Structure, Properties, and Biological Relevance of the DNA and RNA G-Quadruplexes: Overview 50 Years after Their Discovery
N. G. Dolinnaya1*, A. M. Ogloblina2, and M. G. Yakubovskaya2
1Lomonosov Moscow State University, Department of Chemistry, 119991 Moscow, Russia; E-mail: dolinnaya@hotmail.com2Blokhin Cancer Research Center, Institute of Carcinogenesis, Russian Academy of Medical Sciences, 115478 Moscow, Russia
* To whom correspondence should be addressed.
Received June 15, 2016
G-quadruplexes (G4s), which are known to have important roles in regulation of key biological processes in both normal and pathological cells, are the most actively studied non-canonical structures of nucleic acids. In this review, we summarize the results of studies published in recent years that change significantly scientific views on various aspects of our understanding of quadruplexes. Modern notions on the polymorphism of DNA quadruplexes, on factors affecting thermodynamics and kinetics of G4 folding–unfolding, on structural organization of multiquadruplex systems, and on conformational features of RNA G4s and hybrid DNA–RNA G4s are discussed. Here we report the data on location of G4 sequence motifs in the genomes of eukaryotes, bacteria, and viruses, characterize G4-specific small-molecule ligands and proteins, as well as the mechanisms of their interactions with quadruplexes. New information on the structure and stability of G4s in telomeric DNA and oncogene promoters is discussed as well as proof being provided on the occurrence of G-quadruplexes in cells. Prominence is given to novel experimental techniques (single molecule manipulations, optical and magnetic tweezers, original chemical approaches, G4 detection in situ, in-cell NMR spectroscopy) that facilitate breakthroughs in the investigation of the structure and functions of G-quadruplexes.
KEY WORDS: G-quadruplex thermodynamics and kinetics, multiquadruplexes, G-quadruplex-specific ligands and proteins, RNA G-quadruplexes, hybrid DNA–RNA G-quadruplexes, telomeric G-quadruplexes, G-quadruplexes in oncogene promotersDOI: 10.1134/S0006297916130034
The non-canonical DNA structures formed by conformational rearrangement of the double-stranded genome regions with specific base sequences are currently considered as a novel set of regulatory elements. Four-stranded G-rich helical structures called the G-quadruplexes (G4s) are among the most amazing and actively investigated non-canonical forms of DNA. G-quadruplexes discovered more than half a century ago as a curious phenomenon of guanosine gel formation now are perceived in the scientific mind as an important structural element of the genome. Currently, G4 formed by RNA molecules have become the subject of intensive studies [1, 2].
Nucleic acid sequences forming G4 (G4-motifs) have been found in G-rich repeats of eukaryotic genomes such as telomeric DNA, micro- and minisatellite sequences, as well as in regulatory regions of genomes, specifically in oncogene promoters. Data have been obtained by novel experimental approaches indicating that G4 play an important role in regulation of key cellular processes [3, 4], such as replication [5, 6], chromosome end protection [7], transcription [8], mutagenesis [9], genome damage repair [10], DNA recombination [11], and epigenetic processes [12, 13], as well as posttranscriptional events: translation [14], RNA splicing, alternative polyadenylation of mRNA [15], and others.
Many reviews and even books have been dedicated to the different aspects of G4 formation, their topology, factors affecting stability and polymorphism of these structures, and interaction of G4s with small-molecule ligands [3, 16-18]. However, the number of publications examining these non-canonical forms of nucleic acids has grown steadily in the last decade, which has been related mainly with progress in investigation of their biological functions. Interest in G4s is also because their formation is associated with widespread human diseases (cancer, cardiovascular diseases, diabetes, neurodegenerative disorders) [19, 20]. In recent years, G4s have been actively used as components of nanostructures and low-cost switches that offer promise for applications in nanotechnology and biology [21-23].
Unlike in previous review articles that considered specific issues related to G4s, in this review we summarize all aspects of the quadruplex studies. Data from recent papers that significantly change our notions on the structural polymorphism, thermodynamics and kinetics of G4 folding–unfolding, as well as factors affecting the equilibrium between DNA duplexes and non-canonical four-stranded DNAs, and the structural arrangement of multiquadruplexes are analyzed. In addition to these aspects, new information on the structure and stability of G4s in telomeric DNA and oncogene promoters, and on conformational features of RNA G4 and of hybrid DNA–RNA quadruplexes is reviewed. Also we summarized data on various classes of ligands recognizing G-quadruplexes, on G4-specific proteins and antibodies, and on the existence of G4s in vivo. In this review, we present the current state of nucleic acid G-quadruplex science including a short history of the development of our notions on their structure and properties. Special attention was paid to novel experimental approaches that provided the breakthrough in investigation of G-quadruplexes and their visualization in biological objects.
STRUCTURAL FEATURES OF NUCLEIC ACID G-QUADRUPLEXES AND FACTORS
AFFECTING THEIR STABILITY AND CONFORMATIONAL DIVERSITY
G4s are formed via intra- or intermolecular interactions of DNA or RNA molecules containing tracts of oligoG (G-tracts). The central part of a quadruplex (core) consists of G-tetrads, in which four guanine residues from different strands or different G-tracts (in the case of intramolecular G4) are linked through a system of Hoogsteen hydrogen bonds (more precisely H-bonds formed with participation of Watson–Crick and Hoogsteen faces of guanine bases). Stacking interactions of planar G-tetrads as well as additional interactions of sugar-phosphate backbone fragments [24] form the specific G4 structure (Fig. 1). It should be mentioned that the quadruplex-like structures are formed even during self-association of guanosine monophosphates. This is related with the specific properties of guanine residues, which have several mutually corresponding electron donor and electron acceptor centers and exhibit enhanced capability for stacking interactions.
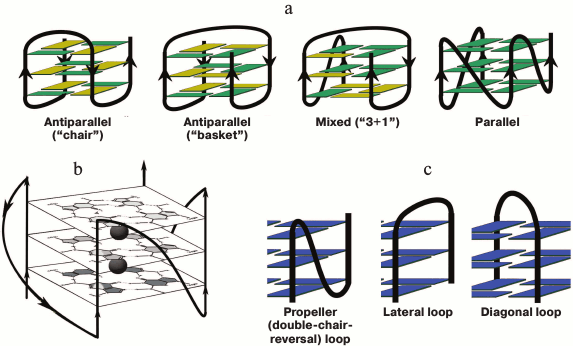
Fig. 1. Schematic representation of intramolecular G-quadruplexes differing in strand orientation in the quadruplex core; guanosines in syn-conformation are presented in yellow, and in anti-conformation – in green (a). Parallel-stranded G4 with presented structure of G-tetrads and potassium-binding sites (black circles) (b). Types of loops that connect G-tracts (c).
In in vitro studies, G-quadruplexes were formed by intermolecular interactions of two, three, or four oligonucleotide molecules containing G-tracts, as well as by intramolecular folding of one oligonucleotide molecule (Fig. 1a). A base sequence capable of intramolecular folding into a G4 is usually denoted as G2-5L1G2-5L2G2-5L3G2-5, where G2-5 are G-tracts containing from two to five consecutive guanosine residues, and Ln are oligonucleotide linkers connecting these tracts. Until recently, the quadruplex core was considered as several right-handed G-tetrads [16]. Only in 2015, a left-handed G-quadruplex analog of Z-DNA was discovered using X-ray analysis and NMR spectroscopy, which was formed during intramolecular folding of the d(T(GGT)4TG(TGG)3TGTT) oligonucleotide under near-physiological conditions [25].
Considerable structural diversity is characteristic for DNA G4s due to such factors as the number of forming molecules, length of G-tracts, mutual orientation of strands, base sequence (composition) and length of G4 loops, availability of oligonucleotide fragments flanking the G4-motif, type of cations in the medium, and others [26]. G4s can adopt manifold topologies characterized by different orientation of the four strands in the quadruplex stem. The structure is parallel if all the strands point in the same direction. In antiparallel G4 two of the four strands point in the same direction (two types are identified – “chair” and “basket” differing in the type of loops), and in mixed antiparallel–parallel (3 + 1) G4 three strands have the same orientation (Fig. 1a). Classification of DNA G-quadruplexes on the basis of the quadruplex twist angles is presented in [27]. Despite the fact that loops do not involve in G-tetrad formation, they play a key role in the overall folding and stability of G4; quadruplex loops represent elements of secondary structure and are the main sources of tensions and limitations in the quadruplex structure. Three types of loops are characteristic for G4s (Fig. 1c): propeller (double-chair-reversal) ones connecting G-tracts forming neighboring edges of the four-stranded core (arbitrarily they can be defined as loops connecting “upper” and “lower” G-tetrads); no inversion of the nucleic acid chain direction occurs in this case; lateral (side) loops connecting neighboring G-tracts (change of the chain direction occurs); diagonal loops connecting G-tracts forming opposite edges of the G-quadruplex (change of the chain direction occurs). Formation of one type of loop or another depends on the number of G-tetrads, number of nucleotide units between the G-tracts, and their composition, and it is directly related to the strand orientation in the quadruplex.
Specific coordination of mono- and bivalent cations inside the G4 as a stabilizing factor of such non-canonical structures of nucleic acids. Considering that G-quadruplexes are formed due to association of four poly(oligo)anions, electrostatic interactions are their main stabilizing factor. However, the ions concentrated around the G-quadruplex helix that neutralize the charges of phosphate groups in the sugar-phosphate backbone contribute only slightly to G4 formation [28]. Unlike other types of structured nucleic acids, G4 specifically bind (coordinate) ions of mono- and bivalent metals within the highly electronegative central channel along the axis of the G4 stem [29]; the cations lose their hydration shell in the process. That is why the ionic radius is a significant factor ensuring the ability of ions to stabilize quadruplexes. Information on location of cations inside the G-quadruplex structure and on the dynamic of their binding to DNA was obtained with the NMR spectroscopy and X-ray study. The mechanism of a cation migration into the G4 core was elucidated using molecular dynamics simulation; it was shown that migration occurs predominately via the terminal G-tetrads, and the quadruplex loops affect the process. Location of internal cations depends on their size and charge. The distance between the electronegative oxo-groups of guanine residues in the G-tetrad decreases when a cation enters the quadruplex channel, due to electrostatic attraction, which prevents the release of cation into bulk solution [30].
Among monovalent cations, the maximum stabilizing effect has been observed for K+, which is located between two consecutive G-tetrads forming eight coordination bonds with the guanine carbonyl groups (Fig. 1b). The spaces between the lateral or diagonal loops of the quadruplex and the terminal G-tetrad are alternative sites for binding potassium ions [30]. Sodium ions, which usually localize in the tetrad plane and form four coordination bonds with the guanine residues, exhibit less pronounced stabilizing effect [31]. In addition to the ionic radius, the difference in the hydration free energy of the ions is suggested as an explanation for the different effect of K+ and Na+ on the G4 stability.
Ammonium ions were used as NMR probes for identification of coordination sites of monovalent ions with DNA. It was shown that NH4+ could be localized inside the quadruplex between neighboring G-tetrads the same as potassium ions [32]. According to other data [33], ammonium ions are too large to be embedded into the inner cavity of the quadruplex without partial opening of G-tetrads. Hence, the type of binding of these ions with G4 depends on the rigidity of the four-stranded structure defined by the temperature, and it is controlled by steric limitations of G-tetrads and the structure of the loops [33]. It was shown using NMR spectroscopy that thallium and rubidium ions are coordinated within the G-quadruplex structure like K+. Thermodynamic stability of G4 increases with increasing concentration of monovalent ions in the medium [34, 35]. It must be noted that Li+ and Cs+ ions have practically no stabilizing effect on the four-stranded DNA structure because their ionic radius either prevents their entry into the quadruplex channel or (if it is too small) remaining there by forming coordination bonds with the guanine residues [28]. As recently shown, increasing the concentration of ions that do not stabilize quadruplexes (Cs+ or trimethylammonium ion) at constant concentration of K+ could result in collapse of G4; this effect was not observed for any other DNA structure [35].
Ions of bivalent metals also affect the stability and conformation transitions between different forms of G4. Some of them (Ba2+, Sr2+) have sizes that allow entering the central quadruplex cavity [36] and stabilizing G4 significantly more strongly than monovalent ions. For example, the binding constant of intramolecular antiparallel G4 with barium cation is 10-fold higher than with the potassium cation [30, 37]. The pronounced stabilizing ability of strontium ions has been explained by their strong interaction with the guanine carbonyl groups and effective screening of the phosphate group charges [38]. Bivalent lead ions also stabilize quadruplexes efficiently because, as shown by X-ray analysis, they are located between the planes of neighboring G-tetrads and can replace K+ and Na+ in the quadruplex structure. Ca2+ and Na+ can coexist in the G4 core, exchanging their positions easily because their ionic radii are similar. The dependence of the G-quadruplex stabilizing ability of bivalent metal ions on concentration is bell-shaped: 2-5 mM salt solutions initiate G4 formation, while when concentrations are above 10 mM in combination with elevated temperature, some ions (Ca2+, Co2+, Mn2+, Zn2+, Ni2+, and Mg2+) can cause the quadruplex unfolding. Probably this is due to specific interactions of bivalent ions with electron acceptor groups of the guanines responsible for the formation of Hoogsteen hydrogen bonds. Comparative ranking of the degree of G4 stabilization by the mono- and bivalent metal ions is as follows: Sr2+ > Ba2+ > K+ > Ca2+ > Na+, NH4+ > Rb+ > Mg2+ > Li+ ≥ Cs+ [22]. The cations, especially the bivalent ones, initiate conformational transitions between the different G4 folding topologies. Thus, conversion of antiparallel G4 into parallel one occurs under the action of Ca2+ [39]. A similar effect is observed for telomeric G4s when a small amount of KCl is added to Na+ solution. It was shown recently that the G4 with only two G-tetrad layers instead of the usually observed three-tetrad structure becomes the predominant form at extremely low concentration of potassium or strontium ions [22]. According to molecular dynamics simulation, the cation does not initiate the quadruplex formation, but rather stabilizes the preformed structure [30].
Factors affecting topology of DNA quadruplexes. The type of loops connecting G-tracts, their length, and base sequence represent such factors [40-42]. The two- or three-tetrad G4 with three single-nucleotide loops has parallel topology and no other, and all the loops are of propeller type [43, 44]. In the presence of K+, the structure with parallel orientation of all the strands is characteristic for the quadruplex with two single-nucleotide loops regardless of the length and base sequence of the third loop [26, 42, 45]. At the same time, in the presence of Na+ the topology of G4s formed by the same oligonucleotides is much more variable and depends to a greater degree on the G4-motif sequence than in K+ solution [42]. The G-quadruplex with three dinucleotide loops can adopt either parallel or antiparallel folding topology, but the parallel one is more energetically favorable [46]. The parallel-stranded structure prevails in G4 with only one single-nucleotide loop. The type of nucleotide (A, T, G, or C) in this loop does not affect the G-quadruplex topology [42]. It was shown using CD spectroscopy that within the library of intramolecular DNA quadruplexes with loop lengths varying from one to three nucleotides, the G4 topology shifts from parallel to antiparallel or mixed when the total length of all loops is larger than five nucleotide residues [26]. The fact that the propeller loops contain no more than three nucleotides [26] imposes steric limitations on the number of G-tetrad layers in the intramolecular quadruplex (that is on its “height”) [47, 48]. The number of guanosines in the G-tract does not necessary correspond to the number of G-tetrads [47]. Thus, it was shown by NMR spectroscopy that oligonucleotide containing G-tract made from 15 guanosine residues formed a parallel intramolecular G-quadruplex consisting of three G-tetrad layers and three propeller loops containing guanosine residues [48].
There is a correlation between the type of G4-motif folding and N-glycosidic syn- and anti-conformations of guanosines forming the quadruplex core. Four possible conformation sets in the GpG dinucleotide sequence within G4 are considered: syn–anti, anti–anti, anti–syn, and syn–syn. They differ significantly in their energy parameters due to the different geometry of stacking contacts between guanines. The 5′-terminal GpG sequence that assume syn–anti and syn–syn conformations are also considered in [49]. In these dinucleotides, the 5′-end hydroxyl group forms H-bonds with N3 of the guanine in syn-conformation. When all four strands in the G4 are oriented in parallel to each other, all the guanosines are in anti-conformation, which is most favorable energetically. In the case of G4 with other topologies, the syn–anti conformation of guanosines correlates with the mutual orientation of the strands changing both inside the G-tetrad cycle and along the quadruplex axis (Fig. 1a). Four grooves are located on the G4 surface with the width (narrow, medium, and wide) and distribution of the phosphate group charges in the sugar-phosphate backbone depending on the topology of the four-stranded structures [50, 51]. The parallel intermolecular quadruplex has four almost identical grooves that are of the same size as the minor groove of the DNA duplex. Two narrow and two wide grooves are formed in the G4 with alternating conformations anti–syn–anti–syn in each tetrad, while one wide, one narrow, and two medium grooves are formed in the G4 with alternating anti–anti–syn–syn-conformations in the G-tetrads.
Taking into consideration that there are 26 combinations of different loops and eight possible types of G-tetrads with various sets of syn–anti-conformations of guanosine residues, the number of possible conformations even for the quadruplexes formed via intramolecular folding of one oligonucleotide molecule is very large [52]. Formation of intermolecular structures further increases conformational diversity of G4s. Additional conformation possibilities emerge due to introduction of A-, C-, or T-tetrads into the quadruplex core, as well as due to formation of more complex planar pentads, hexads, heptads, and octads stabilized by H-bonds [53, 54] that contain other bases along with guanines.
Quadruplex associates. Some G4s, especially parallel ones, are prone to inter-quadruplex interactions [55]. It was shown by NMR spectroscopy that two intramolecular parallel G-quadruplexes in K+ solution stuck together due to interactions of 5′-end G-tetrads (“blunt” end interaction) [43, 56]. Models of quadruplex multimerization via stacking of 3′-3′- and 3′-5′-end G-tetrads have been described [57]. Both intramolecular [43] and intermolecular DNA quadruplexes can interact with each other; moreover, the contact between the G4 subunits can be formed not only by the G-tetrads, but also by heptads and octads with large hydrophobic surface [53, 54] that contain different bases in addition to four guanine residues. Another type of dimeric G4s is formed similarly to the assembly of DNA duplexes via “sticky” ends. “Sticking” together of two G-quadruplexes can occur due to guanosine residues from each subunit participating in formation of new G-tetrad at the two quadruplex subunit junction. This is what happens during intermolecular assembly of G4 by four molecules of the d(GGGT) oligonucleotide. The G-tracts of the initial G4 slide along each other providing the dimer formation. The design of oligonucleotide constructs for formation of G-quadruplex structures of higher order has been described [58].
LOCATION OF G-QUADRUPLEX-FORMING SEQUENCES IN GENOMES AND
TRANSCRIPTOMES OF VARIOUS ORGANISMS
The main strategy for genome analysis involves correct determination of sequences that can be assigned to G4-motifs. The set of sequences that were shown experimentally to form G-quadruplexes is considered as a basis set [59]. Algorithms for searching and marking of G4-forming sequences have been developed in the last decade due to accumulation of a large amount of data on sequencing of genomes of mammals and bacteria [60, 61]. The distribution of G4-motifs revealed by the QuadParser algorithm (GnL1-7GnL1-7GnL1-7Gn, where n ≥ 3, L = A, T(U), G, C) in the genomes of different organisms was investigated in several works [62-64]. It was shown that predominantly the eukaryotic genomes are enriched with G4-motifs. They are distributed non-randomly primarily away from the regions involved in nucleosome formation and in non-methylated regions [65]. High frequency of G4-motifs is observed within telomeric DNA [66] and micro(mini)satellite repeats [67, 68], in long terminal repeats (LTRs) of retrotransposons [69], in genes of ribosomal RNA, in regulatory regions of the genome such as gene promoters [70]; predominately in the coding (sense) strand [71], in origins of replication [5, 72, 73], in immunoglobulin switch regions and breakpoint regions of chromosome translocation [74], recombination hotspots [75], intronic sequences [76, 77], CpG islands, enhancers, and insulators [78], in mitochondrial DNA [79], as well as in various regions of the transcriptome including sites of alternative mRNA processing, splicing [80], 5′- and 3′-untranslated mRNA regions [2], telomeric non-coding RNA [81], pre-microRNA [82], and long non-coding RNA [83].
It was shown using the computational methods that G4-motifs are evolutionarily conserved [84]. The density of G4-forming sequences is significantly higher in the non-coding regions compared with coding ones [85]. There are more than 376,000 G4-motifs in the human genome according to the QuadParser algorithm. More than 40% of genes encoding human proteins contain at least one G4-motif in the promoter region [62]. On average, the distribution density of such sequences along the human genome is 0.153 per 1000 base pairs (bp), while their content in promoter regions is 1.48. The maximum on the distribution probability curve of G4-motifs in human, mouse, rat, yeast, and E. coli genomes corresponds to a site located at approximately 50 bp from the transcription start site (TSS). On the other hand, more than 800 G4-motifs are localized in regions removed from the TSS of 22,049 human genes by up to 5000 bp [86, 87]. The fact that G4-forming sequences were evolutionarily conserved support the role of DNA G-quadruplexes in key cellular processes [88]. The gene regulatory regions of warm-blooded animals are more enriched with G4 motifs [80], with the human genome containing the highest number. Moreover, the presence of G4-motifs has been observed equally in regulatory sites of early and late genes. In general, they precede oncogenes and less often tumor cell suppressor genes [89]. The G4-motifs are mostly associated with genes encoding proteins responsible for regulatory functions rather than “housekeeping” proteins. Homology between G4-motifs of vertebrates and yeasts in the mitochondrial and nuclear genomes was revealed by comparative in silico analysis [85]. Interestingly, there is one order of magnitude more G4-motifs in the mitochondrial DNA from S. cerevisiae than in the nuclear genome [90]. In plants, quadruplex-forming sequences acting as translation repressors were found in 5′-untranslated regions (UTR) of mRNA [91]. Retrotransposon LTRs, which are highly abundant in plants, contain a large number of G4-motifs [69].
