Knockdown of Minichromosome Maintenance Proteins Inhibits Foci Forming of Mediator of DNA-Damage Checkpoint 1 in Response to DNA Damage in Human Esophageal Squamous Cell Carcinoma TE-1 Cells
Jinzhong Yu1#, Ruijie Wang2#*, Jinfeng Wu3, Zhongqin Dang1, Qinsheng Zhang1, and Bo Li1
1Henan Province Hospital of TCM, Department of Gastroenterology, 450002 Zhengzhou, Henan, China2Henan University of Chinese Medicine, 450008 Zhengzhou, Henan, China; E-mail: wangruijie8954@126.com
3Shenzhen Luohu People’s Hospital, Department of Gastroenterology, Shenzhen, 518001 Guangdong, China
# These authors contributed equally to this work.
* To whom correspondence should be addressed.
Received May 11, 2016; Revision received July 1, 2016
Esophageal squamous cell carcinoma (ESCC) has a high morbidity in China and its treatment depends greatly on adjuvant chemotherapy. However, DNA damage repair in cancer cells severely affects the outcome of treatment. This study investigated the potential mechanism regarding mediator of DNA-damage checkpoint 1 (MDC1) and minichromosome maintenance proteins (MCMs) during DNA damage in ESCC. Recombinant vectors of MDC1 and MCMs with tags were constructed and transfected into human ESCC cell line TE-1. Immunoprecipitation and mass spectrometry were performed to screen the MCMs interacting with MDC1, and direct interaction was confirmed by glutathione S-transferase (GST) pull-down assay in vitro. MCM2 and MCM6 were knocked down by shRNAs, after which chromatin fraction and foci forming of MDC1 upon bleomycin-induced DNA damage were examined. The results showed that MCM2/3/5/6 were immunoprecipitated by antibodies against the tag of MDC1 in TE-1 nuclei, and the GST pull-down assay indicated the direct interaction. Knockdown of MCM2 or MCM6 reduced the chromatin fraction of MDC1 according to Western blot results. Moreover, knockdown of MCM2 or MCM6 could significantly inhibit foci forming of MDC1 in TE-1 nuclei in response to bleomycin-induced DNA damage (p < 0.001). This study indicates the direct interaction between MDC1 and MCMs in TE-1 nuclei. Downregulation of MCMs can inhibit chromatin fraction and foci forming of MDC1 in TE-1 cells upon DNA damage, which suggests MCMs and MDC1 as potential targets to improve the outcome of chemotherapy in ESCC.
KEY WORDS: esophageal squamous cell carcinoma (ESCC), DNA damage repair, mediator of DNA-damage checkpoint 1 (MDC1), minichromosome maintenance protein (MCM)DOI: 10.1134/S0006297916100205
Esophageal squamous cell carcinoma (ESCC) is one of the main types of esophageal carcinoma, arising from the epithelial cells that line the esophagus. It is common in developing countries and has a relatively high frequency in China [1], accounting for 90% of all esophageal carcinoma cases [2]. Risk factors for ESCC include tobacco, alcohol, hot drinks, and poor diets. The current general therapeutic methods rely on surgery, like endoscopic mucosal resection for early stages of ESCC, and require the assistance of chemotherapy with or without radiation therapy for some more severe cases [3]. Adjuvant chemotherapy with cisplatin, vindesine, and bleomycin has been applied on esophageal carcinoma patients [4], which exploits the effects of inducing DNA damage to accelerate cancer cell death [5].
Many proteins respond to DNA damage and participate in the DNA damage repair process, among which mediator of DNA damage checkpoint 1 (MDC1) plays important roles in early stages. Upon DNA damage, MDC1 not only itself is recruited to the double-strand break site, but also helps to recruit other proteins related to DNA damage repair [6]. It was reported that the downregulation of MDC1 accelerates DNA damage-induced cell apoptosis [7]. Minichromosome maintenance (MCM) complex, a heterohexamer composed of MCM2-7, is recruited to DNA replication sites after the anchor of origin recognition complex (ORC) [8], thus it is also required for DNA damage repair [9]. These DNA repair molecules respond to DNA damage and constitute a network to enable cells to survive.
Like normal cells, cancer cells exhibit ability for DNA damage repair and have developed their specific DNA damage repair mechanisms [10]. However, DNA damage repair in cancer cells severely affected the outcome of chemotherapy in various diseases including ESCC [11, 12]. Thus, it is imperative to find means for effective modulation of DNA damage repair to suppress the survival of ESCC cells during chemotherapy.
