Microbiota Induces Expression of Tumor Necrosis Factor in Postnatal Mouse Skin
D. V. Yuzhakova1,2, M. V. Shirmanova1, A. A. Bocharov3, I. V. Astrakhantseva2, E. A. Vasilenko2, E. N. Gorshkova2, M. S. Drutskaya3, E. V. Zagaynova1, S. A. Nedospasov2,3,4,5, and A. A. Kruglov2,4,5*
1Nizhny Novgorod State Medical Academy, 603005 Nizhny Novgorod, Russia2Lobachevsky State University of Nizhny Novgorod, 603950 Nizhny Novgorod, Russia; E-mail: andrey_krugloff@mail.ru
3Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, 119991 Moscow, Russia
4Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119991 Moscow, Russia
5Lomonosov Moscow State University, Faculty of Biology, 119991 Moscow, Russia
* To whom correspondence should be addressed.
Received July 1, 2016; Revision received August 9, 2016
Tumor necrosis factor (TNF) is a pleiotropic cytokine that regulates many important processes in the body. TNF production in a physiological state supports the structure of lymphoid organs and determines the development of lymphoid cells in hematopoiesis. However, chronic TNF overexpression leads to the development of various autoimmune disorders. Sites of TNF production in the naïve state remain unclear due to the lack of in vivo models. In the present study, we used TNF-2A-Kat reporter mice to monitor the expression of TNF in different tissues. Comparative analysis of tissue fluorescence in TNF-2A-Kat reporter mice and wild type mice revealed constitutive expression of TNF in the skin of naïve adult mice. In the skin of TNF-2A-Kat reporter mouse embryos, no statistically significant differences in the expression of TNF compared to wild type animals were observed. Furthermore, we established that local depletion of microflora with topical antibiotics leads to a reduction in the fluorescence signal. Thus, we assume that the skin microflora is responsible for the expression of TNF in the skin of mice.
KEY WORDS: tumor necrosis factor (TNF), TNF-2A-Kat reporter mice, in vivo, fluorescent imagingDOI: 10.1134/S0006297916110080
Abbreviations: AMP, antimicrobial peptides; IFN, interferon; IL, interleukin; TNF, tumor necrosis factor.
Skin is not only a barrier tissue, but it is also a complex ecosystem
colonized by various microorganisms including bacteria, fungi, and
viruses [1]. The skin plays an important role in
the maintenance of immune homeostasis provided by Langerhans’
cells, T- and B-lymphocytes, granulocytes, keratinocytes, and mast
cells [1]. Symbiotic microorganisms of the skin
participate in the “tuning” of the immune system,
particularly in educating T lymphocytes to respond to pathogens bearing
similar antigens [2]. During infection or injury,
the immune response in the skin assures the development of inflammatory
reaction and can itself modulate the composition and the quantity of
commensal microflora colonizing the skin [3].
Tumor necrosis factor (TNF) belongs to the TNF superfamily of cytokines. TNF exerts its functions in the form of a homotrimer that is recognized by TNF-R1 and TNF-R2 receptors and induces signals important for the development of inflammation and immune response to pathogens. However, overexpression of TNF can lead to the development of different immunopathologies. TNF plays a role as the main molecular mediator of septic shock [4]. Moreover, local overexpression of TNF is pathologic and leads to the development of a large number of autoimmune diseases such as rheumatoid arthritis, ankylosing spondylitis, psoriasis, etc. [5]. Thus, TNF demonstrates both protective and pathogenic properties, which makes studies of its expression in various tissues especially important for therapy. Some of the factors inducing the expression of TNF are the ligands produced by microorganisms, such as lipopolysaccharide, peptidoglycan, lipoteichoic acid, and flagellin. Thus, one possible mechanism of TNF induction in barrier tissues, such as skin or intestine, is the surface colonization by commensal microflora.