False negative and false positive G4-motifs. Numerous regions have been elucidated in recent years in the human genome with primary structure deviating from the GnL1-7GnL1-7GnL1-7Gn pattern (where n ≥ 3, L = A, T(U), G, C), which nevertheless form quadruplex structures [92]. The G4-motif within minisatellite DNA that folds into a quadruplex with 9-nt middle loop is considered as a false negative [67]; the dominating G4 formed in the promoter of the human Bcl-2 gene assumes parallel topology with a 13-nt loop [87]. According to other data, the average middle loop of a stable G4 can contain up to 21 nucleotide residues [93]. Moreover, recently the evidence was presented for occurrence of bulges (that is the incorporation of non-guanine bases in G-tracts) in different G4 contexts. It was shown using NMR spectroscopy and other methods that single or multiple bulges in the quadruplex core can vary in length, nucleotide composition, and location [94]. There are also data of false-positive G4-motifs that obey the classical pattern but do not form quadruplexes. Thus, G-rich sequences in 5′-UTR of human mRNA were not able to fold into quadruplexes due to the availability of C-tracts in flanking regions, which hybridized with the G-tracts forming alternative secondary structures [14]. To overcome these limitations, other more accurate tools to predict G4 propensity of a given DNA or RNA sequences are needed. More general criteria should be taken into account, for example, the effect of neighboring sequences, which could cover wider functional regions of genomes and transcriptomes [95, 96]. Development and testing of a radically different algorithm, G4Hunter, was suggested in 2016. This algorithm takes into consideration G-richness and G-skewness (G/C asymmetry between the complementary strands of the double helix) of a given sequence [97]. To validate the G4Hunter, a large dataset was analyzed, and G4-forming potential for human mitochondrial DNA fragments was evaluated using the combination of biophysical methods. Analysis of genomes of various organisms using the developed algorithm showed that the number of sequences capable of forming stable G4s in the human genome is significantly (2-10-fold) higher than estimated before.
G4-motifs at the ends of eukaryotic linear chromosomes (telomeres). It is known that telomeric DNA consists of tandem repeats of G·C-rich non-coding sequences with a 150-250-nt single-stranded G-rich overhang at the 3′-end. In human cells as well as in the cells of all vertebrates, telomeric DNA comprises a (TTAGGG)/(CCCTAA) repeat with size of several thousands of base pairs. Similar G·C-rich repeats are characteristic for many phylogenetically distant species; the typical telomeric sequence in insects contains the (TTAGG)/(CCTAA) repeat, in plants – (TTTAGGG)/(CCCTAAA), in Tetrahymena – (TTGGGG/CCCCAA), and in the lower eukaryote (ciliate) Oxytricha nova – (TTTTGGGG/CCCCAAAA).
G4-motifs in bacterial and viral genomes. The distribution of G4-motifs in the genomes of these species is still poorly understood. The implication of G4s in virology only begins to be realized. In the E. coli genome, they also are mainly located in regulatory regions, which suggests participation of G4s in regulation of transcription [98]. The search for G-quadruplexes and elucidation of their functions in viral genomes has mostly focused on the HIV retrovirus. Location of G4-motifs near recombination hotspots, central polypurine tract at the 3′-end of the pol gene, and in HIV-1 LTR promoter indicates participation of G4s in recombination, replication, and regulation of HIV-1 promoter activity [99-101]. It has been suggested that G4s formed close to the origins of replication within DNA-containing Epstein–Barr virus play an important role in DNA replication and metaphase chromosome attachment [102]. Moreover, it was shown recently that G4 clusters were responsible for regulation of virus-encoded nuclear antigene 1 mRNA translation [103]. The G4-motifs presented in the non-coding regulatory DNA region of the SV40 virus can form unusual structures with a C-tetrad integrated into the quadruplex core. It was suggested that these quadruplexes play an important role in the regulation of replication as well as early and late transcription of the viral genome. The G4-forming sequences and their potential to form quadruplex structures were analyzed in the genomes of all known human papillomaviruses [104]. Participation of G4s in transcription, replication, and alternative splicing required for production of viral proteins has been discussed for at least some of them. The role of G-quadruplexes in the life cycle of viruses, such as HIV, as well as applications of G4-containing DNA aptamers as antiviral agents, diagnostics, and innovation tools have been discussed [43, 105].
METHODS USED FOR G4 ANALYSIS in vitro; THERMODYNAMICS AND
KINETICS OF QUADRUPLEX FOLDING–UNFOLDING
To characterize G-quadruplexes formed in a certain genome locus, each identified G4-motif is investigated in vitro under different experimental conditions close, to a lesser or greater degree, to the intracellular ones. The methods commonly used for the description of G4 topology and determination of thermodynamic and kinetic parameters of G-quadruplex folding and unfolding include CD [106, 107] and UV-spectroscopy [108, 109], differential scanning calorimetry [110, 111], gel electrophoresis [29, 52], X-ray crystallography [25, 112], chemical and enzymatic probing [101, 113, 114], fluorescence spectroscopy, fluorescence resonance energy transfer (FRET) [34, 46, 115, 116], including single molecule FRET [41, 117, 118], Raman spectroscopy [119], electron paramagnetic resonance [120, 121], hydrodynamic and chromatographic techniques [52], molecular modelling and molecular dynamics simulation [30, 122-124], mass-spectrometry [125], and NMR spectroscopy [56, 126, 127]. The detailed characteristic of several methods used for G4 investigation have been described [128].
A large amount of data is available in the literature on the kinetics and thermodynamics of G4 formation that were obtained using traditional approaches [128]. However, unlike G-quadruplex structure, these aspects have not been studied systematically. Also, correlation between the thermodynamic stability, kinetics, and G4 structures has not been considered. Typically, the thermodynamic parameters of individual G4 folding–unfolding were investigated and usually by different methods [129]. An algorithm for predicting quadruplex stability was developed based on non-systematic data [130]. Spontaneous folding of single-stranded sequences containing G-tracts into quadruplexes is a thermodynamically favorable process [128]. Unlike other non-canonical structures, G-quadruplexes are formed under conditions close to physiological, and their melting temperature (Tm) can exceed 70-80°C. Of critical importance for the stabilization and unfolding paths of G4s, not found in DNA and RNA duplexes, are the cations within the central quadruplex channel. The ΔG° and ΔH° values of intramolecular G4 formation from 22-26-nt sequences (3-4 G-tetrads) in 100 mM K+ vary in the range from –2 to –8 kcal/mol and from –30 to –80 kcal/mol, respectively; the stability of G-quadruplexes in Na+ solution is significantly lower [31]. Unlike G4, the stability of DNA- and RNA-duplexes does not depend on the type of monovalent cations. Analysis of the thermodynamic data has shown that intramolecular G4 are not more stable than other intramolecular complexes of nucleic acids [128]. Hairpin DNA structures exhibit significantly higher Tm value than G4s of comparable length under equal conditions [131]. It was also shown that in most cases the energy of G4 formation is not sufficient to separate the strands of double helix (>20 bp) in inner DNA sites such as promoter regions [128]. However, special properties have been assigned to intramolecular parallel G4 formed by the d((GGGT)3GGG) sequence; folding of this G4 structure is much more energetically favorable than formation of the DNA duplex in the presence of complementary strand [132]. Moreover, the replacement of guanosine in the first G-tract (at the second position) by an apurinic site does not prevent folding of the analogous sequence into G4, although it results in significant destabilization of the structure [133]. It was shown by molecular dynamics simulation that the vacant place in the formed G-triad is occupied by a water molecule. The unique binding pocket in G4 can be used as a specific target for low molecular weight ligands and nucleic acids metabolites. The G4 containing more than one vacant site (in one or different G-tetrads) is also reasonably stable [134]. Furthermore, G4 with a single apurinic site can be formed by other sequences, such as human telomeric repeats [135]. It is interesting that “excess” of guanosines in some G-tracts can cause their sliding relative to each other during formation of quadruplex structures. It was shown that this process stabilized the folded state; the Tm value of G4 increased significantly due to favorable entropic changes [136].
Factors affecting thermodynamic parameters of G4 folding–unfolding. Among these factors, the cation composition and ionic strength of the buffer solution, number of interacting molecules, their primary structure, and length of G-tracts [109], as well as the presence of nucleotide fragments flanking G4-motif [137] have been studied. The stability of G4 depends strongly on the type of loops, their length, and base composition/sequence [93, 138]. Replacement of only one base in a one- or two-nucleotide loop of a quadruplex and in the G4-flanking fragments can affect G4 stability [42, 137, 139]. In particular, the replacement of T or C by A in the one-nucleotide loop of parallel G4 with (1:3–9:1) loops causes significant destabilization of the quadruplex structure [42]; according to NMR-spectroscopy data, the adenine residue is pushed out of the G4 groove, unlike the thymine residue, which is in the quadruplex groove [140]. It was shown that independent of the orientation of DNA strands, the thermodynamic stability of intramolecular G4 decreased with increasing length of loops [26, 93, 139]. Indeed, each additional nucleotide in a propeller loop decreases the Tm value by 2°C [42, 93]. This dependence is not connected with the base sequence in the G4 loops; this was shown using a set of three-tetrad intramolecular G-quadruplexes with partially randomized loop sequences [26]. In the case when the total length of all loops exceeds five nucleotide residues, its further increase affects the thermodynamic stability only insignificantly. On the other hand, H-bonds or stacking contacts in long quadruplex loops [141], as well as stacking interactions of loop bases with terminal G-tetrads, could provide favorable contribution of enthalpy to the thermodynamics of G4 formation. The stability of G4 also increases when the bases in the long loop can hybridize with the bases of the 5′-end sequence flanking the G4-motif [142].
It is known that the G4 formation is accompanied by the release of water molecules [143]. The thermodynamic contribution of dehydration and capture of counter ions was estimated in [50]. It was shown that water affected thermodynamic stability and polymorphism of G4s [110, 144]. The decrease in water activity caused by addition of various alcohols or polyethylene glycol (PEG) into aqueous solution, results in stabilization of the quadruplex [145]; PEG is usually used as molecular crowding agent to create cell-mimicking conditions. It should be noted that the solvation effects observed for intramolecular quadruplexes and DNA double helices are diametrically opposite; decrease in water activity destabilized B-DNA, but increased the stability of G4 [131]. However, the effect of PEG causing dehydration of quadruplexes on their stability is not so unambiguous. As shown in 2016, the G4 stability in the presence of monovalent ions increased after addition of molecular crowding agents. In contrary, PEG stabilized only parallel G4s, but had no effect on stability or even destabilized antiparallel or mixed G4 structures in the presence of bivalent ions [143]. According to a suggested hypothesis, Sr2+, Ba2+, and Pb2+ compensate to some degree the effect of dehydration accompanying formation of G4 structure, especially in the case of G-quadruplexes with antiparallel orientation of strands.
The most detailed and comprehensive information is available on the thermodynamics and kinetics of intramolecular G4 formation by oligonucleotides containing four G-tracts from human telomeric DNA repeats [109, 128, 130]. The thermodynamic data for oligonucleotide models with identical or similar primary structure were summarized in a review published in 2010 [129], where the presented ΔG° values varied over a wide range (3.4-14.8 kcal/mol). The reasons for the observed parameter variation were associated with polymorphism of telomeric DNA quadruplexes. It is now recognized that most G4-motifs can form G-quadruplexes of different architecture that are energetically similar and at dynamic equilibrium. Various types of G4 conformations can be formed at different rates, which results in non-identical sets of conformers under variant conditions of sample preparation and using different investigation methods [146]. But even under the same conditions the identical telomeric sequence can fold into many different quadruplex structures [109], and the formed G4 can convert into other conformations that are separated by only slight energy barriers (2 kcal/mol) [147]. It was shown that individual conformers could be almost identical in thermodynamic, electrophoretic, and hydrodynamic properties, but exhibit different CD and NMR spectra as well as kinetic stability [52].
Controversial opinions have been presented in the papers regarding whether the intramolecular folding–unfolding of the single G4 is a simple one-step process. It has been advocated in some publications that G4 melting proceeds according to a two-state model [111]. However, most researchers believe this process proceeds via intermediate stages and does not correspond to an “all-or-none” model [129, 148, 149]. Using CD spectroscopy, fluorescence assay, and FRET, a multistep mechanism of telomeric G4 folding was verified recently, and the kinetics of each step was investigated [150, 151]. According to data of biophysical, computational, and other methods, G4 formation proceeds via thermodynamically and kinetically significant intermediates even during intramolecular folding of the oligonucleotide [149, 152]. These intermediates can comprise hairpin structures whose interaction initiates formation of the quadruplex core, or these can be G-triplexes varying in topology that are formed via separation of one of the G-tracts from the quadruplex [153]. The evidence for the occurrence of these structures stabilized by G-triads was obtained experimentally by NMR and CD spectroscopies, differential scanning calorimetry, and other techniques [134, 149], and the properties of possible G-triplex forms were analyzed by molecular dynamics techniques [154]. It was shown that the number of intermediates determined the kinetics of full-sized G4 folding. The number of these states observed in K+ solution was less than in Na+ buffer [155]. The fundamental difference in the behavior of parallel G4 with all guanosines in anti-conformation, which allowed slippage of G-tracts along each other with formation of intermediates with lower number of G-tetrads in the quadruplex core, and antiparallel G4, in which the syn- or anti-conformation of guanosine residues correlating with the strand directions blocked the slippage process, was specifically emphasized. It is of interest that the number and type of residues attached to 5′- and to 3′-ends of the G4 affect the rate of quadruplex formation, but they do not change its multistep mechanism.
The kinetics of G-quadruplex folding depends on the base sequence of the G4-motif and varies in the range from milliseconds to minutes. It was noted that the time of G4 formation of human telomeric repeats that were capable of very fast intramolecular folding correlated with the time of DNA replication [156]. According to data obtained using various experimental approaches, high rate of telomeric G4 folding (16-50 s–1) was observed at low concentrations of potassium ions (<10 mM), while the rate of their unfolding was low (1.3·10–3 s–1) [128, 155]. Unusually slow melting of intramolecular quadruplexes was also postulated [157]. The statement on the high folding rate of the G-quadruplex structure contradicts the results of theoretical analysis [158], according to which competition between the different conformations of G-quadruplexes including the intermediates (especially DNA-hairpin structures) makes this multidirectional process very slow. In the view of authors, the G4 formation follows a very complex kinetic pathway involving multiple alternative states, and only a small fraction of molecules is folded into the final G4 structure. Data obtained recently by NMR [159] and CD [110] agree with such kinetic pattern.
Regarding intermolecular G4s, the polymorphism of these structures is defined by kinetics limitations even more than in the case of intramolecular G4s. Kinetic models that differed significantly depending on the type of the cation (K+ or Na+) in the environment were suggested in a work of Prislan et al. [160] based on traditional approaches. These models describe the processes of G4 folding–unfolding and structural transitions between the different quadruplex conformations formed by the d(G4T4G3) oligonucleotide with two G-tracts.
Potential of single-molecule techniques for G-quadruplex study. The commonly used experimental methods allowing exploring averaged properties of a multitude of molecules (CD, NMR spectroscopy, X-ray crystallography) are not so informative for the analysis of distinct G4 structures, especially those comprising a small fraction within heterogeneous population. Investigation of behavior of single molecules provides more information for determination of the subpopulations of various structures in polymorphic mixture. For instance, the structure and dynamics of G4 at the single molecule level was investigated using FRET, and the coexistence of several types of quadruplex conformations formed by human telomeric repeats was revealed [117]. In recent years, the original laser and magnetic tweezers were developed for investigation of thermodynamics and kinetics of conformational transitions that occur due to mechanical stretching–relaxation of single-stranded G-rich repeats (telomeric DNA or promoter fragments) sandwiched between long pulling double-stranded DNA (dsDNA) handles immobilized on two optically-trapped or paramagnetic beads [146, 153, 157, 161, 162]. According to [163], unwinding of structured DNA under stretching is physiologically more rational that thermal melting, the latter being impossible in vivo. On the contrary, DNA replication, transcription, and other processes can stretch DNA, which causes local unwinding of the double helix. The mechanical stability of G4s was evaluated using optical tweezers, and it was found to be comparable with the force causing the stop of DNA- and RNA-polymerases. Comparative study of mechanical stability of G4 and the hairpin DNA under physiological conditions using the same technique revealed that unlike the hairpin structures, DNA quadruplexes could serve as an efficient barrier for replication or transcription [164]. It was shown using molecular dynamics simulation that the unfolding event of intramolecular G4s of different topology under mechanical stretching was accompanied by the loss of coordination between the central ion and guanines of the G-tetrads [165], which agreed with experimental data [166].
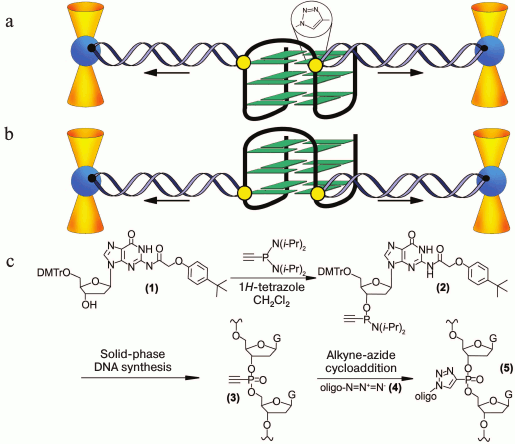
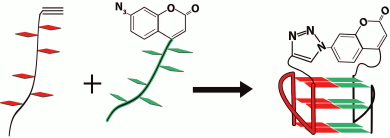
In pioneering work [167], new strategy was developed to discriminate the contribution of loops and G-tetrad stacking to quadruplex stability at the force-induced G4 unfolding. The contributions from G4 elements were determined using optical tweezers technique and click chemistry (Fig. 2). Mechanical unfolding of human telomeric G4 revealed that the loops’ interactions contribute more to the G4 stability than the stacking of G-tetrads.
The use of magnetic tweezers allowed characterizing the equilibrium conformational transitions in intramolecular telomeric G4 at physiological concentrations of potassium ions (~100 mM) [146], which was impossible to do using optical techniques due to the high stability of quadruplexes under these conditions. Three quadruplex conformations were revealed in a timeframe of several minutes, which exhibited different lifetimes and mechanical stabilities. It was shown that the kinetically controlled distribution of human telomer DNA G4 isomers depended on conditions of their formation [168]; in other words, the longer-lived and more thermodynamically stable isomers could be accumulated in a sample with increasing experiment duration. The magnetic tweezers technique was used to study the kinetic properties of G4 formed from a 27-nt G-rich region of the oncogene c-Myc promoter at 100 mM K+. The dominating G4 conformation was shown to melt more slowly by several orders of magnitude than G-quadruplexes of other topologies [157]. It was suggested that this non-canonical structure could play a role of a kinetic barrier in transcription regulation.
Fig. 2. Direct quantification of loop interactions and G-tetrad stacking using the optical tweezers and chemical approaches. Assisted by click chemistry, pulling dsDNA handles were attached via two modified guanines in each of the three G-tetrads. The G4 is presented on the scheme with “upper” or “lower” tetrad covalently bound to the DNA duplexes (a, b). Attachment of the dsDNA fragment carrying an azide group (4) to the guanosine derivative of the G4-motif with an alkyne group in the adjacent phosphate residue (3) was conducted via alkyne-azide cycloaddition (5). The modified guanosine residues after introduction of the alkyne group to the 3′-end phosphate (1) were used as synthons (2) in solid-phase oligonucleotide synthesis. This allowed introducing alkyne-containing residues to predetermined positions of the G-rich oligonucleotide (c). A similar strategy was used for attachment of dsDNA to G-quadruplex loops. The laser beams focused onto beads are presented as yellow cones.
Analysis of the published data indicates that the paths of G4 folding–unfolding and their thermodynamic and kinetic parameters can vary for different quadruplexes and different experimental conditions. It is likely that there is no universal mechanism appropriate for all G4s and sample preparation conditions. But the combination of modern experimental techniques and advanced computational methods could help to elucidate which quadruplex conformations are most represented on biologically significant time scales.
