Inhibition of Chaperonin GroEL by a Monomer of Ovine Prion Protein and Its Oligomeric Forms
S. S. Kudryavtseva1, Y. Y. Stroylova1, I. A. Zanyatkin2, T. Haertle3, and V. I. Muronetz1,2*
1Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119234 Moscow, Russia; E-mail: vimuronets@belozersky.msu.ru2Lomonosov Moscow State University, Faculty of Bioengineering and Bioinformatics, 119234 Moscow, Russia
3Institut National de la Recherche Agronomique, 44000 Nantes, France
* To whom correspondence should be addressed.
Received June 17, 2016; Revision received July 29, 2016
The possibility of inhibition of chaperonin functional activity by amyloid proteins was studied. It was found that the ovine prion protein PrP as well as its oligomeric and fibrillar forms are capable of binding with the chaperonin GroEL. Besides, GroEL was shown to promote amyloid aggregation of the monomeric and oligomeric PrP as well as PrP fibrils. The monomeric PrP was shown to inhibit the GroEL-assisted reactivation of the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The oligomers of PrP decelerate the GroEL-assisted reactivation of GAPDH, and PrP fibrils did not affect this process. The chaperonin GroEL is capable of interacting with GAPDH and different PrP forms simultaneously. A possible role of the inhibition of chaperonins by amyloid proteins in the misfolding of the enzymes involved in cell metabolism and in progression of neurodegenerative diseases of amyloid nature is discussed.
KEY WORDS: amyloid proteins, chaperonin GroEL, prion PrP, oligomeric forms of prion protein, glyceraldehyde-3-phosphate dehydrogenase, reactivation, inhibition of chaperoninDOI: 10.1134/S0006297916100199
Abbreviations: DEAE, diethylaminoethyl; DLS, dynamic light scattering; EDTA, ethylenediaminetetraacetate; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HEPES, 4-(2-hydroxyethyl)-1-piperazinethanesulfonic acid; β-ME, β-mercaptoethanol; MOPS, 3-[N-morpholino]propanesulfonic acid; PBST, phosphate-buffered saline containing 0.05% Tween-20; PrP, prion protein; ThT, thioflavin T.
Despite the large number of publications covering the role of chaperones
in formation, transformation, and destruction of oligomeric and
fibrillary protein structures [1-5], one question that we consider extremely important
has eluded research interest. Can amyloidogenic proteins recognized as
misfolded by chaperones influence the efficacy of chaperone-assisted
refolding of regular proteins? We have previously shown that artificial
protein constructs unable to correctly fold are capable of inhibiting
chaperone activity: glyceraldehyde-3-phosphate dehydrogenase (GAPDH)
bearing point mutations in the intersubunit contact area,
carboxymethylated, and even oxidized GAPDH forms were shown to inhibit
GroEL chaperonin activity [6, 7]. The most evident reason for such action is that a
polypeptide chain unable to form a native conformation will
continuously be recognized as misfolded and therefore will not be able
to leave the inner cavity of the chaperone. We assume that amyloid-like
proteins are able to exert the same action on the chaperone system, and
of the most interest are proteins connected to the progression of
amyloid diseases – α-synuclein, prion protein,
β-amyloid peptide, and their oligomeric forms. Amyloid
neurodegenerative diseases are known to be accompanied by significant
changes in neural tissue metabolism [8-11], which are most commonly found as changes in
energy metabolism, i.e. in glycolysis [12-14]. One possible reason for such changes is that
upon being blocked by amyloid-like proteins, the chaperone system is no
longer able to fulfill its function, i.e. assisted protein folding and
prevention of aggregation. To test this hypothesis, we have chosen an
enzyme playing one of the key roles in glycolysis – GAPDH.
This enzyme is found in all tissue types in relatively high
concentrations (5-10% of cytoplasmic protein content) [15, 16], and the formation of
its quaternary structure is known to be assisted by complex
ATP-dependent chaperonins [17-21]. Bacterial chaperonin GroEL used in the present
work is an example of such proteins, and it closely resembles
cytoplasmic chaperonin TriC from mammals [22, 23]. The possibility of blocking chaperonin function
by an amyloid protein was tested using ovine prion protein and its
oligomeric and fibrillary forms.
MATERIALS AND METHODS
Materials. ATP, 3-phosphoglyceraldehyde, NAD+, Coomassie G-250, DEAE-Sephacel, thioflavin T (ThT), and dithiothreitol (DTT) were purchased from Sigma (USA); EDTA, guanidine hydrochloride, and imidazole – from Panreac (Spain); MgCl2, sodium acetate, protease inhibitor cocktail, HEPES, MOPS, Tris, and KH2PO4 – from Amresco (USA). Recombinant chaperonin GroEL and co-chaperonin GroES were isolated and purified as described previously [7]. GAPDH was isolated from rabbit skeletal muscle as described by Scopes [24].