Progress in molecular analysis methods has revealed and characterized a significantly broader species diversity of skin microbiota than traditional microbiological culture methods [6]. Thus, 16S rDNA analysis of the skin microbiota has shown that the majority of bacteria on the skin belong to four phyla: Actinobacteria, Firmicutes, Bacteroidetes, and Proteobacteria [7]. Interestingly, these four dominating phyla of bacteria constitute also the core of gastrointestinal mucosal microbiota, but their ratio differs considerably: while Actinobacteria are more widespread on skin, Firmicutes and Bacteroidetes are more represented in the digestive tract [7]. A common feature of microbial communities of the digestive tract and the skin is the low level of variety at the level of phyla, but rather great variety at the level of species [8-13]. Molecular analysis of the bacterial diversity demonstrates that the skin microflora profile depends on localization [14-16].
Microbiota colonizes the skin surface and interacts with the host organism through pattern-recognizing receptors, such as Toll-like receptors (TLRs), mannose receptors, and NOD-like receptors. These receptors recognize pathogen-associated molecular patterns, such as lipopolysaccharides of Gram-negative bacteria, mannan, and zymosan from the cell wall of fungi, peptidoglycan and lipoteichoic acid from Gram-positive bacteria, and flagellin. Activation of these receptors on different types of cells triggers the innate immune response, which leads to the secretion of antimicrobial peptides (AMP), cytokines, including TNF, and chemokines [17]. Thus, there is a constant interaction between skin cells of the host organism, immune system cells, and microorganisms that is regulated by AMP, cytokines, chemokines, and other factors [2, 18].
Live imaging techniques are now widely used for studying of functions of the immune system of healthy organisms and in case of pathology. One of the main advantages of in vivo imaging is the possibility to obtain images of individual cells noninvasively under conditions close to physiological. This approach is being intensively developed and is successfully used in pharmacological and medico-biological studies of animal models of human diseases, in particular, to study various pathologies of the immune system [18]. In vivo imaging can be used to analyze different cells of the immune system, such as CD4+ and CD8+ effector T-lymphocytes, dendritic cells, regulatory T-lymphocytes, natural killers (NK), and NK T-cells [19]. Moreover, use of various reporter systems allow analysis of such molecular targets as cytokines, including TNF, interferons (IFN), various interleukins, such as IL-2, IL-4, IL-7, IL-10, IL-12, IL-13, and IL-17, transforming growth factor (TGF-β), and proteins responsible for maturation, development, and functioning of immune system cells, for example, FoxP3, a transcription factor responsible for the development and function of regulatory T-lymphocytes [18-20].
Fluorescent imaging is a powerful tool for studying the dynamics of the immune system using in vivo labeling of cells and molecules [21, 22]. With this approach fluorescent proteins, GFP and its homologs, can be used for visualization [23, 24] (studied cells are transfected with a gene coding for fluorescent protein), or exogenously administrated fluorescent dyes (cells are labeled in vitro with a vital dye such as CFSE, SNARF1, or CMTMR and re-introduced into the organism [25-27]). A radically new approach for in vivo studies using fluorescent tags consists in generation of genetically modified strains of “reporter” animals, in which target molecules are either labeled endogenously, or their expression occurs simultaneously with a reporter protein expression from the same genetic construct. Various proteins with distinct emission spectra can be used as tags – green proteins (GFP, EGFP), red proteins (RFP, Katushka), yellow proteins (YFP, eYFP), and luciferase (LacZ, Luc) coexpressed with target molecules such as IL-2, IL-4, IL-7, IL-10, IL-12p40, IL-13, IL-17, IFN-β1, and IFN-γ, TNF, TGF-β1, transcription factors FoxP3, Blimp1 (inducing maturation of lymphocytes), and GATA3 [20, 22]. As an example of such reporter strains, we can mention a mouse strain IL4-YFP in which CD4+ and CD8+ T-lymphocytes coexpress IL-4 and YFP [28], a mouse strain Yeti, in which IFN-γ is coexpressed with eYFP [29], and Tnfα-luc strain, in which the expression of TNF is monitored using LacZ [30].
The present work has been performed using the TNF-2A-Kat reporter mouse strain. In the immune system cells of these animals, TNF expression is associated with the expression of the red fluorescent protein Katushka (TurboFP635). This model can reveal the expression of TNF using fluorescent imaging and can be used for detailed studies of TNF expression and physiological functions under normal conditions, as well as in cases of TNF-associated pathologies. In this work, we analyzed in vivo the levels of expression of proinflammatory cytokine TNF in the skin of healthy mice without visible signs of inflammation.
