REVIEW: Investigation of Structure of the Ribosomal L12/P Stalk
I. V. Mitroshin, M. B. Garber*, and A. G. Gabdulkhakov
Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; E-mail: garber@vega.protres.ru* To whom correspondence should be addressed.
Received July 6, 2016
This review contains recent data on the structure of the functionally important ribosomal domain, L12/P stalk, of the large ribosomal subunit. It is the most mobile site of the ribosome; it has been found in ribosomes of all living cells, and it is involved in the interaction between ribosomes and translation factors. The difference between the structures of the ribosomal proteins forming this protuberance (despite their general resemblance) determines the specificity of interaction between eukaryotic and prokaryotic ribosomes and the respective protein factors of translation. In this review, works on the structures of ribosomal proteins forming the L12/P-stalk in bacteria, archaea, and eukaryotes and data on structural aspects of interactions between these proteins and rRNA are described in detail.
KEY WORDS: ribosome, stalk, ribosomal proteinsDOI: 10.1134/S0006297916130022
Abbreviations: IF2, translation initiation factor 2; mRNA, messenger RNA; rRNA, ribosomal RNA; tRNA, transport RNA.
The cell organelles responsible for protein biosynthesis were discovered
in the 1950s. These organelles were named ribosomes. The ribosome is a
macromolecular ribonucleoprotein complex. Ribosomal RNA (rRNA)
determines the main structural and functional properties of the
ribosome, but the presence of both rRNA and ribosomal proteins is
necessary for the normal functioning of the ribosome.
Ribosomal structure was originally studied by the methods of ultracentrifugation and electron microscopy. These studies showed that under certain conditions (e.g. the low concentration of magnesium ions) the ribosome dissociates into a small subunit and a large subunit. The large ribosomal subunit has three peripheral stalks: the lateral finger-shaped stalk (the L12 stalk in bacteria and the P stalk in archaea and eukaryotes) on one side of the subunit; the central protuberance that can be called the head of the large subunit in the middle; and the lateral L1 stalk on the other side [1, 2].
During protein biosynthesis, the ribosome interacts with messenger RNA (mRNA), transport RNA (tRNA), translation initiation, elongation, and termination factors, and other ligands [3]. The working cycle of the ribosome consists of three steps: initiation, elongation, and termination. Translation factors promote protein synthesis at each step of ribosomal working cycle. The lateral L12/P stalk of the ribosome promotes interaction between the ribosome and the translation elongation and termination factors, while the bacterial L12 stalk is involved also in translation initiation, increasing the rate of association between the small and large ribosomal subunits [4]. Cryoelectron microscopy and the subsequent reconstruction of ribosomal structure have shown that the L12 stalk undergoes different conformational rearrangements during the ribosomal elongation cycle.
NOMENCLATURE OF RIBOSOMAL PROTEINS
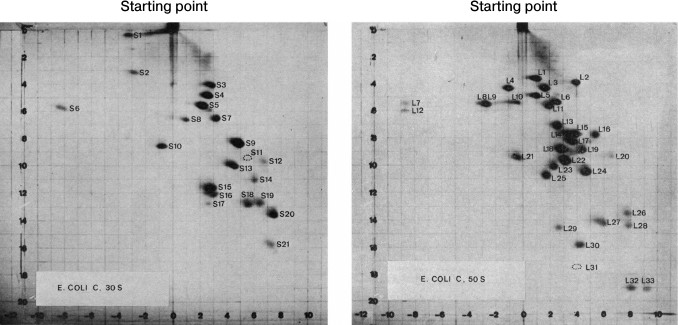
The small and large ribosomal subunits contain many individual proteins. Nearly all are represented by a single copy on the ribosome. The first attempt to systematize ribosomal proteins was based on a standard experimental method: two-dimensional gel electrophoresis. It was the most convenient method for complete separation of ribosomal proteins by molecular size and charge (Fig. 1) [5].
Fig. 1. Separation of the ribosomal proteins of the 30S (a) and the 50S (b) ribosomal subunits from Escherichia coli by 2D polyacrylamide gel electrophoresis under denaturing conditions, and the nomenclature of these proteins. First direction: horizontal; second direction: vertical (top-down). The proteins of the small ribosomal subunit are marked with S (English “small”) and the proteins of the large subunit are marked with L (English “large”). The figures with minor changes were taken from a work by Kaltschmidt and Wittmann [5].
Initially, ribosomal proteins from each type of organisms had their own designations in accordance with their electrophoretic separation. With that nomenclature, the same numbers could be given to nonhomologous proteins of different species. The comparison of amino acid sequences of ribosomal proteins revealed homology between the proteins of different species and demonstrated the evolutionary conservatism of most ribosomal proteins [6]. Hence, it was possible to create a new nomenclature for conservative homologous proteins of bacteria, archaea, and eukaryotes.
With the advent of the models of bacterial, archaeal, and eukaryotic ribosomes obtained by crystallography or cryoelectron microscopy, the absence of a universal nomenclature for ribosomal proteins began to impede the comparative analysis of structure. To solve this problem, a single nomenclature for ribosomal proteins of all domains of life was created in 2014 (table) [7]. The new nomenclature is based on the designation of ribosomal proteins of E. coli because the first ribosomal proteins were isolated from this organism, their amino acid sequences were identified prior to the sequences of other ribosomal proteins, and these proteins have been described most thoroughly in the literature. The prefix “u” (from “universal”) and the numbers of E. coli proteins were assigned to the proteins found in the ribosomes of all domains of life. The prefix “b” (from “bacterial”) was assigned to bacterial proteins that were shown to have no homologs in archaea and eukaryotes. The prefix “a” (from “archaeal”) was assigned to archaeal ribosomal proteins having no homologs in the bacterial and eukaryotic ribosomes. The prefix “e” (from “eukaryotic”) was assigned not only to the eukaryotic ribosomal proteins having no bacterial and archaeal homologous, but also to the homologous archaeal proteins.
Original and revised nomenclatures of ribosomal proteins (the table was
taken with modifications from [7, 8])
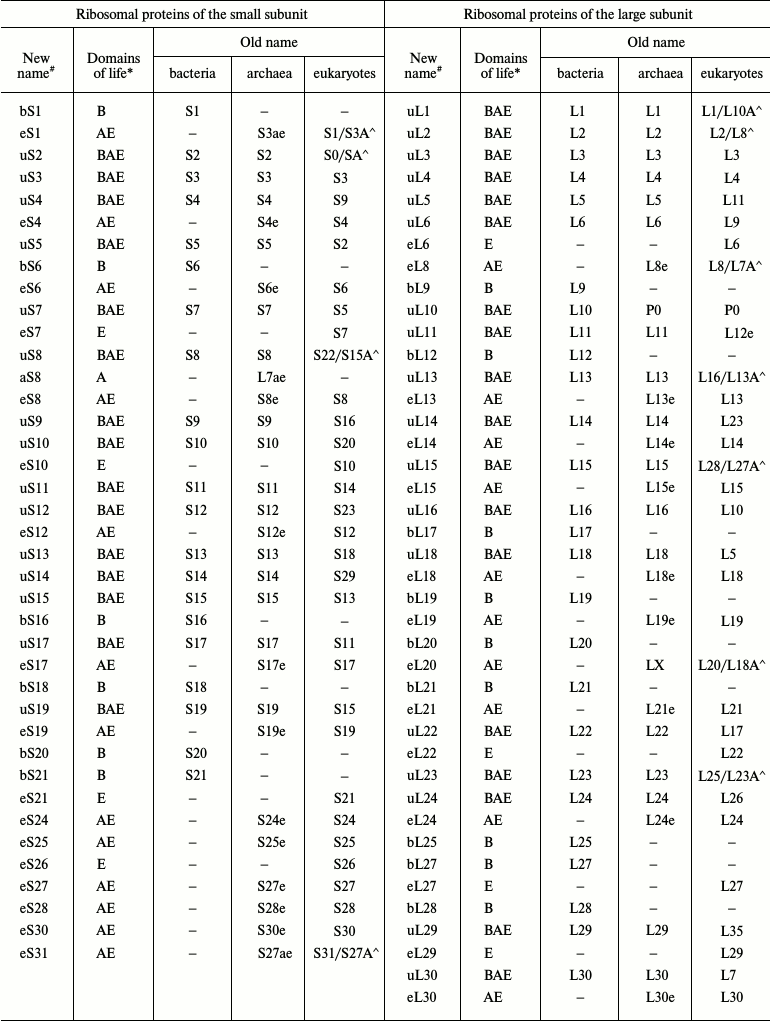
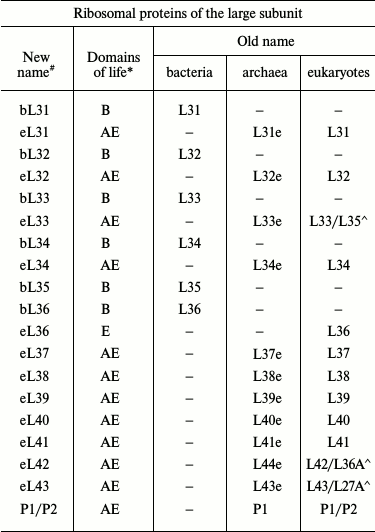
Note: (#) b, bacterial; e, eukaryotic; a, archaeal; u,
universal. (*) B, Bacteria; A, Archaea; E, Eukaryotes. Symbol (^) marks
yeast/human eukaryotic ribosomal proteins.