FACTORS AFFECTING DNA-DUPLEX–G-QUADRUPLEX–i-MOTIF
EQUILIBRIUM
Besides telomeric ends, G4-forming sequences are present in the genome together with complementary strands in the form of double helix. Thus, investigation of thermodynamics and kinetics of the equilibrium between G4 and duplex DNA is important because the double helix is the native context for genomic DNA. The fluorescence assay in a combination with G4-recognizing low molecular weight ligands was used to measure the G4 folding propensity in both single-stranded and double-stranded DNAs [116]. As expected, the ability of G4-motif to form G4 was substantially diminished in the DNA duplexes due to the competition from the Watson–Crick base pairing. Unlike G4 folding sequence in single-stranded DNA, which forms both parallel and antiparallel G4s, dsDNA displays only parallel folding. It was shown in several studies that G4–DNA-duplex equilibrium depended on many factors affecting relative stability of both secondary structures: temperature, availability of fragments flanking G4, primary structure of G-motif, cationic composition, and other factors. One such factor is the length of loops in the quadruplex; the shorter they are, the more stable is the G4 and, consequently, the less stable is the duplex formed in the presence of complementary strand due to the shorter hybridization site [116]. Thus, oligonucleotide with human telomeric repeats d(TTAGGG)4 (the length of loops in the G4 structure are 3 : 3 : 3) forms duplex under physiological conditions in the presence of complementary C-rich strand, while the sequence of the C-myc promoter d(GGGTGGGGAGGGTGGG) predominantly assumes G4 conformation with 1 : 2 : 1 loops. The same factor – change in length of the hybridization site – shifts the equilibrium towards the duplex formations in the case of the G4-motif flanked with nucleotide residues [169]. Similar regularities were observed during monitoring of isothermal G4 unwinding in the presence of complementary strand carrying fluorescent label [170]. It was shown that availability of the sequence neighboring the G-quadruplex accelerated this process approximately two-fold, and the K+ concentration was found to be a key factor controlling the kinetics of G4 unwinding in the presence of a complementary strand. Thus, the G4 formed by the C-myc promoter sequence remains stable in the presence of the complementary strand even at 70°C in 100 mM KCl. If the G4-motifs potentially can fold intramolecularly into imperfect hairpin duplex as in the case of the promoter region of the WNT1 gene, d(GGGCCACCGGGCAGGGGGCGGG), the K+-induced transition into G4 occurs significantly more slowly than the quadruplex formation from the unstructured oligonucleotide [171]. Addition of PEG, which decreases water activity, facilitates G4 assembly even in the absence of cations and shifts the equilibrium from the DNA duplex formed by the telomeric repeats with the complementary strand to G4. The use of complementary probes with enhanced hybridization ability, for example peptide nucleic acids (PNA), on the contrary results in effective G4 unfolding and formation of a heteroduplex (DNA/PNA) [172].
The DNA double helix in vivo becomes temporarily single-stranded during replication, reparation, and transcription, and the open forms can later fold into various non-canonical structures. The conformational rearrangement of duplex DNA into G4, for instance, in gene promotors, can be reached through supercoiling effect. It is known that the transcription machinery can generate negative supercoils behind the moving RNA polymerase. Negative superhelicity stimulates G4 formation [86], because the local unwinding of duplex DNA is facilitated under conditions of topological stress, thus relieving the superhelical constraints. The effect of supercoiling can propagate along the double helix of chromosomal DNA over large distances, initiating conformational rearrangements of genome sites separated by several thousand base pairs from the TSS [86]. That is why not oligonucleotide models but supercoiled plasmids containing inserts of double-stranded G·C-rich DNA with G-tracts on one of the strands and C-tracts on the complementary one are better for modeling in vivo conditions. It was shown using these models that G4 can exist inside a duplex DNA that is the native genomic context [113, 173]. They are formed much more easily in the case of negative supercoiling that is before the transcribed sequence but not behind it [86]. The statistical mechanism of transitions from B-DNA to G4 includes competition between the two states, and the equilibrium distribution between them at a certain level of negative supercoiling is calculated from values of free energy for alternative conformations [174]. Modern techniques of magneto-optical tweezers that combine the nanometer resolution with the possibility to generate the prescribed level of DNA supercoiling by a pair of magnets were used for quantitative evaluation of the relationship between the degree of topological stress and the efficiency of G-quadruplex formation. It was shown that the G4 population increased from 2.4 to 23% with increase in the density of negative supercoiling to –0.05; moreover, the G4 formation efficiency correlated with the readiness of the DNA double helix melting [175]. In addition to supercoiling, the structure of chromatin, binding of proteins specifically recognizing quadruplex structures [4], and the effect of molecular crowding [116] can mediate a shift of equilibrium from DNA duplex to G4 in vivo.
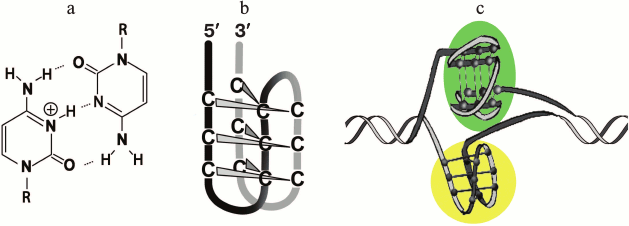
It was shown using oligonucleotide models and plasmid constructs that the intramolecular folding of G-rich sequences into the G4 could be accompanied by the formation of non-canonical four-stranded i-motif by complementary the C-rich strand (Fig. 3) [70, 113, 176, 177]. The enzymatic and/or chemical footprinting experiments are usually used to provide direct insight into the G4 and i-motif folding patterns because these purine-rich/pyrimidine-rich tracts are the nuclease-hypersensitive regions [113]. They are considered as markers of B-form distortions, such as “unwound” regions and various non-canonical structures. It is known that the pH-dependent i-motif structure is formed by two parallel duplexes stabilized by semi-protonated C≡C+ pairs (Fig. 3a) through their intercalation in mutually antiparallel orientations (Fig. 3). The factors that affect the stability of i-motif (pKa value, length of loops, epigenetic modification of cytosine residues, presence of PEG, degree of hydration) were recently analyzed [178, 179]. Oligonucleotides with G-rich and C-rich sequence from the gene promoters were shown in in vitro experiments to fold into G-quadruplexes and i-motifs, respectively, under certain conditions. The competition between two types of four-stranded structures and DNA duplex depended on the cation composition and pH of the medium. Distance-dependent DNA duplex destabilization caused by proximal G4 or i-motif structures was estimated [180]. This rather artificial model comprises a fragment of double helix with protruding single-stranded end, which folds into one of the non-canonical forms. Though the slightly acidic medium stabilized the i-motif [176], the formation of this alternative structure occurred even at neutral pH under conditions of DNA supercoiling [113] or in a molecular crowding environment imitated by adding of special reagents [181, 182] – two physiologically relevant states of the genome.
Fig. 3. Four-stranded i-motif formed by C-rich strand that is complementary to G4-forming sequences. Semi-protonated C≡C+ pairs (a) stabilizing parallel duplexes whose mutual intercalation results in formation of i-motif with the C≡C+ pairs being in different planes (b). Two types of four-stranded structures formed by G/C-rich sequence integrated into DNA plasmid; G4 is shaded with green, and i-motif – with yellow (c).
There are contradicting data on whether an i-motif can be folded in the complementary single-stranded region after a G4 is formed in the opposite strand, or they are mutually exclusive. On one hand, formation of G-quadruplex and i-motif in the promoter regions of the c-Myc gene inserted into the supercoiled plasmid (Fig. 3c) was established using chemical footprinting [113]. It was found that the G4 and i-motif were slightly displaced relative to each other (contain only three common G·C-tracts from the four required); moreover, a longer DNA fragment was used for formation of i-motif, because, unlike G-quadruplex, long loops increased the stability of the four-stranded C-rich structure [70]. On the other hand, it was shown using the laser tweezers-based single-molecule technique and chemical footprinting approach [163] that the fragment of double helix that contained the G·C-rich region (ACAGGGGTGTGGGC)2/(GCCCACACCCCTGT)2 from the insulin-linked polymorphic region (ILRP) formed only one four-stranded structure, but not both, under conditions that favor the formation of G4 and i-motif – pH 5.5, 100 mM K+. Formation of only one of the quadruplexes in this site was attributed to mutual steric hindrances emerging due to the mechanical stretching–relaxation of the dsDNA. A more general conclusion was suggested in a study conducted in 2016, where the formation of G4 and i-motif in different fragments of double-stranded DNA (human telomeres and gene promoter regions) was investigated using the optical tweezers technique and population analysis. It was shown that mutually exclusive formation of one of the quadruplexes was the consequence of steric hindrance in duplex DNA; thus, simultaneous formation of both quadruplexes separated in the complementary strands was observed in the Bcl-2 gene promoter [183]. The variability in formation of four-stranded non-canonical DNA structures governed by appropriate base sequence can facilitate fine-tuning of gene expression [184], because the G4 formation represents the signal for inhibition of transcription, while the i-motif can play a role as the activation signal [177, 184].
Oligonucleotide DNA-models with coexisting duplex and G-quadruplex domains. In recent years, oligonucleotide systems containing G4-duplex hybrids of various topology have attracted considerable attention [185]. Investigation of thermodynamic stability of constructs with contrasting orientations of the duplex stem with respect to the G4 core (coaxial or orthogonal) (Fig. 4) showed that in the case of the structured long loop, the stability of the quadruplex increased; furthermore, the type of the base pair closest to the duplex-G4 junction and the availability of bulges at the boundary of two domains affected significantly the stability of the formed secondary structure [186]. As shown by NMR spectroscopy, stable structures were formed also in the case of multiple duplex stems within the same loop of the parallel G4. Moreover, the multiple duplex inserts could be located in different loops of the same G4, allowing wider variety of the architecture of these structures [187]. The availability of sequences capable of folding into G4 with double helix in one of the loops (with length up to 20 nucleotide units) was analyzed in 2015 [188]. A large number (80,307) of duplex stem-loop-containing G4 motifs organized as clusters was identified in the human genome: in the hotspots of mutations and in the promoter regions of genes associated with oncological diseases and brain tissue pathogenesis. It was suggested that this type of two-domain structure can participate in various biological processes including pathological ones.
Fig. 4. DNA aptamers that have coexisting structures of G4 and duplex localized in one of the G4 loops (base pairs are marked with pink). The construct (a) involves the orthogonal orientation of the duplex and the parallel G4 with no base stacking between two domains. Construct (b) involves coaxial orientation of the duplex and G-quadruplex helices with continual base stacking across the two domains.
In other types of duplex–G4 hybrids, complementary sequences were attached to the 5′- and 3′-ends of a G4-motif [189, 190]. It was shown using the thrombin-binding aptamer as an example that both duplex and antiparallel G-quadruplex domains coexist in intramolecular structure of aptamer. The interrelationship between the duplex and G-quadruplex domains was identified: destruction of one via site-directed modification of oligonucleotide sequence resulted in dissociation of the other [190]. The 3D structure of an aptamer with similar base sequence was investigated by X-ray crystallography [112]. Formation of the duplex domain was not observed when the mutually complementary sequences flanked the parallel G4 because in this case the complementary fragments were separated in space; their existence only destabilized the G4 structure [44].
STRUCTURAL ORGANIZATION OF MULTIQUADRUPLEXES FORMED BY NUCLEIC
ACIDS WITH MULTIPLE G-TRACTS
Simple single-quadruplex models are now replaced by more biologically relevant multiquadruplex models, which provide the possibility for investigation of interactions between G4 subunits [56, 191]. Conformation potential of long single-stranded DNA and RNA with human telomeric repeats, which allow the folding of more than one G4, was mostly investigated [111, 192-194], as well as that of several other sequences from the human genome that contain many G-tracts arranged in tandem, such as gene promoters [195], insulin-linked polymorphic region [193], minisatellite loci CEB25 [67], and microsatellite repeats [196].
Two models were experimentally confirmed for structures with multiple G4s: “beads-on-a-string” [67, 192-194, 197], in which the individual G4s are separated by single-stranded fragment and behave independently from each other, and a model in which G4 are in contact with each other, interacting and forming higher order structures [111]. Furthermore, interaction of neighboring G4 occurs not only via stacking of terminal G-tetrads, but also via loop bases, which results in formation of rigid superstructures. Conflicting opinions have been expressed based on investigation of long telomeric DNA and RNA by X-ray analysis, NMR-spectroscopy, and mass-spectrometry methods regarding the model corresponding to structural organization of sequences with multiple G-tracts [56, 111, 125, 191]. These contradictions could be explained by structural polymorphism of neighboring G-quadruplex units and availability of intermediate and defective structures formed during G4 folding [153, 198]. It was suggested in several papers that both types of structures (“beads-on-a-string” and interacting G4 subunits) could coexist in long telomeric DNA or RNA, and their ratio depended on various factors in the cell.
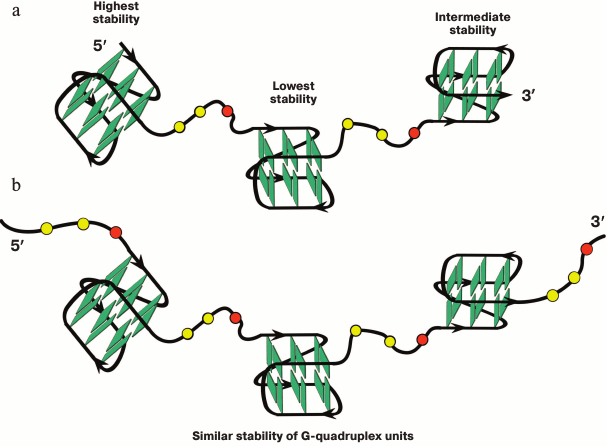
It was shown for simpler models that oligonucleotides containing more than four guanosine tracts formed several G-quadruplexes by using various combinations of G-tracts (position isomers) [52, 199, 200]. It was shown that the sequences capable of forming two individual G4s separated by oligonucleotide linker of various length could fold via different means [199]. Predominately one G-quadruplex is formed if the linker is short (number of nucleotide units ≤3) and is found at different positions along the oligonucleotide chain (entropy gain), while in the case of longer linker only one secondary structure is formed with two G4s, which behave as independent quadruplex units.
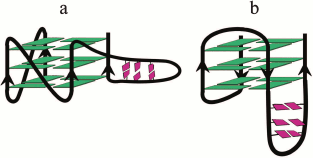
A multistep model is characteristic for unfolding of a G4 ensemble arranged as “beads-on-a-string” [111]. Furthermore, these multiquadruplex structures unfold at lower temperatures than individual G4s [193]. It was shown in 2015 why the stability of the multiquadruplex garland depends on the number of G-quadruplex units [201]. The authors proved that the observed decrease in the stability of the structure formed by d((GGGTTA)4m–1GGG), where m = 2, 3, 4, with increase of the number of G4 units, was related to the fact that the G4 at the 5′-end of the molecule was more thermodynamically stable than the G4 at the 3′-end, whose stability in turn was higher than the stability of the inner quadruplex domains. In other words, the stability of individual G4 in the garland is modulated by the flanking nucleotide sites. Thus, each inner G4 is surrounded by the TTA sequence from both sides, while the terminal G4 – only from one side (Fig. 5a). The validity of this explanation has been confirmed by results of investigation of long G4-motif flanked with trinucleotide sequence TTA: d(TTA-(GGGTTA)4m–1GGG-TTA), where m = 2, 3, 4. It was found that the overall stability of the multiquadruplex structure formed by this oligonucleotide did not depend on the number of G4 units, because in this case both the inner and terminal G4 were flanked with trinucleotide sequences from both sides (Fig. 5b). It is of interest that these results do not change with varying length of the fragment flanking G4.
Fig. 5. Model of multiquadruplex structure with G4 separated by three-nucleotide linkers. a) Tandem G-quadruplexes formed by the d((GGGTTA)11GGG) oligonucleotide in which the individual G4 display different thermodynamic stability due to different position of the flanking single-stranded regions (at 3′-end, at both 5′- and 3′-ends, at 5′-end). b) In contrast, the oligonucleotide d(TTA-(GGGTTA)11GGG-TTA) forms a structure in which each individual G4 neighboring trinucleotide fragment from both sides, thus reducing disparities in the stability of the G4 units. Residues T and A are highlighted by yellow and red circles, respectively.
At the same time, the higher-order G-quadruplex ensembles are significantly more stable than the individual G4s, and the interquadruplex interactions represent the stabilizing factor in this case [196].
SMALL-MOLECULE LIGANDS AND PROTEINS RECOGNIZING AND SPECIFICALLY
BINDING G-QUADRUPLEXES
Conformational features of G4s, which are fundamentally different from the other structured DNAs, represent a basis for molecular recognition of G-quadruplex structures by low molecular weight ligands. Hundreds of small molecules of various chemical nature that are capable of selective interactions with G4 have been produced and investigated in the last two decades [202]. They can recognize either preformed G-quadruplexes [203] or initiate folding of the G4-motifs into such structures. The small molecules that bind and stabilize G4s in telomeric DNA and oncogene promoters are considered as potential antitumor agents [8, 77]. Moreover, G4 ligands are invaluable tools for investigation of the biological role of G-quadruplexes [204], as well as for their detection and isolation from cells [205-207]. Numerous articles and reviews have been devoted to analysis of interactions of G4 ligands of various chemical nature with their polymorphic nucleic acids targets, which is why we limited ourselves to only recently published studies [208-211].
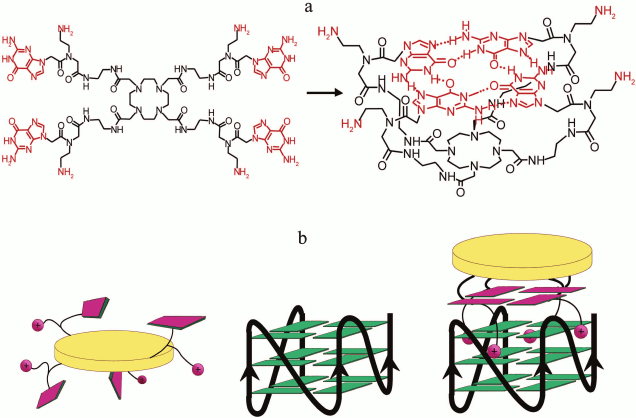
A wide repertoire of chemical approaches has been used for producing compounds with high affinity to G4s [208]. The synthetic G4 ligands include derivatives of anthraquinone (BSU-1051), acridine (BRACO-19, AS1410, RHPS4), quinacridine (BOQ1, NCQ), porphyrin (TMPyP4, NMM) [41], perylene (PIPER), triazine (12459) [212], bis-quinoline (360A, 307A) [213], and others. Furthermore, natural macrocyclic compounds capable of binding to G4 – telomestatin [214], ligands based on natural alkaloids [58], such as berberine [215], daunomycin [216], cryptolepine [217], HXDV, pyridostatin [203, 218], and natural polyamines – have been characterized. Actively investigated duplex DNA minor groove-binding ligands based on benzimidazoles exhibit ability to recognize G4s [219]. Even though some of the investigated ligands demonstrate high selectivity and affinity to G4, several of their properties do not allow considering them as optimal. That is why in recent years numerous compounds have been produced and investigated that in addition to the ability of binding to quadruplex structures provide such advantages as chemical stability and ease of synthesis, enhanced water solubility, ability to penetrate into cells and low toxicity, recognition of G4 with particular topology [220-222], chiral selectivity of interaction with G4 [223, 224], in particular with c Z-G4 [225], induction of conformational transitions in the quadruplex target [226-228], ability of inducing G4 formation (molecular chaperon functions), and high selectivity of the G4–ligand interaction. The selectivity of small-molecule compounds was often assessed by their ability to interfere with biological processes associated with G4 formation [210, 213, 229]. Attempts to evaluate the selectivity of several commonly used G4 ligands by monitoring the quadruplex melting using optimized FRET techniques were reported [34]. Compounds that change their optical (luminescent) properties upon interaction with G4 are of interest [207, 230-233], as well as reactive G4 ligands [234]. Small-molecule compounds exhibiting enhanced affinity to RNA quadruplexes [235-237] that stabilize or disrupt them during interaction [236] have been actively investigated in recent years. A series of new G4 ligands was discovered using molecular docking [122, 238]. The database (http://www. g4ldb.org) comprising a unique collection of more than 800 known G4 ligands was created to facilitate the design of small molecules with predicted affinity to G-quadruplexes [239].
In most ligands, the G4-recognizing site consists of a planar polycyclic aromatic system that binds to the four-stranded helix via interaction of π-electrons of aromatic rings with the terminal G-tetrads. It was found recently that some G4 ligands were capable of discriminating “upper” and “lower” tetrads of the same quadruplex [240, 241]. However, G4s have other sites that demonstrate binding with small molecules: grooves with parameters varying for quadruplexes of different topology, phosphate groups, and loop bases. The ligands recognizing these structural determinants can bind to G4 with a particular secondary structure [242]. Narrow specificity of such ligands is of great importance for their application as novel therapeutic and diagnostic agents [214]. To impart the desired properties to small molecules, one can introduce charged side chains and metal ions into them, and the 3D structure of the ligand can be changed radically as well [243].
Most G4-binding ligands stabilize the G-quadruplex targets. G4 stabilization under the action of a low molecular weight compound is determined by the balance of electrostatic and stacking interactions, hydrogen bonds, and hydrophobic and Van der Waals forces. The electrostatic interactions are the most significant. Energy parameters of G4 binding to ligands and methods for their determination have been described [244]. Two possible mechanisms for G4 stabilization by ligands have been discussed: (i) decrease in the dissociation constant of the formed G4 structure, which results in increase in its lifetime; (ii) increase in the G4 formation constant; in this case, the ligand initiates the quadruplex assembly. For example, the 360A ligand is capable of acceleration the G4 formation 106-fold. Several types of small-molecule ligands are known that cause destabilization or even disruption of quadruplex structures [245]. Some G4-ligands facilitate disturbance of telomere functions, preventing binding of the proteins of shelterin complex to telomeric DNA [66]. Ligands stabilizing G4s can inhibit their unfolding by helicases competing with the proteins for binding to G4 structures [213]. Small molecules are also known to introduce damages into DNA, for example, pyridostatin causing double strand breaks [203]. Such ligands are considered as effective antitumor agents because they affect cell viability, especially in the case, when the DNA repair system is inhibited or mutated [246].