This study investigated the potential mechanism regarding MDC1 and MCMs during DNA damage in ESCC. Immunoprecipitation and mass spectrometry in human ESCC cell line TE-1 and glutathione S-transferase (GST) pull-down assay in vitro were performed to reveal the interaction between MDC1 and MCMs in cell nuclei. We further knocked down MCMs by shRNAs to analyze the involvement of MCMs in the modulation of MDC1 upon DNA damage. This study adds to understanding of the functions of MDC1 and MCMs and provides potential strategies to improve the outcome of chemoradiotherapy in ESCC.
MATERIALS AND METHODS
Cells. Human esophageal squamous carcinoma cell line TE-1 (Institute of Biochemistry and Cell Biology, China) was used in this study. The cells were cultured and passaged in Dulbecco’s modified Eagle Medium (DMEM; Gibco, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) and incubated in a humidified atmosphere with 5% CO2 at 37°C. This study was performed according to the instructions of our institute.
Plasmid construction. The cDNA sequences encoding the complete open reading frames of human MDC1 (BC152556), MCM2 (GenBank Accession No. BC017258), MCM3 (BC003509), MCM5 (BC003656), and MCM6 (BC032374) were amplified by polymerase chain reaction (PCR) and verified by sequencing. The HA and FLAG tag sequences were added to the 3′ end of MDC1 and MCMs by PCR. Then the fusion sequences were inserted into pcDNA3.1 overexpression vectors (Thermo Scientific, USA), and the resulting plasmids were named pcDNA3.1-MDC1-HA, pcDNA3.1-MCM2-FLAG, pcDNA3.1-MCM3-FLAG, pcDNA3.1-MCM5-FLAG, and pcDNA3.1-MCM6-FLAG. The plasmids were purified with Plasmid Purification Kit (Qiagen, China) according to the manufacturer’s instructions.
Cell transfection and treatment. The plasmids were transfected or co-transfected into TE-1 cells in serum-free DMEM using Lipofectamine 2000 (Invitrogen, USA) according to the supplier’s instructions. Before transfection, the cells were adjusted to confluency of about 80%. Plasmids of 1 µg were added to each well of 24-well plates. The cells were collected for further analysis at 48 h post-transfection. The TE-1 cells stably expressing HA-tagged human MDC1 were screened by neomycin (300 µg/ml).
Similar procedures were conducted on TE-1 cells for the transfection of the specific shRNAs for MCM2 and MCM6, sh-MCM2 and sh-MCM6 (Genechem, China), or the negative control (NC) for shRNAs. Bleomycin (2 µM) was added to the medium for a treatment of 1 h to induce DNA damage in TE-1 cells at 48 h post-transfection. Thus, the TE-1 cells were divided into four groups: NC, bleomycin ± NC, bleomycin ± sh-MCM2, and bleomycin ± sh-MCM6.
Chromatin fraction. Chromatin fraction was prepared according to a previous study [8]. Briefly, TE-1 cells stably expressing HA-tagged MDC1 were lysed in buffer containing 10 mM HEPES (pH 7.9), 10 mM KCl, 10 mM MgCl2, 0.34 mM sucrose, 10% glycerol, 0.2% Triton X-100, 1 mM dithiothreitol (DTT), and Protease Inhibitor Cocktail (Roche, Switzerland) for 10 min at 4°C. After centrifugation at 1000 rpm for 5 min, the supernatants were collected, which contained the cytoplasmic fraction. The pellet was resuspended in buffer containing 3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, and Protease Inhibitor Cocktail and lysed for 30 min at 4°C. After centrifugation at 1500 rpm for 5 min, the supernatant was mixed with the cytoplasmic fraction as the non-chromatin fraction.
The remaining pellet was resuspended in buffer containing 50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 1% Nonidet P-40, 1 mM DTT, and Protease Inhibitor Cocktail for 10 min at 4°C, after which it was sonicated and centrifuged at 13,000 rpm for 10 min, and the supernatants were collected as the chromatin fraction.
Immunoprecipitation. To analyze MDC1 and its potential interactive proteins, immunoprecipitation was carried out in nucleoprotein samples and the whole cell extracts (WCE) of TE-1 cells stably expressing HA-tagged MDC1. The nucleoprotein samples were extracted from the TE-1 cells using an EpiQuik Nuclear Extraction Kit (Epigentek, USA) according to the supplier’s instructions. Immunoprecipitation was performed with a Co-Immunoprecipitation Kit (Pierce, USA) according to the supplier’s instructions. Briefly, transfected TE-1 cells were washed with phosphate-buffered saline (PBS) and lysed in cold IP lysate buffer for 5 min, and cell debris was removed by centrifugation. Then the IP lysates were added to resin immobilized with anti-HA antibody (ab18181; Abcam, Great Britain) and incubated at 4°C overnight. Then the resin was washed in PBS three times, and the immunoprecipitates were eluted by elution buffer, after which they were analyzed by SDS-PAGE and Western blot.
