Perfect Hemihedral Twinning in Crystals of the γ-Subunit of Translation Initiation Factor 2 from Sulfolobus solfataricus: Cause and Effect
O. V. Kravchenko*, O. S. Nikonov, N. A. Nevskaya, E. A. Stolboushkina, V. I. Arkhipova, M. B. Garber, and S. V. Nikonov
Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; E-mail: kravchenko.olesya@gmail.com, nikonov@vega.protres.ru, nevskaya@protres.ru* To whom correspondence should be addressed.
Received July 8, 2016; Revision received August 2, 2016
The crystal structure of the γ-subunit of translation initiation factor 2 from the archaeon Sulfolobus solfataricus (SsoIF2γ) has been solved based on perfectly hemihedral twinned data. The protein was cocrystallized with the 10-fold molar excess of GTP analog (GDPCP) over protein. However, no nucleotide was found in the structure, and the model demonstrated the apo form of the protein. Two slightly different molecules in the asymmetric unit of the crystal are related by the non-crystallographic 2-fold axis and form a tightly associated dimer. This dimer is stabilized by an intermolecular hydrophobic core and hydrogen bonds. Lack of GDPCP in the nucleotide-binding pocket of the γ-subunit and significant excess of dimers over monomers in the crystallization solution suggest that these dimers are the building blocks of the crystal. Contrary to SsoIF2γ monomers, these dimers are able to crystallize in two oppositely oriented slightly different crystal domains, thus forming a twinned crystal. Comparison of crystallization conditions for the twinned and untwinned crystals of apo SsoIF2γ showed that stabilization of the dimers in the solution may be caused by higher sodium salt concentration. Since amino acid residues involved in intermolecular contacts in the dimer are responsible for binding of the γ- and α-subunits within SsoIF2, increase in sodium salt concentration may prevent functioning of SsoIF2 in the cell.
KEY WORDS: translation initiation factor 2, Sulfolobus solfataricus, crystal structure, hemihedral twinningDOI: 10.1134/S0006297916100187
In Eukaryotes and Archaea, the transfer of initiator Met-tRNAiMet (tRNAi) to the small ribosomal subunit is mediated through translation initiation factor 2 (e/aIF2). This factor belongs to the family of GTP-binding proteins, which undergo conformational changes under the control of GTP and GDP binding [1, 2]. Like other GTP-binding proteins, it cycles between two distinct forms: an active, GTP-bound form, and an inactive, GDP-bound form. In the active state, e/aIF2·GTP forms a ternary complex with Met-tRNAiMet and incorporates into a translation preinitiation complex with the small ribosomal subunit. After recognition of the start codon and GTP hydrolysis, this factor in its GDP-bound form loses affinity for tRNAi and dissociates from the initiation complex. Recycling of the factor requires dissociation of protein-bound GDP. In eIF2, this is a slow process accelerated by guanine nucleotide-exchange factor eIF2B [3]. In contrast to eukaryotes, a guanine nucleotide-exchange factor has not been identified in the genome of the archaeon Sulfolobus solfataricus. Besides, aIF2 from S. solfataricus (SsoIF2) has similar affinities for GTP and GDP [4], and thus the GDP/GTP exchange appears to occur spontaneously.
The e/aIF2 molecule is composed of three subunits (α, β, γ) connected by hydrogen bonds and van der Waals interactions. The largest, γ-subunit (e/aIF2γ) forms the core of the heterotrimer and has GTPase activity. It is connected with the α- and β-subunits, which have no contact with each other. The structure of the γ-subunit has three domains. Domain I (G domain) incorporates the nucleotide binding pocket and contains seven β-strands and five α-helices. Two flexible regions, switch 1 and switch 2, govern the transition of the γ-subunit from the GTP-bound state to the GDP-bound state and vice versa. Domains II and III are β-barrels and act together as a rigid body [5, 6].