The recombinant allelic variant of ovine prion protein ovPrP-VRQ (full-length amino acid sequence lacking in the signal N- and C-terminal peptides and containing additional S22 and mutations V136, R154, and Q171) was expressed and purified as described previously [25].
Preparation of various forms of prion protein PrP. Lyophilized PrP was dissolved in 20 mM sodium acetate buffer, pH 4.0, prior to each experiment and then was transferred into 20 mM MOPS buffer, pH 7.5, using a Sephadex G-25 desalting column. Solution containing 0.5 mg/ml of prion protein was incubated for 1 h at 60°C and pH 7.5 to yield oligomers. Prion protein that had been stored for 5 months and then exposed to similar conditions formed a homogeneous population of particles with a hydrodynamic diameter of about 90 nm. These particles were called “protofibrils”, as their size was larger than for the oligomers, but smaller than for PrP fibrils.
Amyloid fibrils were prepared as previously described [26]. Guanidine hydrochloride used as a chaotropic agent was added to samples containing 2 mg/ml of prion protein in 100 mM Na-acetate buffer, pH 4.0, to a final concentration of 1 M. Samples were incubated at 37°C for 3 days with vigorous stirring.
Denaturation and reactivation of GAPDH using chaperonin complex GroEL14/GroES7. A solution containing 120 µM of GAPDH (calculated per monomer) was incubated with 4 M guanidine hydrochloride, 10 mM KH2PO4, 1 mM EDTA, 2 mM mercaptoethanol, pH 7.5, for 15 min until complete disappearance of the enzyme activity to denature the enzyme.
GAPDH reactivation was initiated by 200-fold dilution of denatured enzyme to a final concentration of 0.6 µM calculated per monomer (concentration of guanidine hydrochloride was reduced to 0.02 M at the same time). GAPDH was spontaneously reactivated in buffer containing 10 mM KH2PO4, 1 mM EDTA, 5 mM mercaptoethanol, 1.5 mM NAD+, pH 7.5. In the case of chaperonin-dependent reactivation of GAPDH, 2 mM ATP, 2 mM Mg2+, 0.6 µM chaperonin GroEL14, and 1.2 µM co-chaperonin GroES7 were further added into the incubation mixture. During reactivation, aliquots were taken from the reaction mixture for determination of the enzymatic activity of GAPDH.
Determination of enzymatic activity of GAPDH. GAPDH activity was determined using a Shimadzu UV-1601 UV/VIS spectrophotometer at 25°C by measuring the rate of NADH formation at 340 nm in the oxidation reaction of 3-phosphoglyceraldehyde. GAPDH activity was measured at 25°C in pH 9.0 buffer containing 50 mM glycine, 50 mM potassium phosphate, 0.5 mM EDTA, 0.5 mM NAD+, 1 mM 3-phosphoglyceraldehyde, and 0.8-2 µg GAPDH isolated from rabbit muscle.
ELISA modified for study of protein–protein interactions. A 100-µl portion of solution with 2 µg/ml concentration of the first protein (complex GroEL14/GroES7 or one of the three forms of PrP: native PrP, PrP oligomers, PrP fibrils) in phosphate-buffered saline (PBS) was adsorbed in a 96-well plate. The plate was incubated for 1 h at 20°C with stirring. After that, the wells were washed with PBS. In the next step, the second protein (possible partner) was added to the wells at the same concentration (or PBS in case of controls) and incubated under similar conditions. Then plate was washed with PBST (phosphate-buffered saline with 0.05% Tween-20) and incubated with primary monoclonal antibodies (6C5 against denatured GAPDH and F99/97.6.1, specific against the PrP epitope Q220YQRES225) for 1 h at 20°C with stirring. After washing the plate with PBST, samples were incubated with secondary horseradish peroxidase-conjugated anti-mouse antibodies (1 µg/ml in PBST) under the given conditions. Then the plate was washed, and solution containing o-phenylenediamine and hydrogen peroxide was added to the wells according to the standard protocol. The optical density of wells was determined at λ = 492 nm using a State Fax 2100 plate photometer.
Dynamic light scattering (DLS). Dynamic light scattering was measured using a Zetasizer Nano-ZS device (Malvern Instruments, Great Britain) equipped with 173° optics for detection of scattered intensity. The laser wavelength was 532 nm. Numerical distribution of particles was used to interpret the results. Each distribution shown in the graph is represented as the average of five measurements carried out for 75 s. Polydispersity index was calculated as the ratio of the peak width at half-height to the mean squared.