In the proposed nomenclature, the ribosomal L12/P stalk proteins were given new designations. The bacterial protein L10 and its archaeal and eukaryotic homologs P0 were designated as uL10; the bacterial protein L12 was designated as bL12. Archaea and eukaryotes were shown to have no homologs of the bacterial protein L12 but contained its functional analogs named eP1/P2 in eukaryotes and aP1 in archaea. Accordingly, this lateral stalk in archaea and eukaryotes is now designated as a P stalk. The bacterial and archaeal proteins L11 and their eukaryotic analog L12e were designated as uL11.
The more detailed nomenclature will be used hereafter in this review. For example, the prefixes “b”, “a”, and “e” denoting bacterial, archaeal, and eukaryotic proteins, respectively, will be used for differentiating between the “universal” proteins uL10 and uL11 in addition to the prefix “u”.
COMPONENTS OF RIBOSOMAL L12/P STALK
Bacterial proteins bL10, bL11, and bL12. The ribosomal proteins bL10, bL11, and bL12, together with a fragment of domain II of 23S rRNA, form a characteristic morphological stalk of the bacterial ribosome called the L12 stalk. Two-step treatment of the large ribosomal subunit of E. coli with 1 M NH4Cl and 50% ethanol at 0 and 37°C allows complete removal of these proteins from the ribosome [8].
Ribosomal protein bL12. The ribosomal protein bL12 was one of the first proteins isolated from ribosomes. It is the only protein of the large ribosomal subunit that is present in several copies [9]. The ribosomes of E. coli contain also an acetylated variant of this protein designated as ribosomal protein L7 according to the original nomenclature [5]. bL7 is an exact copy of ribosomal protein bL12; the only difference is that bL7 is acetylated at the N-terminal serine residue [10]. Because of their similarity, these proteins have been previously mentioned in the literature as protein bL7/L12. The bL7/bL12 protein ratio in a cell is not constant and depends on a cell growth phase. In the early logarithmic growth phase of E. coli cells, the level of protein bL7 in the ribosomes is minimal. An increasing amount is observed during log-to-stationary transition of the growth phase [11, 12].
During translation initiation, protein bL12 is needed for recognition of translation initiation factor 2 (IF2) in a complex with GTP as a component of the 30S preinitiation complex. This interaction results in a higher rate of association between the small and large ribosomal subunits [4].
The translation and error rates during protein biosynthesis on ribosomes depend on the presence of protein bL12 [13, 14]. The removal of this ribosomal protein from the ribosome impedes the binding of elongation factors EF1A and EF2 with the ribosome [15] and affects other factor-dependent functions, e.g. the binding of aminoacyl-tRNA with the A-site of the ribosome, translocation and, consequently, GTP hydrolysis [16, 17].
The protein bL12 has unique properties among bacterial ribosomal proteins. In addition to the fact that bL12 is a multicopy protein, its isoelectric point is in the acidic region (pH 4.8) [18]. In aqueous solutions, isolated protein bL12 exists only as a dimer [19] or a tetramer [20].
The ribosomal protein bL12 consists of two domains and a long flexible linker [21-23]. The C-terminal domain of protein bL12 (bL12CTD) is responsible for interaction with translation factors [24, 25], while the N-terminal domain of bL12 (bL12NTD) is responsible for dimerization and binding with ribosomal protein bL10 [21]. The N- and C-terminal domains of bL12 are connected to each other with a flexible linker providing mobility of the protein molecule [26]. The length of this linker influences not only the mobility of the two domains relative to each other, but also the binding of elongation factors, GTP hydrolysis on the ribosome, and translation rate and accuracy [27, 28]. The removal of this region inactivates the bL12 protein [28].
In 1980, the C-terminal domain of the E. coli protein bL12 was crystallized and its structure was determined at 2.6 Å resolution [29]. It was the first crystal structure of a ribosomal protein. Much later, the spatial structure of the full-sized protein bL12 from Thermotoga maritima was obtained at 2.0 Å resolution [30]. The bL12NTD contains two short helices: α1 and α2. The long α3 hinge helix is a linker that separates the N-terminal domain from the globular C-terminal domain. This helix is formed by 20 mainly hydrophobic amino acid residues. The C-terminal domain of protein bL12 has dense packing and consists of a three-stranded antiparallel β-sheet surrounded by three α-helices from one side [30].
In 2004, the spatial structure of protein bL7 dimer, the N-acetylated variant of the protein bL12 from E. coli, was determined using solution nuclear magnetic resonance (NMR) [26]. The protein bL7 has an elongated conformation. This structure of the protein bL7 differs greatly from the crystal structure of the protein bL12 from T. maritima in the linker region. The flexible linker in the structure of each bL7 monomer has no particular structural packing. The structure of bL7 dimer determined by NMR revealed how the protein dimerizes in solution (Fig. 2). Dimerization of protein bL7 occurs through the contact between two antiparallel V-shaped α-α-hairpins of the N-terminal domain, which form a symmetrical four-helix bundle.
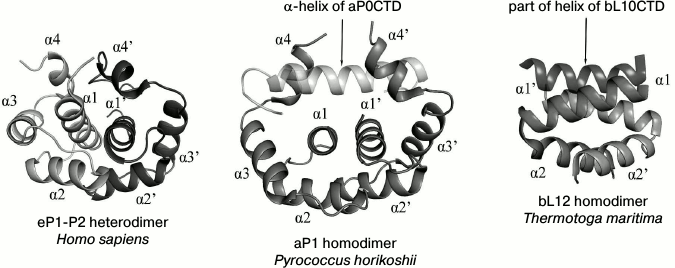
Fig. 2. Structural comparison of dimerization of eukaryotic proteins eP1/P2, archaeal protein aP1, and bacterial protein bL12 (the figure with minor modifications was taken from [34]).
The model of a molecular switch between the two states of the protein was proposed based on the determined protein structures of bL12 from T. maritima and bL7 dimer from E. coli. This model suggests that the region near the linker of protein bL12 plays the role of a molecular switch: the molecule adopts either a “closed” compact conformation, when the elongation factor is bound to the ribosome, or an “open” elongated conformation after GTP hydrolysis and dissociation of the elongation factor from the ribosome [26].
The bL12 dimers are bound to ribosomal protein bL10 with the formation of a strong ribosomal protein complex bL10–bL12 in solution. The thermal stability of bL10 and bL12 in the complex increases compared to the individual states [31]. The bL10–bL12 complex from E. coli remains stable in the presence of 6 M urea at pH 4.6. Hence, when the ribosomal proteins were systematized by two-dimensional electrophoresis, the protein complex bL10–bL12 was preserved under the denaturing conditions was erroneously taken as an individual protein, which was named L8 [32].
For the complex of ribosomal proteins bL10–bL12 from E. coli, the ratio of proteins bL10 and bL12 was shown to be 1 : 4 by the methods of isotope dilution [33], equilibrium ultracentrifugation, and quantitative analysis of protein staining on the electrophoregram [31]. Therefore, it was believed for a long time that the bacterial complex of ribosomal proteins bL10–bL12 can exist only as a pentamer. The determination of the crystal structure of ribosomal complex bL10–bL12NTD from T. maritima changed the notion of the ratio of proteins bL10 and bL12. In this structure of the complex, six molecules of the N-terminal domain of protein bL12 formed a heptameric complex with one molecule of protein bL10 [34]. Hence, it was suggested that the difference between protein ratios in the bL10–bL12 complexes depended on the nature of the organism from which the proteins were isolated, and that the extra amino acid sequence in the C-terminal domain of protein bL10 of thermophilic bacteria could be a binding site for a third dimer of bL12 [35]. Results obtained by mass spectrometry confirmed this assumption. It was shown that the bL10–bL12 complexes from mesophilic bacteria are pentameric, while those from thermophilic bacteria are solely heptameric [35, 36].