Several crystal structures of nucleotide-free, inactive GDP-bound and active GTP-bound forms of SsoIF2γ have been solved [7-11]. These proteins were crystallized under various conditions, and their crystals belonged to different space groups. Structures of the active form of the protein were obtained by cocrystallization of SsoIF2γ with nonhydrolyzable GTP analogs, GDPNP and GDPCP, reliably localized on electron density maps. Recently, structures of two complexes between SsoIF2γ mutants (D19A and H97A) and GTP were determined [11]. The mutations lowered the rate of GTP hydrolysis; in the crystal, GTP was tightly bound within the nucleotide-binding pocket in a position similar to that of GDPNP in the corresponding structures formed by wild type SsoIF2γ. All these structures of the apo and nucleotide-bound forms of SsoIF2γ were determined using untwinned data.
At the same time, the structures of the intact SsoIF2 and of the SsoIF2αγ heterodimer in complex with GDPNP and Met-tRNAiMet were solved by our group using perfect hemihedral twinned crystals [12, 13]. In both these cases, our attempts to obtain untwinned crystals failed, and the reason for twinning remained unexplained. It is known that a hemihedrally twinned crystal consists of two crystal domains related to each other by a specific symmetry operation [14, 15]. For the description of hemihedrally twinned crystals, two parameters are necessary: a description of the orientation of the different domains relative to each other (twin law) and the fractional contribution of each component. The twin law can be expressed as a matrix that transforms the hkl indices of one crystal domain into the other [14, 16]. If the fractional contribution of each domain is close to 0.5, we are dealing with perfect hemihedral twinning.
Here we present the crystal structure of SsoIF2γ solved using perfect hemihedral twinned crystals. In spite of the 10-fold molar excess of the analog of GTP, GDPCP, over the protein in the crystal solution, the nucleotide was not found in the structure, and the model demonstrated the apo form of the protein. In the asymmetric unit of the crystal, two conformationally similar but not identical molecules related by a 2-fold non-crystallographic symmetry axis formed a tightly associated dimer. The significant excess of dimers over monomers in the crystallization solution and lack of GDPCP binding by the γ-subunit suggested that these dimers are formed in solution and then are used as the building blocks of the crystal. This results in appearance of the perfect hemihedral twinning.
Analysis of the crystallization conditions showed that the number of the dimers in the crystallization solution that inhibit GTP binding increases with sodium salt concentration. Since amino acid residues involved in intermolecular contact in the dimer are responsible for binding of the γ- and α-subunits within SsoIF2, high salt concentration may prevent functioning of SsoIF2 in the cell.
MATERIALS AND METHODS
Crystallization. Cloning and expression of the SsoIF2γ gene and purification of the protein was performed as described previously [9]. The GTP analog GDPCP was purchased from Jena Bioscience. Crystallization experiments were performed at 22°C using the hanging drop vapor diffusion technique [17]. A 1.5-µl aliquot of the protein solution (40 mg/ml SsoIF2γ dialyzed into 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 5 mM β-mercaptoethanol) was mixed with 1.5 µl of precipitation solution (15% sodium malonate monohydrate, 50 mM glycine, pH 4.5). Then 1.0 µl solution of 13 mM CdCl2 and 13 mM GDPCP was added to reach 10-fold molar excess of the nucleotide over the protein. The drops were set up on siliconized cover slides and equilibrated against 0.5 ml of the reservoir solution: 19% sodium malonate monohydrate, pH 4.5, 63.3 mM glycine. Crystals were obtained within 10 days. Cryoprotection of the crystals was achieved by a short-time soaking in cryo solution (15% sodium malonate monohydrate, 50 mM glycine, 3.2 mM CdCl2, 15% ethylene glycol (v/v)). After this, the crystals were instantly frozen in liquid nitrogen at 100 K.
Data collection, structure determination, and refinement. Diffraction data were collected at the X12 beamline of DESY (Hamburg, Germany) equipped with a MAR CCD 165 detector. The data were processed and merged with the XDS package [18] (version May 8, 2007). Analysis of the results showed that the P31 and P3121 space groups were the most probable. First, the higher of these groups of symmetry was tested. Neither Matthew’s coefficient [19] nor the Stanley factor [20] detected twinning for this space group. The initial phases were obtained by the molecular replacement method with Phaser [21] using all known SsoIF2γ structures as search models. The best result was obtained with the apo form (PDB ID 2plf). The asymmetric unit of the cell contained one molecule. The model was subjected to several rounds of crystallographic refinement and map calculation with PHENIX [22] and manual model inspection and modification with Coot [23], but we could not refine the structure to an Rfree value less than 32%.