Thioflavin T fluorescence. Thioflavin T (ThT) is a small molecule dye that fluoresces upon binding to β-sheets, and it is used to study amyloid structures [27, 28]. A solution of 0.6 µM of each form of the prion protein (native PrP, PrP oligomers, “protofibrils”, fibrils) in 10 mM potassium phosphate buffer, pH 7.5, containing 1 mM EDTA, 5 mM β-mercaptoethanol, 1.5 mM NAD+, 2 mM ATP, and 2 mM Mg2+ was incubated with GroEL14/GroES7 complex (0.6 µM GroEL14 and 1.2 µM GroES7) for 2 h. Then the solution of ThT was added to the samples to final concentration of 6 µM, and the mixture was incubated for 15 min at 20°C. Fluorescence emission spectra of ThT were recorded at 20°C in the range of 450-600 nm (excitation at 435 nm) using a FluoroMax 3 spectrofluorometer (Horiba Jobin Yvon, France) in 2-ml quartz fluorimetric cuvette.
RESULTS AND DISCUSSION
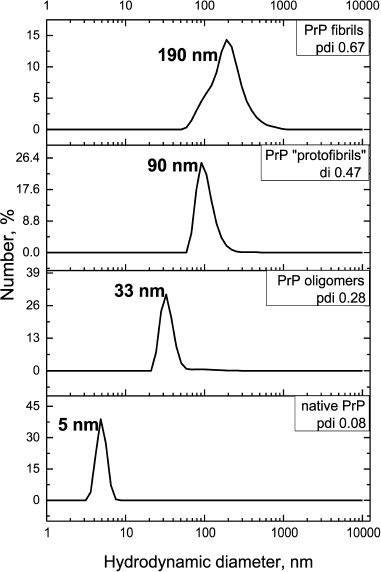
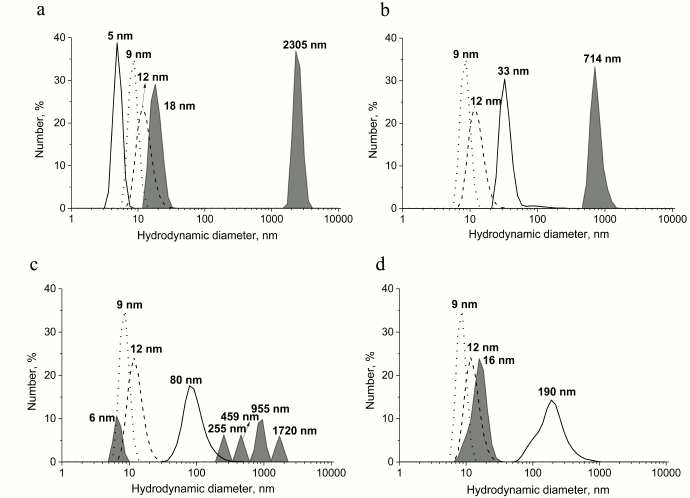
To study the possibility of inhibiting GroEL chaperonin with amyloid proteins, we prepared different forms of prion protein (PrP): a monomeric form, oligomers, “protofibrils”, and fibrils. To yield oligomers, PrP was incubated in 20 mM MOPS, pH 7.5, at 60°C for 1 h. “Protofibrils” were produced under the same conditions from lyophilized protein stored for 5 months. PrP fibrils were formed upon incubating the protein with 1 M guanidine hydrochloride in 100 mM sodium acetate buffer, pH 4.0, for 3 days under intensive stirring (see “Materials and Methods”). Monomeric PrP was not subject to any treatment after isolation and purification, but the conservation of its state was checked before each experiment by DLS. Figure 1 shows the results of particle size measurement of all the PrP preparations. PrP monomers, oligomers, protofibrils, and fibrils were shown to have hydrodynamic diameters of 5, 33, 90, and 190 nm, respectively. Particle polydispersity was shown to increase together with particle size: monomeric PrP shows as a narrow monodisperse peak (polydispersity index PDI = 0.08), PrP oligomers show moderate polydispersity (PDI = 0.28), while protofibrils and fibrils are detected as wide polydisperse peaks (PDI of 0.47 and 0.67, respectively). PrP fibril formation under the reported conditions was confirmed previously using various methods [26].
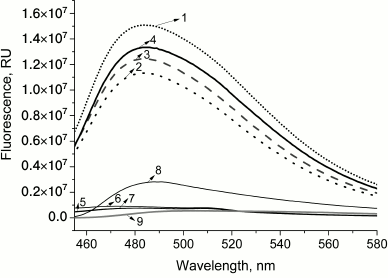
PrP fibrillization is also evidenced by the increased ThT fluorescence (Fig. 2, curve 8). Figure 3 shows the ELISA data for GroEL14/GroES7 complex binding to different forms of PrP.