Ribosomal protein bL10. Ribosomal protein bL10 acts as a bridge between bL12 dimers and the ribosome. The C-terminal part of protein bL10 from mesophilic bacteria contains two independent binding sites for bL12 dimers, whereas the N-terminal part of protein bL10 interacts with 23S rRNA [22].
The crystal structure of a complex of ribosomal protein bL10 with dimers of the N-terminal domain of protein bL12 from T. maritima was determined at 2.3 Å resolution [34]. Protein bL10 consists of two domains: the N-terminal RNA-binding domain, and the C-terminal domain to which protein bL12 is bound. The N-terminal domain is densely packed and contains an α/β motif. The C-terminal domain of bL10 is formed by a long and flexible C-terminal α-helix (helix α8). The α8 helix bends twice, forming three segments of 10 amino acid residues each. Each segment binds one bL12NTD dimer; therefore, three nearly identical elements can be distinguished in the region of contact between proteins bL10 and bL12 [34]. Between the N- and C-terminal domains of bL10 there is the so-called “rotation center”. This “rotation center” provides high mobility of helix α8 with the bL12NTD dimers relative to the RNA-binding domain, which is necessary for the functional activity of the lateral L12 stalk [34].
The site of bL10 binding to the ribosome is on the surface of the large ribosomal subunit. Chemical probing was used to determine the main binding region for protein bL10, which is in domain II of 23S RNA and includes helices H42-44 [37]. The site of bL10 binding to 23S rRNA was localized by superimposing the known structure of the bacterial protein bL10 from T. maritima at the conservative RNA-binding domain on the structure of two N-terminal α-helices of the archaeal protein aL10, which was determined within the 50S ribosomal subunit from the archaeon Haloarcula marismortui [34, 38]. Based on these data, the maximum number of contacts were revealed between helices α1 and α2 of protein bL10 and helix H42 of 23S rRNA, which is in good agreement with the chemical probing data [34, 37]. It should be noted that most contacts between bL10 and 23S rRNA are in the sugar-phosphate backbone of rRNA. Spatial packing of rRNA probably plays a key role in recognition of the uL10 binding site in the ribosomes of all organisms [34].
The genes of bacterial ribosomal proteins bL10 and bL12 are localized in the same operon. Ribosomal protein bL10 as a component of the bL10–bL12 complex is a translational repressor of its operon and is bound to mRNA upstream from the initiation codon of the bL10 gene [39, 40].
The analysis of rRNA showed that the crystal structure of the large ribosomal subunit of Deinococcus radiodurans contained a consensus motif in helix H42 of 23S rRNA around the GTPase center, the so-called “kink-turn motif”, which probably makes a major contribution to rRNA recognition by protein bL10 [41, 42]. The mRNA of the rplJ gene from E. coli might contain the same motif [43]. It is supposed that mRNA and rRNA interact with protein bL10 in a similar way.
Ribosomal protein bL11 is bound to 23S rRNA close to the bL10 binding site. The binding of proteins bL10 and bL11 is cooperative [44]. The cooperative effect can increase the affinity of bL10 to rRNA 100-fold [43]. Cooperative binding of bL10 and bL11 is probably determined by conformational changes in the rRNA structure [34]. Although mRNA and rRNA showed the same affinity for protein bL10 in experiments in vitro, the 100-fold cooperative effect of bL10 and bL11 binding to rRNA ensures almost complete occupation of ribosomes by the bL10–bL12 protein complex [43].
Ribosomal protein bL11. Ribosomal protein bL11 is a necessary component of the ribosomal 50S subunit, being located at the base of the L12 stalk. The bL11-lacking mutant strains of Bacillus megaterium are viable, but their growth is more than twice slower compared to cells of the wild type strain [45]. The function of bL11 on the ribosome is like that of bL12: it is involved in the interaction between the ribosome and elongation factors EF1A and EF2, termination factors RF1 and RF2, and it promotes the association of ribosomal subunits and stimulates GTP hydrolysis [46-48].
Spatial structures of the full-sized protein bL11 from T. maritima have been determined in the free state by the solution NMR technique [49] and in complex with the 23S rRNA fragment by X-ray structure analysis at 2.6 Å resolution [50]. Also, the structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with the antibiotic thiostrepton, where bL11 was visualized, was determined at 3.3 Å resolution [51].
Ribosomal protein bL11 consists of N- and C-terminal globular domains connected with a short linker. The α1 helix of the N-terminal domain of bL11 contains a conservative proline residue (Pro22, numbering for T. maritima), which interacts with thiazole antibiotics (e.g. thiostrepton and micrococcin). The linker between the bL11 domains is formed by a consensus motif of three amino acid residues (Thr72-Pro73-Pro74). The structure of the C-terminal domain (bL11CTD) has a characteristic feature, namely, an elongated disordered loop (84-96 a.a.). This loop participates in RNA–protein interaction and is susceptible to conformational changes during rRNA binding [49, 50].
Protein bL11 interacts via the C-terminal domain with the large ribosomal RNA. Filter-binding assay has shown that the dissociation constant for the protein and rRNA is 1.2·10–9 M [52]. The C-terminal domain of bL11 is bound with the minor groove of 23S rRNA formed by helices H43-44. The RNA-binding surface of bL11 is formed by the α5 helix, the N-terminal part of the α3 helix, as well as by the α3-β4 and α4-α5 loops located on each side of the α5 helix. The α5 helix is localized along the minor groove of 23S rRNA, forming the greatest number of contacts. The α3-β4 and α4-α5 loops, when binding to rRNA, adopt an ordered structure that repeats the surface of the minor groove of rRNA [50]. More than half of the hydrogen bonds of the complex are formed between the sugar-phosphate backbone of rRNA and the main chain of bL11CTD. This fact indicates that the spatial packing of 23S rRNA plays the key role in recognition of the bL11 binding site. Biophysical experiments have shown that bL11 stabilizes the tertiary structure of rRNA via the C-terminal domain [53]. The N-terminal domain of bL11 (bL11NTD) is involved in interaction with translation factors.
The ribosomal complex of protein bL11 with 23S rRNA is a target for thiazole antibiotics [54]. When binding with the ribosome, thiostrepton blocks and micrococcin stimulates GTP hydrolysis on EF2 [55]. The main thiostrepton/micrococcin binding site is localized in a slot between the α1 helix of bL11NTD and the 1067/1095 region of rRNA. Thus, thiostrepton blocks the functionally important structural rearrangements of bL11 through formation of a stable bL11–rRNA–thiostrepton complex.
Proteins of eukaryotic and archaeal P stalk. Eukaryotic and archaeal ribosomes contain a lateral P stalk with structural organization analogous to that of the bacterial L12 stalk. The eukaryotic ribosomal P stalk is formed by two types of P proteins and protein eL11. Protein eL10 is a P protein of the first type (previously denoted as P0) [56]. The proteins of the second type are small acidic proteins eP1/P2 of about 11 kDa [57]. The P proteins form a pentameric complex eL10–eP1/P2, where two heterodimers eP1/P2 are bound to protein eL10. The eukaryotic P stalk is a necessary component of the ribosome, and protein eL10 is vitally important for cell growth [58].
In archaea, the P stalk consists of ribosomal proteins aL11, aL10, and aP1. As concerns amino acid sequence, the archaeal proteins aL10 and aP1 are much more like eukaryotic proteins eL10 and eP1/P2 than to their bacterial analogs [59].
The archaeal P stalk is rather stable. Proteins aL10 and aP1 form a strong protein complex aL10–aP1, which, like the bacterial complex bL10–bL12, is not destroyed in 6 M urea at pH 4.6. After the large ribosomal subunit has been treated with a high-concentration NH4Cl and ethanol solution, protein aL10 is not completely removed from the ribosome as it interacts with rRNA with higher affinity than protein bL10 [60].
Ribosomal proteins eP1/P2 and aP1. The eukaryotic eP1/P2 proteins are very similar to each other both functionally and structurally. The number of groups and subgroups of eP1/P2 proteins varies between different species of organisms [61, 62]. In contrast to bacterial protein bL12 from E. coli, which is represented on the ribosome by one more N-acetylated copy, each type of eP1/P2 is encoded by a separate gene [61]. In solution, the eP1/P2 proteins are present as a stable heterodimer [63, 64].
The archaeal protein aP1 is encoded by a single gene in archaea and is not subject to modifications, although it is structurally similar to the eukaryotic proteins eP1/P2. In solution, as well as on the ribosome, protein aP1 is present as a dimer [60, 65].