This problem may be caused by twinned data, and consequently a lower true symmetry of the crystal. Analysis of data in space group P31 confirmed the perfect twinning with twin law k, h, –l. In this case, two molecules were found in the asymmetric unit by molecular replacement. Taking into account twinned data, the refinement of the structure in space group P31 gave Rwork of 20.1%, and Rfree of 22.0% at 2.5 Å resolution and a good interpretive electron density map. Data collection and refinement statistics are given in Table 1. All residues are within the allowed regions of the Ramachandran plot. Structural data and the coordinates were deposited in the Protein Data Bank (PDB ID 5dsz).
Table 1. Data collection and refinement
statistics
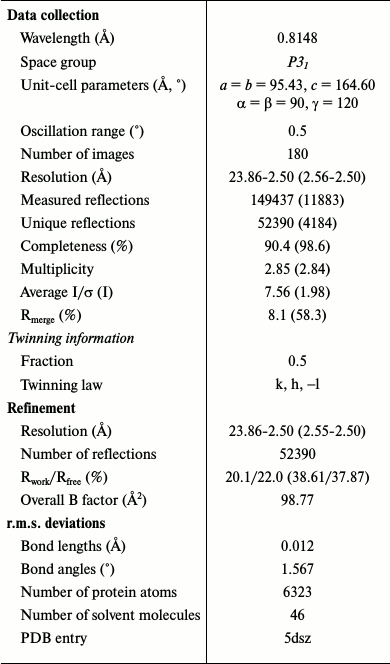
Note: Values in parentheses are for the highest resolution shell.
RESULTS AND DISCUSSION
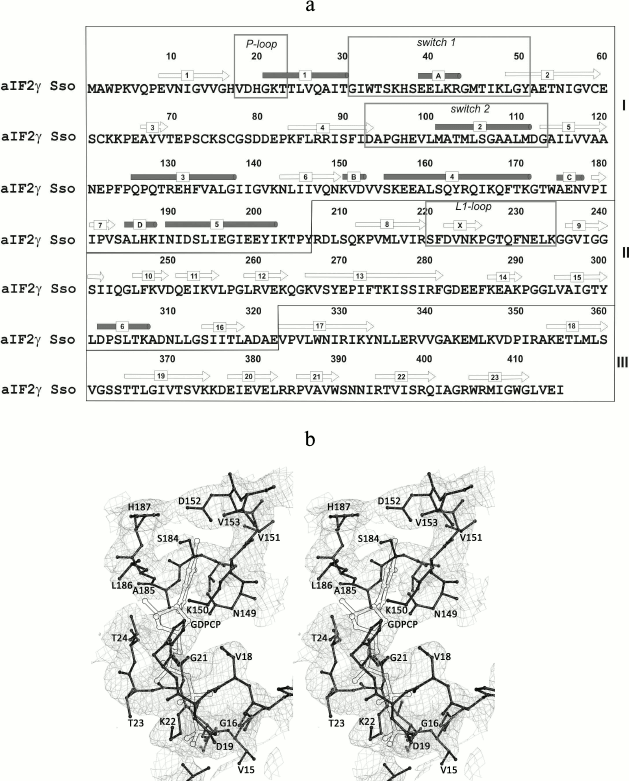
Overall description of the structure. Though the protein was cocrystallized with the 10-fold molar excess of the GTP analog GDPCP, as in all previously solved structures of nucleotide-bound SsoIF2γ [9, 10], no nucleotide was found in the structure. The model demonstrates the apo form of the protein, and similar to other apo forms of SsoIF2γ the nucleotide-binding pocket is occupied by amino acid residues (18-20) of the P-loop (Fig. 1).
