISCUSSION
In the work of Dmitriev et al. [10], it was suggested that factor eIF4B is not required for the scanning of capped β-globin mRNA. They attributed this to the fact that the leader sequence of this mRNA is poorly structured, although they allowed the possibility of intermixture of eIF4B in these experiments. In the present study, the absolute need for eIF4B for the formation of 48S initiation complexes on β-globin mRNA was demonstrated. The reason is not clear; one can only assume that eIF4B together with eIF4A and eIF4F provide proper recognition of the cap structure by the 43S preinitiation complex.
Pestova and Kolupaeva [8] showed that the addition of four CAA triplets to the 5′-end of uncapped β-globin mRNAs greatly enhanced the 48S complex formation up to the level of the natural mRNA. The experiments were carried out in a system with a full set of factors, and the role of individual factors was not explored. Here we have shown that the 48S complexes on mRNA with the (CAA)4-5′-UTRglobin leader were formed in the absence of eIF4A and eIF4B, but not eIF4F.
Summarizing, all of the uncapped leader sequences studied in the present work revealed the same mode of scanning. First, the 48S complex formation is strictly dependent on eIF4F, and second, it is substantially independent of free eIF4A and eIF4B. eIF4F is the only conventional ATP-hydrolyzing initiation factor within the initiating ribosomal complex and thus the only one capable of providing the energy-dependent unidirectional scanning. Formation of initiating 48S complexes because of eIF4F-dependent scanning apparently implies the involvement of ATP and, hence, the energy dependence of the process. However, as shown earlier, the uncapped omega leader permits efficient eIF4F/ATP-independent scanning of the 5′-UTR [13]. To explain this fact, one can speculate that the omega RNA can provide some kind of IRES-like type of translation initiation. In favor of this hypothesis, one can adduce the following arguments. The IRES-dependent mechanism of initiation implies the binding of the ribosomal preinitiation complex directly to a specific binding site within the viral RNA sequence in the vicinity of the initiation codon. It is worth noting that the omega RNA is capable of 5′-end-independent internal translation initiation [13] and, therefore, may contain a binding site within its sequence for recruiting the ribosomal preinitiation complex. Another hallmark of viral IRESs is a highly specific well-ordered secondary and tertiary structure. The omega RNA also has a unique compact and stable structure [14, 21, 22]. Moreover, mutations in the omega sequence, which result in disordering of its overall native structure [14], concurrently alter the mode of its scanning: the scanning becomes eIF4F-dependent (the present work). The synthetic (CAA)n leader does not require either the presence of eIF4F for its scanning [8]. Since the (CAA)n motif was identified as the core regulatory element in the structure [14] and function [23] of the omega RNA, this allows us to consider these leaders as a counterparts with similar features.
Another known example of eIF4F-independent scanning is the poly(A)25 leader sequence [9]. In this case, the mechanism of ATP-independent, diffusional movement (“phaseless wandering”) of the ribosomal subunit along the leader sequence seems to be most probable [24]. It is likely that the poly(A) leader sequence can bind the ribosomal 43S preinitiation complex at random internal sites within the sequence and may thus allow the complex to perform energy-independent diffusional movement along the 5′-UTR until the complex is fixed at the initiation codon [9]. It should be noted that a short distance between the 5′-end and the initiation codon in the poly(A)25 leader sequence probably makes it possible that the 43S complex successfully reaches the start codon using the mechanism of the energy-independent diffusional movement. Longer leader sequences (“obelin” leader, above 50 nt), as shown in the present work, require eIF4F, and their mode of scanning is energy-dependent. Evidently, in the case of long distances, the energy of ATP hydrolysis is required for unidirectional movement of the ribosomal complex, and only a “phaseless wandering” scenario is hardly possible because of very low probability for the ribosomal preinitiation complex to reach the start codon in such a manner.
Here we have demonstrated that free initiation factors eIF4A and eIF4B are dispensable for all cap-independent leader sequences used in this study. eIF4A is thought to unwind secondary structures in the 5′-UTR of mRNAs to facilitate scanning of the 40S ribosomal subunit towards the start codon, and eIF4B greatly stimulates helicase activity of eIF4A. It is reasonable to assume that 5′-UTRs lacking stable stem-loop structures would not require eIF4A and eIF4B for their scanning. In [25] it was shown that the requirement for eIF4A in translation is directly proportional to the degree of the secondary structure of the 5′-UTR of mRNA. Computer modeling of the secondary structure of all the leader sequences studied has not revealed any conventional stem-loop structures. It is therefore not surprising that these leaders bypass the dependence of translation initiation on eIF4A and eIF4B.
Acknowledgements
We would like to thank A. S. Spirin for his support, constant interest, and valuable advices, V. A. Kolb for reading the manuscript and critical comments, and E. Z. Alkalaeva for providing us with the construct encoding (CAA)4-5′-UTRglobin leader sequence.
The work was supported by the Russian Foundation for Basic Research (projects Nos. 15-04-015-25a and 16-34-00304 mol-a) and by the grant of the program of Presidium of the Russian Academy of Sciences “Molecular and Cell Biology”.
REFERENCES
1.Jackson, R. J., Hellen, C. U. T., and Pestova, T.
V. (2010) The mechanism of eukaryotic translation initiation and
principles of its regulation, Nat. Rev. Mol. Cell Biol.,
11, 113-127.
2.Kozak, M. (1978) How do eukaryotic ribosomes select
initiation regions in messenger RNA? Cell, 15,
1109-1123.
3.Kozak, M. (1989) The scanning model for
translation: an update, J. Cell. Biol., 108, 229-241.
4.Kozak, M. (1980) Role of ATP in binding and
migration of 40S ribosomal subunits, Cell, 22,
459-457.