MATERIALS AND METHODS
Laboratory animals. In this work, we used wild type mice of the C57Bl/6 strain and transgenic TNF-2A-Kat reporter mouse strain, generated based on the C57Bl/6 genetic background, bearing the coding sequence of red fluorescent protein Katushka under the control of regulatory sequences of the TNF gene [31].
Fluorescent imaging. In vivo and ex vivo fluorescent imaging was done using an IVIS-Spectrum system (Caliper Life Sciences, USA) in epiluminescence registration mode. Fluorescence was excited at 605/615 nm, and fluorescence signal was acquired at 660/610 nm. For in vivo imaging, animals were first shaved, treated further with depilatory cream, and narcotized with 2.5% isoflurane. For quantitative analysis, average fluorescence intensity of an area of interest was defined using the LivingImage software (Caliper Life Sciences).
Microflora depletion. Local depletion of skin microflora was done with Neosporin®, which is a mix of bacitracin zinc, neomycin sulfate, and polymyxin B sulfate. Neosporin was applied during 9 days at 20 mg/cm2 on the depilated area of 2.5 cm2 at the left shoulder of TNF-2A-Kat reporter mice and wild type mice once in 24 h. To analyze the dynamics of TNF expression in TNF-2A-Kat reporter mice during the depletion, in vivo fluorescent images of animals before the first application of the drug and 24 h after each application during 9 days were taken.
Statistical analysis. Statistically significant differences of values were determined using the StatSoft STATISTICA 10 software using the Mann–Whitney U-test. Differences were considered statistically significant if p ≤ 0.05.
RESULTS
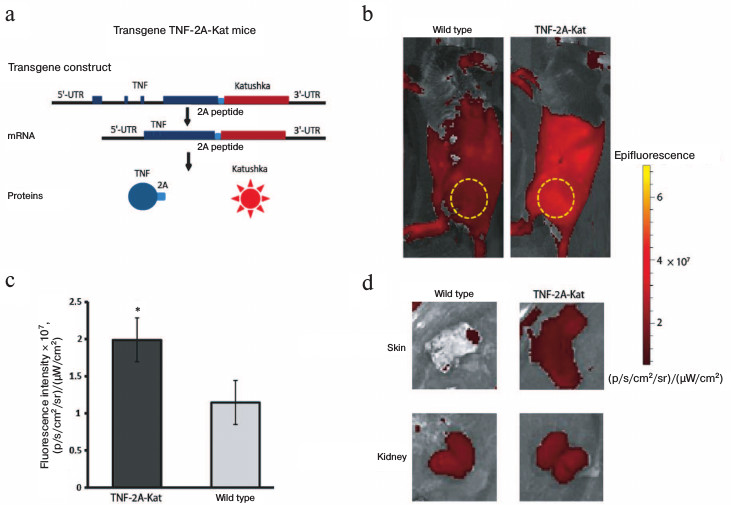
Constitutive expression of TNF in the skin of adult mice. First, we analyzed the florescence of different organs in TNF-2A-Kat mice and in wild type mice to determine the sites of TNF production in naïve mice (Fig. 1a). For this purpose, we used vital visualization of animals and analyzed the fluorescence of different organs (Fig. 1). Comparison of different skin areas of reporter and control mice revealed statistically significant increase in fluorescence signal in reporter mice (Fig. 1, b-d). It should be mentioned that the most intensive fluorescent signal in certain areas results from the autofluorescence of internal organs, since the analysis of isolated skin samples ex vivo confirmed the increased fluorescence in this organ (Fig. 1d). These data show that the TNF gene is constitutively expressed in skin cells in the absence of any visible signs of inflammation.
Fig. 1. Increased fluorescence intensity in the skin of adult TNF-2A-Kat mice in vivo. a) Schematic representation of TNF-2A-Kat transgenic mice allowing visualization of TNF production in vivo. b) In vivo fluorescence images of TNF-2A-Kat reporter mouse and a wild type mouse. The yellow circle designates the area of fluorescent signal used for quantification. c) Fluorescence signal analysis of surface tissues from TNF-2A-Kat reporter mice (n = 7) and from wild type mice (n = 6). Average value ± standard deviation. * Statistically significant difference between groups “TNF-2A-Kat” and “wild type”, p = 0.0034. d) In vivo fluorescent images of the skin fragment and kidneys of a TNF-2A-Kat reporter mouse and a wild type mouse.