Fig. 1. Amino acid sequence of SsoIF2γ (a) and a fragment of the electron density map (b). a) Elements of the SsoIF2γ structure referred to in the text are boxed; domains I, II, and III are shown with roman numerals; secondary structure elements are numbered. b) Stereo view showing that the electron density of the nucleotide-binding pocket is occupied by residues 18-20 of the P-loop of SsoIF2γ. The position of the nucleotide in the structure of the SsoIF2γ complex with the GTP analog (GDPCP) is shown with double lines. The 2Fo-Fc electron density map is contoured at the 1.0 σ level. All figures were prepared with PyMOL (DeLano, W. L. (2002) http://www.pymol.org).
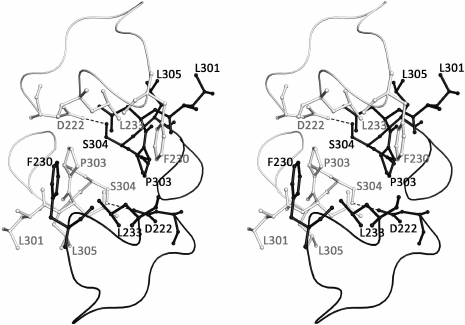
The asymmetric unit of the SsoIF2γ crystal contains two monomers (A and B) related by a non-crystallographic 2-fold symmetry axis. Molecules A and B have similar but not identical conformations: superposition of 401 Cα atoms visible in both molecules yields an r.m.s. deviation of 1.16 Å. The most marked differences are observed in the switch 1 and switch 2 regions, in the β17-β18 and β22-β23 loops and hairpin β12-β13 (Fig. 1a). The superposition of two molecules excluding these regions yields an r.m.s. deviation of 0.71 Å for 365 Cα atoms. The relative orientation of three domains in both molecules is very close (r.m.s. deviation for 102 atoms of domain I and 173 atoms of domains II and III is equal to 0.48 Å). In the crystal, two monomers form a tightly associated dimer and contact each other through the intermolecular hydrophobic core formed by F230, L233, and P303 of each molecule. Two short hydrogen bonds between S304 and D222 of two monomers stabilize the dimer. Loops 224-233 and 306-310 protect the contact region from the solvent (Fig. 2).
Fig. 2. Stereo view of the contact region between two monomers of SsoIF2γ in the dimer. Monomer A is shown in black, monomer B – in gray. Two intermolecular hydrogen bonds between S304 and D222 are shown by dotted lines. The intermolecular hydrophobic core is formed by F230, L233, and P303 of each molecule. Loops 224-233 and 306-310 protect the contact region from the solvent.
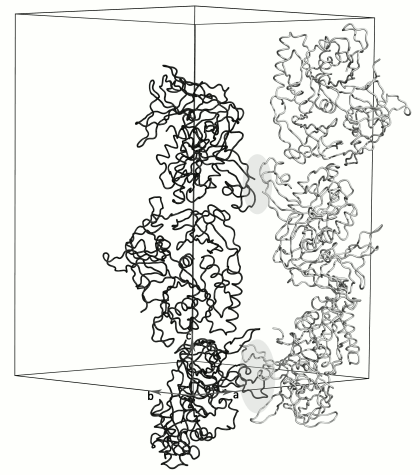
Molecules in the crystal are packed into long antiparallel strands parallel to the 3-fold crystallographic screw axis (Fig. 3). Each of the neighboring strands is formed by one type of the SsoIF2γ monomers (A or B). Molecules of the strands are related by two non-crystallographic 2-fold axes. One of them relates monomers in the dimer within the asymmetric unit of the crystal; the other relates the regions containing the N-terminus and the zinc finger of monomers from two dimers. The last contact is very weak: there is no intermolecular hydrophobic core, and only two intermolecular hydrogen bonds are formed between A2 and E82.
Fig. 3. Crystal packing of dimers along the 3-fold crystallographic screw axis. Each oppositely directed crystal strand contains monomers of only one type A (black) or B (gray). The A and B molecules of neighboring strands are related by two different 2-fold non-crystallographic axes perpendicular to the plane of the figure. These regions (light gray ovals) alternate along the crystal strands. Strands formed by molecules A transform to strands formed by molecules B in accordance with twin low k, h, –l.