Structurally, the archaeal aP1 and eukaryotic eP1/P2 can be divided into N- and C-terminal domains and a flexible linker connecting the domains (by analogy with bacterial protein bL12). Like in bL12, the N-terminal domain is responsible for dimerization of these proteins and for their binding with the ribosomal subunit via protein a/eL10 [66, 67].
During protein biosynthesis, archaeal protein aP1 delivers translation factors to the ribosome. The C-terminal part of the protein directly interacts with elongation factors aEF2 and aEF1α, as well as with initiation factor aIF5B homologous to the bacterial initiation factor IF2 [68, 69]. As shown by surface plasmon resonance, protein aP1 binds elongation factor aEF2 irrespective of whether the factor is in a complex with GTP or GDP. Moreover, the protein complex aL10·(aP1)6 can bind several aEF2 molecules, promoting an increase in the rate of aEF2 delivery to the GTPase-binding center and high rate of GTP hydrolysis [69].
The first 10 a.a. residues of eukaryotic protein eP1/P2 are important for heterodimerization and formation of a pentameric complex [70]. The C-terminal domain of eP1/P2 is responsible for interaction between the ribosome and translation factors [71].
The spatial structure of the full-sized heterodimer eP1/P2 from Homo sapiens has been determined by the solution NMR technique [72]. The N-terminal domains of proteins eP1 and eP2 consist of four α-helices and are rather compact (Fig. 2). The heterodimer of the eP1 and eP2 N-terminal domains is asymmetrical and is formed due to highly conservative hydrophobic residues of the α1, α2, and α4 helices of eP1 and eP2. The primary point of contact is between the α1 helices of eP1 and eP2. The C-terminal part has no specific structure. Thus, the C-terminal “tail” of the eP1–eP2 dimer can be at a distance up to 125 Å from the N-terminal domain. Due to the elongated C-terminal “tail” of eP1/P2, translation elongation factors seem to be easily delivered to the GTPase-binding center [72].
The structure of the full-sized archaeal protein aP1 is unknown, but the crystal structures of dimers of the N-terminal domain of aP1 (in complex with aL10; 2.1 Å resolution) [67] and the C-terminal domain of aP1 (in complex with elongation factor aEF1α; 2.3 Å resolution) from Pyrococcus horikoshii have been determined [73]. The N-terminal domain of the archaeal protein aP1 is formed by four α-helices (Fig. 2). Dimerization of aP1 occurs due to hydrophobic interactions between the α1 and α2 helices of the two monomers. In contrast to the compact C-terminal domain of bacterial bL12, the C-terminal domain of archaeal aP1 (aP1CTD) is unstructured in the free state from the elongation factor [74]; however, during the interaction with translation factors, aP1CTD is structured with the formation of a long α-helix [73].
The structures of the archaeal dimer of the aP1 N-terminal domain and the eukaryotic heterodimer of the eP1 and eP2 N-terminal domains show similar packing of polypeptide chains, as opposed to the bacterial dimer of the bL12 N-terminal domain (Fig. 2). The basic structural difference between the aP1NTD dimer and the eP1/P2NTD heterodimer is the α4 helix. The α4 helix of aP1NTD in the structure of the archaeal complex aL10·(aP1)6 adopts an “open” conformation, which allows it to bind with the short helix of aL10. The α4 helix in the eukaryotic dimer eP1/P2NTD adopts a “closed” conformation but, quite probably, can adopt also the “open” conformation and, in this state, promote the binding of the eP1/P2 heterodimer with the C-terminal part of eukaryotic protein eL10 [66].
The α3 helix of eukaryotic protein eP1 has a strongly hydrophobic surface and, on the contrary, the α3 helix of eP2 has a hydrophilic surface. Neither the eP1 nor the eP2 α3 helix is involved in the dimerization process. The asymmetry of eP1/P2 suggests that the eP1/P2 heterodimers in the pentameric complex are arranged in the following order: eP2–eP1 : eP1–eP2 [66].
In archaeal ribosomes, aL10 and aP1 are present as an aL10–aP1 complex in ratio 1 : 4 or 1 : 6, as determined by mass spectrometry [36, 59]. The ribosomes of hyperthermophilic archaea contain only the heptameric complex aL10·(aP1)6, while mesophilic archaea were shown to have two ribosome populations with either pentameric or heptameric aL10–aP1 complex. The ratio of the ribosomes with pentameric and heptameric complexes varies through the cell life cycle. At the initial stage of cell growth, ribosomes contain mainly the pentameric complex aL10·(aP1)4. When the cells pass into the stationary growth phase, the aL10 to aP1 ratio changes and the ribosomes with the heptameric complex aL10·(aP1)6 become predominant [36].
The activity of GTP hydrolysis and polyphenylalanine synthesis in archaeal ribosomes containing the trimeric complex aL10·(aP1)2 have 55% of the activity of ribosomes with the heptameric complex. The activity of ribosomes with the pentameric complex is about 95%. Thus, the pentameric and heptameric complexes aL10–aP1 are not much different in availability for translation factors. The third dimer of the archaeal protein aP1 seems to be necessary for proper functioning of archaeal the ribosome at temperatures close to the optimal growth temperature (e.g. for P. horikoshii, it is 95°C) [59].
Ribosomal proteins a/eL10. Eukaryotic protein eL10 contains two structural elements for binding of two eP1–eP2 heterodimers [75]. By removing C-terminal amino acid residues, it has been shown that the first and second binding sites for the eP1–eP2 heterodimer are located in the region of 205-230 and 240-255 a.a., respectively (numbering for Bombyx mori) [75, 76].
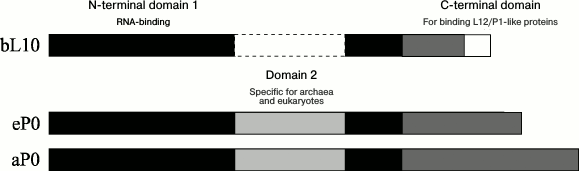
The archaeal and eukaryotic ribosomal proteins a/eL10, like bacterial protein bL10, are intermediates between the archaeal aP1, the eukaryotic eP1/P2, and the ribosome. The archaeal and eukaryotic proteins a/eL10 consist of three domains (Fig. 3). The conservative (in all domains of life) N-terminal domain 1 of protein a/eL10 is an RNA-binding domain; it is responsible for the attachment of the ribosomal complex aL10–aP1 or eL10–eP1/P2 to the large ribosomal subunit. The second (specific domain) has been found only as a component of proteins a/eL10 of archaea and eukaryotes and is absent in the bacterial analog [77, 78]. The C-terminal helical domain is a place of attachment for the two eP1–eP2 heterodimers in the case of eukaryotic P stalk and two or three aP1 homodimers in the case of archaeal P stalk.
Fig. 3. Scheme of polypeptide chain sequences of ribosomal proteins uL10. The N-terminal domains 1 and 2 are rendered in black and light gray, respectively; the C-terminal domain is gray. The white rectangle with a solid line denotes extra amino acid sequences of the thermophilic bacterial proteins bL10.
In 2010, the crystal structures of the archaeal ribosomal complex of aL10 with dimers of the N-terminal domain of aP1 from P. horikoshii (2.1 Å resolution) [67] and the two-domain N-terminal fragment of the archaeal ribosomal protein aL10 (aL10NTF) from Methanococcus jannaschii (1.6 Å resolution) [77] have been determined.
The N-terminal domain 1 of archaeal protein aL10 consists of two parts corresponding to amino acid residues 1-111 and 192-206 (numbering for M. jannaschii) [67, 77]. The second domain of aL10 is an insert in the first and contains amino acid residues 115-188. The two domains are connected with a linker consisting of two oppositely directed β-strands. Domain 2 can shift relative to domain 1 by 13 Å [77]. The C-terminal domain of protein aL10 is twofold longer (Fig. 3) than the bacterial bL10CTD and different in structure. It contains three independent α-helices connected with a short 6-a.a. linker. Each helix of the C-terminal domain of aL10 is bound to one dimer of protein aP1 [67].
The spatial structure of isolated eukaryotic protein eL10 has not been determined. A structural model of eukaryotic protein eL10 in complex with N-terminal domains of the eP1–eP2 heterodimer was suggested based on high homology between archaeal and eukaryotic proteins a/eL10 and the crystal structure of the archaeal protein aL10 in complex with dimers of the aP1 N-terminal domains [79]. This model predicted the structure of only the N-terminal domain 1 and the C-terminal domain of eL10 and, therefore, determination of the structure of its domain 2 is still relevant.