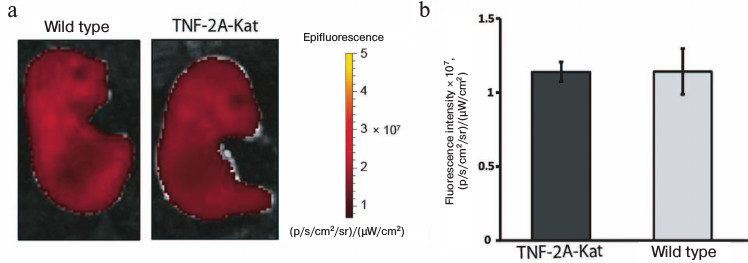
TNF expression in the skin of late embryos is not increased. The increased Katushka expression in adult mice can be caused either by the constitutive expression of TNF throughout the lifetime of an animal, or by response to external stimuli, such as colonization of the skin by microbiota. Since external stimuli starts acting on the skin after birth, we analyzed late embryos of wild type and TNF-2A-Kat mice. To this end, embryos were isolated at the 17th day of pregnancy, and their fluorescence images were taken (Fig. 2). The fluorescence intensity of the skin fragment of TNF-2A-Kat reporter embryos was on average (1.139 ± 0.067)·107 (p/s/cm2/sr)/(µW/cm2), while for wild type embryos the average value was (1.141 ± 0.154)·107 (p/s/cm2/sr)/(µW/cm2). Thus, the fluorescence of superficial tissues of TNF-2A-Kat reporter embryos was similar to that of wild type embryos, which demonstrates that contact with the environment might be necessary for TNF induction.
Fig. 2. In vivo skin fluorescence of late embryos of TNF-2A-Kat mice. a) In vivo fluorescent images of TNF-2A-Kat reporter embryo and a wild type embryo at 17th day of embryogenesis. b) Quantification of tissue fluorescence signal of TNF-2A-Kat reporter embryos (n = 5) and wild type embryos (n = 13). Average value ± standard deviation.
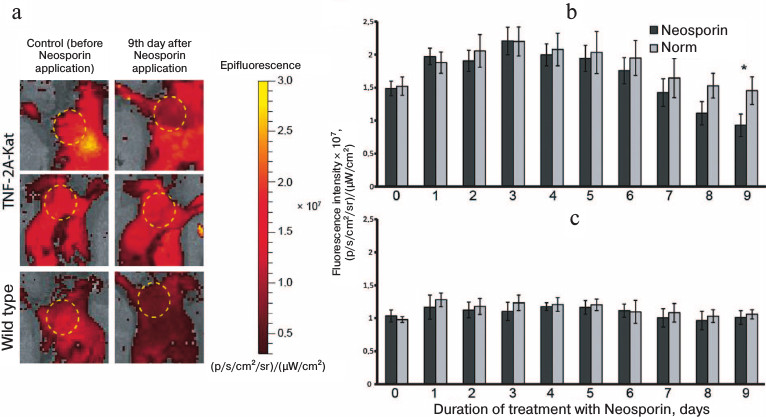
Local decrease in TNF expression in adult mice during local administration of antibiotics. After birth, microorganisms start to colonize the skin, resulting in the maturation of the skin immune system [1]. Taking this into account, we assumed that the skin colonization by microflora leads to the induction of TNF expression. Thus, as the next step, we depleted microflora by local administration of antibiotics and analyzed whether the production of TNF is affected in adult TNF-2A-Kat reporter mice. Figure 3 shows the results of in vivo fluorescence imaging of animals with local depletion of skin microflora. As a control, we used in vivo images of animals before the first application of Neosporin and images of the skin at the right shoulder that was not treated with the drug.
Fig. 3. Results of fluorescence imaging of mice during local microflora depletion. a) In vivo fluorescent images of TNF-2A-Kat reporter mice before application of Neosporin and 9 days after the start of the treatment. The upper panel shows the antibiotic-treated TNF-2A-Kat mouse skin fragment, middle panel shows the control non-treated region of the same animal, and the lower panel shows the antibiotic-treated wild type mouse skin fragment. b, c) Quantification of the skin fluorescence signal in the regions of TNF-2A-Kat mice and wild type mice, respectively, treated with Neosporin. Average value ± standard deviation. * Statistically significant difference between the region of normal skin and the region treated with Neosporin, p < 0.05.