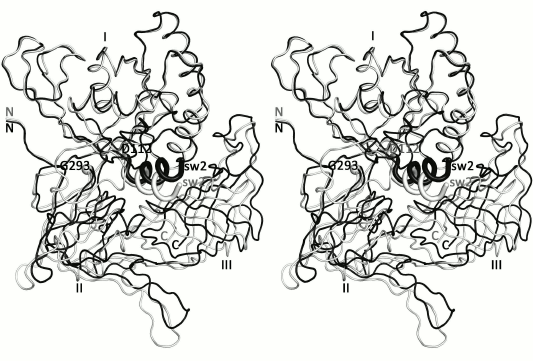
Previously it was shown that the structures of SsoIF2γ in the apo and nucleotide-bound forms differ in position of domains II and III relative to domain I. In all known archaeal IF2γ structures, domain II and III act as a rigid body [5-7, 10, 24]. Superposition of the presented structure onto the structure of the GTP-bound form [10] reveals extra feature characterizing transition of SsoIF2γ from the apo form to the active state. Superposition shows that such transition is accompanied by rotation of helix α2 (switch 2) and domains II and III as a rigid body by about 13° around an axis, approximately passing through the Cα atoms of D112 and G293 (Fig. 4). Such rotation is provided by the interdomain hydrophobic core formed by M101, A102, M104, L105, G107, M358, V396, and M407, which stabilizes the relative position of helix α2 and domain III and retains it during functional transition. Thus, binding of GTP induces a conformational rearrangement of switch 2 of SsoIF2γ and changes the shape of the molecule.
Fig. 4. Stereo view of the presented apo structure (gray) superimposed onto the GDPCP-bound form [10] of SsoIF2γ (black) with least-squares minimization of differences in Cα atom coordinates of domain I. It is seen that under transition of SsoIF2γ from the apo to active state, helix α2 of switch 2 (sw2) moves integrally with domains II and III.
The reason for twinning. Crystals of apo SsoIF2γ belonging to space group P3121 [9] have no twinning. Their molecules are packed into antiparallel strands similar to that of twinned crystals, but all strands are formed by identical molecules (molecule A is identical to molecule B). Therefore, each strand can be transformed to another by a symmetry operation x, y, z → y, x, –z. If molecules A and B are not identical, this symmetry operation is inhibited, and crystallographic 2-fold axis is replaced by a non-crystallographic one. The position of the 3-fold screw axis and origin of coordinates are also changed. This results in the space group P31 and appearance of two crystal domains related by twin low k, h, –l. Because of this, our structure could not be properly refined in the P3121 space group (see “Materials and Methods”).
Building blocks in a twinned crystal. The complex of SsoIF2γ with GTP must be formed in crystallization solution before the crystal nucleation, because this process in the crystal is connected with the change in the shape of the molecule and demands a shift of a significant molecular mass and a rearrangement of crystal contacts. Such conformational changes in the apo form of the protein may result in destruction of the crystal structure, as it happened with the apo form of the SsoIF2βγ from Pyrococcus furiosus in the time of soaking it in solution containing GTP [24].
In the present study, the nucleotide/protein ratio in the crystallization solution was equal to 10/1, as in all previously solved nucleotide-bound forms of SsoIF2γ [9, 10]. However, the resulting structure possesses no nucleotide, and crystals of the apo form are not destroyed in the presence of the GTP analog. According to the structural data, this may be caused by formation of stable dimers (similar to those observed in the crystal structure) in the crystallization solution. In such dimers domains II of the two monomers are connected by an intermolecular hydrophobic core and hydrogen bonds. The main axes of inertia of the molecules in the dimer are oppositely directed, as well as movement of switch 2 and domains II and III under interaction with GTP. If forces of the intermolecular contact in the dimer exceed the protein–nucleotide interactions, SsoIF2γ cannot bind GTP.
To check the existence of such dimers in the solution experimentally, we used the dynamic light scattering (DLS) method, which measures average molecular weight of the protein in liquid medium. Experiments were performed in conditions (Table 2) corresponding to twinned (this study) and untwined crystals of apo SsoIF2γ [9]. This revealed that a dynamic equilibrium between monomers and dimers of SsoIF2γ exists in both crystallization solutions, but the proportion of monomers and dimers is different. An average molecular weight of the protein SsoIF2γ was 79.4 ± 5.8 kDa in the solution producing the twinned crystals, while that for untwined crystals was equal to 55.8 ± 3.3 kDa. Based on these data, the calculated ratio of dimers to monomers was 10 times higher in the solution producing the twinned crystals.