Mass spectrometry. The proteins potentially interacting with MDC1 were isolated with anti-HA antibody by the above-mentioned procedures. Then the protein samples were denatured in 8 M urea, reduced in 10 mM tris-(2-carboxyethyl)-phosphine hydrochloride (Pierce), alkylated with 55 mM iodoacetamide (Sigma-Aldrich, China), and digested by trypsin (Gibco). The samples were desalted, eluted, and then chromatographed using an Agilent 1200 device (Agilent Technologies, USA). Mass spectrometry was performed on an LTQ mass spectrometer (Thermo Scientific). Data were collected by RawXtract 1.9.9 [13] and searched in the International Protein Index database [14].
GST pull-down assay. In vitro GST pull-down was performed to test the direct interaction between MDC1 and MCMs. GST-tagged MDC1 proteins and His-tagged MCMs were expressed respectively using recombinant vectors pGEX-6p-1 and pET-32a (Novagen, China) in BL21 (DE3) (Takara, China) and purified with the corresponding purification kits (Bio-Rad, USA). GST pull-down was performed with GST Protein Interaction Pull-Down Kit (Pierce) according to the supplier’s instruction, with GST protein as a negative control. The pull-down products were tested by Western blot.
Immunofluorescence. Foci forming of MDC1 in TE-1 nuclei were analyzed by immunofluorescence after the transfected cells were screened for the green fluorescence protein on shRNAs by flow cytometry. Briefly, the four groups of TE-1 cells, namely, NC, bleomycin ± NC, bleomycin ± sh-MCM2, and bleomycin ± sh-MCM6, were fixed with 4% paraformaldehyde for 10 min, permeabilized in 0.2% Triton X-100 (Solarbio, China) for 5 min, and incubated in the specific primary antibodies against MDC1 (ab50003; Abcam) diluted in 1% normal goat serum (Solarbio) for 2 h at room temperature. After washing in PBS, the cells were incubated in goat anti-mouse secondary antibodies (Alexa Fluor 594-conjugated, ab150116; Abcam) for 1 h at room temperature. 4′,6-Diamidino-2-phenylindole (DAPI; Sigma-Aldrich) was used to stain the nuclei. In each group, 20 fields were randomly selected. Fluorescence signals were observed under a fluorescence microscope (Bio-Rad) and analyzed with ImageJ 1.49 software (National Institutes of Health, Bethesda, MD, USA).
Western blot. TE-1 cell lysates prepared with Protein Extraction Reagent (Thermo Scientific), the chromatin fraction, immunoprecipitates, and GST pull-down products were analyzed by Western blot. The samples were separated by SDS-PAGE and transferred to polyvinylidene fluoride membranes. After blocking in normal goat serum for 2 h at room temperature, the membranes were incubated in the specific primary antibodies for FLAG tag (ab18230), His tag (ab184607), MCM2 (ab6153), MCM6 (ab167521), ORC2 (ab68348), or MDC1 (ab50003) overnight at 4°C. GAPDH (ab8245) and TATA box-binding protein (TBP, ab818) were used as internal controls. After washing in PBS, the membranes were incubated in goat anti-mouse secondary antibodies (horseradish peroxidase-conjugated, ab6789) for 1 h at room temperature. Positive signals were developed with 3,3′-diaminobenzidine Kit (Sangon Biotech, China) and analyzed with the ImageJ 1.49 software.
Statistical analysis. All experiments were performed in triplicate, and results are represented as the mean ± standard deviation. Comparison between groups was conducted by one-way analysis of variance and t-test with SPSS 20 (IBM, USA). p < 0.05 was considered as statistically significant differences.
RESULTS
MDC1 interacts with MCMs in TE-1 cells. After immunoprecipitation with the HA tag of MDC1 in the chromatin fraction of transfected TE-1 cells, mass spectrometry results showed that four MCMs were identified in immunoprecipitates of MDC1, which might be the MCMs interacting with MDC1 in TE-1 cell nuclei (table). However, not all the MCMs composing the MCM complex were found, thus MDC1 could at least interact with MCM2, MCM3, MCM5, and MCM6 in TE-1 cell nuclei.
Mass spectrometric data for MCMs in TE-1 nuclei immunoprecipitated with
antibody against the HA tags
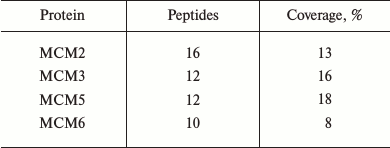
Note: MCM, minichromosome maintenance protein.