Structural information on ligand–G4 complexes has been obtained using NMR spectroscopy [126] and X-ray crystallography [247]. It was stated in a review devoted to structural analysis of such complexes [16] that the planar aromatic site of the ligand did not intercalate into the quadruplex core upon its binding, but interacted with terminal G-tetrads. Positively charged groups such as protonated nitrogen of the acridine ring can use the central electronegative channel of the G-quadruplex to enhance interaction of the ligand with G4 structures. The possibility of structural adaptation of the quadruplex to the ligand was mentioned, for example reorganization of loops to increase the surface area of G-tetrads (to hexads and octads) via incorporation of additional bases or base pairs of the loops into the plane of terminal tetrads. The X-ray crystallography data show that the type of G4 loops defining the accessibility of quadruplex grooves significantly affects the selectivity of the ligand [247].
It must be noted that all the intramolecular G4s cocrystallized with small-molecule ligands display parallel topology [16, 215]. This is likely related with the dehydration of the sample during crystallization (it is known that dehydration induces formation of parallel G4) and constraints imposed by the crystal lattice. Interaction of the ligand with G-quadruplexes of different conformations was also characterized using molecular dynamics simulation [248]. As of now, structures of approximately 25 G4–ligand complexes have been deposited in the Protein Data Bank (PDB) [248].
Despite the obvious desire to understand better the mechanism of molecular recognitions of G4 by aromatic ligands, only a few reports are available on investigation of thermodynamics of the complex formation. The thermodynamic and structural characteristics of binding of six different ligands with human telomeric DNA were compared in a paper in 2015 [226]. The analysis of experimental data obtained from isothermal calorimetry, CD, fluorescent spectroscopy, gel electrophoresis, and molecular modeling allowed estimation of dominating forces (specific interactions, changes in solvation and conformational state of the quadruplex) that controlled the binding of ligands to G4. Moreover, the existence of intermediate structures during G4 formation was taken into consideration for the first time. It was found that the parameters differed significantly for the poorly selective and quadruplex-specific (highly selective) ligands.
Investigation of binding of aromatic ligands with multimeric G4s showed that the ligand was usually incorporated between two G4 subunits forming a sandwich-like structure due to stacking interactions with terminal G-tetrads of neighboring quadruplexes [249, 250]. Furthermore, the distance between the individual quadruplexes in the multimeric system affected the efficiency of formation of these complexes [251]. However, it was shown recently that one of the enantiomers of the chiral nickel-containing ligand was not incorporated between the two quadruplex units, but rather covered them from above, corresponding to two G-tetrads in size. Furthermore, its binding with the G-quadruplex dimer was 200-fold more efficient than with the individual G4 [252].
Metal complexes occupy a very important place among a huge number of organic molecules capable of binding to DNA quadruplexes, and they have been investigated thoroughly in the last ten years. The G4-ligands based on metal complexes combine advantages of organic compounds and coordinating action of a metal ion. They are relatively easy to produce and exhibit unusual optical, magnetic, and catalytic properties, which makes them appealing for creation of libraries of the G4-selective ligands, G-quadruplex optical probes, and cleaving agents. Exhaustive classification of binding of metal complexes to G4s (macrocyclic ligands, non-macrocyclic polydentate ligands, bulky metal assemblies with flat surfaces, supramolecular chiral cylinders, luminescent metal complexes, and others) was presented in reviews [253, 254] together with their structural peculiarities, additional geometric and electronic possibilities for the specific binding to G4s, and mechanisms of their interactions with DNA quadruplexes. Several types of metal complexes were designed to introduce damage and breaks into DNA. For example, a method for determination of strand orientation in G4s using the reactive metal complex Zn-TTAP based on phthalocyanine was developed [255]. This method is based on ligand-induced photocleavage of the DNA strand between guanosines along the quadruplex axis, predominately after the guanosine belonging to the “upper” G-tetrad.
Alkylating G4 ligands are now considered as a class of chemotherapeutic agents [234]. Targeted DNA modification induced by these compounds can prevent correct translation of genetic information, causing death of tumor cells. In addition to medical application, the covalent modification of G4 using reactive ligands has been suggested as an effective strategy for detection and monitoring of G4 formation in the entire genome and transcriptome. Marking of G4-sites can be performed via affinity isolation of the respective DNA fragments with covalently bound highly specific ligands [256]. It is important for reactive molecules to minimize non-specific nucleophilic attack and make the alkylation process reversible, so that the DNA fragment isolated from the genome would not carry covalently bound small-molecule ligand on it. The electrophilic intermediate quinone methide was suggested as an alkylating agent attached to the G4 ligand, which is capable of reversible alkylation and can form adducts with DNA that are stable in aqueous solutions [257]. The fact that quinone methide can be easily produced from suitable water-soluble precursors via mild activation (photochemical, reductive, oxidative, thermal, and other) signifies its advantage [258]. Recently developed chemical strategies for isolation and intracellular localization of G-quadruplexes based on modified G4 ligands were summarized in a review [256]. The developed approaches allow a biotin residue to be introduced into a ligand, facilitating further affinity isolation of the DNA quadruplex on streptavidin-coated magnetic beads [205], transfer a fluorophore from the ligand to G4 by chemical reactions stimulated by short-distance effects, or introduce the quinone methide precursor into the small molecule for its further chemical modification. Considering, that the availability of bulky substituents could affect the transport of the ligand into the cell, as well as affinity and selectivity of their interaction with G4, the strategy based on introduction of low-volume alkyne residues into the ligand seems more promising. These groups can be transferred from the ligand to G4 via, for example, reaction between the nucleophilic group in the quadruplex loop and an electrophilic center on the linker connecting the alkyne with the ligand [256]. Small molecules containing alkyne functions can be immobilized via the cycloaddition technique onto beads carrying azide groups, or they can be attached to fluorescent labels directly in the cell. Fluorogenic G4 ligands were used for optical visualization of G-quadruplex aptamers introduced into lung cancer cells [259]. The efficiency of capture of G-rich oligonucleotides was monitored by confocal microscopy, and the intracellular localization of the quadruplexes thus formed was revealed. It was shown that parallel G4s were observed mostly in cell lysosomes, while antiparallel structures were found in mitochondria. These findings are important from the point of view of drug delivery to specialized cell organelles using G-quadruplex aptamers.
An innovative strategy for the synthesis of “smart” G4 ligands was developed recently based on template-mediated assembly of water-soluble compounds imitating G-tetrad (Template-Assembled Synthetic G-Quartet, TASQ); moreover, this process occurs only in the presence of a G-quadruplex target. For example, a novel molecule was designed and produced [260] with a structure that could rearrange when it interacts with a G4, but not interacts with duplex DNA. The “smart” ligand forms artificial G-tetrad via template-mediated intramolecular reaction, which binds to the terminal G-tetrad of the quadruplex, stabilizing it in the process (Fig. 6). The metal (terbium) coordination in the G4 channel facilitates organization of the active closed state, and the precisely located positive charges on the side substituents enhance affinity of the ligand to G4 even further. Application of these compounds as artificial receptors for testing the interaction of terminal G-tetrads in the quadruplex with low molecular weight ligands was demonstrated [261]. The new generation of G-quadruplex biomimetic ligands is also capable of playing a role of “smart” fluorescent labels [262]. The ligand fluorescence is quenched in the inactive open state due to intramolecular photoinduced electron transfer, while the fluorescence is restored in the closed state induced by the self-assembly of the artificial G-tetrad. The first such ligand – PyroTASQ based on pyrene – was shown to be an effective G4-specific probe for in vitro studies [262]. However, it showed a tendency for aggregation in intracellular medium. The specifically designed N-TASQ with pyrene replaced by a naphthalene residue was found to be suitable for intracellular studies. N-TASQ was used for the direct detection of RNA G4s in the non-pretreated (fixation, permeabilization, etc.) human and mouse cells [263].
Fig. 6. “Smart” G4 ligand capable of artificial G-tetrad formation via template-mediated intramolecular reaction; the terminal G-tetrad of G4 used as a template in the process. a) Chemical structure of the “smart” ligand, which rearranges itself into active (closed) state generating an artificial G-tetrad. b) Schematic representation of the mode of “smart” ligand action with G4-binding domain of the molecule presented by a yellow circle and the guanine residues forming a new G-tetrad as pink rectangles.
Cellular and viral proteins. The availability of functionally important proteins capable of specific binding to G4 is one of most significant factors supporting the hypothesis of biological relevance of these structures. Most regulatory functions of G4s in cells are mediated by the binding of proteins that modulate their thermodynamic stability and conformation. As of now, numerous proteins have been identified that specifically bind to DNA quadruplexes [264-266] and to the RNA-analogs of G4 [267, 268]. Affinity chromatography (for example, using biotinylated G4-forming oligonucleotides that are linked to streptavidin-coated beads) was applied for isolation of target proteins followed by the mass-spectral analysis (LC-MS/MS) [269, 270]. Identification and characterization of many cellular RNA G4-binding proteins (nuclear ribonucleoproteins, ribosomal proteins, splicing factors) using developed screening system was described in a recent paper [271]. The selected proteins were characterized by MALDI TOF mass spectrometry, and their dissociation constants with RNA G4 (nanomolar range) were determined with the surface plasmon resonance technique.
The proteins recognizing G4s belong to different classes and perform different functions in the cell: regulation of telomere length, ribosome assembly [272], transcription, translation, splicing, and alternative splicing [20, 269, 273-276], DNA replication and reparation [277, 278], replicative bypass base damaged sites [279], reverse transcription [206], conjugation of homologous chromosomes during meiosis, DNA duplex unwinding, changing DNA topology, and other processes of nucleic acid metabolism. The G4-binding proteins are classified into three functional groups: (i) telomere-related proteins, such as proteins of shelterin complex [267, 280, 281]; (ii) G4-unwindig or cleaving enzymes, such as DNA- and RNA-helicases (molecular motors that unwind structured nucleic acids and thus are involved in numerous biological processes), nucleases [273, 282, 283]; (iii) proteins that stabilize G-quadruplexes and prone their folding (molecular chaperons), for example, nucleolin (multifunctional nuclear protein) and nucleophosmin [269, 276, 284]. The proteins and enzymes with enhanced affinity to G-quadruplexes in comparison with double- and single-stranded DNA not only discriminate such non-canonical structures, but can also recognize their topology [269].
Despite intensive investigation of protein–G4 complexes, the molecular mechanisms for recognition of G-quadruplexes by protein molecules remain poorly understood, and investigations in this area are fragmentary and unsystematic. It was established using high-resolution methods that protein–nucleic acid binding is affected by the G4 loops [112] and duplex–G-quadruplex junction [189, 285]. It was shown by molecular modeling that the cavities in the HIV-1 integrase and RecA protein matched exactly the G4 size [55], and the domain of the nucleophosmin fit the G4 groove [286]. It was shown recently using NMR spectroscopy that two peptide molecules with sequences corresponding to prion protein fragments were bound to the terminal G-tetrads of RNA G4 via hydrophobic interactions (favorable change of water entropy) [287]. An 18-unit G4-binding amino acid sequence of RNA-helicase (RHAU) was identified in 2015 using the same method; it covered the end G-tetrad of parallel DNA G4 and enveloped the quadruplex core due to electrostatic interactions between the three positively charged amino acids and the negatively-charged phosphate groups [288]. Interestingly, this binding model closely resembles one for most G4-specific small-molecule ligands. Investigation of the crystal structure of a complex of arginine- and glycine-rich peptide fragment of the FMRP protein with RNA containing G-quadruplex and duplex domains with 2.8 Å resolution elucidated the molecular principles explaining the specificity of protein–nucleic acid interactions and identified a wide network of contacts between the peptide and RNA, which included not only the G-tetrad and boundary between the G4 and the duplex domain, but were extended into the RNA duplex domain [289]. It must be noted that FMRP is a well-characterized regulatory RNA-binding protein that plays a central role in the development of a series of human neurological disorders including autism. It was suggested that the interaction of this protein with RNA quadruplexes directly affected regulation of translation of hundreds of key mRNAs in neuronal cells, and the disruption of these interactions was considered as one of the major mechanisms initiating pathological changes causing mental disorders [20]. The role of aromatic amino acids – tyrosine and phenylalanine – in the C-terminal domain of proteins associated with liposarcoma and Ewing sarcoma in recognition of telomeric DNA- and RNA quadruplexes was investigated [290]. It was shown that phenylalanine residues were extremely important for specific binding to G4. Thus, the replacement of three Phe by Ala residues prevented interaction of the peptide with the G-quadruplex structure. Also, it was possible to discriminate telomeric DNA G4s and RNA G4s by replacing the Phe residues with Tyr ones. This substituted peptide recognizes and binds predominately RNA quadruplexes due to the higher hydrophilicity of tyrosine in comparison with phenylalanine. It was shown using model structures containing modifications in the loops of DNA- and RNA quadruplexes (apurinic/apyrimidinic sites, 2′-O-methyl derivatives, or nucleosides with locked ribose ring (LNA, Locked Nucleic Acids)) that the Tyr residues recognized primarily the ribose 2′-hydroxyl groups.
According to the modern paradigm, the G4s and recognizing proteins are key players in many very important metabolic processes. In particular, G4 structures are considered as an endogenous source of chromosomal instability [291] associated with numerous human diseases [292, 293]. Accumulated data indicate that if four-stranded non-canonical DNAs are not resolved by proteins, they can interfere with transcription and replication processes [294, 295], as well as reparation and homologous DNA recombination. This results in double stranded DNA breaks, genome rearrangements, and expansion of G-rich repeats [296-298]. Moreover, genomic damages are greater the higher is thermodynamic stability of the G4 [139]. At the same time, it has been mentioned that stable G4s are not the only reason for defective replication, and alternative mechanisms could exist that explain this phenomenon. For example, the DNA polymerase δ stalls near telomeric G4-motifs independent of the quadruplex structure formation on the lagging strand [299]; investigation at the cellular level showed that these sequences were generally difficult to replicate. Unlike the high-fidelity replicative DNA polymerases, the specialized DNA polymerases from eukaryotic cells, which can insert nucleosides opposite certain types of point damages, can prevent genomic instability in the vicinity of G4. Indirect indications exist that some of them, for example Pol κ, Pol η, and Rev1 (members of Y-family), can ensure efficient replication progress through DNA quadruplexes [300]. The structural features of the most investigated Rev1 explain why this polymerase operates predominately on G-rich templates.
Helicases that prevent chromosomal instability caused by G4-forming sequences occupy a specific place among the proteins specifically binding and unwinding G-quadruplex structures [282, 298]. The mechanism and kinetics of G4 resolving by Pif1 family of replicases conserved from bacteria to humans have been discussed in [301-303]. Interestingly, these helicases are activated in the process of G4 resolving and more readily unwind double-stranded DNA during replication. Unlike the highly processive Pif1 family of replicases that remain bound to their DNA substrate, G4-resolvase 1 (also known as DHX36 or RHAU), which is capable of strongly binding and resolving DNA G4 and RNA G4, is non-processive, because it dissociates from the substrate after each act of ATP hydrolysis [304]. The unwinding of intramolecular G4 by the ATP-dependent DNA-helicase BLM from the RecQ family is realized via different mechanisms depending of the G-quadruplex DNA surrounding (single- or double-stranded), as has been shown by FRET [305]. The technique based on fluorescence methods allowed monitoring the unwinding of different G-quadruplexes by DNA-helicases in real time [34]; G4 binding with small-molecule quadruplex-stabilizing ligand inhibited the enzyme activity, the inhibition degree depending on the G4 structure and the ligand type. Different efficiency of denaturation of G4s with different folding topologies by replication protein A (RPA) was revealed by single-molecule FRET [306]. G4 stabilizing factors, such as short loops (less than three nucleotide units) and more than three G-tetrad layers, helped to preserve the G-quadruplex conformation even at the physiologically relevant protein concentration, which increases the functional significance of these structures [307].
The effect of G-quadruplexes on functioning of G4-binding proteins was investigated in several recent studies. Thus, it was shown using single molecule technique that telomeric DNA G4, which modulated telomerase activity in vitro, could affect the binding of other proteins involved into telomere biology [118, 308]. And vice versa, proteins binding to telomeric ends controlled DNA G4 formation in the cell. Data are available on the effect of G4 on translocation of the telomerase catalytic subunit (TERT) from the nucleus to mitochondria, which plays an important role in cellular apoptosis induced by the G-quadruplex-binding ligands [309]. It was established that G4s of different topology could inhibit topoisomerase I-mediated nicking of double-stranded DNA [310, 311], thus affecting the degree of DNA supercoiling. The specialized low-fidelity DNA polymerases hPol η and hPol ε from the Y and B families, respectively, which are capable of replicative bypassing base-damaged sites, retain various activity towards G4-containing substrates depending on the degree of specific binding with G-quadruplexes [279]. It was shown that in 20% of human genes G4 was a target for two components of the TFIIH transcriptional factor (XPD and XPB) [283]; furthermore, XPD acted as DNA G4-helicase, and XPB as G4-binding protein. Considering that both these components play a major role in coordinating the system of DNA damage identification and reparation mechanisms related to local melting of the double helix, it has been suggested that G4 can regulate these processes. A mechanism of G4-mediated regulation of gene expression and other processes of DNA metabolism has been discussed [312]. This mechanism is based on interaction of DNA G4s with yeast transcription coactivator Sub1 and its human homolog PC4.
In recent years, our understanding of the biological relevance of G-quadruplexes in prokaryotes [10, 98] and viruses [313] has increased. It was shown that G4s are specifically recognized by various viral proteins such as Epstein–Barr nuclear antigen I [103], coronavirus SARC-unique domain (SUD) existing exclusively in highly pathogenic strains [314], and nucleolin, which stabilizes G4 structures formed by LTR promoter and silences HIV-1 viral transcription [276]. Moreover, it was established that the G-rich DNA sequence in the U3 region of the HIV-1 virus that is capable of folding into G4 contains three binding sites of the Sp1 transcriptional factor, which is involved in regulation of the viral gene expression [100]. The G-quadruplex aptamers targeting HIV-1 proteins that were considered as potential inhibitors of the viral gene replication in cells and viral integration in vitro [315] demonstrated antiviral activity [316].
G4-specific antibodies. G-quadruplexes are immunogenic like any other non-canonical DNA form. Antibodies are described in the literature that exhibit 1000-fold higher affinity to the four-stranded DNA in comparison with duplex structures; furthermore, some of them are highly specific to parallel G4, while others bind either to parallel or antiparallel G-quadruplex structures. Moreover, antibodies were isolated and characterized that display the ability to discriminate with 100-fold specificity parallel G4s differing only in the structure of their loops [317]. Usually antibodies are produced from a huge library of bacteriophages, with each bacteriophage displaying a unique antibody on its surface (phage display). Bacteriophages carrying G4-recognizing antibodies are isolated via binding with immobilized DNA quadruplexes [317]. Antibodies of different specificity were successfully used as tools for quantitative detection of DNA- and RNA quadruplexes in cells using different versions of immunofluorescence assay [318-320].
Antibodies to G-quadruplexes were also used to demonstrate that four-stranded DNAs modulates gene expression in vivo. The presence of such antibodies in human colon carcinoma cells caused significant changes in the expression of a large group of genes (from the investigated set consisting of more than 18,000 genes) whose regulatory regions are enriched with G4-motifs [321].
POLYMORPHISM OF TELOMERIC G-QUADRUPLEXES
Intramolecular G-quadruplexes formed by single-stranded telomeric repeats in in vitro studies represent a unique set of diverse topologies [148, 322]. The G4s formed by human telomeric repeats are the best studied [57, 323], and this elevated interest is because they can act as suppressors of tumor growth [66, 324].
Telomeres are specialized nucleoprotein structures at the ends of eukaryotic chromosomes, which protect the cells from chromosome fusions and DNA damage responses. It is known that the telomeric DNA in somatic cells progressively shorten with each round of cell division due to inability of DNA-polymerase to replicate the 3′-end completely and due to nucleolytic processing. After reaching a critical length (the Hayflick limit), the replication ceases and the cells enter a senescence phase that precedes apoptosis. In contrast, a telomere length maintenance mechanism is induced in tumor cells via activation of telomerase – an enzyme of ribonucleoprotein nature capable of synthesizing G-rich repeats at the 3′-end of telomeric DNA according to the reverse-transcription mechanism or via alternative lengthening of telomeres based on recombination (ALT). Increase in telomerase activity is observed in more than 85% of tumor cells, and the ALT mechanism is realized in 15% of tumors. Since telomerase activity is either completely absent or significantly lowered (as in stem and germ cells) in the absolute majority of cells in the organism, inhibition of this enzyme would facilitate deceleration of tumor growth without toxic effects on the organism [325].