5.Vassilenko, K. S., Alekhina, O. M., Dmitriev, S.
E., Shatsky, I. N., and Spirin, A. S. (2011) Unidirectional constant
rate motion of the ribosomal scanning particle during eukaryotic
translation initiation, Nucleic Acids Res., 39,
5555-5567.
6.Sonenberg, N. (1993) Remarks on the mechanism of
ribosome binding to eukaryotic mRNAs, Gene Expr., 3,
317-323.
7.Haghighat, A., and Sonenberg, N. (1997) eIF4G
dramatically enhances the binding of eIF4E to the mRNA 5′-cap
structure, J. Biol. Chem., 272, 21677-21680.
8.Pestova, T. V., and Kolupaeva, V. G. (2002) The
roles of individual eukaryotic translation initiation factors in
ribosomal scanning and initiation codon selection, Genes. Dev.,
16, 2906-2922.
9.Shirokikh, N. E., and Spirin, A. S. (2008) Poly(A)
leader of eukaryotic mRNA bypasses the dependence of translation on
initiation factors, Proc. Natl. Acad. Sci. USA, 2105,
10738-10743.
10.Dmitriev, S. E., Terenin, I. M., Dunaevsky, Y.
E., Merrick, W. C., and Shatsky, I. N. (2003) Assembly of 48S
translation initiation complexes from purified components with mRNAs
that have some base pairing within their 5′-untranslated regions,
Mol. Cell. Biol., 23, 8925-8933.
11.Gudkov, A. T., Ozerova, M. V., Shiryaev, V. M.,
and Spirin, A. S. (2005) 5′-poly(A) sequence as an effective
leader for translation in eukaryotic cell-free systems, Biotechnol.
Bioeng., 91, 468-473.
12.Shaloiko, L. N., Granovsky, I. E., Ivashina, T.
V., Ksenzenko, V. N., Shirokov, V. A., and Spirin, A. S. (2004)
Effective non-viral leader for cap-independent translation in a
eukaryotic cell-free system, Biotechnol. Bioeng., 88,
730-739.
13.Agalarov, S. Ch., Sakharov, P. A., Fattakhova, D.
Kh., Sogorin, E. A., and Spirin, A. S. (2014) Internal translation
initiation and eIF4F/ATP-independent scanning of mRNA by eukaryotic
ribosomal particles, Sci. Rep., 4, doi:
10.1038/srep04438.
14.Agalarov, S. C., Sogorin, E. A., Shirokikh, N.
E., and Spirin, A. S. (2011) Insight into the structural organization
of the omega leader of TMV RNA: the role of various regions of the
sequence in the formation of a compact structure of the omega RNA,
Biochem. Biophys. Res. Commun., 404, 250-253.
15.Kopeina, G. S., Afonina, Z. A., Gromova, K. V.,
Shirokov, V. A., Vasiliev, V. D., and Spirin, A. S. (2008) Step-wise
formation of eukaryotic double-row polyribosomes and circular
translation of polysomal mRNA, Nucleic Acids Res., 36,
2476-2478.
16.Alkalaeva, E. Z., Pisarev, A. V., Frolova, L. Y.,
Kisselev, L. L., and Pestova, T. V. (2006) In vitro
reconstitution of eukaryotic translation reveals cooperativity between
release factors eRF1 and eRF3, Cell, 125, 1125-1136.
17.Sakharov, P. A., Sokolov, A. S., and Agalarov, S.
C. (2015) Nonhydrolyzable ATP analog 5′-adenylyl-imidodiphosphate
(AMP-PNP) does not inhibit ATP-dependent scanning of leader sequence of
mRNA, Biochemistry (Moscow), 80, 45-49.
18.Hartz, D., McPheeters, D. S., Traut, R., and
Gold, L. (1988) Extension inhibition analysis of translation initiation
complexes, Methods Enzymol., 164, 419-425.
19.Gould, P. S., Bird, H., and Easton, A. J. (2005)
Translation toeprinting assays using fluorescently labeled primers and
capillary electrophoresis, Biotechniques, 38,
397-400.
20.Sleat, D. E., Gallie, D. R., Jefferson, R. A.,
Bevan, M. W., Turner, P. C., and Wilson, T. M. A. (1987)
Characterization of the 50-leader sequence of tobacco mosaic virus RNA
as general enhancer of translation in vitro, Gene,
60, 217-225.
21.Kovtun, A. A., Shirokikh, N. E., Gudkov, A. T.,
and Spirin, A. S. (2007) The leader sequence of tobacco mosaic virus
RNA devoid of Watson–Crick secondary structure possesses a
cooperatively melted, compact conformation, Biochem. Biophys. Res.
Commun., 358, 368-372.
22.Shirokikh, N. E., Agalarov, S. C., and Spirin, A.
S. (2010) Chemical and enzymatic probing of spatial structure of the
omega leader of tobacco mosaic virus RNA, Biochemistry (Moscow),
75, 405-411.
23.Gallie, D. R., and Walbot, V. (1992)
Identification of the motifs within the tobacco virus 5′-leader
responsible for enhancing translation, Nucleic Acids Res.,
20, 4631-4638.
24.Sarabhai, A., and Brenner, S. (1967) A mutant
which reinitiates the polypeptide chain after chain termination, J.
Mol. Biol., 27, 145-162.
25.Svitkin, Y. V., Pause, A., Haghighat, A.,
Pyronnet, S., Witherell, G., Belsham, G. J., and Sonenberg, N. (2001)
The requirement for eukaryotic initiation factor 4A (elF4A) in
translation is in direct proportion to the degree of mRNA
5′-secondary structure, RNA, 7, 382-394.