Fig. 1. Hydrodynamic diameters of forms of PrP. From bottom to top: native PrP, 0.5 mg/ml in 20 mM MOPS buffer, pH 7.5; PrP oligomers formed by incubating at 60°C for 1 h in 20 mM MOPS buffer, pH 7.5; PrP “protofibrils” formed from stored for a long time protein preparation by incubating at 60°C for 1 h in 20 mM MOPS buffer, pH 7.5; PrP fibrils formed by incubation of solution with 2 mg/ml concentration of protein in 100 mM Na-acetate buffer, pH 4.0, in the presence of 1 M guanidine hydrochloride at 37°C for 3 days. The numerical distribution of the particles is shown.
Fig. 2. Emission spectra of thioflavin T fluorescence in the presence of different forms of PrP and complex GroEL14/GroES7 after coincubation. Curves: 1) chaperonin complex GroEL14/GroES7 after incubation with native prion protein PrPc (frequent dotted line); 2) chaperonin complex GroEL14/GroES7 after incubation with PrP oligomers (rare dotted line); 3) chaperonin complex GroEL14/GroES7 after incubation with PrP “protofibrils” (dashed line); 4) chaperonin complex GroEL14/GroES7 after incubation with PrP fibrils (solid line); 5) native prion protein PrPc; 6) PrP oligomers; 7) PrP “protofibrils”; 8) PrP fibrils; 9) chaperonin GroEL14/GroES7 complex. PrP forms at a concentration of 0.6 µM (calculated per monomer) were incubated in the presence of 0.6 µM GroEL14, 1.2 µM GroES7 and in medium containing 10 mM KH2PO4, 1 mM EDTA, 5 mM β-ME, 1.5 mM NAD+, 2 mM ATP, 2 mM Mg2+, pH 7.5.
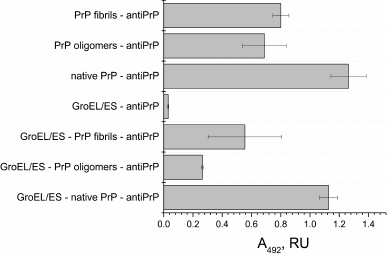
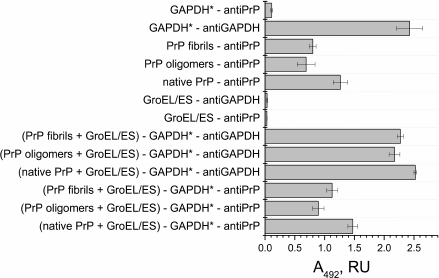
Fig. 3. ELISA of binding of chaperonin complex GroEL14/GroES7 with different forms of prion protein PrP. A 0.2-µg sample of the complex GroEL14/GroES7 or one of the three forms of PrP (native PrP, PrP oligomers, or PrP fibrils) in PBS was added in wells of a 96-well plate. After incubation for 1 h with stirring and washing with PBS, the wells with GroEL14/GroES7 complex were filled with one of the four forms of PrP in the same concentration and incubated under similar conditions. The following steps of immunoassay were performed according to standard protocol using the primary monoclonal antibodies (F99/97.6.1, specific against the PrP epitope Q220YQRES225) and secondary horseradish peroxidase-conjugated anti-mouse antibodies.
PrP-specific antibodies were shown to interact with the various PrP preparations and GroEL14/GroES7 chaperonin preincubated with different forms of PrP. One should expect ELISA signal to be diminished upon PrP oligomer binding to the chaperonin inner cavity due to the screening of PrP antigenic determinants, but this effect was not found in the case of PrP monomers. This observation leads to a hypothesis that chaperonin is not only capable of binding monomeric PrP, but also of causing its aggregation or fibrillization.
This hypothesis was confirmed by recording ThT fluorescence spectra in presence of different PrP forms and GroEL14/GroES7 complexes upon simultaneous incubation (Fig. 2). For monomeric, oligomeric, and protofibrillar preparations of PrP, and also for chaperonin, ThT fluorescence intensity remains basically unchanged (Fig. 2, curves 5-7 and 9), whereas in the case of PrP fibrils a 4-fold increase in fluorescence intensity is observed, indicating the presence of amyloid structures and confirming its fibrillary nature (Fig. 2, curve 8). Upon incubation of chaperonin with any of the studied PrP forms, ThT fluorescence was found to increase by multiple orders of magnitude (i.e. from 7.5·105 to 1.5·107 in case of PrP monomers; Fig. 2, curves 1-4). These data suggest the existence of chaperonin-stimulated amyloid conversion of not only already formed oligomers and fibrils, but also PrP monomers. It is also possible that amyloid aggregation (and/or fibrillization) of monomeric PrP bound by chaperonin is the explanation for the above-mentioned interaction with specific antibodies, as they are only capable of binding with solvent-exposed PrP aggregate antigens, while monomeric PrP bound to the inner cleft of chaperonin would be effectively screened from this interaction.