It was shown that local administration of the antibiotic decreases the skin fluorescence signal of the studied region of TNF-2A-Kat reporter mice compared to the intact tissue fragment (Fig. 3, a and b). Statistically significant difference in fluorescence intensity was detected in the normal skin fragment compared to the skin fragment treated with Neosporin for 9 days (Fig. 3b). Nine days after the start of the treatment, the intensity of fluorescence signal in the zone of drug application was (9.31 ± 1.68)·106 (p/s/cm2/sr)/(µW/cm2), whereas the signal in a normal tissue from the opposite shoulder and the signal before the first application of the drug were (1.45 ± 1.68)·107 and (1.5 ± 1.1)·107 (p/s/cm2/sr)/(µW/cm2), respectively. In general, the intensity of fluorescence of the skin fragment after 9 days of the treatment with Neosporin was about 1.6 times lower than the control values. In wild type mice, no changes in fluorescence signal during treatment with Neosporin were detected (Fig. 3, a and c).
Thus, the decrease in the intensity of fluorescence signal in TNF-2A-Kat reporter mice after local administration of Neosporin for local microflora depletion indicates that skin microflora participates in maintenance of a certain constitutive level of TNF expression in healthy adult TNF-2A-Kat reporter mice.
DISCUSSION
In the present work, we compared for the first time the skin fluorescence intensity in healthy adult mice of TNF-2A-Kat reporter strain and wild type mice or embryos. These results show constitutive expression of TNF, one of the main proinflammatory cytokines, in the skin in absence of visible signs of inflammation. Such constitutive expression was not detected in embryos. The level of TNF expression, determined by the reporter protein fluorescence, decreased after treatment of certain regions of the skin with antibiotics. In total, these data demonstrate that expression of TNF in the skin is triggered by the skin microbiota and can represent one way of interaction between the immune system and microbiota.
Skin is a complex organ, where keratinocytes, skin epithelial cells, T lymphocytes, and dendritic cells are actively interacting [1]. Our data show a global signal from the skin and do not allow deducing a contribution of each cell type. Further analysis of cells using flow cytometry methods and microscopy will determine which type of cells produce TNF in the normal state and during various skin pathologies.
The skin microflora is composed of many different species [2]. Many studies indicate that some species of bacteria can stimulate both proinflammatory and anti-inflammatory signals on barrier surfaces [1]. In this work, we have shown that complete microflora depletion in one region of the skin of TNF-2A-Kat mice using a broad-spectrum antibiotic leads to a decrease in fluorescence signal, indicating the decrease in TNF expression. Thus, microflora participates in the induction of TNF in the skin, but it is not yet clear which bacteria induce TNF and which, to the contrary, can cause its repression.
After birth, different commensal microorganisms colonize the skin in a strictly regulated manner [1]. We have found that embryos and newborn TNF-2A-Kat mice do not demonstrate the increased fluorescence signal of the skin. The analysis of fluorescence throughout the whole lifetime will reveal at which period TNF is induced in the skin and what consequences for maturation of the skin immune system it has.
Unlike intestinal microflora, controlled by external factors, the composition of skin microflora is regulated by the host organism [7]. The role of TNF in the control of skin microflora composition remains unclear. Our data show the constitutive expression of TNF in adult mice, which can be important for the control of skin microflora composition. Further detailed analysis of skin microflora using deep-sequencing methods will allow addressing this question.
Production of high amounts of TNF leads to the development of autoimmune psoriasis [32]. However, the molecular mechanisms of this process and the kinetics of TNF expression during the development of psoriasis are still not known. In future, using this model system, it will be interesting to analyze the mechanisms governing the activation of TNF production during psoriasis or skin neoplasia.
Acknowledgements
Authors are grateful to R. V. Zvartsev and R. Sh. Kazarian for the technical help with the project.
This work was done with financial support from the Ministry of Education and Science (project No. 14.Z50.31.0008, S. A. Nedospasov) and the Russian Science Foundation (project No. 14-15-00139, A. A. Kruglov).