Folding of the single-stranded overhang at the chromosome ends into G-quadruplex structure can effectively block the undesired extension of telomeric DNA via preventing its hybridization with telomerase RNA, which serves as a template for synthesis of telomeric sequences [326]. Quadruplex formation also prevents realization of the alternative lengthening of telomeres due to inhibition of early recombination stages [18, 326]. It was shown that the recombination proteins attached to DNA only after unwinding of the telomeric G-quadruplexes by helicases. That is why stabilization of quadruplex structures in human telomeric DNA by a small-molecule ligand is a promising antitumor strategy [66]. The use of G4-stabilizing ligands can also cause telomere dysfunction independent of the telomeric DNA lengthening with telomerase. For example, telomestatin can remove the telomere shelterin protein TRF2, which results in damage of telomeric DNA in glioma stem cells and, consequently, slowing the growth of glioblastoma – a malignant brain tumor. It was found that the non-glioma cells were insensitive to the treatment with G4-stabilizing ligand [327]. Moreover, it was suggested that G-quadruplexes could be involved in capping and telomere protection from degradation or end fusion. The G-quadruplex function as a protective mechanism of chromosome ends, postulated many decades ago, was clearly shown for yeast telomeres, at least when all other forms of capping were defective [326].
The telomeric G4s are formed via intramolecular folding of tandem G-rich repeats, although it has been mentioned in earlier studies that in some ciliates the telomeric 3′-overhangs form intermolecular G-quadruplexes, which link together the macronuclear chromosomes.
The use of G-quadruplexes of telomeric DNA as targets of antitumor reagents requires knowledge of the detailed G4 structure under conditions mimicking intracellular ones. The issue is complicated by the fact that the structure and stability of the most investigated G-quadruplexes formed by the four human telomeric DNA repeats, d(TTAGGG), strongly depend on the presence of flanking nucleotide residues, DNA concentration [119], environmental conditions such as ionic composition, availability of G4-specific ligands [228], the degree of DNA hydration controlled by PEG addition [328], on sample pretreatment, and methods of investigation [119]. Moreover, it must be taken in consideration that the telomeric repeats form intermediate hairpin and G-triplex structures in addition to the final G4 [299].
It was shown that the topology of G-quadruplex formed by the d(AGGG(TTAGGG)3) oligonucleotide and by oligomers with similar base sequence differ significantly in the crystal state and in solution. According to the X-ray data, d(AGGG(TTAGGG)3) in the presence of K+ folds into parallel intramolecular G-quadruplex with propeller-type loops (PDB ID: 1KF1). At the same time, antiparallel G4 and different variants of mixed (parallel–antiparallel) forms of G-quadruplexes with various loop structures were identified in K+ solution [47]; furthermore, the ratio between them was affected by the 3′-end nucleotides attached to the oligonucleotide. However, the parallel intramolecular G-quadruplex, like the one identified in the crystal state, was observed under conditions of lower water activity (dehydration) caused by addition of ethanol, DMSO, acetonitrile, or high molecular weight PEG of different length [191, 329]. Because PEG was usually added to the oligonucleotide solution to adjust the condition to intracellular molecular crowding, until recently the intramolecular parallel structure of the G4 formed by the human telomeric repeats was considered as a physiologically significant one [191, 329]. However, as shown recently, the presence of PEG not so much imitates the high concentration of biopolymers as it facilitates dehydration of the sample, changing the water structure the same way as other cosolvents [18, 330]. The PEG-initiated transition of the quadruplex structure from the more compact mixed form into the hydrodynamically less compact parallel one cannot be explained by the excluded volume effect, but rather can be explained by the higher affinity of PEG to the parallel G4 conformation, as shown in [331]. Moreover, the use of other compounds instead of PEG more suitable for mimicking the natural biological environment and creating high concentration of biopolymers such as 40% polysaccharide (Ficoll 70) or bovine serum albumin (300 mg/ml) did not facilitate stabilization of the G4 parallel form [194, 332, 333]. The fact that it was precisely dehydration and not the excluded volume effect that induced formation of the parallel quadruplex was shown in 2013 [164]. In that study, the structural and mechanical properties of human telomeric quadruplex were investigated at the single molecule level using optical tweezers.
Either increase in the cosolvent amount or high DNA concentration (6-9 mM) can cause conversion of antiparallel G4 into the parallel one [334]. The oligonucleotide itself can play a role of the agent mimicking molecular crowding in such concentrated solutions. Despite differences in the action mechanism, both these factors result in the dehydration effect, that is decrease in water activity [335]. The concentration dependence of the G4 topology can provide an explanation why different experimental methods, which require DNA samples with concentrations differing by orders of magnitude, reveal different types of quadruplex folding for the same oligonucleotide. It is of interest that G4 stabilized with sodium ions maintains antiparallel topology independent of the DNA concentration [119].
It is commonly accepted that the structural polymorphism of telomeric repeats is much less pronounced in solution containing Na+ than in K+ solution. For example, the d(AGGG(TTAGGG)3) oligonucleotide in Na+ solution forms exclusively the antiparallel three-tetrad G4 with “basket”-type conformation. However, formation of the antiparallel intramolecular structure differing from the standard quadruplex by the nature of hydrogen bonds in G-tetrads and by the loop locations was shown recently by NMR spectroscopy for longer oligomers containing nucleotide units flanking the G4 core [127]. Different conformations of G-quadruplexes formed by telomeric sequences and their dynamics in Na+ solutions was investigated using single-molecule FRET [336]. This method allows characterization of nucleic acids structural polymorphism and the kinetics of conformational transitions on a time scale from milliseconds to minutes. In addition to the unfolded state, at least three different telomeric quadruplexes were identified under dynamic equilibrium. These data change our perspective on the limited conformational options of telomeric G4s in Na+ solutions and indicate the polymorphic nature of the telomeric repeats under these conditions (see reviews on telomere structure in [57, 281]). In a study published in 2016, conformational potential of telomeric DNA was investigated under conditions of low cation concentration, and it was shown that the conversion of the antiparallel G4 into the parallel one occurred when a minor amount of KCl was added to the Na+ solution [22].
Taking into consideration the fact that the level of K+ in mammalian cells (approximately 100-150 mM) is significantly higher than the content of Na+, the G4 structure formed in the presence of K+ seems more relevant physiologically. The conformation of d(AGGG(TTAGGG)3 in live X. laevis oocytes and ex vivo in cellular extract of these oocytes where the natural conditions of molecular crowing were realized was investigated in 2013 [194]. It must be noted that the recently developed in-cell NMR technique was used for in vivo studies, which allowed evaluating of the nucleic acids structure in the eukaryotic intracellular environment. It was shown using this approach and using fluorescence spectroscopy that d(AGGG(TTAGGG)3) adapted the G-quadruplex structure with mixed topology (3 + 1). The same structure is formed in diluted K+ solution (but not in the presence of Na+). Investigation of a fragment of telomeric DNA containing 8 and 12 G-tracts revealed that the conformation of the quadruplex units in the long strand was different from the conformation of the isolated G4. The end and inner quadruplexes exist predominantly as a two-tetrad antiparallel “basket” and a structure with mixed topology with three G-tetrad layers arranged as “beads-on-a-string” [194]. The unusual two-tetrad antiparallel G4 binding only one cation in the quadruplex cavity was documented with a combination of physicochemical methods at extremely low concentration of potassium or strontium ions [22].
The proof that telomeres fold into G-quadruplexes of various types under in vivo conditions allows new comprehension of the data according to which not all G4 conformations actively inhibit telomere lengthening by telomerase, including human telomerase [18]. In particular, parallel tetramolecular G4s are good substrates of the enzyme from Tetrahymena; the parallel intramolecular quadruplexes are also lengthened by telomerase. At the same time, the intramolecular antiparallel quadruplexes (that is the ones that have been postulated) are resistant to lengthening under the action of telomerase [194]. These are precisely the G4 formed by the single-stranded telomere ends that are capable of inhibiting telomerase acting as tumor suppressors.
Structure and stability of G-quadruplexes in elongated telomeric DNA. In the last decade, conflicting reports have been published on the structural arrangement and stability of G-quadruplexes in such DNA in vitro. Data of CD spectroscopy and of electrophoretic studies indicate that the long telomeric sequences predominantly fold into intramolecular rather than intermolecular quadruplexes. Experimental confirmations were obtained for both the “beads-on-a string” model in which the individual G4s were independent [194], and for the model in which the contiguously stacked G-quadruplex units affected each other [111, 193, 337]. The results of optical tweezers investigation demonstrated that only 5% of higher order G4 structures were formed by the DNA fragment containing 24 d(TTTAGGG) repeats, the length comparable with the total single-stranded human telomeric overhang, while the clear majority of the quadruplexes behaved as “beads-on-a-string” [162]. The contradictions related to the arrangement of multiquadruplex telomeric systems were partially explained in a work published in 2016 [31]. It was found that realization of one model or another depended on the ionic composition of the medium. The individual G4s practically do not affect each other in the K+ solutions, while Na+ facilitated interaction of quadruplex subunits. These are precisely the interactions that make the quadruplex structure formed by the d((GGGTAA)7GGG) oligonucleotide more stable in the Na+-containing solutions than in the presence of KCl.
There is some disagreement in the published data on the number of formed G4 units in the long telomeric sequences. Thermodynamic studies revealed that more than 90% of the oligonucleotides d(TTAGGG)12 and d((TTAGGG)12TT) fold into three adjacent G-quadruplexes, in other words into the structure with the maximum possible numbers of G4s, and only 10% form the two-quadruplex structure [111]. Nevertheless, it has been mentioned that in the case of very long telomeric repeats (~200 nucleotide residues), formation of the maximum amount of G4s becomes energetically unfavorable [111]. The direct visualization of the d(TTAGGG)16 telomeric repeats with single-molecule atomic force microscopy [197] supported the “beads-on-a-string” model, where the G4 were separated by single-stranded DNA regions, which in turn created more flexible structure with the number of quadruplexes not at the maximum level possible. Such only partial folding of the long telomeric DNA provides the possibility of binding of the telomere protective protein POT1 with single-stranded sites. In addition to the proteins of shelterin complex, a whole series of telomere processing enzymes requires free 3′-end for binding and further functioning. The minimal size of the single-stranded binding site (6-12 nucleotides) for these enzymes was determined in in vitro experiments [338]. Considering that G-quadruplexes are formed more readily at the 3′-end of DNA [197], and that the probability of folding into quadruplex structures decreases on moving towards the 5′-end of the telomeric DNA (from 55% to 21.8, 14.5, and 7.9% for the 1st, 2nd, 3rd, and 4th G4 position, respectively), the length of the single-stranded 3′-end region can vary from 0 to 18 nucleotides (three d(TTAGGG) repeats) in the case of human telomeric DNA [197]. It was noted that the size of single-stranded site as a result of coordinated action of G4 formation and its unwinding by helicases affected telomere lengthening by telomerase or DNA polymerase (ALT mechanism) [338].
G-quadruplexes with modified nucleotide residues. The effect of guanine analogs (2-aminopurine, 8-bromo- and 8-methyl guanine, hypoxanthine, and others), substituted or modified ribose residues (LNA), and modified non-guanine bases in the G4-motif on the folding topology and stability of intra- and intermolecular G-quadruplexes [339-341], as well as on the functioning of proteins recognizing G4 structures [342] was explored in several recent works and reviews. It was mentioned that certain types of DNA damages could act as modulators of G4 formation. The effect of natural modifications of bases (oxidative damages, apurinic sites, alkylated derivatives, and others) localized either in the loops or in the quadruplex core of human telomeric G-quadruplexes has been discussed in detail [343-345]. As a rule, the analogs of bases that replace guanines in the G-tetrad significantly destabilize G4. The loop modifications change the structure of the quadruplex and its stability less dramatically, and the effect depends on the type of damage and its location in the base sequence. For example, apurinic sites in loops of DNA G4 facilitate formation of propeller loops and stabilize the parallel G-quadruplexes [346]. And 8-oxoA either slightly destabilizes or stabilizes G4 depending on localization of the oxidized base. It must be mentioned that the reparation of natural damages in telomeric G-quadruplexes remains poorly understood.
G-QUADRUPLEXES FORMED IN PROMOTER REGIONS OF EUKARYOTIC
GENES
It was shown using bioinformatics approaches that more than 40% of genes encoding human proteins contained at least one G4-motif in a promoter region. The G-motifs located in the promoters of human oncogenes and the genes of proteins expressed in tumor cells or involved in tumor angiogenesis, unlimited replication, deregulation of apoptosis, and metastasizing are the most popular study objects: c-Myc [113, 138, 140, 157], Bcl-2 [87], c-Kit-1, c-Kit-2 [142, 347, 348], c-Myb, WT1 [349], VEGF [114, 350], VEGFR-2 [351], RET [352], Rb, HIF-1α, KRAS, PDGF-A [353], and PDGFR-β [354], transcription factors Nrf2 [227], SRC [203], as well as the promoter of HIV-1 genes [99, 355], 3′-proximal promoter region of the human tyrosine hydroxylase gene (TH) associated with neurological disorders [356], promoter of the human telomerase reverse transcriptase gene (hTERT) [357, 358]. Under certain conditions, the minisatellite repeats from the ILPR, which is localized 363 bp upstream from the insulin encoding sequence, can also be assigned to this category [359].
Oligonucleotides containing four consecutive G-tracts, sometimes with flanking regions, were usually used for in vitro investigations of the secondary structures formed by G4-motifs from gene promoters [45]. As shown using NMR and CD spectroscopy and by chemical probing, these oligonucleotide models usually folded in K+ solutions into parallel intramolecular quadruplexes [8, 227, 350], although conformational heterogeneity of the promoter G4s was reported in some publications. The common features of the parallel G4s dominating in the population include three G-tetrads connected by two single-nucleotide loops (first and third) and the middle loop variable in length [323, 350, 360]. For example, loops 1 : 2 : 1 are characteristic for the c-Myc quadruplex, and for the G4 VEGF – 1 : 4 : 1 [350]. Despite the indicated common features of promoter quadruplexes, it has been suggested that each G4 can assume a unique structure due to interaction of the nucleotides from the middle loop with flanking sequences [350]. Conformation of G4s from the gene promoter regions was also investigated by X-ray crystallography [361]. It was established that a dichotomy between the structure in crystal state and in solution, which was characteristic for telomeric quadruplexes, was not extended to the other eukaryotic G-quadruplexes.
Considering that many G-motifs in promoter regions contain more than four G-tracts, they can form several discrete quadruplexes depending on what combination of G-tracts is used for in vitro studies, its length, and availability or absence of flanking regions. The various G4 conformations can be at dynamic equilibrium with each other [138, 350]. The set of quadruplex conformers formed in the c-Myc promoter as well as the effect of loops and flanking nucleotides on thermodynamic stability and kinetics of the G4 folding was evaluated [138]. The dependence of the results on the selected sequence can be clearly illustrated using the promoter region P1 of the Bcl-2 gene as an example. The 39-nt G-rich sequence (Pu39) contains six G-tracts from 3-5 consecutive guanosine residues. Investigation of the conformational potential of the set of oligonucleotides (shorter than Pu39) with partly overlapping sequences containing four consecutive G-tracts showed that they could form various quadruplex structures, which could bind G4-specific ligands with different degree of selectivity [362, 363]. Moreover, these G-quadruplexes were not parallel but had mixed topology (3 + 1), which excluded them from the common trend. The MidG4 formed by four central G-tracts was the most stable. However, when the full-sized Pu39 was used as a model, the prevailing structure into which it folded comprised an intramolecular parallel quadruplex with two single-nucleotide loops and a middle 13-nt loop. The G-tracts I, II, IV, and V participated in formation of this quadruplex, as shown by chemical footprinting and NMR spectroscopy, and the guanosine residues of G-tract III were included in the middle loop [87]. Moreover, the full-sized parallel G4 was found to be even more thermodynamically stable than MidG4. The presence of several topologically different quadruplexes in the same regulatory region can be important for fine tuning of gene expression mediated by the binding of different G4s with different proteins. According to recent data, in the close vicinity of the P1 promoter of the Bcl-2 gene there is another 28-nt G4-forming sequence P1G4, which can fold under physiological conditions into two parallel G4s that are in a state of equilibrium with each other. It was shown using chemical footprinting and NMR-spectroscopy that one of the quadruplexes contained two single-nucleotide loops and 12-nt middle loop, while the other had unusual conformation with three single-nucleotide loops and 11-nt site protruding from the G4 core [364]. The long loops in both quadruplexes assume the hairpin duplex conformation, which differentiates them from other parallel G4s and can be specifically recognized by small molecules. The authors suggested that the presence of Pu39 and P1G4 in the neighboring regions of the P1 promoter provides an additional mechanism for regulation of the Bcl-2 gene transcription. Furthermore, the P1G4 sequence plays a critical role in the repression of transcriptional activity [364].
The occurrence of various G4s formed in gene promoters was discussed in a review published in 2010 [70], and their classification was presented. Four classes of intramolecular quadruplexes were suggested: (i) single G4, (ii) pair of different G4 separated by ~30 base pairs, (iii) tandem of two G4s interacting with each other, (iv) multiple G4s formed by overlapping G4-motifs. Evidence of the interactions between quadruplexes formed by sequences from promoter regions with multiple G-tracts has been presented in several reports [358, 365]. Formation of G4 multimers can be a characteristic feature of the promoter G4s, because it is exactly the parallel G4s that demonstrate a tendency for “sticking” to each other. The interaction between two individual G-quadruplexes formed by the promoter region of the hTERT gene containing 12 G-tracts was investigated in most detail (Fig. 7a). It must be mentioned that controversial opinions have been expressed in the published data on the number of G4s formed by this sequence. On one hand, arguments have been presented in favor of the existence of three parallel G4s interacting with each other via stacking interactions of the end G-tetrads [358]. On the other hand, the formation of a two-quadruplex structure has been reported, in which the components formed a unified structure; the formation of parallel G4 with four consecutive G-tracts (I-IV) and formation of a quadruplex with unusual conformation formed by two pairs of consecutive G-tracts (V, VI and XI, XII) separated by 26-nt loop (tracts VII-X) was shown by CD spectroscopy, Taq-polymerase inhibition, chemical footprinting, and optical tweezers methods (Fig. 7b). This loop likely forms a stable hairpin, which can explain high stability of this atypical G-quadruplex [195]. Direct evidence of interaction between two G4s obtained with optical tweezers was reported in 2016 [195]. This approach was modified in such a way to break down the entire structure into individual components and to monitor sequentially the dynamics of conformational transitions in each G4-component without disrupting contacts between the G-quadruplexes. To target the desired domain of the biomolecule, the reaction of alkyne-azide cycloaddition to the alkyne group attached between two analyzed G4s and 3'- and 5'-ends of the construct was used. This allowed targeted addition of dsDNA handles with a terminal azide group and separate monitoring mechanical folding–unfolding of one of the G-quadruplex components, leaving the other part of the biomolecule free of stress (Fig. 7c). It was the authors’ opinion that this approach provided a significant advantage over the use of truncated mutants, in which a part of the structure is deleted. The developed technique for the analysis of high order G-quadruplex structures provided the possibility to determine hierarchy of the unfolding of the analyzed system (the less stable G4 at the 5′-end of the molecule unfolded first, followed by the more stable 3′-end G4 with the structured middle loop), and it was possible to estimate the stabilization energy of π–π interactions between the terminal G-tetrads in the interquadruplex interface (2 kcal/mol) [195]. It should be noted that the G4-multimers in this work are considered as elements of the tertiary structure, which is uncharacteristic for genomic DNA unlike in proteins and RNA molecules. Usually conclusions on the spatial organization of multiplex G4 are based on knowledge of the structure of quadruplex units in its composition. It was shown in a study devoted to direct evidence of interaction between two G4s formed by the human ILPR sequence (d(ACAGGGGTGTGGGG)4) [359] that this principle was not obeyed. The data were obtained by chemical footprinting and optical tweezers techniques indicating that the conformation adapted by the intramolecular quadruplexes could not be automatically predicted based on the structure of individual G4, and the characterized interquadruplex interactions differed from those observed in telomeric sequences. The availability of “tertiary” interactions between the neighboring G4s can produce an important biological impact. The extended rigid quadruplex structures can cause more serious errors in the replication processes, create new targets for G4-specific ligands and cellular proteins, and ensure fine-tuning of the gene expression level.
Fig. 7. Evaluation of the energy of interaction between two quadruplexes formed by the promoter region of the hTERT gene containing 12 G-tracts (a) using optical tweezers (b). The reaction of alkyne-azide cycloaddition was used for mechanical unfolding–assembly of the target domain of the biomolecule; the alkyne group placed between two analyzed G4 allowed targeted addition of the dsDNA handles containing the azide group. Stretching–relaxation of different combinations of dsDNA handles (indicated by green arrows) immobilized on optically trapped beads to monitor the dynamics of conformational transitions and their energy in the G-quadruplex tandem and in each G4-component separately (c). Laser beams focused onto the beads are presented as yellow cones.