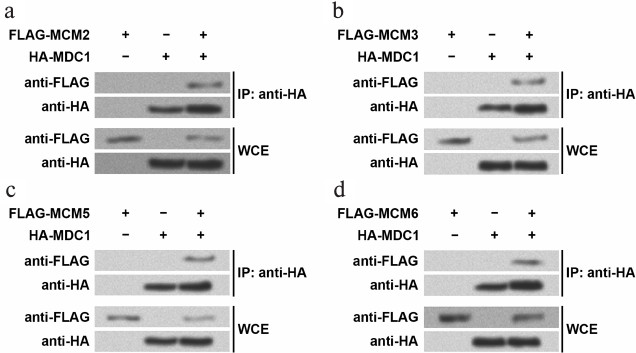
The mass spectrometry results were confirmed by Western blot following immunoprecipitation (Fig. 1). In the TE-1 cells overexpressing HA-tagged MDC1 and one of the four FLAG-tagged MCMs, the antibodies against HA and FLAG were used to detect the existence of these proteins in immunoprecipitates. For example, FLAG-tagged MCM2 or HA-tagged MDC1 could be detected in WCE of TE-1 cells overexpressing the two fusion proteins (Fig. 1a), indicating successful overexpression. In the nuclear immunoprecipitates with anti-HA, FLAG-tagged MCM2 and HA-tagged MDC1 could be found only in the cells overexpressing both fusion proteins, while FLAG-tagged MCM2 could not be detected in nuclei of cells only expressing one fusion protein. Similar results were also obtained when investigating MCM3, MCM5, or MCM6 (Fig. 1, b-d). Thus, this implied that MDC1 interacted with the four MCMs in TE-1 cell nuclei.
Fig. 1. Immunoprecipitates examined by Western blot. HA-tagged MDC1 and one of the four MCMs with FLAG tag were overexpressed in TE-1 cells, and then the chromatin fractions were immunoprecipitated with the antibody against HA tags. Western blot was performed with the antibodies against HA tags or FLAG tags to detect MDC1 or MCM2 (a), MCM3 (b), MCM5 (c), MCM6 (d). Whole cellular extract (WCE) was examined to confirm the overexpression of fusion proteins in TE-1 cells. MCM, minichromosome maintenance protein; MDC1, mediator of DNA-damage checkpoint 1; IP, immunoprecipitation.
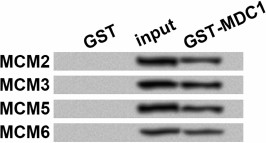
The possibility that MDC1 interacted with MCMs via association of other proteins could not be excluded just based on the immunoprecipitation results, thus we performed in vitro GST pull-down assay using fusion proteins from prokaryotic expression. The results showed that in input and the pull-down samples, GST-tagged MDC1 could pull down MCM2/3/5/6, which were detected with the antibody against their His tags (Fig. 2), while none of these MCMs could be detected in GST negative control. Thus, MDC1 might directly interact with the four MCMs in TE-1 cells.
Fig. 2. Western blot results for glutathione S-transferase (GST) pull-down assay. GST pull-down was performed to evaluate the direct interaction in vitro between MDC1 (GST tag) and MCM2, MCM3, MCM5, or MCM6 (His tag), all from prokaryotic expression. Western blot was performed with the antibody against His tags. Input samples were used as positive controls. GST protein was used as a negative control. MCM, minichromosome maintenance protein; MDC1, mediator of DNA-damage checkpoint 1.
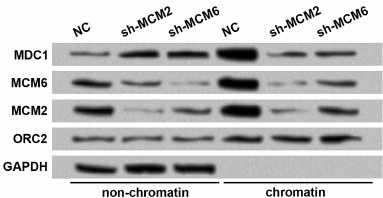
Knockdown of MCMs reduces chromatin fraction and foci forming of MDC1 in TE-1 cells in response to DNA damage. Since MDC1 plays roles in DNA damage repair, and it could interact with MCMs based on the aforementioned results, we wondered whether MCMs were involved in the function of MDC1. First, MCM2 and MCM6 were downregulated to evaluate whether MCM levels could affect the chromatin fraction of MDC1. Western blot showed that transfection of shRNAs successfully induced the knockdown of MCM2 or MCM6 in both non-chromatin and chromatin fractions of TE-1 cells (Fig. 3). With the downregulation of MCM2 or MCM6, the non-chromatin level of MDC1 increased, and the chromatin level of MDC1 decreased, implying that inhibiting MCMs affected the chromatin fraction of MDC1 in TE-1 cells.
Fig. 3. Protein levels of MDC1 in non-chromatin and chromatin fractions of TE-1 cells with MCM2 or MCM6 knockdown. TE-1 cells were transfected with shRNAs for MCM2 or MCM6 (sh-MCM2 or sh-MCM6). NC, negative control for shRNAs. Origin recognition complex 2 (ORC2) and GAPDH were used as controls. MCM, minichromosome maintenance protein; MDC1, mediator of DNA-damage checkpoint 1.