The region of archaeal and eukaryotic proteins a/eL10 corresponding to domain 2 ensures specific interaction between the ribosome and the eukaryotic or archaeal, but not bacterial, translation factors. The removal of this domain reduces to 40% the factor-dependent GTP hydrolysis and the level of polyphenylalanine synthesis by hybrid ribosomes of E. coli with eukaryotic and archaeal translation factors [67].
The N-terminal domain 1 of archaeal protein aL10 interacts with helix H42 of 23S rRNA domain II, similar to the binding of bacterial protein bL10 with 23S rRNA, and the major point of contact between the archaeal protein aL10 and rRNA is also located in the sugar-phosphate backbone of RNA (“kink-turn motif”) [34].
The first 20 a.a. of eukaryotic protein eL10 are needed for binding with 26/28S rRNA [76]. Between amino acids 40 and 70, there is an arginine-rich region providing an extra contact with rRNA [80]. In complex with proteins eP1/P2, the affinity of eL10 to rRNA increases [81]. It is most probable that eP1/P2 additionally functions as a modulator during the binding of eL10 with the ribosome.
Ribosomal proteins a/eL11. Eukaryotic ribosomal protein eL11 is a functional analog of the archaeal and bacterial proteins a/bL11. Chemical cross-linking of neighboring molecules has shown that eukaryotic protein eL11 interacts with eukaryotic elongation factors eEF1α and eEF2 [82]. Protein eL11 binds with 26/28S rRNA in the region equivalent to the binding region for the archaeal/bacterial proteins a/bL11 on 23S rRNA.
It should be noted that the spatial structure of isolated eukaryotic protein eL11 has not been determined. It is only known about the model of eL11 as a component of the yeast ribosome as a polyalanine chain [83].
Archaeal protein aL11 is homologous to bacterial protein bL11 [65]. The spatial structure of archaeal protein aL11 has been partially visualized only within the 50S subunit of the ribosome from H. marismortui [84]. Protein aL11 consists of two domains connected with a short linker. Due to the low quality of electron density maps for the region of the N-terminal domain, the structural details of the latter are poorly discernible [84].
The archaeal aL11 is bound via the C-terminal domain to helices H43-44 of 23S rRNA domain II. The binding sites of archaeal aL10 and aL11 are located on rRNA side-by-side. It is important to note that archaeal protein aL11 stimulates binding of archaeal aL10 with RNA only at high temperatures (70°C); at low temperatures (37°C), stimulation is not observed [85]. The RNA-binding surface of protein aL11 is formed by the α5 helix and the α3-α4 and α4-α5 loops located on both sides of the α5 helix. The α5 helix of the C-terminal domain forms an extensive network of interactions with 23S rRNA, being localized along the minor groove of rRNA [84].
The N-terminal domain of archaeal aL11 promotes delivery of translation elongation and termination factors to the GTPase-binding center of the archaeal ribosome. It has been shown that the archaeal ribosome is sensitive to the peptide antibiotics thiostrepton and micrococcin. It is most probable that archaeal protein aL11 can be a target for this class of antibiotics [86].
INTERCHANGEABILITY OF LATERAL STALK IN BACTERIA, ARCHAEA, AND
EUKARYOTES
After it had been shown that the L12/P stalk is a functionally important morphological element of ribosomes in bacteria, archaea, and eukaryotes, the first attempts were made to create a hybrid ribosome under in vitro conditions. In 1981, the first eukaryotic hybrid ribosome was reconstructed. After the removal of proteins eL10, eL11, and eP1/P2 from the eukaryotic ribosome by extraction with NH4Cl–ethanol mixture, the respective bacterial proteins were added to the “vacant” ribosome. However, the resulting hybrid ribosome was inactive [87]. In addition, the aL10–aP1 protein complex of the archaeal ribosome was replaced by the bacterial complex bL10–bL12 and, on the contrary, the bL10–bL12 complex of the bacterial ribosome was replaced by the archaeal complex aL10–aP1. Electron microscopy showed that the hybrid ribosomes had an L12/P stalk formed by the bL10–bL12 or aL10–aP1 complexes, but the functional activity of such ribosomes was not tested [88].
The first active hybrid bacterial ribosome from E. coli with eukaryotic P stalk of the rat ribosome was obtained in Hachimori laboratory [89]. This substitution changes the specific binding of bacterial EF2 into binding of eukaryotic eEF2 and stimulates GTPase activity in the latter. The level of GTP hydrolysis on eEF2 in the hybrid ribosome is comparable with the level of GTP hydrolysis on eEF2 in the eukaryotic 80S ribosome [68]. At the same time, the activity of the hybrid ribosome during polyphenylalanine synthesis in the presence of eukaryotic elongation factors eEF1α and eEF2 is at the same level as in the eukaryotic ribosome [68, 76].
The archaeal P-stalk proteins, like their eukaryotic homologs, can substitute for bacterial L12-stalk proteins [68]. As a result of such substitution, the reconstructed hybrid ribosome becomes available both for the archaeal and eukaryotic elongation factors but not for the bacterial elongation factors. The activity of GTP hydrolysis and polyphenylalanine synthesis in the hybrid ribosome containing the archaeal lateral stalk proteins in the presence of eukaryotic elongation factors in comparable with those for the hybrid ribosome where the bacterial L12 stalk is replaced by its eukaryotic analog. It should be noted that polyphenylalanine synthesis and GTPase activity of the hybrid ribosome with the archaeal P stalk are the same as in eukaryotic and archaeal elongation factors [68].
The L12-stalk proteins of human mitochondria have a high degree of homology with the respective bacterial proteins. The mitochondrial L12 stalk can substitute for the analogous stalk on the ribosome of E. coli with the formation of a functionally active hybrid ribosome [90]. The resultant hybrid ribosome with the mitochondrial L12 stalk has a high activity of polyphenylalanine synthesis in the presence of bacterial or mitochondrial elongation factors.
Thus, these experiments have demonstrated the key role of the L12/P-stalk proteins in the specific recognition of translation factors. The uL10–L12-like complex (but not the uL11 proteins) is responsible for specific interaction between the ribosome and the translation factors. It is important to note that the change in specific recognition of the elongation factors caused by the replacement of lateral stalk proteins by their homologs (or analogs) on the ribosome is accompanied by structural changes in 23S/28S rRNA regions around the sarcin–ricin loop and helices H43-44 [91].
CRYSTALLOGRAPHIC STUDIES OF THE L12/P STALK IN RIBOSOMES
The X-ray structure analysis made it possible to determine not only the significant morphological features of the ribosome, but also the internal structure of the ribosome, the tertiary structures of ribosomal RNA within the ribosome, and the localizations and structures of ribosomal proteins. The first ribosome crystals suitable for the X-ray structure analysis were obtained as early as in the late 1980s. However, the crystal structure of the 50S ribosomal subunit from the archaeon H. marismortui was determined for the first time at high resolution only in 2000 [38]. This structure contained 2711 out of 2923 nucleotide residues of 23S rRNA, the whole 5S rRNA, and the structures for 27 out of 31 ribosomal proteins. The electron density for the proteins aL1, aL10, aL11, and aP1 was absent, though previously they have been localized within this ribosomal subunit using electron microscopy and low-resolution diffraction data [92].
In 2009, the L12 stalk structure was determined within the bacterial 70S ribosome in complex with EF2 from T. thermophilus [93]. Elongation factor EF2 directly interacts with bL11 and the C-terminal domain of bL12. Due to low quality of electron density map for the region of the lateral stalk, the structure of protein complex bL10–bL12 was determined as a polyalanine chain. The α8 helix of bL10 is bent compared to its position in the structure of isolated complex bL10–bL12NTD, allowing the C-terminal domain bL12CTD to interact with both the N-terminal domain of bL11 and the G'-domain of EF2. In turn, the N-terminal domain of bL11, together with 23S rRNA nucleotide residues 1067 and 1095, interacts with domain V of elongation factor EF2 [93].
In the eukaryotic 80S ribosome from S. cerevisiae, the P stalk was visualized partially [83]. The proteins of this stalk could be placed into the electron density map only as polyalanine chains. The exception was the conservative N-terminal RNA-binding domain 1 of protein eL10, whose structure was determined fully. The overall packing of protein eL10NTD coincides with the packings of the N-terminal domains of bacterial bL10 and archaeal aL10.
The most complete structure of the archaeal P stalk could be determined after additional refinement of the structure of the 50S ribosomal subunit from the archaeon H. marismortui in 2013 [84]. As a result of revision, not only the P stalk but also some ribosomal components of previously unknown structure were visualized (e.g. the archaea-specific LX protein). Refinement of the large subunit structure made it possible to interprete the electron density map for approximately 2/3 of protein aL10 (the C-terminal domain as a polyalanine chain) and one dimer of the aP1 N-terminal domain (as a polyalanine chain), as well as to supplement the aL11 protein structure.