It is generally accepted that tandem-arranged G-tracts composed of three or more consecutive guanosine residues participate in the formation of intramolecular G4 forming “edges” of the quadruplex core, while the linker sequences connecting G-tracts become the quadruplex loops. However, an exception from this rule was found recently using NMR spectroscopy and X-ray analysis. The sequences of Pu24 and c-Kit87 from the human promoters c-Myc and c-Kit [361], respectively, can fold into unusual G4s. The characteristic feature of such quadruplexes is that the guanosine tract in one of the “edges” is interrupted, despite the presence of four full G-tracts. For example, the structure formed by c-Kit87 investigated in vitro exhibits unique topology like none of the known G4s. Despite the availability of four (GGG) tracts in the c-Kit87 sequence (d(A1G2G3G4A5G6G7G8C9G10C11T12G13G14G15A16G17G18A19G20G21G22)), which suggests formation of the standard parallel G4, the separately positioned residue G10 participates in formation of one of the “edges” of the experimentally detected quadruplex together with only two of the three guanosines (G21 and G22) at the 3′-end of the molecule. The unusual quadruplex structures formed by the 32-nt oligonucleotide d(G1C2G3G4T5G6T7G8G9G10A11A12G13A14G15G16G17A18A19G20A21G22G23G24G25G26A27G28G29C30A31G32) from the KRAS gene promoter were recorded using NMR spectroscopy, CD, and other methods [366]. This oligomer folds in K+ solution into two dimeric quadruplexes at the 5′- and 3′-ends of the molecule, while the central segment A11-A21 only connects two quadruplex units even though it is enriched with G residues. One of the characteristic features of these structures is the fact that the C2 residue that disrupts the G-tract at the 5′-end bulges from the G-quadruplex core. The described dimeric G4s cannot be formed in the genome context; however, it is the authors’ opinion that conditions of molecular crowding can facilitate the intramolecular assembly of the quadruplex. It was shown in earlier studies that oligonucleotides of various lengths from the promoter region of the KRAS gene in KCl solution folded into G4s of different topology, including monomeric forms [367].
Effect of promoter G4 on gene expression. Plasmid constructs with inserted G-rich promoter regions or their fragments are better for mimicking in vivo conditions. Unlike in the single-stranded telomeric ends, the formation of G-quadruplex structure in the promoter regions occurs in competition with preservation of the double helix and is regulated by the level of DNA supercoiling and binding with G4-recognizing proteins. The G4 formation could be energetically favorable only in negatively supercoiled genome fragments (or plasmids imitating them) [113]. The mechanism of regulation of c-Myc gene transcription has been elucidated using plasmid models in a conceptual study [113] where the major role was assigned to G-quadruplexes. Participation of G4 structures formed in promoter regions in inhibition of oncogene expression has been postulated in many recent publications. That is why the use of low molecular weight ligands, which specifically bind and stabilize G4s, is a basis for antitumor strategy [8, 229].
A strong effect of the G4 location in the eukaryotic gene promoter on the level of transcription was shown in recent publications using reporter constructs [368]. Whereas the G-motif in the antisense (template) strand inhibits transcription significantly, its localization in the sense (coding) strand does not affect the efficiency of the process. More complicated regularities related to the position effect of G-motifs in regulatory sites were revealed during investigation of the role of G4s in the expression of bacterial genes [369]. It was established that the degree of transcription inhibition in the case of G4 location in the antisense chain of the promoter site correlated with the G4 stability. However, when the G4-motif was introduced into the antisense strand between the TSS and stop codon, an increase in the level of gene expression was observed. As suggested by the authors, the G4 formed beyond the promoter facilitated double helix unwinding, thus supporting the unwinding activity of RNA polymerase. Enhancement of promoter activity was also observed in the case when G-rich sequence with seven G-tracts was located at the 3′-proximate region of the human TH gene promoter [356]. It was suggested that one of the reasons for promoter activation was the formation of multiple quadruplexes by these G4-motifs, which could differentially regulate the gene expression.
Because gene expression is insensitive to the availability of G4s in the sense strand of the promoter region [368], it follows that the quadruplex structure in the C-rich antisense strand (i-motif) of the promoter region either does not form intracellularly or does not affect the transcription process. However, participation of the i-motif in transcription regulation was not ruled out in other publications. For example, it was shown using the methods of molecular population dynamics and optical tweezers that the small molecules and proteins interacting with i-motif target could modulate transcription of the Bcl-2 oncogene [370]. It was suggested that while the G4 could act as an inhibition signal for the oncogene expression, the i-motif could activate this process [184].
Correlation between point mutations in G4-motifs that affected conformation and stability of quadruplexes and variation in the gene expression profiles of humans was analyzed [371]. In addition to the development of areas affiliated with personalized medicine, these findings have been related to evolutionary aspects. It was implied that the elements of the quadruplex core remained evolutionarily conserved, while loops in G4s were subjected to mutations.
STRUCTURAL FEATURES OF RNA QUADRUPLEXES AND THEIR BIOLOGICAL
RELEVANCE
The same as the DNA sequences, RNA analogs containing G-tracts are capable of folding into quadruplex structures under physiological conditions, with stabilization factors and folding pathways similar to those observed for DNA quadruplexes [372]. As in the case of DNA G4s, the length and base sequence of the loops, concentration of K+ and Na+, and number of G-tetrads are the major factors of RNA quadruplex stability [96, 373]. Considering that molecules of ribonucleic acid are usually single-stranded, RNA G4 formation should occur more easily than in the case of double-stranded DNA, because hybridization with the complementary sequence will not compete with the process of quadruplex assembly. However, it has been mentioned that the nucleotide context is one of the major factors controlling the G4 folding in RNA molecules. Thus, the availability of C-tracts in the vicinity that are capable of hybridization with the G4-motifs interferes with the formation of four-stranded RNA structures [95].
Many G4-motifs have been identified in the human transcriptome. According to data of bioinformatic analysis, they are present in the 5′-UTR of more than 2000 human genes [374], as well as in 3′-untranslated sites [375]. The G4-motifs were found in various regions of mRNA encoding or not encoding proteins, for example, in the open reading frame of certain mRNA [376], in sites of alternative processing of pre-mRNA, as well as in pre-microRNA [82], in long non-coding RNA [83], and in viral RNA. A new approach for detection and mapping of RNA G4s in cellular transcripts has been described [206]. It is based on the monitoring of the reverse transcriptase stops due to formation of RNA G4; it has been shown that this process correlated with the stability of RNA quadruplexes, which was controlled by metal ions and G4-specific ligands. A promising strategy for detection of RNA G4s under different conditions in vitro and in fixed and live cells based on the use of the fluorogenic cyanine dye has been developed [377].
Telomeres – the specialized DNA–protein structures at the ends of eukaryotic chromosomes – for a long time were considered transcriptionally inactive. However, recent studies have shown that telomeric DNA were transcribed in mammalian cells with formation of long non-coding RNA (TERRA) that contain repeats of the 5′-UUAGGG-3′ sequence. These G4-motifs exist as quadruplexes in K+ solutions and in vivo [1, 57, 81, 378]. The fact that the human TERRA can fold into stable G-quadruplex structures was demonstrated using CD and NMR spectroscopy, X-ray crystallography, and mass spectrometry [1, 125, 379]. The formation of parallel G4 from human TERRA molecules in live cells was demonstrated using oligonucleotide probes with light-switchable pyrene groups [1]. Series of oligonucleotides were synthesized for this purpose that contained the 5′-GGGUUAGGG sequence with two G-tracts and with pyrene residues attached to their ends via flexible linkers. The two pyrene molecules were spatially separated in the unstructured state, and their fluorescence signal was weak (monomeric emission). However, the pyrene molecules at the 5′- and 3′-ends of the oligonucleotide probes were brought together during the formation of the bimolecular parallel G4 in the presence of K+, forming the fluorescence-excited excimer state (Fig. 8). The change in color of the excimer fluorescence observed by fluorescence microscopy allowed real time monitoring of G4 formation in HeLa cells incubated with oligonucleotide probes.
Fig. 8. Use of 5′-GGGUUAGGG oligonucleotides with two G-tracts containing pyrene residues at the 5′- and 3′-ends for demonstrating formation of parallel bimolecular RNA G4. In the unstructured state, the pyrene fluorescence signal is weak (monomeric emission). The pyrene residues are brought together during complex formation, forming fluorescence-excited excimer state (shown with green ovals).
Unlike the polymorphic DNA quadruplexes, the RNA G4 adopt only parallel topology [57, 380]; moreover, this is also true for both short oligonucleotide models [57, 81, 380] and long telomeric RNA forming a garland of intramolecular quadruplex structures [192]. It was shown that parallel orientation of G-tracts in the quadruplexes formed by TERRA RNA did not depend on the base sequence of loops, nature of monovalent cations (K+ or Na+), state of the sample (crystalline or in aqueous buffer), and the presence of small-molecule ligands [379, 381]. It is known that all guanosines in the G4 of parallel topology have anti-conformation, while in the antiparallel G4, half of the residues have anti- and another half – syn-conformation. That is why the conformational preferences of nucleosides (syn-anti) can define the mutual arrangement of strands in the G-quadruplex. Since rG residues in oligo- and polyribonucleotides are predominantly in anti-conformation due to the steric hindrance imposed by the 2′-hydroxyl group, and the dG readily assume the syn-conformation also, the parallel topology is characteristic for G-quadruplexes with ribonucleotide residues. Transition of the antiparallel G4 formed by the aptamer to thrombin (5′-d(GGTTGGTGTGGTTGG)) to the quadruplex with parallel topology was achieved via partial or complete replacement of the dG residues by rG. Moreover, this replacement resulted in conversion of the monomolecular G4 into a bimolecular one due to incompatibility of the loop lengths of the quadruplex with the parallel intramolecular folding of the ribo-version of the aptamer [382].
The RNA quadruplexes are usually more stable than the G4s formed by the same DNA sequence [380]. For example, Tm and ΔG (at 310 K) of the RNA quadruplex containing three G-tetrads and single-nucleotide loops in solution with 15 mM concentration of cations are 359 K and 9 kcal/mol, respectively, versus 350 K and 5 kcal/mol for the DNA analog [131]. More likely, the difference in the properties of two nucleic acids is the reason for that, rather than the difference in the folding topology of the quadruplexes [57, 380, 383]. The availability of the 2′-OH-group plays a significant role in organization of the hydration shell in the grooves of the four-stranded structure, in formation of the network of hydrogen bonds, and in conformation of the sugar residues. According to the molecular modeling data, the additional H-bonds formed with participation of the 2′-OH-group and electron acceptor centers such as oxygen atoms of the sugar-phosphate backbone (O3′, O4′, O5′) and polar exocyclic groups of the bases (NH2-group) provide significant contribution to stabilization of G-quadruplexes of parallel topology [381]. Crystallographic studies showed that the sugar residues in the guanosines from the end tetrads had C2′-endo-conformation, while the C3′-endo-conformation was characteristic for the guanosines in the center of the quadruplex core, which was significantly different from the corresponding parameters of the DNA quadruplex [381]. These findings agree with the data of NMR spectroscopy and computer modeling [381]. As in the case of DNA G4s, thermodynamic stability of RNA quadruplexes decreases with increasing loop lengths [96]; furthermore, on one hand this effect is more pronounced, and on the other it is governed by more complex regularities than in the case of DNA quadruplexes [96, 384]. The effects of the number of nucleotide residues in loops, length of G-tracts, and potassium ion concentration on the thermodynamic stability of RNA quadruplexes and the degree of cooperativity of their folding have been characterized [96, 373]; the G4 topology remained invariably parallel. It was shown on the example of the r(GGGGCC)n sequence from the C9orf72 gene transcript that depending on the RNA concentration, length of repeats, and availability of flanking sequences, this type of oligonucleotides could form either very stable parallel intramolecular quadruplexes with propeller-type loops, or parallel tetramolecular RNA G4 [385]. A mechanism was suggested recently according to which G4 formation in the long (GGGGCC) repeat both on the level of DNA and RNA initiates a molecular cascade of neurodegenerative disorders [292].
Some sufficiently long RNA molecules can form several stable intramolecular conformations that coexist with each other [386]. For example, the double helical hairpin structure can compete with formation of RNA G4s [387]. The relative amount of mono- and bivalent metal ions that stabilize competing structures to various degree affect the “quadruplex–hairpin” dynamic equilibrium characterized by NMR spectroscopy. It was shown that the shift of “hairpin duplex–RNA quadruplex” due to potassium ion content regulated maturation of human pre-microRNA [82]. Small-molecule ligands that can unfold or stabilize RNA quadruplexes can also shift the “quadruplex–hairpin” equilibrium [387, 388]. Interestingly, some ligands affect differently the RNA G4s and the DNA G4s. For example, the cationic porphyrin TmPyP4 stabilized the DNA quadruplex formed by the G4-motif in the c-Myc promoter, thus inhibiting transcription of this oncogene. The same ligand exerts the opposite action on RNA G4s [236] unwinding, for example, the extremely stable quadruplex formed by the mRNA that encodes MT3-MMP protein, which results in the mitigation of the suppression effect and increase the level of translation of this mRNA in eukaryotic cells [236]. The reaction of alkyne-azide cycloaddition in situ was used for identification of ligands selectively binding to the TERRA G4 but not to the telomeric DNA quadruplex [235].
Conformational transitions from duplex to G-quadruplex mediated by metal ions, by oligodeoxyribonucleotides of certain primary structure, by proteins, and by other agents represent a moving force for switching the activity of the specially designed ribozymes and DNA-zymes [389-391].
Parallel RNA quadruplexes exhibit a strong tendency to stick together [379, 381]. For example, the oligonucleotide with two telomeric repeats r(UAGGGUUAGGGU) form in K+ or Na+ solutions bimolecular parallel quadruplexes that dimerize via stacking interactions of the 5′-end G-tetrads. The lack of UA residues at the 5′-end of the molecule facilitates formation of dimeric RNA quadruplexes even more. Moreover, as shown by NMR spectroscopy, two additional adenine residues from UUA loops were brought into the plane of the end G-tetrads, thus increasing the surface area of the stacking contact [81]. It has been suggested that the hexad A·(G·G·G·G)·A is stabilized by H-bonds between N1 of adenine and 2′-OH of guanosine [379]. Dimerization of parallel RNA quadruplexes via stacking interactions of G·(A)·G·G·(A)·G hexads was also reported for the G4 formed by the r(GGAGGAGGAGGA) oligonucleotide in the presence of potassium ions. The propensity of RNA G4 to multimerization is an important factor inducing formation of the high-order structures in long RNA sequences [192]. According to mass-spectral analysis, the TERRA forms stable multimeric G4. It was suggested that they could be arranged as “beads-on-a-string” in which each bead comprised either a single RNA G4 or a pair of quadruplexes combined via stacking interactions [192]. These RNA quadruplex dimers could serve as targets for ligands recognizing the G-tetrad planes by intercalating between them or inducing emergence of flat structures with large hydrophobic surface area (such as A·G·A·G·A·G·A·G octads) for more efficient binding [379]. As established with the spectral methods and electrophoretic data, the r(GGGUUGCGGAGGGUGGGC) sequence from the 5′-end region (hTERC) of the human telomerase RNA folded into G4 dimer in K+ solution [392]. It was specifically mentioned that the thermodynamically stable intramolecular parallel RNA quadruplex contained a single nucleotide bulge (the cytidine residue underlined in the presented sequence) in one of the G4 “edges”. Direct experimental evidence for formation of RNA G4s in the full-sized hTERC was also reported [206].
Biological functions of RNA quadruplexes. The studies of recent years have shown that RNA G4s play an important role in cellular processes [275, 393]. They participate in transcription termination and in regulation of different stages of RNA metabolism, for example, posttranscriptional gene expression, which included control over mRNA half lifetime, splicing, and polyadenylation of pre-mRNA [15, 394], as well as translation [372, 393, 395]. Folding of G4-motifs in RNA sequences into quadruplex structure affecting translation can be especially important in plants and other organisms, where cellular ionic conditions, e.g. the concentration of potassium ions, can be changed as a result of stress [373].
Until recently, the G4s formed in different mRNA regions were considered as a network of translation repressors. It was shown that RNA G-quadruplexes localized in 5′-UTR mRNA encoding the NRAS, Bcl-2, Zic-1, TRF2, MT3, and ESR proteins inhibited translation, although the exact mechanism of the inhibition by RNA G4s was not understood. It has been mentioned that the level of translation repression could reach 50% (for most G4-motifs) and even 80%. The RNA quadruplexes formed in the 3′-UTR mRNA [375] and in the mRNA open reading frame also inhibit translation elongation in vitro and decrease protein synthesis in cells [376]. It was shown that G4 localized in the coding regions of mRNA can stimulate reading frame shift in vitro and in cell culture, with this effect correlating with quadruplex stability, which can be modulated by G4-binding small molecules [396].
To inhibit translation of the target mRNA, the following strategy was developed: formation of a bimolecular hybrid DNA–RNA G4 was initiated in the predetermined regions of mRNA containing only two G-tracts using modified oligodeoxyribonucleotides with design allowing targeted addition of missing G-tracts required for G4 formation [397]. It was shown using the mRNA encoding eIF-4E protein, which was actively expressed in various malignant tumors, that formation of G-quadruplexes in the 5′-UTR mRNA and in the protein coding region of the eIF-4E mRNA initiated by phosphorothioate derivatives of oligonucleotides resulted in 30- and 60% inhibition of expression of this protein in human cancer cells, respectively [397]. On the contrary, addition of antisense oligonucleotides (complementary to the G4-motif) can result in the destruction of RNA G4 [398]. The effect of antisense peptide nucleic acids on RNA quadruplex assembly in vitro with relatively high degree of discrimination between the different G4-motifs has been described [399]. This approach – unwinding of RNA G4 by 17-19-nt oligonucleotides – was successfully used for two-fold change in the level of inhibition of specific mRNA translation in human cells [400].
However, it was recently shown that modulation of translation depended on the nucleotide context in which G4-motifs were located. The formed RNA quadruplexes can either inhibit or activate gene expression depending on their location in the 5′-UTR mRNA [393, 401, 402]. It has been suggested that the neighboring base sequences, intracellular environment, and interacting partners can affect the functions of G4 in the cell. Indeed, when the RNA G4 is located in the IRES-element, it facilitates a new mechanism of cap-independent translation initiation [403]. It was reported [404] that RNA G4 formed in the IRES of the mRNA encoding human VEGF protein directly involved in interaction with the 40S ribosome subunit. In the review published in 2015, the role of RNA G4s in regulation of processing and functioning of non-coding RNAs was discussed [20]. These are promising new objects for investigation of G4 functions in the context of various neurodegenerative diseases.
Several publications in recent years have been devoted to investigation of biological functions of the TERRA telomeric repeats and quadruplex structures formed by these non-coding RNAs. It was shown that TERRA was not only accumulated in the cell nucleus, but it was colocalized with telomeric DNA at the chromosome ends (FISH, Fluorescence In situ Hybridization method) and with proteins of telomere shelterin complex (immunofluorescence assay) [1]. Participation of TRF2 in simultaneous binding to TERRA in the form of a G4 and to telomeric DNA as a duplex or G4 (ELISA, Enzyme Linked ImmunoSorbent Assay) provides evidence that TERRA is the key component of the telomere machinery [267]. During the process, one TERRA molecule, which can fold into a quadruplex garland [192], can interact with several TRF2 molecules [267]. It has been suggested that TERRA represses its own transcription and participates in maintenance of telomere structure and formation of heterochromatin [405]. It was shown that TERRA caused changes in gene expression in human cancer cells in vivo. The telomeric RNA that forms G4s inhibits expression of genes associated with the innate immune system, thus affecting tumor malignancy [406].
It is interesting that the TERRA repeats, 5′-UUAGGG-3′, are complementary to the telomerase RNA sequence. It was shown using in vitro reconstructed telomerase and synthetic TERRA molecule that the TERRA repeats hybridized with the enzyme RNA inhibiting its activity. However, the inhibiting action of TERRA upon telomerase can be mitigated in vivo by the heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) involved in the control of telomere length [407]. It has been suggested that the enzymatic elongation of telomeric ends required a balance between the levels of TERRA and hnRNPA1 protein functioning as regulators of enzymatic activity. The role of G-quadruplexes in RNA biology, including TERRA telomeric repeats and telomerase RNA component hTERC, has been discussed in the recent reviews [394, 408].
From the practical point of view, a study in which G4-containing RNA aptamer produced by in vitro selection activated fluorescence of a ligand similar to green fluorescent protein is of great interest [409]. The approach was developed recently based on the replacement of pyrimidine residues in the loops of telomeric G4s with fluorescent 5-benzofuran derivatives of 2′-deoxyuridine and uridine, which allowed the development of switchable fluorescent probes for discrimination of DNA quadruplexes of different topology and parallel RNA G4 [410]. These G4 sensors were also used for estimation of the specificity and affinity of low molecular weight ligands to quadruplexes of various chemical nature and conformation.