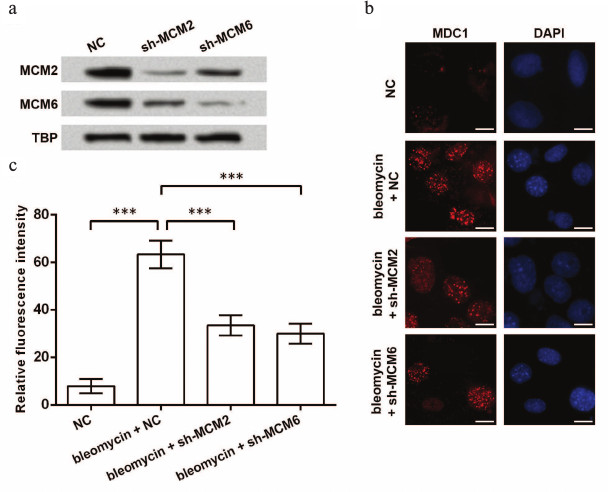
Next, foci forming of MDC1 under the situation of bleomycin-induced DNA damage was investigated. Effective knockdown of MCM2 or MCM6 was examined before bleomycin treatment (Fig. 4a). After the induction of DNA damage, foci forming of MDC1 in TE-1 cell nuclei was detected by immunofluorescence, and the results showed that bleomycin increased foci forming of MDC1, which was then inhibited by knockdown of MCM2 or MCM6 (Fig. 4b). Significant differences in the fluorescence intensity were found between groups (p < 0.001, Fig. 4c). Thus, knockdown of MCMs might also reduce foci forming of MDC1 in TE-1 cells in response to DNA damage.
Fig. 4. Foci forming of MDC1 impacted by knockdown of MCMs. a) Western blot indicating the inhibition of MCM2 and MCM6 in TE-1 nuclei by shRNAs. TATA box-binding protein (TBP) was used as an internal control. b) Immunofluorescence of MDC1 showing foci forming of MDC1 in TE-1 nuclei. TE-1 cells were transfected with shRNAs for MCM2 or MCM6 (sh-MCM2 or sh-MCM6) and then treated with bleomycin (2 µM) for 1 h to induce DNA damage. Bar indicates 10 µm. c) Relative fluorescence intensity in immunofluorescence experiments; *** p < 0.001; NC, negative control for shRNAs; MCM, minichromosome maintenance protein; MDC1, mediator of DNA-damage checkpoint 1.
DISCUSSION
MDC1 and MCMs are key proteins involved in DNA replication and DNA damage repair. In this study, we found that MDC1 could interact with MCM2/3/5/6 in the nuclei of human ESCC cell line TE-1. Knockdown of MCMs suppressed the chromatin fraction of MDC1 and reduced foci forming of MDC1 in TE-1 cells in response to bleomycin-induced DNA damage.
Immunoprecipitation was performed in the nuclear protein of TE-1 cells followed by mass spectrometry to find the proteins interacting with MDC1. MCM2/3/5/6 were found in the immunoprecipitates, and their direct interaction with MDC1 was further confirmed by GST pull-down assay in vitro, through which the bridging of other proteins between MDC1 and MCMs was basically excluded, given that there was no adsorption generated from the effects of electric charges. Thus, it can be assumed that MDC1 directly interacts with MCM2/3/5/6 in TE-1 cell nuclei.
It was noticed that MCM2/3/5/6 were found in the immunoprecipitates of MDC1, while MCM4 and MCM7, the other two proteins composing the MCM heterohexamer, possessed very low sequence coverages in the mass spectrometry results. The heterohexamer is essential for the MCM proteins to execute helicase activities in vivo [15], and the ring structure of MCM complex splits duplex DNAs pumped into the central channel [16], which was shown in a recent study to be a kinked one formed by MCM2-7 double hexamer [17]. Since in each of the hexamers, MCM2/3/5/6 units are arranged adjacent to each other, it is possible that MDC1 interacts with MCM complex via the direct interaction with at least MCM2/3/5/6. Thus, it can be further speculated that MDC1 directly interacts with MCM complex in TE-1 cell nuclei.
When MCM2 or MCM6 was knocked down in TE-1 cells, we found that transfecting sh-MCM2 or sh-MCM6 alone could suppress the protein levels of both MCM2 and MCM6, which might suggest the inhibition of the entire MCM complex, according to similar results in a previous study [9]. The expression of ORC2 was barely altered, and it had higher levels in the chromatin fraction than in the non-chromatin fraction generally. ORC2 is a subunit composing the heterohexameric ORC1-6, whose winged-helix domain face engages MCM complex indirectly during replicative helicase loading [18]. The dephosphorylation of ORC2 promotes chromatin loading of ORC [19]. Based on the prima facie results of this study, knockdown of any MCM protein may suppress the level of MCM complex and its directly interactive protein MDC1, but barely affect proteins or complexes of indirect interaction. The underlying detailed mechanism needs to be determined in future research.