CONCLUSION
The lateral L12/P stalk plays a key role in the interaction between ribosome and translation factors. The structural organization of this stalk is similar in all domains of life, although the comprising ribosomal proteins are different: the proteins of the bacterial L12 stalk show a low homology, both in sequence and in structure, with the respective proteins that form the P stalk in archaea and eukaryotes.
In the last decade, there has been an enormous breakthrough in determination of spatial structures of the ribosome and the isolated proteins of this lateral stalk; however, there are problems thus far with understanding the interactions of these proteins between each other and with the high molecular weight rRNA. Works of recent years devoted to this subject have been considered in this review.
Acknowledgements
This work was supported by the Program of the Presidium of the Russian Academy of Sciences “Molecular and Cell Biology”.
REFERENCES
1.Lake, J. A. (1976) Ribosome structure determined by
electron microscopy of Escherichia coli small subunits, large
subunits and monomeric ribosomes, J. Mol. Biol., 105,
131-159.
2.Boublik, M., Hellmann, W., and Roth, H. E. (1976)
Localization of ribosomal proteins L7L12 in the 50S subunit of
Escherichia coli ribosomes by electron microscopy, J. Mol.
Biol., 107, 479-490.
3.Spirin, A. S. (2011) Molecular Biology:
Ribosomes and Protein Biosynthesis [in Russian], Akademiya,
Moscow.
4.Huang, C., Mandava, C. S., and Sanyal, S. (2010)
The ribosomal stalk plays a key role in IF2-mediated association of the
ribosomal subunits, J. Mol. Biol., 399, 145-153
5.Kaltschmidt, E., and Wittmann, H. G. (1970)
Ribosomal proteins. XII. Number of proteins in small and large
ribosomal subunits of Escherichia coli as determined by
two-dimensional gel electrophoresis, Proc. Natl. Acad. Sci. USA,
67, 1276-1282.
6.Liao, D., and Dennis, P. P. (1994) Molecular
phylogenies based on ribosomal protein L11, L1, L10, and L12 sequences,
J. Mol. Evol., 38, 405-419.
7.Ban, N., Beckmann, R., Cate, J. H. D., Dinman, J.
D., Dragon, F., Ellis, S. R., Lafontaine, D. L. J., Lindahl, L.,
Liljas, A., Lipton, J. M., McAlear, M., Moore, P. B., Noller, H. F.,
Ortega, J., Panse, V. G., Ramakrishnan, V., Spahn, C. M. T., Steitz,
T., Tchorzewski, M., Tollervey, D., Warren, A. J., Williamson, J. R.,
Wilson, D., Yonath, A., and Yusupov, M. (2014) A new system for naming
ribosomal proteins, Curr. Opin. Struct. Biol., 24,
165-169.
8.Highland, J. H., and Howard, G. A. (1975) Assembly
of ribosomal proteins L7, L10, L11, and L12 on the 50S subunit of
Escherichia coli, J. Biol. Chem., 250,
831-834.
9.Hardy, S. J. S. (1975) The stoichiometry of the
ribosomal proteins of Escherichia coli, Mol. Gen. Genet.,
140, 253-274.
10.Terhorst, C., Moller, W., Laursen, R., and
Wittmann-Liebold, B. (1972) Amino acid sequence of a 50S ribosomal
protein involved in both EF-G and EF-T dependent GTP-hydrolysis,
FEBS Lett., 28, 325-328.
11.Ramagopal, S., and Subramanian, A. R. (1974)
Alteration in the acetylation level of ribosomal protein L12 during
growth cycle of Escherichia coli, Proc. Natl. Acad. Sci.
USA, 71, 2136-2140.
12.Gordiyenko, Y., Deroo, S., Zhou, M., Videler, H.,
and Robinson, C. V. (2008) Acetylation of L12 increases interactions in
the Escherichia coli ribosomal stalk complex, J. Mol.
Biol., 380, 404-414.
13.Pettersson, I., and Kurland, C. G. (1980)
Ribosomal protein L7/L12 is required for optimal translation,
Biochemistry, 77, 4007-4010.
14.Kirsebom, L. A., and Isaksson, L. A. (1985)
Involvement of ribosomal protein L7/L12 in control of translational
accuracy, Proc. Natl. Acad. Sci. USA, 82, 717-721.
15.Sander, G., Marsh, R. C., Voigt, J., and
Parmeggiani, A. (1975) A comparative study of the 50S ribosomal subunit
and several 50S subparticles in EF-T- and EF-G-dependent activities,
Biochemistry, 14, 1805-1814.
16.Koteliansky, V. E., Domogatsky, S. P., and
Gudkov, A. T. (1978) Dimer state of protein L7/L12 and EF-G-dependent
reactions on ribosomes, FEBS J., 90, 319-323.
17.Donner, D., Villems, R., Liljas, A., and Kurland,
C. G. (1978) Guanosinetriphosphatase activity dependent on elongation
factor Tu and ribosomal protein L7/L12, Proc. Natl. Acad. Sci.
USA, 75, 3192-3195.
18.Brot, N., and Weissbach, H. (1981) Chemistry and
biology of E. coli ribosomal protein L12, Mol. Cell.
Biochem., 63, 47-63.
19.Wong, K.-P., and Paradies, H. H. (1974) Shape
properties of proteins L7 and L12 from E. coli ribosomes,
Biochem. Biophys. Res. Commun., 61, 178-184.
20.Kar, E. G., and Aune, K. C. (1981) Solution
behavior of proteins L7/L12 from the 50S ribosomal subunit of
Escherichia coli, Biochemistry, 20, 4638-4646.
21.Gudkov, A. T., and Behlke, J. (1978) The
N-terminal sequence protein of L7/L12 is responsible for its
dimerization, FEBS J., 90, 309-312.
22.Gudkov, A. T., Tumanova, L. G., Gongadze, G. M.,
and Bushuev, V. N. (1980) Role of different regions of ribosomal
proteins L7 and L10 in their complex formation and in the interaction
with the ribosomal 50S subunit, FEBS Lett., 109,
34-38.
23.Gudkov, A. T., Gongadze, G. M., Bushuev, V. N.,
and Okon, M. S. (1982) Proton nuclear magnetic resonance study of the
ribosomal protein L7/L12 in situ, FEBS Lett., 138,
229-232.
24.Olson, H. M., Tewari, D. S., Traut, R. R., and
Glitz, D. G. (1986) Localization of two epitopes of protein L7/L12 to
both the body and stalk of the large ribosomal subunit, J. Biol.
Chem., 261, 6924-6932.
25.Oleinikov, A. V., Jokhadze, G. G., and Traut, R.
R. (1998) A single-headed dimer of Escherichia coli ribosomal
protein L7/L12 supports protein synthesis, Proc. Natl. Acad. Sci.
USA, 95, 4215-4218.
26.Bocharov, E. V., Sobol, A. G., Pavlov, K. V.,
Korzhnev, D. M., Jaravine, V. A., Gudkov, A. T., and Arseniev, A. S.
(2004) From structure and dynamics of protein L7/L12 to molecular
switching in ribosome, J. Biol. Chem., 279,
17697-17706.
27.Dey, D., Oleinikov, D., Dey, A. V., and Traut, R.
R. (1995) The hinge region of Escherichia coli ribosomal protein
L7/L12 is required for factor binding and GTP hydrolysis,
Biochimie, 77, 925-930.
28.Bubunenko, M. G., Chuikov, S. V., and Gudkov, A.
T. (1992) The length of the interdomain region of the L7/L12 protein is
important for its function, FEBS J., 313, 232-234.
29.Leijonmarck, M., Eriksson, S., and Liljas, A.
(1980) Crystal structure of a ribosomal component at 2.6 Å
resolution, Nature, 286, 824-826.
30.Wahl, M. C., Bourenkov, G. P., Bartunik, H. D.,
and Huber, R. (2000) Flexibility, conformational diversity and two
dimerization modes in complexes of ribosomal protein L12, EMBO
J., 19, 174-186.
31.Gudkov, A. T., Tumanova, L. G., Venyaminov, S.
Y., and Khechinashvilli, N. N. (1978) Stoichiometry and properties of
the complex between ribosomal proteins L7 and L10 in solution, FEBS
Lett., 93, 215-218.
32.Pettersson, I., Hardy, S. J. S., and Liljas, A.
(1976) The ribosomal protein L8 is a complex of L7/L12 and L10, FEBS
Lett., 64, 135-138.