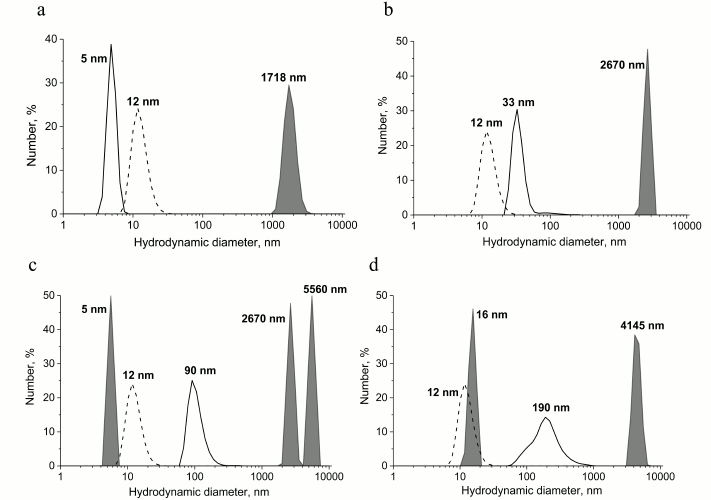
Interaction of various PrP forms with GroEL chaperonin and the subsequent changes in its aggregation state was also investigated using the dynamic light scattering technique. Data shown in Fig. 4 indicate that upon incubation of monomeric and oligomeric PrP with GroEL chaperonin, large aggregates with observed hydrodynamic diameters of 1718 and 2670 nm, respectively, are formed (Fig. 4, a and b). Aggregates of similar size and even larger ones are formed upon interaction of PrP protofibrils and fibrils with GroEL (Fig. 4, c and d), but in these cases smaller soluble particles are also formed (6 nm in case of protofibrils and 16 nm in case of PrP fibrils); therefore, it is possible that chaperonin is capable of cleaving PrP monomers from preformed protofibrils. The emergence of 16-nm-sized particles could result from formation of chaperonin with small fibril fragments resulting in conformational changes and hydrodynamic diameter of chaperonin, but this hypothesis requires further study.
Fig. 4. Hydrodynamic diameter of preparations of various forms of PrP after their incubation with chaperonin complex GroEL14/GroES7. The populations of particles that are complexes of chaperonins with different forms of PrP are in gray, and the control of individual chaperonin complex (dotted line) and different forms of PrP (solid line) are shown separately on the plots. a) Incubation of 0.6 µM native PrP with 0.6 µM GroEL14/GroES7 complex; b) incubation of 0.6 µM PrP oligomers with 0.6 µM GroEL14/GroES7 complex; c) incubation of 0.6 µM PrP “protofibrils” with 0.6 µM GroEL14/GroES7 complex; d) incubation of 0.6 µM PrP fibrils with 0.6 µM GroEL14/GroES7 complex. PrP forms at concentration of 0.6 µM (counting on monomer) were incubated in the presence of 0.6 µM GroEL14, 1.2 µM GroES7, and in medium containing 10 mM KH2PO4, 1 mM EDTA, 5 mM β-ME, 1.5 mM NAD+, 2 mM ATP, 2 mM Mg2+, pH 7.5.
Having demonstrated effective binding of various PrP forms to a GroEL chaperonin, we proceeded to study the influence of such mechanism inhibiting chaperonin functional activity to its capability of reactivating denatured form of GAPDH. According to the ELISA data, all PrP forms remain bound to the GroEL/GroES complex after incubation with denatured GAPDH (Fig. 5), whereas the later also binds to the chaperonin complex. Therefore, it is possible that one of the chaperonin rings binds to various PrP forms absorbed on plate walls, while another ring is still capable of binding denatured GAPDH.
Fig. 5. ELISA of binding of chaperonin complex GroEL14/GroES7 with different forms of prion protein PrP. A 0.2-µg sample of the complex GroEL14/GroES7 or one of the three forms of PrP (native PrP, PrP oligomers, PrP fibrils) in PBS was added into wells of a 96-well plate. After incubation for 1 h with stirring and washing with PBS, the wells with complex GroEL14/GroES7 were filled with one of the four forms of PrP in the same concentration and incubated under similar conditions. Following steps of immunoassay were performed according to the standard protocol using primary monoclonal antibodies (6C5, specific against denatured GAPDH, and F99/97.6.1, specific against the PrP epitope Q220YQRES225) and secondary horseradish peroxidase-conjugated anti-mouse antibodies.
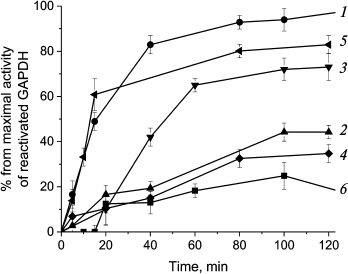
We hypothesized that preincubating GroEL/GroES chaperonin complex with various PrP forms might influence its activity of reactivating denatured GAPDH. The data obtained for chaperone-mediated and spontaneous reactivation of GAPDH that was treated with guanidine hydrochloride and then diluted 200-fold is shown at Fig. 6 (curves 1 and 6, respectively).