Knockdown of MCM2 or MCM6 increased the non-chromatin fraction and decreased the chromatin fraction of MDC1 in TE-1 cells, which might reflect suppressed function of MDC1 by the inhibition of MCM complex. Similar results have been found in H2A histone family member X (H2AX), whose phosphorylation in response to DNA damage is a signal for the recruitment of DNA-damage-response proteins to the damaged regions [20]. MDC1 directly binds to phosphorylated H2AX [21], and knockdown of H2AX inhibits the chromatin fraction of MDC1 [22]. Thus, it is inferred from this study that the inhibition of the entire MCM complex may suppress the chromatin fraction of MDC1, possibly due to reduced DNA replication and reduced requirements for DNA-damage-response proteins including MDC1 in TE-1 cell nuclei.
It should be clarified that MCMs are significant cell cycle modulators whose depletion triggers G1/S arrest, as found in Drosophila S2 cells and human non-small cell lung carcinomas [23, 24]. Besides, an early study found that the effect of bleomycin was weaker in S-phase cells than in G2/M-phase cells, possibly due to the varied cellular uptake of bleomycin [25]. When knocking down MCMs in TE-1 cells, the DNA damage repair process is hindered, and it is also possible that MCM knockdown leads to cell cycle arrest and reduce bleomycin-sensitive cells. Moreover, according to the treatment method in this study, knockdown of MCMs generates more profound effects than bleomycin treatment. An aggregated influence of bleomycin effectiveness, DNA damage repair, and DNA replication changes is to be assessed in future research.
Existing reports have shown the downregulation of MDC1 has multiple benefits in controlling tumor cells. For example, knockdown of MDC1 generates defective radiation-induced apoptosis in human lung adenocarcinoma cells [26], and it enhances radiosensitivity of nasopharyngeal cancer cells [27] and ESCC cells [28]. Its downregulation induces cell cycle arrest, high apoptotic rate, and increased sensitivity to adriamycin in cervical cancer cells [29]. In addition, MCMs have been revealed as therapeutic targets for a variety of tumors such as non-small cell lung cancer [30] and leukemia [31] as well as ESCC [32, 33]. In this study, MDC1 foci forming was induced by bleomycin treatment in TE-1 cells, which was consistent with its functions during DNA damage [34]. Its inhibited chromatin fraction and foci forming by suppressing MCM complex imply the possibility of modulating DNA replication or damage repair in ESCC cells via the suppression of MDC1 and its regulators such as MCMs, thus improving the effects of chemotherapy. Further research is necessary to verify the efficient regulation of MDC1 and MCMs in multiple ESCC cells.
In summary, this study suggests that MDC1 directly interacts with MCM2/3/5/6, and that knockdown of MCMs reduces the chromatin fraction and foci forming of MDC1 in response to DNA damage in ESCC cell line TE-1. These results reveal MDC1 and MCMs as potential targets to improve the outcome of chemotherapy in ESCC.
Acknowledgements
This work was supported by the Key Project of Science and Technology of Henan Education Ministry (13A360579), by the Doctoral Foundation of Henan University of Chinese Medicine (BSJJ2012-21), and by the Hospital-grade Research Project of Henan Province Hospital of TCM (2015).
REFERENCES
1.Wu, C., Hu, Z., He, Z., Jia, W., Wang, F., Zhou,
Y., Liu, Z., Zhan, Q., Liu, Y., Yu, D., Zhai, K., Chang, J., Qiao, Y.,
Jin, G., Liu, Z., Shen, Y., Guo, C., Fu, J., Miao, X., Tan, W., Shen,
H., Ke, Y., Zeng, Y., Wu, T., and Lin, D. (2011) Genome-wide
association study identifies three new susceptibility loci for
esophageal squamous-cell carcinoma in Chinese populations, Nat.
Genet., 43, 679-684.
2.Torre, L. A., Bray, F., Siegel, R. L., Ferlay, J.,
Lortet-Tieulent, J., and Jemal, A. (2015) Global cancer statistics,
2012, Cancer J. Clin., 65, 87-108.
3.Van Heijl, M., Van Lanschot, J. J., Koppert, L. B.,
Van Berge Henegouwen, M. I., Muller, K., Steyerberg, E. W., Van Dekken,
H., Wijnhoven, B. P., Tilanus, H. W., Richel, D. J., Busch, O. R.,
Bartelsman, J. F., Koning, C. C., Offerhaus, G. J., and Van der Gaast,
A. (2008) Neoadjuvant chemoradiation followed by surgery versus surgery
alone for patients with adenocarcinoma or squamous cell carcinoma of
the esophagus (CROSS), BMC Surg., 8, 21.