REFERENCES
1.Belkaid, Y., and Tamoutounour, S. (2016) The
influence of skin microorganisms on cutaneous immunity, Nat. Rev.
Immunol., 16, 353-366.
2.Grice, E. A., and Segre, J. A. (2011) The skin
microbiome, Nat. Rev. Microbiol., 9, 244-253.
3.Borkowski, A. W., and Gallo, R. L. (2011) The
coordinated response of the physical and antimicrobial peptide barriers
of the skin, J. Invest. Dermatol., 131, 285-287.
4.Kruglov, A. A., Tumanov, A. V., Grivennikov, S. I.,
Shebzukhov, Y. V., Kuchmiy, A. A., Efimov, G. A., Drutskaya, M. S.,
Scheller, J., Kuprash, D. V., and Nedospasov, S. A. (2011) Modalities
of experimental TNF blockade in vivo: mouse models, Adv. Exp.
Med. Biol., 691, 421-431.
5.Taylor, P. C., and Feldmann, M. (2009) Anti-TNF
biologic agents: still the therapy of choice for rheumatoid arthritis,
Nat. Rev. Rheumatol., 5, 578-582.
6.Fierer, N., Hamady, M., Lauber, C. L., and Knight,
R. (2008) The influence of sex, handedness, and washing on the
diversity of hand surface bacteria, Proc. Natl. Acad. Sci. USA,
105, 17994-17999.
7.Bashan, A., Gibson, T. E., Friedman, J., Carey, V.
J., Weiss, S. T., Hohmann, E. L., and Liu, Y. Y. (2016) Universality of
human microbial dynamics, Nature, 534, 259-262.
8.Eckburg, P. B., Bik, E. M., Bernstein, C. N.,
Purdom, E., Dethlefsen, L., Sargent, M., Gill, S. R., Nelson, K. E.,
and Relman, D. A. (2005) Diversity of the human intestinal microbial
flora, Science, 308, 1635-1638.
9.Dewhirst, F. E., Chen, T., Izard, J., Paster, B.
J., Tanner, A. C., Yu, W. H., Lakshmanan, A., and Wade, W. G. (2010)
The human oral microbiome, J. Bacteriol., 192,
5002-5017.
10.Zaura, E., Keijser, B. J., Huse, S. M., and
Crielaard, W. (2009) Defining the healthy “core microbiome”
of oral microbial communities, BMC Microbiol., 9,
259.
11.Bik, E. M., Long, C. D., Armitage, G. C., Loomer,
P., Emerson, J., Mongodin, E. F., Nelson, K. E., Gill, S. R.,
Fraser-Liggett, C. M., and Relman, D. A. (2010) Bacterial diversity in
the oral cavity of 10 healthy individuals, ISME J., 4,
962-974.
12.Bik, E. M., Eckburg, P. B., Gill, S. R., Nelson,
K. E., Purdom, E. A., Francois, F., Perez-Perez, G., Blaser, M. J., and
Relman, D. A. (2006) Molecular analysis of the bacterial microbiota in
the human stomach, Proc. Natl. Acad. Sci. USA, 103,
732-737.
13.Pei, Z. H., Bini, E. J., Yang, L. Y., Zhou, M.
S., Francois, F., and Blaser, M. J. (2004) Bacterial biota in the human
distal esophagus, Proc. Natl. Acad. Sci. USA, 101,
4250-4255.
14.Gao, Z., Tseng, C. H., Pei, Z., and Blaser, M. J.
(2007) Molecular analysis of human forearm superficial skin bacterial
biota, Proc. Natl. Acad. Sci. USA, 104, 2927-2932.
15.Grice, E. A., Kong, H. H., Renaud, G., Young, A.
C., Bouffard, G. G., Blakesley, R. W., Wolfsberg, T. G., Turner, M. L.,
and Segre, J. A. (2008) A diversity profile of the human skin
microbiota, Genome Res., 18, 1043-1050.
16.Costello, E. K., Lauber, C. L., Hamady, M.,
Fierer, N., Gordon, J. I., and Knight, R. (2009) Bacterial community
variation in human body habitats across space and time, Science,
326, 1694-1697.