HYBRID DNA–RNA G-QUADRUPLEXES
Hybrid DNA–RNA G-quadruplexes are also functionally important. The possibility of association between telomeric DNA and TERRA via formation of hybrid DNA–RNA quadruplex with parallel topology was investigated for the first time in [411]. Formation of such structures in vitro was established by several independent and complementary methods using oligonucleotide models. To prove occurrence of the hybrid G4s in the cellular medium, telomeric DNA- and RNA-oligonucleotides labeled with different fluorophores (FAM and Cy3) at the 3′-ends were transfected into HeLa cells. The observed change in the color of signal occurring during colocalization of fluorophores served as evidence that they were brought together in the hybrid bimolecular G4. If the DNA- and RNA-probes were not interacting, there would be two different fluorescence signals. The additional brilliant evidence of the hybrid telomeric G4 formation in live cells was based on the use of a chemical approach [411]. The DNA probe (12-mer with two telomeric repeats) contained an azidocoumarin residue at the 5′-end, and the 12-nt RNA – a 5′-alkyne group. Azidocoumarin was selected as a pro-fluorophore with quenched fluorescence by the electron-rich α-nitrogen of the azido group. However, the synthesis of the triazole ring by alkyne-azide cycloaddition is accompanied by the emergence of a strong fluorescence signal due to disappearance of the quenching effect (Fig. 9). Hence, the fluorescence enhancement could indicate formation of the hybrid parallel G4 in which the cycloaddition reaction was initiated due to bringing the reactive groups at the ends of RNA- and DNA-oligonucleotides into proximity. Bright blue fluorescence was observed (using a fluorescence microscope) following addition of the cellular medium containing copper catalyst to the HeLa cells incubated with the oligonucleotides. It was established that the simple molecular colocalization of oligomers did not initiate the reaction of cycloaddition. The fact that this method was successfully used to demonstrate the formation of hybrid RNA–DNA G4 in solution in vitro agrees with the experimental data with the cells [411]. The formation of hybrid G4 between the natural telomeric RNA (TERRA) and oligodeoxyribonucleotide containing telomeric repeats with fluorescence label at both ends was observed under conditions close to native ones. The probe was designed in a way that it can form a hairpin structure in the absence of target and, hence, the FAM fluorescence was quenched by the DABCYL quencher located at the other end of the molecule. Formation of the hybrid quadruplex with telomeric RNA forced the DNA-hairpin to open, which resulted in fluorescence enhancement because the fluorophore and quencher were separated. This effect was observed after penetration of the DNA beacon into HeLa cells, confirming intracellular formation of the telomeric hybrid G4 [411]. It was shown in the same work that quadruplexes containing telomeric DNA and RNA could contribute to the protection of telomeric ends. It is known that telomeric single-stranded DNA forms a quadruplex garland. Formation of G4 structures of higher order is also characteristic for telomeric RNA [381]. Intermolecular interaction of the DNA and RNA end repeats can lead to formation of hybrid G4, which is located at the interface of the structured telomeric DNA sequences and TERRA and can provide protective effect creating steric hindrance [411]. Moreover, it is known that the telomeric protein of shelterin complex TRF2 binds simultaneously DNA G4 and RNA G4, thus bringing them closer together in the cell [267].
Fig. 9. Evidence of formation of DNA–RNA hybrid telomeric G4 in HeLa cells using the alkyne-azide cycloaddition reaction. The 12-nt DNA with two telomeric repeats (G residues highlighted with green color) contained an azidocoumarin residue at the 5′-end, where fluorescence was quenched by the electron-rich α-nitrogen in the azide group, and the 12-nt RNA (G residues marked with red) containing the 5′-alkyne group. Synthesis of a triazole ring on addition of copper catalyst was accompanied by the emergence of strong fluorescence signal due to disappearance of the quenching effect.
The hybrid DNA–RNA G4s can be a predominant form of quadruplexes also in the transcription process. They are formed from the non-template DNA sequence and the RNA transcript [412]. It was shown that the DNA–RNA G4s modulated transcription under in vitro and in vivo conditions, and it was precisely these structures that comprised the major form of G4 that were involved in the transcription process. The fact that the RNA transcript is involved in the formation of hybrid G4 was confirmed by demonstrating that the effect of transcription inhibition was achieved on addition of GTP participating in the RNA synthesis, and its replacement by the 7-deazaGTP, which was not capable of G-tetrad formation restored the transcription level. These results indicate that the intermolecular DNA–RNA G4s are more involved in the regulation of transcription and related processes than the intramolecular DNA quadruplexes formed by the template strand of the double helix [412]. The fact that only two G-tracts on the non-template DNA strand participate in formation of hybrid G4 seems especially important, because as was shown with bioinformatics techniques these motifs are more abundant in the transcribed regions of the genome than the intramolecular DNA quadruplex formation motifs containing four and more G-tracts on the template strand [413]. The hybrid quadruplex motifs, which are evolutionarily conserved [413], are abundant in the genomes of warm-blooded animals [414] and occur in more than 97% of human genes [412]. The folding mechanism of hybrid G4s associated with transcription allows controlling their formation and, consequently, the transcription level. For example, the presence of C-rich oligonucleotides complementary to the RNA transcript prevents formation of the hybrid G4, and the quadruplex-specific ligands stabilize these structures [414].
According to some published data [415], quadruplex hybrids can be involved also in regulation of DNA synthesis, at least in mitochondria, where the transcription machine generates RNA primers required for initiation of replication. The DNA of the non-template strand containing fragments with G-rich conserved sequence 5′-GGGGGAGGGGGGG (two G-tracts) from human mitochondrial DNA and the formed transcript fold into a stable hybrid G4 with parallel topology close to the origin of replication. The 3′-end of the primer in this quadruplex is removed from the template strand and is not accessible to the action of the mitochondrial replication machinery, which explains the mechanism of negative regulation of the DNA replication. Only after unwinding of the G4 with helicase (such as, for example, evolutionarily conserved Pif1) the released primer can be used for initiation of the DNA replication. Participation of hybrid quadruplexes in immunoglobulin gene recombination and in neurodegenerative diseases caused by expansion of G-rich repeats in the C9orf72 [416] and FMR1 [417] genes has also been discussed.
EVIDENCE FOR DNA AND RNA QUADRUPLEX OCCURRENCE IN CELL CULTURES
AND in vivo
Over several decades, the possibility of formation of biologically relevant G-quadruplexes in live cells that were unlike the other non-canonical DNA structures stable under physiological condition has been discussed among scientists. The discovery of ligands specific to G4 structures provided a tool for investigation of the properties of such structures, as well as for development of methods for their isolation and identification in cells [205]. The chromatin immunoprecipitation analysis was used to determine the localization of G4s in the genome. The chromatin fragments were fractionated depending on their affinity to quadruplex-binding ligands followed by analysis of selected samples using sequencing or template analysis on microchips. Identification of G4s in cellular cultures was also performed using monoclonal antibodies highly specific to these structures [320].
The complexity of G4 testing in live cells and ambiguity of interpretation of the results is related to many factors. Numerous proteins and enzymes existing in vivo can interact with non-canonical DNAs, thus stabilizing or destabilizing these structures [418], as well as cleaving them. The genome regions containing G4-motifs can be included in nucleosomes, where they are completely shielded by histone proteins. Rearrangement of the genome G4-motif region into the G-quadruplex structure depends on the factors affecting stability of the double helix: the degree of methylation and local topological stress of the DNA [419]. It should be also taken into account that in vitro experiments are commonly conducted in dilute solutions, i.e. under conditions that are significantly different from the molecular crowding characteristic of in vivo systems. Moreover, small-molecule ligands can shift the equilibrium towards quadruplex structure formation by stabilizing their structure, which can skew the real distribution pattern of these non-canonical structures in the cell DNA. Nevertheless, direct evidence of the G4 existence in the gene promoter region was reported as early as 2002. However, this and similar studies were based on the biological effects of G4-stabilizing ligands introduced into the cell. The discovery of G-quadruplex structures at the telomeres in macronuclei of the Stylonychia lemnae ciliate using fluorescent antibodies was an important breakthrough in this research area. It was shown later that the G4 formation depended on the phase of the cell cycle and was controlled by proteins binding to the telomeric ends. Only in studies of recent years the possibility to visualize the loci in mammalian cells (including human cells), which adopt the G4 structures, and to elucidate their role in the cascade of biological events has been achieved.
Two independent lines of evidence demonstrated the genome-wide distribution of G4 structures in human cancer cells [203]. In the first approach, a highly selected G4-binding small-molecule ligand, pyridostatin, was used to target polymorphic G4s regardless of their topology. Pyridostatin was shown to promote growth arrest in cells and modulate the expression of the genes containing G4-motifs by inducing replication- and transcription-dependent DNA damage. It was suggested that the ligand prevented the polymerase movement along the double helix by stabilizing the G4s, which resulted in a DNA break caused by the physical forces acting on a substrate or by endonuclease activity. A chromatin immunoprecipitation sequencing analysis of the DNA damage marker provided the genome distribution of pyridostatin-induced damaged loci. A chemical approach (reaction of alkyne-azide cycloaddition of fluorescent group to pyridostatin) was used in the independent series of experiments to visualize the ligand molecules in the cell nuclei as fluorescent signals. The regioselective reaction of the alkyne group attached to pyridostatin with the azide group on the fluorescent dye Alexa Fluor was initiated in cellulo by a copper catalyst. Loci with folded G-quadruplex structures in human cell DNA were revealed by high-resolution microscopy, and it was shown that they were located predominately in the regions away from the chromosome ends. The hPif1 helicase, known for its high selectivity towards G4 and regulation of these structures formation and processing in the genome, was used as a second reference point. Distribution of helicase signals in the cell nuclei in comparison with the fluorescently labeled pyridostatin was investigated on the human osteosarcoma cell lines expressing human helicase fused to green fluorescent protein. This analysis showed that the helicase was bound to G4-motifs in the genome even without the ligand treatment. The general distribution pattern of fluorescent signals was comparable with that observed for the labeled small molecules. In an independent experiment with cells fixed by formaldehyde “freezing” all biological processes prior to the pyridostatin treatment significant overlapping between the genomic targets of pyridostatin and helicase was observed. These data indicate the existence of the preformed G4 in human cells, formation of which was not initiated by addition of the G4-binding ligand.
Not all the sites with high content of G4-motifs were labeled with the fluorescent ligand. It is likely that additional mechanisms facilitating G4 folding should be realized. Negative supercoiling of DNA in a certain locus or activity of replication or transcription processes that are accompanied by local melting of the double helix and short-lived release of single-stranded regions for reading by polymerase could serve as examples of such mechanisms. The ability of G4-motif to form quadruplex and bind the ligand could also depend on whether the G-rich regions are on the transcribed or non-transcribed strand of the gene [368] or, in the case of replication, on the leading or lagging DNA strand. It also must be taken in consideration that certain cellular effects of G4-specific ligands might be caused by their interaction with RNA G4s [420].
Direct quantitative visualization of DNA G4 structures in human tumor cells using quadruplex-specific antibody has been described [318]. The visualization was achieved via amplification of the fluorescent signal in an antibody sandwich. The loss of signals on pre-incubation of the antibody with excess of preformed G4 oligonucleotides as well as during incubation of the cells with fluorescent antibodies in the absence of the primary G4-binding antibody indicated specificity of the signal. The colored loci in the nucleus also disappeared during treatment of the cells with DNase, but not with RNase. Location of the signals at the ends of metaphase chromosomes confirms the presence of G4s at the telomeric ends, and the scattering of the signals in other chromosome loci indicates that most G-quadruplex structures (approximately 75%) can be formed in promoter regions and in other DNA regions despite the presence of competing complementary strand (Fig. 10). It was shown in this excellent work that formation of G4s in cell nuclei was modulated depending on the phase of the cell cycle; maximum level was observed in the phase of active DNA replication (S-phase), which was 4.8-fold higher than the level of G4 detection in the resting phase. According to these data, addition of small-molecule ligands stabilizing endogenous DNA quadruplexes resulted in an increase (2.9-fold) of the amount of detected G4 targets in the cell nucleus, which indicated the shift of the duplex–G4 equilibrium towards formation of quadruplexes, and this opened new possibilities for targeted regulation of cellular processes such as inhibition of oncogene expression.
Fig. 10. Visualization of endogenous DNA G4 structures in metaphase chromosomes isolated from HeLa cancer cells by immunofluorescence microscopy (using G4-specific antibody BG4). The fluorescent foci are concentrated both within the non-telomeric region (a-c) and at telomeres (d and e). The symmetric appearance of the foci in the sister chromatids (e) indicates formation of G-quadruplexes in the same locations on the newly replicated DNA. This figure is partly adapted from Fig. 3 in [318].
These data were corroborated by the work of another scientific group [421] where highly specific G4-antibodies were also used. The increase in the number of detected G4s in nuclei of human cells was observed under the action of two G4-stabilizing ligands – telomestatin and TMPyP4. Intracellular optical visualization of DNA G4s using fluorogenic ligands was described [259]. Very recently, a report was published [422] where direct evidence of the existence of DNA quadruplexes in cell mitochondria was presented. Fluorescent G4-specific ligands based on carbazole derivatives that can migrate from the nucleus to mitochondria of malignant cells were used for visualization of quadruplexes by confocal microscopy. It should be noted that in the case of normal cells they accumulated in the cytoplasm. The developed compounds were shown to stabilize G4s and, hence, to inhibit expression of mitochondrial genes acting as antitumor agents.
In addition to DNA G4s, the ribonucleotide analogs of quadruplexes and their role in RNA biology have aroused considerable and growing interest [394]. Direct evidence of formation of RNA quadruplexes in human cells was presented in [319], in which the detection of G-quadruplexes was conducted in the cell cytoplasm by G4-specific BG4. Prior to that it was shown by ELISA that this antibody recognized the major structural determinant of G4s effectively binding to both DNA and RNA quadruplexes. The endogenous G4s were visualized by fluorescence microscopy in normal, immortalized, and malignant human cells. The fluorescence signals were amplified using a sandwich of labeled antibodies; BG4 served as a primary antibody in the process. Both the intranuclear signals assigned to the DNA quadruplexes and the fluorescence signals (with longer exposure) distributed in the cytoplasm of all the investigated cell lines and corresponded to RNA G4s were identified using the described approach. Cell staining with the CellMask green fluorescent dye, which revealed cell boundaries, corroborated cytoplasmic location of the primary BG4 antibody. When fixed permeabilized cells were treated with RNase A prior to addition of the BG4 antibody, the quadruplex signals in the cytoplasm disappeared, which indicated the presence of RNA G4s. Along with the immunofluorescence assay, low molecular weight ligands were used to provide evidence of the occurrence of endogenous RNA quadruplexes in human cells. For example, carboxypyridostatin, which was shown by chemical approaches (alkyne-azide cycloaddition in situ) to bind more effectively to RNA G4 than to the DNA analogs [235], increased (approximately 2.4-fold) the number of G4s only in the cell cytoplasm, not affecting the number of DNA quadruplexes with nuclear localization. Recently, RNA G4 were found in live cells using a new generation of quadruplex-specific compound, N-TASQ, which combined functions of the “smart” ligand and “smart” fluorescence probe [263]. These compounds behave as intracellular molecular sensors targeting only the preformed G4s. They use G4-quadruplexes as a template for self-assembly of the artificial G-tetrad, formation of which causes fluorescence enhancement. N-TASQ exhibits unique properties because, unlike the quadruplex-specific antibodies, it does not require fixation and permeabilization of the cells. It was shown that under the conditions used the cytoplasmic RNA G4s (in free state and more likely in the form of complex ribonucleoprotein subcellular ensembles) served as targets of the N-TASQ, and they were revealed as intensive discrete loci in human breast cancer and osteosarcoma cells as well as in murine melanoma. Moreover, the cells were not pretreated in the process. It has been suggested that N-TASQ penetrates easily through the cytoplasmic membrane, accumulating in the cytoplasm under non-denaturing physiological conditions.
It was shown [423] that G4-specific BG4 antibody could be used to stain DNA G4 structures in human tissues using immunohistochemistry. A significantly elevated number of G4-positive nuclei in tissues of liver and stomach cancer patients in comparison with healthy tissues was observed. To identify malignant tissue transformation, the G4-binding fluorescent ligand 3,6-bis(1-methyl-4-vinylpyridinium) carbazole diiodate was also used [424]. It was found that the number of signals increased depending on the degree of cell transformation, making this approach promising for early diagnostics of oncological diseases.
The G-quadruplex structures were identified by immunoelectron microscopy in samples from different organisms: ciliates, flatworms, fruit flies, and mammals [425]. It was found that the loci stained by the 1H6 monoclonal antibodies, which were specific to DNA G4s of various topologies, were present in heterochromatin of all samples. It was found that the sites of 1H6 antibody binding in the polytene chromosomes from drosophila salivary glands were colocalized with heterochromatic proteins. It was also observed that the somatic cells in different organisms displayed more intensive staining than pluripotent stem cells. These findings indicate a conserved role of G4s in nuclear organization and cell differentiation. It is the authors’ opinion that the G-quadruplex structures are present in postmitotic cells as well as in mitotic chromosomes. They can exist independently of DNA replication, transcription, or recombination stimulating genetic instability.
The studies conducted in recent years have shown that G4s exist in genomic DNA in all phases of the cell cycle. These non-canonical structures are diverse. On one hand, they can perform regulatory functions in the cell, inhibit oncogene expression, block undesired elongation of telomeric DNA, control the level of negative supercoiling in the genome, and serve as targets of antitumor therapeutics. Recently, the role of G-quadruplexes in embryo development, which is the most controlled process in vertebrate biology, was established [426]. On the other hand, G4 formation causes genome instability (genomic inversions, recombination, mutations, deletions, and others) associated with oncological diseases and neurological disorders. In this review, we have presented state-of-the-art knowledge on these amazing structures in all investigated aspects. The most important information on the structure and biological functions of G-quadruplexes was obtained using novel research techniques, and their major features were emphasized in this review.
Acknowledgements
This work was supported by the Russian Foundation for Basic Research (project No. 16-04-00575_A).
REFERENCES
1.Xu, Y., Suzuki, Y., Ito, K., and Komiyama, M.
(2010) Telomeric repeat-containing RNA structure in living cells,
Proc. Natl. Acad. Sci. USA, 107, 14579-14584.
2.Huppert, J. L., Bugaut, A., Kumari, S., and
Balasubramanian, S. (2008) G-quadruplexes: the beginning and end of
UTRs, Nucleic Acids Res., 36, 6260-6268.
3.Bochman, M. L., Paeschke, K., and Zakian, V. A.
(2012) DNA secondary structures: stability and function of G-quadruplex
structures, Nat. Rev. Genet., 13, 770-780.
4.Lipps, H. J., and Rhodes, D. (2009) G-quadruplex
structures: in vivo evidence and function, Trends Cell
Biol., 19, 414-422.
5.Valton, A. L., Hassan-Zadeh, V., Lema, I.,
Boggetto, N., Alberti, P., Saintome, C., Riou, J. F., and Prioleau, M.
N. (2014) G4 motifs affect origin positioning and efficiency in two
vertebrate replicators, EMBO J., 33, 732-746.
6.Foulk, M. S., Urban, J. M., Casella, C., and Gerbi,
S. A. (2015) Characterizing and controlling intrinsic biases of lambda
exonuclease in nascent strand sequencing reveals phasing between
nucleosomes and G-quadruplex motifs around a subset of human
replication origins, Genome Res., 25, 725-735.
7.Paeschke, K., Juranek, S., Simonsson, T., Hempel,
A., Rhodes, D., and Lipps, H. J. (2008) Telomerase recruitment by the
telomere end binding protein-beta facilitates G-quadruplex DNA
unfolding in ciliates, Nat. Struct. Mol. Biol., 15,
598-604.
8.Balasubramanian, S., Hurley, L. H., and Neidle, S.
(2011) Targeting G-quadruplexes in gene promoters: a novel anticancer
strategy? Nat. Rev. Drug Discov., 10, 261-275.
9.Kruisselbrink, E., Guryev, V., Brouwer, K.,
Pontier, D. B., Cuppen, E., and Tijsterman, M. (2008) Mutagenic
capacity of endogenous G4 DNA underlies genome instability in
FANCJ-defective C. elegans, Curr. Biol., 18,
900-905.
10.Beaume, N., Pathak, R., Yadav, V. K., Kota, S.,
Misra, H. S., Gautam, H. K., and Chowdhury, S. (2013) Genome-wide study
predicts promoter-G4 DNA motifs regulate selective functions in
bacteria: radioresistance of D. radiodurans involves G4
DNA-mediated regulation, Nucleic Acids Res., 41,
76-89.
11.Cahoon, L. A., and Seifert, H. S. (2009) An
alternative DNA structure is necessary for pilin antigenic variation in
Neisseria gonorrhoeae, Science, 325, 764-767.
12.Tarsounas, M., and Tijsterman, M. (2013) Genomes
and G-quadruplexes: for better or for worse, J. Mol. Biol.,
425, 4782-4789.
13.Schiavone, D., Guilbaud, G., Murat, P.,
Papadopoulou, C., Sarkies, P., Prioleau, M. N., Balasubramanian, S.,
and Sale, J. E. (2014) Determinants of G quadruplex-induced epigenetic
instability in REV1-deficient cells, EMBO J., 33,
2507-2520.
14.Beaudoin, J. D., and Perreault, J. P. (2010)
5′-UTR G-quadruplex structures acting as translational
repressors, Nucleic Acids Res., 38, 7022-7036.
15.Beaudoin, J. D., and Perreault, J. P. (2013)
Exploring mRNA 3′-UTR G-quadruplexes: evidence of roles in both
alternative polyadenylation and mRNA shortening, Nucleic Acids
Res., 41, 5898-5911.
16.Haider, S. M., Neidle, S., and Parkinson, G. N.
(2011) A structural analysis of G-quadruplex/ligand interactions,
Biochimie, 93, 1239-1251.
17.Bryan, T. M., and Baumann, P. (2010)
G-Quadruplex DNA: Methods and Protocols, Methods in Molecular
Biology Series, Humana Press, p. 276.
18.Yatsunyk, L. A., Bryan, T. M., and Johnson, F. B.
(2012) G-ruption: the third international meeting on G-quadruplex and
G-assembly, Biochimie, 94, 2475-2483.
19.Wu, Y., and Brosh, R. M., Jr. (2010) G-quadruplex
nucleic acids and human disease, FEBS J., 277,
3470-3488.
20.Simone, R., Fratta, P., Neidle, S., Parkinson, G.
N., and Isaacs, A. M. (2015) G-quadruplexes: emerging roles in
neurodegenerative diseases and the non-coding transcriptome, FEBS
Lett., 589, 1653-1668.