4.Roth, J. A., Pass, H. I., Flanagan, M. M., Graeber,
G. M., Rosenberg, J. C., and Steinberg, S. (1988) Randomized clinical
trial of preoperative and postoperative adjuvant chemotherapy with
cisplatin, vindesine, and bleomycin for carcinoma of the esophagus,
J. Thorac. Cardiovasc. Surg., 96, 242-248.
5.Liu, M., Hales, B. F., and Robaire, B. (2014)
Effects of four chemotherapeutic agents, bleomycin, etoposide,
cisplatin, and cyclophosphamide, on DNA damage and telomeres in a mouse
spermatogonial cell line, Biol. Reprod., 90, 72.
6.Coster, G., and Goldberg, M. (2010) The cellular
response to DNA damage: a focus on MDC1 and its interacting proteins,
Nucleus, 1, 166-178.
7.Solier, S., and Pommier, Y. (2011) MDC1 cleavage by
caspase-3: a novel mechanism for inactivating the DNA damage response
during apoptosis, Cancer Res., 71, 906-913.
8.Mendez, J., and Stillman, B. (2000) Chromatin
association of human origin recognition complex, Cdc6, and
minichromosome maintenance proteins during the cell cycle: assembly of
prereplication complexes in late mitosis, Mol. Cell. Biol.,
20, 8602-8612.
9.Han, X., Aslanian, A., Fu, K., Tsuji, T., and
Zhang, Y. (2014) The interaction between checkpoint kinase 1 (Chk1) and
the minichromosome maintenance (MCM) complex is required for DNA
damage-induced Chk1 phosphorylation, J. Biol. Chem., 289,
24716-24723.
10.Costantino, L., Sotiriou, S. K., Rantala, J. K.,
Magin, S., Mladenov, E., Helleday, T., Haber, J. E., Iliakis, G.,
Kallioniemi, O. P., and Halazonetis, T. D. (2014) Break-induced
replication repair of damaged forks induces genomic duplications in
human cells, Science, 343, 88-91.
11.Okumura, H., Natsugoe, S., Matsumoto, M., Mataki,
Y., Takatori, H., Ishigami, S., Takao, S., and Aikou, T. (2005) The
predictive value of p53, p53R2, and p21 for the effect of
chemoradiation therapy on oesophageal squamous cell carcinoma, Br.
J. Cancer, 92, 284-289.
12.Okumura, H., Uchikado, Y., Setoyama, T.,
Matsumoto, M., Owaki, T., Ishigami, S., and Natsugoe, S. (2014)
Biomarkers for predicting the response of esophageal squamous cell
carcinoma to neoadjuvant chemoradiation therapy, Surg. Today,
44, 421-428.
13.McDonald, W. H., Tabb, D. L., Sadygov, R. G.,
MacCoss, M. J., Venable, J., Graumann, J., Johnson, J. R., Cociorva,
D., and Yates, J. R. (2004) MS1, MS2, and SQT-three unified, compact,
and easily parsed file formats for the storage of shotgun proteomic
spectra and identifications, Rapid Commun. Mass Spectrom.,
18, 2162-2168.
14.Peng, J., Elias, J. E., Thoreen, C. C.,
Licklider, L. J., and Gygi, S. P. (2003) Evaluation of multidimensional
chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for
large-scale protein analysis: the yeast proteome, J. Proteome
Res., 2, 43-50.
15.Schwacha, A., and Bell, S. P. (2001) Interactions
between two catalytically distinct MCM subgroups are essential for
coordinated ATP hydrolysis and DNA replication, Mol. Cell,
8, 1093-1104.
16.Takahashi, T. S., Wigley, D. B., and Walter, J.
C. (2005) Pumps, paradoxes and ploughshares: mechanism of the MCM2-7
DNA helicase, Trends Biochem. Sci., 30, 437-444.
17.Li, N., Zhai, Y., Zhang, Y., Li, W., Yang, M.,
Lei, J., Tye, B. K., and Gao, N. (2015) Structure of the eukaryotic MCM
complex at 3.8 Å, Nature, 524, 186-191.
18.Bleichert, F., Botchan, M. R., and Berger, J. M.
(2015) Crystal structure of the eukaryotic origin recognition complex,
Nature, 519, 321-326.
19.Lee, K. Y., Bae, J. S., Yoon, S., and Hwang, D.
S. (2014) Dephosphorylation of Orc2 by protein phosphatase 1 promotes
the binding of the origin recognition complex to chromatin, Biochem.
Biophys. Res. Commun., 448, 385-389.