33.Pettersson, I., and Liljas, A. (1979) The
stoichiometry and reconstruction of a stable protein complex from
Escherichia coli ribosomes, FEBS Lett., 98,
139-144.
34.Diaconu, M., Kothe, U., Schlunzen, F., Fischer,
N., Harms, J. M., Tonevitsky, A. G., Stark, H., Rodnina, M. V., and
Wahl, M. C. (2005) Structural basis for the function of the ribosomal
L7/12 stalk in factor binding and GTPase activation, Cell,
121, 991-1004.
35.Ilag, L. L., Videler, H., McKay, A. R., Sobott,
F., Fucini, P., Nierhaus, K. H., and Robinson, C. V. (2005) Heptameric
(L12)6/L10 rather than canonical pentameric complexes are
found by tandem MS of intact ribosomes from thermophilic bacteria,
Proc. Natl. Acad. Sci. USA, 102, 8192-8197.
36.Gordiyenko, Y., Videler, H., Zhou, M., McKay, A.
R., Fucini, P., Biegel, E., Muller, V., and Robinson, C. V. (2010) Mass
spectrometry defines the stoichiometry of ribosomal stalk complexes
across the phylogenetic tree, Mol. Cell. Proteom., 9,
1774-1783.
37.Rosendahl, G., and Douthwaite, S. (1993)
Ribosomal proteins L11 and L10. (L12)4 and the antibiotic
thiostrepton interact with overlapping regions of the 23S rRNA backbone
in the ribosomal GTPase centre, J. Mol. Biol., 234,
1013-1020.
38.Ban, N., Nissen, P., Hansen, J., Moore, P. B.,
and Steitz, T. A. (2000) The complete atomic structure of the large
ribosomal subunit at 2.4 Å resolution, Science,
289, 905-920.
39.Climie, S. C., and Friesen, J. D. (1987) Feedback
regulation of the rplJL-rpoBC ribosomal protein operon of
Escherichia coli requires a region of mRNA secondary structure,
J. Mol. Biol., 198, 371-381.
40.Johnsen, M., Christensen, T., Dennis, P. P., and
Fiil, N. P. (1982) Autogenous control: ribosomal protein L10–L12
complex binds to the leader sequence of its mRNA, EMBO J.,
1, 999-1004.
41.Harms, J., Schluenzen, F., Zarivach, R., Bashan,
A., Gat, S., Agmon, I., Bartels, H., and Yonath, A. (2001) High
resolution structure of the large ribosomal subunit from a mesophilic
eubacterium, Cell, 107, 679-688.
42.Klein, D. J., Schmeing, T. M., Moore, P. B., and
Steitz, T. A. (2001) The kink-turn: a new RNA secondary structure
motif, EMBO J., 20, 4214-4221.
43.Iben, J. R., and Draper, D. E. (2008) Specific
interactions of the L10–(L12)4 ribosomal protein
complex with mRNA, rRNA, and L11, Biochemistry, 10,
2721-2731.
44.Dijk, J., Garrett, R. A., and Muller, R. (1979)
Studues on the binding of the ribosomal protein complex L7/12–L10
and protein L11 to the 5′-one third of 23S RNA: a functional
centre of the 50S subunit, Nucleic Acids Res., 6,
2717-2729.
45.Stark, M. J. R., Cundliffe, E., Dijk, J., and
Stoeffler, G. (1980) Functional homology between E. coli
ribosomal protein L11 and B. megaterium protein BM-L11, Mol.
Gen. Genet., 15, 11-15.
46.Tate, W. P., Dognin, M. J., Noah, M.,
Stoffler-Meilicke, M., and Stoffler, G. (1984) The
NH2-terminal domain of Escherichia coli ribosomal
protein L11, J. Biol. Chem., 259, 7317-7324.
47.Schrier, P. I., and Moller, W. (1975) The
involvement of 50S ribosomal protein L11 in the EF-G dependent GTP
hydrolysis of E. coli ribosomes, FEBS Lett., 54,
130-134.
48.Kazemie, M. (1976) Binding of aminoacyl-tRNA to
reconstituted subparticles of Escherichia coli large ribosomal
subunits, Eur. J. Biochem., 67, 373-378.
49.Ilin, S., Hoskins, A., Ohlenschläger, O.,
Jonker, H. R. A., Schwalbe, H., and Wohnert, J. (2005) Domain
reorientation and induced fit upon RNA binding: solution structure and
dynamics of ribosomal protein L11 from Thermotoga maritima,
ChemBioChem, 6, 1611-1618.
50.Wimberly, B. T., Guymon, R., McCutcheon, J.
P.,White, S. W., and Ramakrishnan, V. (1999) A detailed view of a
ribosomal active site: the structure of the L11–RNA complex,
Cell, 97, 491-502.
51.Harms, J. M., Wilson, D. N., Schluenzen, F.,
Connell, S. R., Stachelhaus, T., Zaborowska, Z., Spahn, C. M. T., and
Fucini, P. (2008) Translational regulation via L11: molecular switches
on the ribosome turned on and off by thiostrepton and micrococcin,
Mol. Cell, 30, 26-38.
52.Bausch, S. L., Poliakova, E., and Draper, D. E.
(2005) Interactions of the N-terminal domain of ribosomal protein L11
with thiostrepton and rRNA, J. Biol. Chem., 280,
29956-29963.
53.Xing, Y., and Draper, D. E. (1996) Cooperative
interactions of RNA and thiostrepton antibiotic with two domains of
ribosomal protein L11, Biochemistry, 35, 1581-1588.
54.Thompson, J., Cundliffe, E., and Stark, M. (1979)
Binding of thiostrepton to a complex of 23S rRNA with ribosomal protein
L11, FEBS J., 98, 261-265.
55.Cundliffe, E., Dixon, P., Stark, M., Stoffler,
G., Ehrlich, R., Stoffler-Meilicke, M., and Cannon, M. (1979) Ribosomes
in thiostrepton-resistant mutants of Bacillus megaterium lacking
a single 50S subunit protein, J. Mol. Biol., 132,
235-252.
56.Uchiumi, T., Albert, J. W., and Traut, R. R.
(1987) Topography and stoichiometry of acidic proteins in large
ribosomal subunits from Artemia salina as determined by
crosslinking, Proc. Natl. Acad. Sci. USA, 84,
5580-5584.
57.Van Agthoven, A., Kriek, J., Amons, R., and
Moller, W. (1978) Isolation and characterization of the acidic
phosphoproteins of 60S ribosomes from Artemia salina and rat
liver, Eur. J. Biochem., 91, 553-565.
58.Santos, C., and Ballesta, J. P. (1994) Ribosomal
protein P0, contrary to phosphoproteins P1 and P2, is required for
ribosome activity and Saccharomyces cerevisiae viability, J.
Biol. Chem., 269, 15689-15696.
59.Maki, Y., Hashimoto, T., Zhou, M., Naganuma, T.,
Ohta, J., Nomura, T., Robinson, C. V., and Uchiumi, T. (2007) Three
binding sites for stalk protein dimers are generally present in
ribosomes from archaeal organism, J. Biol. Chem., 282,
32827-32833.
60.Casiano, C., Matheson, A. T., and Traut, R. R.
(1990) Occurrence in the archaebacterium Sulfolobus solfutaricus
ribosomal protein complex corresponding to Escherichia coli
(L7/L12)4–L10 and eukaryotic
(P1)2/(P2)2–P0, J. Biol. Chem.,
265, 18757-18761.
61.Szick, K., Springer, M., and Bailey-Serres, J.
(1998) Evolutionary analyses of the 12-kDa acidic ribosomal P-proteins
reveal a distinct protein of higher plant ribosomes, Proc. Natl.
Acad. Sci. USA, 95, 2378-2383.
62.Bailey-Serres, J.,Vangala, S., Szick, K., and
Lee, C.-H. K. (1997) Acidic phosphoprotein complex of the 60S ribosomal
subunit of maize seedling roots, Plant Physiol., 114,
1293-1305.
63.Tchorzewski, M., Boldyreff, B., Issinger, O. G.,
and Grankowski, N. (2000) Analysis of the protein–protein
interactions between the human acidic ribosomal P-proteins: evaluation
by the two hybrid system, Int. J. Biochem. Cell Biol.,
32, 737-746.
64.Nusspaumer, G., Remacha, M., and Ballesta, J. P.
(2000) Phosphorylation and N-terminal region of yeast ribosomal protein
P1 mediate its degradation, which is prevented by protein P2, EMBO
J., 19, 6075-6084.