Fig. 6. Inhibition of GroEL14/GroES7-dependent reactivation of GAPDH with different forms of prion protein. Curves: 1) chaperonin-dependent reactivation of GAPDH; 2) chaperonin-dependent reactivation of GAPDH in the presence of 0.6 µM native PrP; 3) chaperonin-dependent reactivation of GAPDH in the presence of 0.6 µM PrP oligomers; 4) chaperonin-dependent reactivation of GAPDH in the presence of 0.6 µM PrP “protofibrils”; 5) chaperonin-dependent reactivation of GAPDH in the presence of 0.6 µM PrP fibrils; 6) spontaneous reactivation of GAPDH in the absence of GroEL14/GroES7. After denaturation in 4 M guanidine hydrochloride, GAPDH reactivation was performed by 200-fold dilution of the denatured enzyme to a final concentration of 0.6 µM in the presence of 0.6 µM GroEL14, 1.2 µM GroES7, and in medium containing 10 mM KH2PO4, 1 mM EDTA, 5 mM β-ME, 1.5 mM NAD+, 2 mM ATP, 2 mM Mg2+, pH 7.5.
Preincubation of chaperonin with native PrP and PrP protofibrils was found to suppress chaperone-mediated GAPDH reactivation almost completely (Fig. 6, curves 2 and 4), while PrP fibrils had almost no effect (Fig. 6, curve 5), and PrP oligomers had an inhibitory effect only for the first 60 min of incubation (curve 3). One should note that as PrP monomer to chaperonin molar ratio was equal to 1 : 1 for all PrP forms, the larger size of prion aggregates should lead to decreased probability of inhibiting chaperonin function. It is evident that PrP fibrils consisting of hundreds of monomers should not be able to inhibit chaperonin activity due to high particle-to-particle ratio, and the observed reactivation data support this assumption. However, one should remember that smaller fragments cleaved from PrP fibrils by chaperones and other possible agents would still be able to inhibit chaperonin function.
To investigate the probable causes of different action on chaperonin-mediated reactivation of GAPDH exerted by PrP forms, we studied size distribution of particles formed in each case using the DLS technique (Fig. 7). First, we found that upon simultaneous incubation of PrP fibrils, denatured GAPDH, and large chaperonin aggregates with size exceeding 6000 nm (these cannot be measured with DLS) are formed and precipitated. The resulting supernatant mainly contains 16-nm-sized particles that are considered to be chaperonin complexed with its protein substrates. PrP oligomers preincubated with chaperonin slow chaperonin-mediated GAPDH reactivation, but in this case, the formation of larger PrP aggregates (>1000 nm) is observed. In the case of PrP monomers and protofibrils, chaperone activity is effectively blocked, possibly due to the presence of not only large aggregates, but also smaller particles including monomers.
Fig. 7. Hydrodynamic diameters of preparations of the various forms of PrP after their incubation with chaperonin complex GroEL14/GroES7 and GAPDH. The populations of particles that are complexes of chaperonins with different forms of PrP and GAPDH are in gray, and the control of individual chaperonin complex (dashed dotted line), GAPDH (dotted line), and different forms of PrP (solid line) are shown separately on the graphs. a) Incubation of 0.6 µM native PrP with 0.6 µM GroEL14/GroES7 complex and 0.6 µM GAPDH; b) incubation of 0.6 µM PrP oligomers with 0.6 µM GroEL14/GroES7 complex and 0.6 µM GAPDH; c) incubation of 0.6 µM PrP “protofibrils” with 0.6 µM GroEL14/GroES7 complex and 0.6 µM GAPDH; d) incubation of 0.6 µM PrP fibrils with 0.6 µM GroEL14/GroES7 complex and 0.6 µM GAPDH. PrP forms at concentration 0.6 µM (counting on monomer) were incubated in the presence of 0.6 µM GroEL14, 1.2 µM GroES7, 0.6 µM GAPDH, and in medium containing 10 mM KH2PO4, 1 mM EDTA, 5 mM β-ME, 1.5 mM NAD+, 2 mM ATP, 2 mM Mg2+, pH 7.5.
Therefore, we have shown that one of the amyloid-like proteins, ovine prion protein PrP, is capable of binding to GroEL chaperonin and blocking its capability of restoring the functional state of a substrate, GAPDH. One should specifically note that a PrP/chaperonin complex loses its ability for reactivating GAPDH, and the latter is known to fold in the inner cavity of chaperonin. In the case of other protein substrates, i.e. lactate dehydrogenase, this effect could have been attributed to PrP screening of apical chaperonin domains, which are known to be centers of reactivation for many proteins. Also, not only GroEL interaction with different PrP forms was found, but also the ability of chaperonin to stimulate amyloid PrP aggregation. Obviously, the discovered inhibition of chaperonin functional activity by PrP is a first step in the investigation of the role of different amyloid-like proteins in chaperone-mediated inhibition of enzymes participating in energy metabolism in neural tissue. However, the similarity of bacterial chaperonin GroEL to some complex mammalian chaperonins (i.e. TriC), the key role of GAPDH in glycolysis, and the presence of different forms of amyloid-like proteins (α-synuclein, prion protein, and β-amyloid peptide) in neural tissue allows suggests the involvement of the described mechanism into development and progression of amyloid neurodegenerative diseases.