20.Celeste, A., Difilippantonio, S.,
Difilippantonio, M. J., Fernandez-Capetillo, O., Pilch, D. R.,
Sedelnikova, O. A., Eckhaus, M., Ried, T., Bonner, W. M., and
Nussenzweig, A. (2003) H2AX haploinsufficiency modifies genomic
stability and tumor susceptibility, Cell, 114,
371-383.
21.Stucki, M., Clapperton, J. A., Mohammad, D.,
Yaffe, M. B., Smerdon, S. J., and Jackson, S. P. (2005) MDC1 directly
binds phosphorylated histone H2AX to regulate cellular responses to DNA
double-strand breaks, Cell, 123, 1213-1226.
22.Shi, H., Zhu, S., Liu, Z., and Su, J. (2013) The
impact to MDC1 and 53BP1 after silence H2AX in esophageal carcinoma
ECA109, Basic Clin. Med., 33, 808-813.
23.Crevel, G., Hashimoto, R., Vass, S., Sherkow, J.,
Yamaquchi, M., Heck, M. M. S., and Cotterill, S. (2007) Differential
requirements for MCM proteins in DNA replication in Drosophila
S2 cells, PLoS One, 2, e833.
24.Zhang, X., Teng, Y., Yang, F., Wang, M., Hong,
X., Ye, L. G., Gao, Y. N., and Chen, G. Y. (2015) MCM2 is a therapeutic
target of lovastatin in human non-small cell lung carcinomas, Oncol.
Rep., 33, 2599-2605.
25.Olive, P. L., and Banath, J. P. (1993) Detection
of DNA double-strand breaks through the cell cycle after exposure to
X-rays, bleomycin, etoposide and 125IdUrd, Int. J. Radiat.
Biol., 64, 349-358.
26.Lou, Z., and Chen, J. (2004) Use of siRNA to
study the function of MDC1 in DNA damage responses, Methods Mol.
Biol., 281, 179-187.
27.Wang, Z., Zeng, Q., Chen, T., Liao, K., Bu, Y.,
Hong, S., and Hu, G. (2015) Silencing NFBD1/MDC1 enhances the
radiosensitivity of human nasopharyngeal cancer CNE1 cells and results
in tumor growth inhibition, Cell Death Dis., 6,
e1849.
28.Liu, Z. K., Zhu, S. C., Su, J. W., Wang, Y. X.,
Yang, J. J., Li, J., and Shen, W. B. (2010) Short hairpin RNA-mediated
MDC1 gene silencing enhances the radiosensitivity of esophageal
squamous cell carcinoma cell line ECA109, J. South Med. Univ.,
30, 1830-1834.
29.Yuan, C., Bu, Y., Wang, C., Yi, F., Yang, Z.,
Huang, X., Cheng, L., Liu, G., Wang, Y., and Song, F. (2012) NFBD1/MDC1
is a protein of oncogenic potential in human cervical cancer, Mol.
Cell. Biochem., 359, 333-346.
30.Toyokawa, G., Masuda, K., Daigo, Y., Cho, H. S.,
Yoshimatsu, M., Takawa, M., Hayami, S., Maejima, K., Chino, M., Field,
H. I., Neal, D. E., Tsuchiya, E., Ponder, B. A., Maehara, Y., Nakamura,
Y., and Hamamoto, R. (2011) Minichromosome Maintenance Protein 7 is a
potential therapeutic target in human cancer and a novel prognostic
marker of non-small cell lung cancer, Mol. Cancer, 10,
65.
31.Ha, S. A., Shin, S. M., Namkoong, H., Lee, H.,
Chio, G. W., Hur, S. Y., Kim, T. E., and Kim, J. W. (2004)
Cancer-associated expression of minichromosome maintenance 3 gene in
several human cancers and its involvement in tumorigenesis, Clin.
Cancer Res., 10, 8386-8295.
32.Kato, H., Miyazaki, T., Fukai, Y., Nakajima, M.,
Sohda, M., Takita, J., Masuda, N., Fukuchi, M., Manda, R., Ojima, H.,
Tsukada, K., Asao, T., and Kuwano, H. (2003) A new proliferation
marker, minichromosome maintenance protein 2, is associated with tumor
aggressiveness in esophageal squamous cell carcinoma, J. Surg.
Oncol., 84, 24-30.
33.Huang, B., Hu, B., Su, M., Tian, D., Guo, Y.,
Lian, S., Liu, Z., Wu, X., Li, Q., Zheng, R., and Gao, Y. (2011)
Potential role of minichromosome maintenance protein 2 as a screening
biomarker in esophageal cancer high-risk population in China, Hum.
Pathol., 42, 808-816.
34.Goldberg, M., Stucki, M., Falck, J.,
D’Amours, D., Rahman, D., Pappin, D., Bartek, J., and Jackson, S.
P. (2003) MDC1 is required for the intra-S-phase DNA damage checkpoint,
Nature, 421, 952-956.