17.Braff, M. H., Bardan, A., Nizet, V., and Gallo,
R. L. (2005) Cutaneous defense mechanisms by antimicrobial peptides,
J. Invest. Dermatol., 125, 9-13.
18.Massoud, T. F., and Gambhir, S. S. (2003)
Molecular imaging in living subjects: seeing fundamental biological
processes in a new light, Genes Dev., 17, 545-580.
19.Germain, R. N., Miller, M. J., Dustin, M. L., and
Nussenzweig, M. C. (2006) Dynamic imaging of the immune system:
progress, pitfalls and promise, Nat. Rev. Immunol., 6,
497-507.
20.Shebzukhov, Y. V., Kuchmiy, A. A., Kruglov, A.
A., Zipp, F., Siffrin, V., and Nedospasov, S. A. (2014) Experimental
applications of TNF-reporter mice with far-red fluorescent label,
Methods Mol. Biol., 1155, 151-162.
21.Luker, G. D., and Luker, K. E. (2008) Optical
imaging: current applications and future directions, J. Nucl.
Med., 49, 1-4.
22.Kuchmiy, A. A., Efimov, G. A., and Nedospasov, S.
A. (2012) Methods for in vivo molecular imaging, Biochemistry
(Moscow), 77, 1339-1353.
23.Chudakov, D. M., Matz, M. V., Lukyanov, S., and
Lukyanov, K. A. (2010) Fluorescent proteins and their applications in
imaging living cells and tissues, Physiol. Rev., 90,
1103-1163.
24.Chudakov, D. M., Belousov, V. V., Zaraisky, A.
G., Novoselov, V. V., Staroverov, D. B., Zorov, D. B., Lukyanov, S.,
and Lukyanov, K. A. (2003) Kindling fluorescent proteins for precise
in vivo photolabeling, Nat. Biotechnol., 21,
191-194.
25.Stoll, S., Delon, J., Brotz, T. M., and Germain,
R. N. (2002) Dynamic imaging of T cell–dendritic cell
interactions in lymph nodes, Science, 296, 1873-1876.
26.Mempel, T. R., Henrickson, S. E., and Von
Andrian, U. H. (2004) T-cell priming by dendritic cells in lymph nodes
occurs in three distinct phases, Nature, 427,
154-159.
27.Bousso, P., Bhakta, N. R., Lewis, R. S., and
Robey, E. (2002) Dynamics of thymocyte-stromal cell interactions
visualized by two-photon microscopy, Science, 296,
1876-1880.
28.Kumar, S., Skeen, M. J., Adiri, Y., Yoon, H.,
Vezys, V. D., Lukacher, A. E., Evavold, B. D., Ziegler, H. K., and
Boss, J. M. (2005) A cytokine promoter/yellow fluorescent protein
reporter transgene serves as an early activation marker of lymphocyte
subsets, Cell Immunol., 237, 131-140.
29.Stetson, D. B., Mohrs, M., Reinhardt, R. L.,
Baron, J. L., Wang, Z. E., Gapin, L., Kronenberg, M., and Locksley, R.
M. (2003) Constitutive cytokine mRNAs mark natural killer (NK) and NK
T-cells poised for rapid effector function, J. Exp. Med.,
198, 1069-1076.
30.Kravchenko, V. V., Kaufmann, G. F., Mathison, J.
C., Scott, D. A., Katz, A. Z., Grauer, D. C., Lehmann, M., Meijler, M.
M., Janda, K. D., and Ulevitch, R. J. (2008) Modulation of gene
expression via disruption of NF-kappaB signaling by a bacterial small
molecule, Science, 321, 259-263.
31.Kuchmiy, A. A., Kruglov, A. A., Galimov, A. R.,
Shebzukhov, Yu. V., Chudakov, D. M., Lukyanov, S. A., and Nedospasov,
S. A (2011) New line of transgenic reporter mice for studying the
expression of tumor necrosis factor, Russ. Immunol. J.,
5, 205-214.
32.Tracey, D., Klareskog, L., Sasso, E. H., Salfeld,
J. G., and Tak, P. P. (2008) Tumor necrosis factor antagonist
mechanisms of action: a comprehensive review, Pharmacol. Ther.,
117, 244-279.