65.Casiano, C., and Traut, R. R. (1991) Protein
topography of Sulfolobus solfataricus ribosomes by cross-linking
with 2-iminothiolane, J. Biol. Chem., 266,
21578-21583.
66.Lee, K.-M., Yu, C. W.-H., Chiu, T. Y.-H., Sze,
K.-H., Shaw, P.-C., and Wong, K.-B. (2012) Solution structure of the
dimerization domain of the eukaryotic stalk P1/P2 complex reveals the
structural organization of eukaryotic stalk complex, Nucleic Acids
Res., 40, 3172-3182.
67.Naganuma, T., Nomura, N., Yao, M., Mochizuki, M.,
Uchiumi, T., and Tanaka, I. (2010) Structural basis for translation
factor recruitment to the eukaryotic/archaeal ribosomes, J. Biol.
Chem., 285, 4747-4756.
68.Nomura, T., Nakano, K., Maki, Y., Naganuma, T.,
Nakashima, T., Tanaka, I., Kimura, M., Hachimori, A., and Uchiumi, T.
(2006) In vitro reconstitution of the GTPase-associated centre
of the archaebacterial ribosome: the functional features observed in a
hybrid form with Escherichia coli 50S subunits, Biochem.
J., 396, 565-571.
69.Nomura, N., Honda, T., Baba, K., Naganuma, T.,
Tanzawa, T., Arisaka, F., Noda, M., Uchiyama, S., Tanaka, I., Yao, M.,
and Uchiumi, T. (2012) Archaeal ribosomal stalk protein interacts with
translation factors in a nucleotide-independent manner via its
conserved C-terminus, Proc. Natl. Acad. Sci. USA, 109,
3748-3753.
70.Naganuma, T., Shiogama, K., and Uchiumi, T.
(2007) The N-terminal regions of eukaryotic acidic phosphoproteins P1
and P2 are crucial for heterodimerization and assembly into the
ribosomal GTPase-associated center, Genes Cells, 12,
501-510.
71.Uchiumi, T., Traut, R. R., and Kominami, R.
(1990) Monoclonal antibodies against acidic phosphoproteins P0, P1, and
P2 of eukaryotic ribosomes as functional probes, J. Biol. Chem.,
265, 89-95.
72.Lee, K.-M., Yusa, K., Chu, L.-O., Yu, C. W.-H.,
Oono, M., Miyoshi, T., Ito, K., Shaw, P.-C., Wong, K.-B., and Uchiumi,
T. (2013) Solution structure of human P1•P2 heterodimer provides
insights into the role of eukaryotic stalk in recruiting the
ribosome-inactivating protein trichosanthin to the ribosome, Nucleic
Acids Res., 41, 8776-8787.
73.Ito, K., Honda, T., Suzuki, T., Miyoshi, T.,
Murakami, R., Yao, M., and Uchiumi, T. (2014) Molecular insights into
the interaction of the ribosomal stalk protein with elongation factor
1α, Nucleic Acids Res., 42, 14042-14052.
74.Grela, P., Bernado, P., Svergun, D., Kwiatowski,
J., Abramczyk, D., Grankowski, N., and Tchorzewski, M. (2008)
Structural relationships among the ribosomal stalk proteins from the
three domains of life, J. Mol. Evol., 67, 154-167.
75.Baba, K., Tumuraya, K., Tanaka, I., Yao, M., and
Uchiumi, T. (2013) Molecular dissection of the silkworm ribosomal stalk
complex: the role of multiple copies of the stalk proteins, Nucleic
Acids Res., 41, 3635-3643.
76.Hagiya, A., Naganuma, T., Maki, Y., Ohta, J.,
Tohkairin, Y., Shimizu, T., Nomura, T., Hachimori, A., and Uchiumi, T.
(2005) A mode of assembly of P0, P1, and P2 proteins at the
GTPase-associated center in animal ribosome: in vitro analyses
with P0 truncation mutants, J. Biol. Chem., 280,
39193-39199.
77.Kravchenko, O., Mitroshin, I., Nikonov, S.,
Piendl, W., and Garber, M. (2010) Structure of a two-domain N-terminal
fragment of ribosomal protein L10 from Methanococcus jannaschii
reveals a specific piece of the archaeal ribosomal stalk, J. Mol.
Biol., 399, 214-220.
78.Santos, C., Remacha, M., and Ballesta, J. P. G.
(2004) Ribosomal P0 protein domain involved in selectivity of
antifungal sordarin derivatives, Antimicrob. Agents Chemother.,
48, 2930-2936.
79.Choi, A., Wong, E., Lee, K.-M., and Wong, K.-B.
(2015) Structures of eukaryotic ribosomal stalk proteins and its
complex with trichosanthin, and their implications in recruiting
ribosome-inactivating proteins to the ribosomes, Toxins,
7, 638-647.
80.Shimizu, T., Nakagaki, M., Nishi, Y., Kobayashi,
Y., Hachimori, A., and Uchiumi, T. (2002) Interaction among silkworm
ribosomal proteins P1, P2 and P0 required for functional protein
binding to the GTPase-associated domain of 28S rRNA, Nucleic Acids
Res., 30, 2620-2627.
81.Uchiumi, T., and Kominami, R. (1997) Binding of
mammalian ribosomal protein complex P0•P1•P2 and protein L12
to the GTPase-associated domain of 28S ribosomal RNA and effect on the
accessibility to anti-28S RNA autoantibody, J. Biol. Chem.,
272, 3302-3308.
82.Uchiumi, T., Kikuchi, M., Terao, K., Iwasaki, K.,
and Ogata, K. (1986) Cross-linking of elongation factor 2 to rat-liver
ribosomal proteins by 2-iminothiolane, Eur. J. Biochem.,
156, 37-48.
83.Ben-Shem, A., De Loubresse, S., Melnikov, N. G.,
Jenner, L., Yusupova, G., and Yusupov, M. (2011) The structure of the
eukaryotic ribosome at 3.0 Å resolution, Science,
334, 1524-1529.
84.Gabdulkhakov, A., Nikonov, S., and Garber, M.
(2013) Revisiting the Haloarcula marismortui 50S ribosomal
subunit model, Acta Crystallogr. Sect. D Biol. Crystallogr.,
69, 997-1004.
85.Shcherbakov, D., Dontsova, M., Tribus, M.,
Garber, M., and Piendl, W. (2006) Stability of the “L12
stalk” in ribosomes from mesophilic and (hyper)thermophilic
Archaea and Bacteria, Nucleic Acids Res., 34,
5800-5814.
86.Beauclerk, A. A. D., Hummel, H., Holmes, D. J.,
Bock, A., and Cundliffe, E. (1985) Studies of the GTPase domain of
archaebacterial ribosomes, FEBS J., 151, 245-255.
87.Sanchez-Madrid, F., Vidales, F. J., and Ballesta,
J. P. G. (1981) Functional role of acidic ribosomal poteins.
Interchangeability of proteins from bacterial and eukaryotic cells,
Biochemistry, 20, 3263-3266.
88.Stoffler-Meilicke, M., and Stoffler, G. (1991)
The binding site of ribosomal protein L10 in Eubacteria and
Archaebacteria is conserved: reconstitution of chimeric 50S subunits,
Biochimie, 73, 797-804.
89.Uchiumi, T., Hori, K., Nomura, T., and Hachimori,
A. (1999) Replacement of L7/L12–L10 protein complex in
Escherichia coli ribosomes with the eukaryotic counterpart
changes the specificity of elongation factor binding, J. Biol.
Chem., 274, 27578-27582.
90.Han, M.-J., Cimen, H., Miller-Lee, J. L., Koc,
H., and Koc, E. C. (2011) Purification of human mitochondrial ribosomal
L7/L12 stalk proteins and reconstitution of functional hybrid ribosomes
in Escherichia coli, Protein Expr. Purif., 78,
48-54.
91.Uchiumi, T., Honma, S., Endo, Y., and Hachimori,
A. (2002) Ribosomal proteins at the stalk region modulate functional
rRNA structures in the GTPase center, J. Biol. Chem.,
277, 41401-41409.
92.Ban, N., Nissen, P., Hansen, J., Capel, M.,
Moore, P. B., and Steitz, T. A. (1999) Placement of protein and RNA
structures into a 5 Å-resolution map of the 50S ribosomal
subunit, Nature, 400, 841-847.
93.Gao, Y.-G., Selmer, M., Dunham, C. M.,
Weixlbaumer, A., Kelley, A. C., and Ramakrishnan, V. (2009) The
structure of the ribosome with elongation factor G trapped in the
posttranslocational state, Science, 326, 694-699.