Table 2. Initial concentrations (mM) of
components in crystallization drops before equilibration
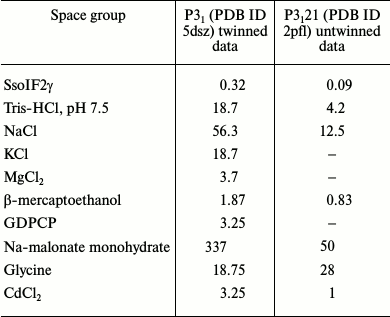
To understand the biochemical reasons for the preferable formation of dimers in the solution used in the presented study, we compare the crystallization conditions in the cases of twinned and untwinned data. Initial concentrations of components in the crystallization drops before the equilibration are shown in Table 2 for twinned (PDB ID 5dsz) and untwinned (PDB ID 2pfl) crystals. As seen, these conditions differ considerably in salt concentration. First, this concerns the concentration of sodium ions (Na-malonate and NaCl), which is considerably higher in the case of twinned crystals. It is known [25] that in high salt concentration the role of the intermolecular hydrophobic core increases. In our case, interactions between monomers in the dimer becomes stronger and cannot be destroyed by GDPCP binding.
Therefore, we conclude that two conformationally similar but not identical molecules of SsoIF2γ are able to form stable dimers in solution before the crystallization event, and these dimers can serve as the building blocks of crystals with perfect hemihedral twinning.
Comparison of the presented structure of SsoIF2γ with the structures of αγ-heterodimer [7] and intact protein [12] shows that the contact region between monomers in the dimer coincides with the contact region between the α- and γ-subunits. High salt concentration forces affinity of SsoIF2γ molecules to each other, in contrast to electrostatic interactions between subunits. This may prevent the formation of the intact SsoIF2 and disrupt normal functioning of the factor in the cell.
Acknowledgements
We are indebted to Dr. Alexei Kazakov (Institute for Biological Instrumentation of the Russian Academy of Sciences, Pushchino, Russia) for assistance with DLS measurements.
This work was supported by the Russian Foundation for Basic Research (projects Nos. 14-04-00571, 14-04-00406) and by the Program of the Russian Academy of Sciences on Molecular and Cell Biology.
REFERENCES
1.Vetter, I. R., and Wittinghofer, A. (2001) The
guanine nucleotide-binding switch in three dimensions, Science,
294, 1299-1304.
2.Grant, B. J., Gorfe, A. A., and McCammon, J. A.
(2010) Large conformational changes in proteins: signaling and other
functions, Curr. Opin. Struct. Biol., 20, 142-147.
3.Kyrpides, N. C., and Woese, C. R. (1998) Archaeal
translation initiation revisited: the initiation factor 2 and
eukaryotic initiation factor 2B alpha-beta-delta subunit families,
Proc. Natl. Acad. Sci. USA, 95, 3726-3730.
4.Pedulla, N., Palermo, R., Hasenohrl, D., Blasi, U.,
Cammarano, P., and Londei, P. (2005) The archaeal eIF2 homologue:
functional properties of an ancient translation initiation factor,
Nucleic Acids Res., 33, 1804-1812.
5.Schmitt, E., Blanquet, S., and Mechulam, Y. (2002)
The large subunit of initiation factor aIF2 is a close structural
homologue of elongation factors, EMBO J., 21,
1821-1832.
6.Roll-Mecak, A., Alone, P., Cao, C., Dever, T. E.,
and Burley, S. K. (2004) X-ray structure of translation initiation
factor eIF2γ: implications for tRNA and eIF2alpha binding, J.
Biol. Chem., 279, 10634-10642.
7.Yatime, L., Mechulam, Y., Blanquet, S., and
Schmitt, E. (2006) Structural switch of the γ-subunit in an
archaeal aIF2αγ heterodimer, Structure, 14, 119-128.