Acknowledgements
This work was supported by the Russian Science Foundation (project No. 16-14-10027).
REFERENCES
1.Nussbaum-Krammer, C., Mogk, A., Nillegoda, N. B.,
Szlachcic, A., Guilbride, D. L., Saibil, H. R., Mayer, M. P., and
Bukau, B. (2015) Human Hsp70 disaggregase reverses
Parkinson’s-linked α-synuclein amyloid fibrils, Mol.
Cell, 59, 781-793.
2.Landreh, M., Rising, A., Presto, J., Jornvall, H.,
and Johansson, J. (2015) Specific chaperones and regulatory domains in
control of amyloid formation, J. Biol. Chem., 290,
26430-26436.
3.Sablon-Carrazana, M., Fernandez, I., Bencomo, A.,
Lara-Marinez, R., Rivera-Marrero, S., Dominguez, G., Perez-Perera, R.,
Jimenez-Garcia, L. F., Altamirano-Bustamante, N. F., Diaz-Delgado, M.,
Vedrenne, F., Rivillas-Acevedo, L., Pasten-Hidalgo, K., De Lourdes
Segura-Valdez, M., Islas-Andrade, S., Garrido-Magana, E.,
Perera-Pintado, A., Prats-Capote, A., Rodriguez-Tanty, C., and
Altamirano-Bustamante, M. M. (2015) Drug development in conformational
diseases: a novel family of chemical chaperones that bind and stabilise
several polymorphic amyloid structures, PLoS One, 10,
1-24.
4.Wisniewski, T., and Sadowski, M. (2008) Preventing
beta-amyloid fibrillization and deposition: beta-sheet breakers and
pathological chaperone inhibitors, BMC Neurosci.,
9 (Suppl. 2), S5.
5.Xi, D., Dong, X., Deng, W., and Lai, L. (2011)
Dynamic behavior of small heat shock protein inhibition on amyloid
fibrillization of a small peptide (SSTSAA) from RNase A, Biochem.
Biophys. Res. Commun., 416, 130-134.
6.Polyakova, O. V., Roitel, O., Asryants, R. A.,
Poliakov, A. A., Branlant, G., and Muronetz, V. I. (2005) Misfolded
forms of glyceraldehyde-3-phosphate dehydrogenase interact with GroEL
and inhibit chaperonin-assisted folding of the wild-type enzyme,
Protein Sci., 14, 921-928.
7.Naletova, I. N., Muronetz, V. I., and Schmalhausen,
E. V. (2006) Unfolded, oxidized, and thermoinactivated forms of
glyceraldehyde-3-phosphate dehydrogenase interact with the chaperonin
GroEL in different ways, Biochim. Biophys. Acta, 1764,
831-838.
8.Tillement, L., Lecanu, L., and Papadopoulos, V.
(2011) Alzheimer’s disease: effects of β-amyloid on
mitochondria, Mitochondrion, 11, 13-21.
9.Mamelak, M. (2012) Sporadic Alzheimer’s
disease: the starving brain, J. Alzheimer’s Dis.,
31, 459-474.
10.Yao, J., Rettberg, J., Klosinski, L., Cadenas,
E., and Brinton, R. (2011) Shift in brain metabolism in late onset
Alzheimer’s disease: implications for biomarkers and therapeutic
interventions, Mol. Asp. Med., 32, 247-257.
11.Rama Rao, K. V., and Norenberg, M. D. (2012)
Brain energy metabolism and mitochondrial dysfunction in acute and
chronic hepatic encephalopathy, Neurochem. Int., 60,
697-706.
12.Vlassenko, A. G., Vaishnavi, S. N., Couture, L.,
Sacco, D., Shannon, B. J., Mach, R. H., Morris, J. C., Raichle, M. E.,
and Mintun, M. A. (2010) Spatial correlation between brain aerobic
glycolysis and amyloid-β (Aβ) deposition, Proc. Natl.
Acad. Sci. USA, 107, 17763-17767.
13.Vaishnavi, S. N., Vlassenko, A. G., Rundle, M.
M., Snyder, A. Z., Mintun, M. A., and Raichle, M. E. (2010) Regional
aerobic glycolysis in the human brain, Proc. Natl. Acad. Sci.