8.Yatime, L., Mechulam, Y., Blanquet, S., and
Schmitt, E. (2007) Structure of an archaeal heterotrimeric initiation
factor 2 reveals a nucleotide state between GTP and GDP states,
Proc. Natl. Acad. Sci. USA, 104, 18445-18450.
9.Nikonov, O., Stolboushkina, E., Nikulin, A.,
Hasenohrl, D., Blasi, U., Manstein, D. J., Fedorov, R., Garber, M., and
Nikonov, S. (2007) New insights into the interactions of the
translation initiation factor 2 from archaea with guanine nucleotides
and initiator tRNA, J. Mol. Biol., 373, 328-336.
10.Nikonov, O., Stolboushkina, E., Arkhipova, V.,
Kravchenko, O., Nikonov, S., and Garber, M. (2014) Conformational
transitions in the γ-subunit of the archaeal translation
initiation factor 2, Acta Crystallogr. D Biol. Crystallogr.,
D70, 658-667.
11.Dubiez, E., Aleksandrov, A., Lazennec-Schurdevin,
C., Mechulam, Y., and Schmitt, E. (2015) Identification of a second
GTP-bound magnesium ion in archaeal initiation factor 2, Nucleic
Acids Res., 43, 2946-2957.
12.Stolboushkina, E., Nikonov, S., Nikulin, A.,
Blasi, U., Manstein, D., Federov, R., Garber, M., and Nikonov, O.
(2008) Crystal structure of the intact archaeal translation initiation
factor 2 demonstrates very high conformational flexibility in the
alpha- and beta-subunits, J. Mol. Biol., 382,
680-691.
13.Stolboushkina, E., Nikonov, S., Zelinskaya, N.,
Arkhipova, V., Nikulin, A., Garber, M., and Nikonov, O. (2013) Crystal
structure of the archaeal translation initiation factor 2 in complex
with a GTP analogue and Met-tRNAf(Met), J. Mol. Biol.,
425, 989-998.
14.Yeates, T. O. (1997) Detecting and overcoming
crystal twinning, Methods Enzymol., 276, 344-358.
15.Parsons, S. (2003) Introducing to twinning,
Acta Crystallogr. D Biol. Crystallogr., 59,
1995-2003.
16.Dauter, Z. (2003) Twinned crystals and anomalous
phasing, Acta Crystallogr. D Biol. Crystallogr.,
59, 2004-2016.
17.Davies, D., and Segal, D. (1971) Protein
crystallization: microtechniques involving vapor diffusion, Methods
Enzymol., 22, 266-269.
18.Kabsch, W. (1993) Automatic processing of
rotationdiffraction data from crystals of initially unknown symmetry
and cell constants, J. Appl. Crystallogr., 26,
795-800.
19.Matthews, B. W. (1968) Solvent content of protein
crystals, J. Mol. Biol., 33, 491-497.
20.Stanley, E. (1972) The identification of twins
from intensity statistics, J. Appl. Crystallogr., 5,
191-194.
21.McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P.
D., Winn , M. D., Storoni, L. C., and Read, R. J. (2007) Phaser
crystallographic software, J. Appl. Crystallogr., 40,
658-674.
22.Afonine, P. V., Grosse-Kunstleve, R. W., Echols,
N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C.,
Urzhumtsev, A., Zwart, P. H., and Adams, P. D. (2012) Towards automated
crystallographic structure refinement with phenix.refine, Acta
Crystallogr. D Biol. Crystallogr., 68, 352-367.
23.Emsley, P., Lohkamp, B., Scott, W. G., and
Cowtan, K. (2010) Features and development of Coot, Acta
Crystallogr. D Biol. Crystallogr., 66, 486-501.
24.Sokabe, M., Yao, M., Sakai, N., Toya, S., and
Tanaka, I. (2006) Structure of archaeal translational initiation factor
2 betagamma-GDP reveals significant conformational change of the
beta-subunit and switch 1 region, Proc. Natl. Acad. Sci.
USA, 103, 13016-13021.
25.Date, M. S., and Dominy, B. N. (2013) Modeling
the influence of salt on the hydrophobic effect and protein fold
stability, Commun. Comput. Phys., 13, 90-106.