USA, 107, 17757-17762.
14.Jayasena, T., Poljak, A., Braidy, N., Smythe, G.,
Raftery, M., Hill, M., Brodaty, H., Trollor, J., Kochan, N., and
Sachdev, P. (2015) Upregulation of glycolytic enzymes, mitochondrial
dysfunction and increased cytotoxicity in glial cells treated with
Alzheimer’s disease plasma, PLoS One, 10,
e0116092.
15.Rodacka, A. (2013) Properties and functional
diversity of glyceraldehyde-3-phosphate dehydrogenase, Postepy Hig.
Med. Doswiadczalnej, 67, 775-789.
16.Seidler, N. W. (2013) Basic biology of GAPDH,
Adv. Exp. Med. Biol., 985, 1-36.
17.Naletova, I. N., Popova, K. M., Eldarov, M. A.,
Kuravsky, M. L., Schmalhausen, E. V., Sevostyanova, I. A., and
Muronetz, V. I. (2011) Chaperonin TRiC assists the refolding of
sperm-specific glyceraldehyde-3-phosphate dehydrogenase, Arch.
Biochem. Biophys., 516, 75-83.
18.Bulatnikov, I. G., Polyakova, O. V., Asryants, R.
A., Nagradova, N. K., and Muronetz, V. I. (1999) Participation of
chaperonin GroEL in the folding of D-glyceraldehyde-3-phosphate
dehydrogenase. An approach based on the use of different oligomeric
forms of the enzyme immobilized on sepharose, J. Protein Chem.,
18, 79-87.
19.Li, X. L., Lei, X. D., Cai, H., Li, J., Yang, S.
L., Wang, C. C., and Tsou, C. L. (1998) Binding of a burst-phase
intermediate formed in the folding of denatured
D-glyceraldehyde-3-phosphate dehydrogenase by chaperonin 60 and
8-anilino-1-naphthalenesulphonic acid, Biochem. J., 331
(Pt. 2), 505-511.
20.Viet, M. H., Ngo, S. T., Lam, N. S., and Li, M.
S. (2011) Inhibition of aggregation of amyloid peptides by beta-sheet
breaker peptides and their binding affinity, J. Phys. Chem. B,
115, 7433-7446.
21.Markossian, K. A., Golub, N. V., Chebotareva, N.
A., Asryants, R. A., Naletov, I. N., Muronetz, V. I., Muranov, K. O.,
and Kurganov, B. I. (2010) Comparative analysis of the effects of
alpha-crystallin and GroEL on the kinetics of thermal aggregation of
rabbit muscle glyceraldehyde-3-phosphate dehydrogenase, Protein
J., 29, 11-25.
22.Kiselev, G. G., Naletova, I. N., Sheval, E. V.,
Stroylova, Y. Y., Schmalhausen, E. V., Haertle, T., and Muronetz, V. I.
(2011) Chaperonins induce an amyloid-like transformation of ovine prion
protein: the fundamental difference in action between eukaryotic TRiC
and bacterial GroEL, Biochim. Biophys. Acta, 1814,
1730-1738.
23.Ricci, C., Ortore, M. G., Vilasi, S., Carrotta,
R., Mangione, M. R., Bulone, D., Librizzi, R., Spinozzi, F., Burgio,
G., Amenitsch, H., and San Biagio, P. L. (2016) Stability and
disassembly properties of human naïve Hsp60 and bacterial GroEL
chaperonins, Biophys. Chem., 208, 68-75.
24.Scopes, R. K., and Stoter, A. (1982) Purification
of all glycolytic enzymes from one muscle extract, Methods
Enzymol., 90 (Pt. E), 479-490.
25.Rezaei, H., Marc, D., Choiset, Y., Takahashi, M.,
Hui Bon Hoa, G., Haertle, T., Grosclaude, J., and Debey, P. (2000) High
yield purification and physico-chemical properties of full-length
recombinant allelic variants of sheep prion protein linked to scrapie
susceptibility, Eur. J. Biochem., 267, 2833-2839.
26.Breydo, L., Makarava, N., and Baskakov, I. V.
(2008) Methods for conversion of prion protein into amyloid fibrils,
Methods Mol. Biol., 459, 105-115.
27.Ban, T., Hamada, D., Hasegawa, K., Naiki, H., and
Goto, Y. (2003) Direct observation of amyloid fibril growth monitored
by thioflavin T fluorescence, J. Biol. Chem., 278,
16462-16465.
28.Voropai, E. S., Samtsov, M. P., Kaplevskii, K.
N., Maskevich, A. A., Stepuro, V. I., Povarova, O. I., Kuznetsova, I.
M., Turoverov, K. K., Fink, A. L., and Uverskii, V. N. (2003) Spectral
properties of thioflavin T and its complexes with amyloid fibrils,
Appl. Spectrosc., 70, 868-874.