21.Yatsunyk, L. A., Mendoza, O., and Mergny, J. L.
(2014) “Nano-oddities”: unusual nucleic acid assemblies for
DNA-based nanostructures and nanodevices, Acc. Chem. Res.,
47, 1836-1844.
22.Largy, E., Marchand, A., Amrane, S., Gabelica,
V., and Mergny, J. L. (2016) Quadruplex turncoats: cation-dependent
folding and stability of quadruplex-DNA double switches, J. Am.
Chem. Soc., 138, 2780-2792.
23.Rajendran, A., Endo, M., Hidaka, K., Tran, P. L.,
Mergny, J. L., and Sugiyama, H. (2013) Controlling the stoichiometry
and strand polarity of a tetramolecular G-quadruplex structure by using
a DNA origami frame, Nucleic Acids Res., 41,
8738-8747.
24.Kogut, M., Kleist, C., and Czub, J. (2016)
Molecular dynamics simulations reveal the balance of forces governing
the formation of a guanine tetrad – a common structural unit of
G-quadruplex DNA, Nucleic Acids Res., 44, 3020-3030.
25.Chung, W. J., Heddi, B., Schmitt, E., Lim, K. W.,
Mechulam, Y., and Phan, A. T. (2015) Structure of a left-handed DNA
G-quadruplex, Proc. Natl. Acad. Sci. USA, 112,
2729-2733.
26.Bugaut, A., and Balasubramanian, S. (2008) A
sequence-independent study of the influence of short loop lengths on
the stability and topology of intramolecular DNA G-quadruplexes,
Biochemistry, 47, 689-697.
27.Reshetnikov, R. V., Kopylov, A. M., and Golovin,
A. V. (2010) Classification of G-quadruplex DNA on the basis of
the quadruplex twist angle and planarity of G-quartets, Acta
Naturae, 2, 72-81.
28.Kim, B. G., Shek, Y. L., and Chalikian, T. V.
(2013) Polyelectrolyte effects in G-quadruplexes, Biophys.
Chem., 184, 95-100.
29.Gaynutdinov, T. I., Neumann, R. D., and Panyutin,
I. G. (2008) Structural polymorphism of intramolecular quadruplex of
human telomeric DNA: effect of cations, quadruplex-binding drugs and
flanking sequences, Nucleic Acids Res., 36,
4079-4087.
30.Reshetnikov, R. V., Sponer, J., Rassokhina, O.
I., Kopylov, A. M., Tsvetkov, P. O., Makarov, A. A., and Golovin, A. V.
(2011) Cation binding to 15-TBA quadruplex DNA is a multiple-pathway
cation-dependent process, Nucleic Acids Res., 39,
9789-9802.
31.Saintome, C., Amrane, S., Mergny, J. L., and
Alberti, P. (2016) The exception that confirms the rule: a higher-order
telomeric G-quadruplex structure more stable in sodium than in
potassium, Nucleic Acids Res., 44, 2926-2935.
32.Sket, P., Virgilio, A., Esposito, V., Galeone,
A., and Plavec, J. (2012) Strand directionality affects cation binding
and movement within tetramolecular G-quadruplexes, Nucleic Acids
Res., 40, 11047-11057.
33.Podbevsek, P., Sket, P., and Plavec, J. (2008)
Stacking and not solely topology of T3 loops controls rigidity and
ammonium ion movement within
d(G4T3G4)2 G-quadruplex,
J. Am. Chem. Soc., 130, 14287-14293.
34.De Rache, A., and Mergny, J. L. (2015) Assessment
of selectivity of G-quadruplex ligands via an optimised FRET melting
assay, Biochimie, 115, 194-202.
35.Kim, B. G., Evans, H. M., Dubins, D. N., and
Chalikian, T. V. (2015) Effects of salt on the stability of a
G-quadruplex from the human c-MYC promoter, Biochemistry,
54, 3420-3430.
36.Zhang, D., Huang, T., Lukeman, P. S., and
Paukstelis, P. J. (2014) Crystal structure of a DNA/Ba2+
G-quadruplex containing a water-mediated C-tetrad, Nucleic Acids
Res., 42, 13422-13429.
37.Liu, W., Zhu, H., Zheng, B., Cheng, S., Fu, Y.,
Li, W., Lau, T. C., and Liang, H. (2012) Kinetics and mechanism of
G-quadruplex formation and conformational switch in a G-quadruplex of
PS2.M induced by Pb2+, Nucleic Acids Res., 40,
4229-4236.
38.Martin-Hidalgo, M., and Rivera, J. M. (2011)
Metallo-responsive switching between hexadecameric and octameric
supramolecular G-quadruplexes, Chem. Commun. (Camb.), 47,
12485-12487.
39.Gray, R. D., and Chaires, J. B. (2011) Linkage of
cation binding and folding in human telomeric quadruplex DNA,
Biophys. Chem., 159, 205-209.
40.Sattin, G., Artese, A., Nadai, M., Costa, G.,
Parrotta, L., Alcaro, S., Palumbo, M., and Richter, S. N. (2013)
Conformation and stability of intramolecular telomeric G-quadruplexes:
sequence effects in the loops, PLoS One, 8, e84113.
41.Tippana, R., Xiao, W., and Myong, S. (2014)
G-quadruplex conformation and dynamics are determined by loop length
and sequence, Nucleic Acids Res., 42, 8106-8114.
42.Guedin, A., De Cian, A., Gros, J., Lacroix, L.,
and Mergny, J. L. (2008) Sequence effects in single-base loops for
quadruplexes, Biochimie, 90, 686-696.
43.Do, N. Q., Lim, K. W., Teo, M. H., Heddi, B., and
Phan, A. T. (2011) Stacking of G-quadruplexes: NMR structure of a
G-rich oligonucleotide with potential anti-HIV and anticancer activity,
Nucleic Acids Res., 39, 9448-9457.
44.Ogloblina, A. M., Bannikova, V. A., Khristich, A.
N., Oretskaya, T. S., Yakubovskaya, M. G., and Dolinnaya, N. G. (2015)
Parallel G-quadruplexes formed by guanine-rich microsatellite repeats
inhibit human topoisomerase I, Biochemistry (Moscow), 80,
1026-1038.
45.Qin, Y., and Hurley, L. H. (2008) Structures,
folding patterns, and functions of intramolecular DNA G-quadruplexes
found in eukaryotic promoter regions, Biochimie, 90,
1149-1171.
46.Kwok, C. K., Sherlock, M. E., and Bevilacqua, P.
C. (2013) Effect of loop sequence and loop length on the intrinsic
fluorescence of G-quadruplexes, Biochemistry, 52,
3019-3021.
47.Lim, K. W., Amrane, S., Bouaziz, S., Xu, W., Mu,
Y., Patel, D. J., Luu, K. N., and Phan, A. T. (2009) Structure of the
human telomere in K+ solution: a stable basket-type
G-quadruplex with only two G-tetrad layers, J. Am. Chem. Soc.,
131, 4301-4309.
48.Sengar, A., Heddi, B., and Phan, A. T. (2014)
Formation of G-quadruplexes in poly-G sequences: structure of a
propeller-type parallel-stranded G-quadruplex formed by a
G15 stretch, Biochemistry, 53, 7718-7723.
49.Cang, X., Sponer, J., and Cheatham, T. E., 3rd.
(2011) Explaining the varied glycosidic conformational, G-tract length
and sequence preferences for anti-parallel G-quadruplexes, Nucleic
Acids Res., 39, 4499-4512.
50.Olsen, C. M., Lee, H. T., and Marky, L. A. (2009)
Unfolding thermodynamics of intramolecular G-quadruplexes: base
sequence contributions of the loops, J. Phys. Chem. B,
113, 2587-2595.
51.Chen, B., Liang, J., Tian, X., and Liu, X. (2008)
G-quadruplex structure: a target for anticancer therapy and a probe for
detection of potassium, Biochemistry (Moscow), 73,
853-861.
52.Dailey, M. M., Miller, M. C., Bates, P. J., Lane,
A. N., and Trent, J. O. (2010) Resolution and characterization of the
structural polymorphism of a single quadruplex-forming sequence,
Nucleic Acids Res., 38, 4877-4888.
53.Borbone, N., Amato, J., Oliviero, G.,
D’Atri, V., Gabelica, V., De Pauw, E., Piccialli, G., and Mayol,
L. (2011) d(CGGTGGT) forms an octameric parallel G-quadruplex via
stacking of unusual G(:C):G(:C):G(:C):G(:C) octads, Nucleic Acids
Res., 39, 7848-7857.
54.D'Atri, V., Borbone, N., Amato, J., Gabelica, V.,
D'Errico, S., Piccialli, G., Mayol, L., and Oliviero, G. (2014)
DNA-based nanostructures: the effect of the base sequence on octamer
formation from d(XGGYGGT) tetramolecular G-quadruplexes,
Biochimie, 99, 119-128.
55.Kuryavyi, V., Cahoon, L. A., Seifert, H. S., and
Patel, D. J. (2012) RecA-binding pilE G4 sequence essential for pilin
antigenic variation forms monomeric and 5′-end-stacked dimeric
parallel G-quadruplexes, Structure, 20, 2090-2102.
56.Adrian, M., Ang, D. J., Lech, C. J., Heddi, B.,
Nicolas, A., and Phan, A. T. (2014) Structure and conformational
dynamics of a stacked dimeric G-quadruplex formed by the human CEB1
minisatellite, J. Am. Chem. Soc., 136, 6297-6305.
57.Phan, A. T. (2010) Human telomeric G-quadruplex:
structures of DNA and RNA sequences, FEBS J., 277,
1107-1117.
58.Phan, A. T., and Do, N. Q. (2013) Engineering of
interlocked DNA G-quadruplexes as a robust scaffold, Nucleic Acids
Res., 41, 2683-2688.
59.Todd, A. K., and Neidle, S. (2011) Mapping the
sequences of potential guanine quadruplex motifs, Nucleic Acids
Res., 39, 4917-4927.
60.Yadav, V. K., Abraham, J. K., Mani, P.,
Kulshrestha, R., and Chowdhury, S. (2008) QuadBase: genome-wide
database of G4 DNA-occurrence and conservation in human, chimpanzee,
mouse and rat promoters and 146 microbes, Nucleic Acids Res.,
36, D381-385.
61.Kikin, O., Zappala, Z., D’Antonio, L., and
Bagga, P. S. (2008) GRSDB2 and GRS_UTRdb: databases of quadruplex
forming G-rich sequences in pre-mRNAs and mRNAs, Nucleic Acids
Res., 36, D141-148.
62.Huppert, J. L., and Balasubramanian, S. (2007)
G-quadruplexes in promoters throughout the human genome, Nucleic
Acids Res., 35, 406-413.
63.Todd, A. K., and Neidle, S. (2008) The
relationship of potential G-quadruplex sequences in cis-upstream
regions of the human genome to SP1-binding elements, Nucleic Acids
Res., 36, 2700-2704.
64.Huppert, J. L. (2008) Hunting G-quadruplexes,
Biochimie, 90, 1140-1148.
65.Maizels, N., and Gray, L. T. (2013) The G4
genome, PLoS Genet., 9, e1003468.
66.Neidle, S. (2010) Human telomeric G-quadruplex:
the current status of telomeric G-quadruplexes as therapeutic targets
in human cancer, FEBS J., 277, 1118-1125.
67.Amrane, S., Adrian, M., Heddi, B., Serero, A.,
Nicolas, A., Mergny, J. L., and Phan, A. T. (2012) Formation of
pearl-necklace monomorphic G-quadruplexes in the human CEB25
minisatellite, J. Am. Chem. Soc., 134, 5807-5816.
68.Sawaya, S., Bagshaw, A., Buschiazzo, E., Kumar,
P., Chowdhury, S., Black, M. A., and Gemmell, N. (2013) Microsatellite
tandem repeats are abundant in human promoters and are associated with
regulatory elements, PLoS One, 8, e54710.
69.Lexa, M., Kejnovsky, E., Steflova, P.,
Konvalinova, H., Vorlickova, M., and Vyskot, B. (2014)
Quadruplex-forming sequences occupy discrete regions inside plant LTR
retrotransposons, Nucleic Acids Res., 42, 968-978.
70.Brooks, T. A., Kendrick, S., and Hurley, L.
(2010) Making sense of G-quadruplex and i-motif functions in oncogene
promoters, FEBS J., 277, 3459-3469.
71.Du, Z., Zhao, Y., and Li, N. (2008) Genome-wide
analysis reveals regulatory role of G4 DNA in gene transcription,
Genome Res., 18, 233-241.
72.Besnard, E., Babled, A., Lapasset, L., Milhavet,
O., Parrinello, H., Dantec, C., Marin, J. M., and Lemaitre, J. M.
(2012) Unraveling cell type-specific and reprogrammable human
replication origin signatures associated with G-quadruplex consensus
motifs, Nat. Struct. Mol. Biol., 19, 837-844.
73.Cayrou, C., Coulombe, P., Puy, A., Rialle, S.,
Kaplan, N., Segal, E., and Mechali, M. (2012) New insights into
replication origin characteristics in metazoans, Cell Cycle,
11, 658-667.
74.Katapadi, V. K., Nambiar, M., and Raghavan, S. C.
(2012) Potential G-quadruplex formation at breakpoint regions of
chromosomal translocations in cancer may explain their fragility,
Genomics, 100, 72-80.
75.Mani, P., Yadav, V. K., Das, S. K., and
Chowdhury, S. (2009) Genome-wide analyses of recombination prone
regions predict role of DNA structural motif in recombination, PLoS
One, 4, e4399.
76.Kuryavyi, V., and Patel, D. J. (2010) Solution
structure of a unique G-quadruplex scaffold adopted by a guanosine-rich
human intronic sequence, Structure, 18, 73-82.
77.Zizza, P., Cingolani, C., Artuso, S., Salvati,
E., Rizzo, A., D’Angelo, C., Porru, M., Pagano, B., Amato, J.,
Randazzo, A., Novellino, E., Stoppacciaro, A., Gilson, E., Stassi, G.,
Leonetti, C., and Biroccio, A. (2016) Intragenic G-quadruplex structure
formed in the human CD133 and its biological and translational
relevance, Nucleic Acids Res., 44, 1579-1590.
78.Du, Z., Zhao, Y., and Li, N. (2009) Genome-wide
colonization of gene regulatory elements by G4 DNA motifs, Nucleic
Acids Res., 37, 6784-6798.
79.Dong, D. W., Pereira, F., Barrett, S. P.,
Kolesar, J. E., Cao, K., Damas, J., Yatsunyk, L. A., Johnson, F. B.,
and Kaufman, B. A. (2014) Association of G-quadruplex forming sequences
with human mtDNA deletion breakpoints, BMC Genomics, 15,
677.
80.Huppert, J. L. (2010) Structure, location and
interactions of G-quadruplexes, FEBS J., 277,
3452-3458.
81.Martadinata, H., and Phan, A. T. (2013) Structure
of human telomeric RNA (TERRA): stacking of two G-quadruplex blocks in
K+ solution, Biochemistry, 52, 2176-2183.
82.Mirihana Arachchilage, G., Dassanayake, A. C.,
and Basu, S. (2015) A potassium ion-dependent RNA structural switch
regulates human pre-miRNA 92b maturation, Chem. Biol.,
22, 262-272.
83.Jayaraj, G. G., Pandey, S., Scaria, V., and
Maiti, S. (2012) Potential G-quadruplexes in the human long non-coding
transcriptome, RNA Biol., 9, 81-86.
84.Frees, S., Menendez, C., Crum, M., and Bagga, P.
S. (2014) QGRS-Conserve: a computational method for discovering
evolutionarily conserved G-quadruplex motifs, Hum. Genomics,
8, 8.
85.Zybailov, B. L., Sherpa, M. D., Glazko, G. V.,
Raney, K. D., and Glazko, V. I. (2013) G4-quadruplexes and genome
instability, Mol. Biol. (Moscow), 47, 197-204.
86.Zhang, C., Liu, H. H., Zheng, K. W., Hao, Y. H.,
and Tan, Z. (2013) DNA G-quadruplex formation in response to remote
downstream transcription activity: long-range sensing and signal
transducing in DNA double helix, Nucleic Acids Res., 41,
7144-7152.
87.Agrawal, P., Lin, C., Mathad, R. I., Carver, M.,
and Yang, D. (2014) The major G-quadruplex formed in the human BCL-2
proximal promoter adopts a parallel structure with a 13-nt loop in
K+ solution, J. Am. Chem. Soc., 136,
1750-1753.
88.Davis, L., and Maizels, N. (2011) G4 DNA: at risk
in the genome, EMBO J., 30, 3878-3879.
89.Eddy, J., Vallur, A. C., Varma, S., Liu, H.,
Reinhold, W. C., Pommier, Y., and Maizels, N. (2011) G4 motifs
correlate with promoter-proximal transcriptional pausing in human
genes, Nucleic Acids Res., 39, 4975-4983.
90.Capra, J. A., Paeschke, K., Singh, M., and
Zakian, V. A. (2010) G-quadruplex DNA sequences are evolutionarily
conserved and associated with distinct genomic features in
Saccharomyces cerevisiae, PLoS Comput. Biol., 6,
e1000861.
91.Kwok, C. K., Ding, Y., Shahid, S., Assmann, S.
M., and Bevilacqua, P. C. (2015) A stable RNA G-quadruplex within the
5′-UTR of Arabidopsis thaliana ATR mRNA inhibits
translation, Biochem. J., 467, 91-102.
92.Chambers, V. S., Marsico, G., Boutell, J. M., Di
Antonio, M., Smith, G. P., and Balasubramanian, S. (2015)
High-throughput sequencing of DNA G-quadruplex structures in the human
genome, Nat. Biotechnol., 33, 877-881.
93.Guedin, A., Gros, J., Alberti, P., and Mergny, J.
L. (2010) How long is too long? Effects of loop size on G-quadruplex
stability, Nucleic Acids Res., 38, 7858-7868.
94.Mukundan, V. T., and Phan, A. T. (2013) Bulges in
G-quadruplexes: broadening the definition of G-quadruplex-forming
sequences, J. Am. Chem. Soc., 135, 5017-5028.
95.Beaudoin, J. D., Jodoin, R., and Perreault, J. P.
(2014) New scoring system to identify RNA G-quadruplex folding,
Nucleic Acids Res., 42, 1209-1223.
96.Pandey, S., Agarwala, P., and Maiti, S. (2013)
Effect of loops and G-quartets on the stability of RNA G-quadruplexes,
J. Phys. Chem. B, 117, 6896-6905.
97.Bedrat, A., Lacroix, L., and Mergny, J. L. (2016)
Re-evaluation of G-quadruplex propensity with G4Hunter, Nucleic
Acids Res., 44, 1746-1759.
98.Du, X., Wojtowicz, D., Bowers, A. A., Levens, D.,
Benham, C. J., and Przytycka, T. M. (2013) The genome-wide distribution
of non-B DNA motifs is shaped by operon structure and suggests the
transcriptional importance of non-B DNA structures in Escherichia
coli, Nucleic Acids Res., 41, 5965-5977.
99.Amrane, S., Kerkour, A., Bedrat, A., Vialet, B.,
Andreola, M. L., and Mergny, J. L. (2014) Topology of a DNA
G-quadruplex structure formed in the HIV-1 promoter: a potential target
for anti-HIV drug development, J. Am. Chem. Soc., 136,
5249-5252.
100.Piekna-Przybylska, D., Sullivan, M. A., Sharma,
G., and Bambara, R. A. (2014) U3 region in the HIV-1 genome adopts a
G-quadruplex structure in its RNA and DNA sequence,
Biochemistry, 53, 2581-2593.
101.Perrone, R., Nadai, M., Frasson, I., Poe, J.
A., Butovskaya, E., Smithgall, T. E., Palumbo, M., Palu, G., and
Richter, S. N. (2013) A dynamic G-quadruplex region regulates the HIV-1
long terminal repeat promoter, J. Med. Chem., 56,
6521-6530.
102.Norseen, J., Johnson, F. B., and Lieberman, P.
M. (2009) Role for G-quadruplex RNA binding by Epstein–Barr virus
nuclear antigen 1 in DNA replication and metaphase chromosome
attachment, J. Virol., 83, 10336-10346.
103.Murat, P., Zhong, J., Lekieffre, L., Cowieson,
N. P., Clancy, J. L., Preiss, T., Balasubramanian, S., Khanna, R., and
Tellam, J. (2014) G-quadruplexes regulate Epstein–Barr
virus-encoded nuclear antigen 1 mRNA translation, Nat. Chem.
Biol., 10, 358-364.
104.Tluckova, K., Marusic, M., Tothova, P., Bauer,
L., Sket, P., Plavec, J., and Viglasky, V. (2013) Human papillomavirus
G-quadruplexes, Biochemistry, 52, 7207-7216.
105.Musumeci, D., Riccardi, C., and Montesarchio,
D. (2015) G-quadruplex forming oligonucleotides as anti-HIV agents,
Molecules, 20, 17511-17532.
106.Vorlickova, M., Kejnovska, I., Sagi, J.,
Renciuk, D., Bednarova, K., Motlova, J., and Kypr, J. (2012) Circular
dichroism and guanine quadruplexes, Methods, 57,
64-75.
107.Olsen, C. M., and Marky, L. A. (2010)
Monitoring the temperature unfolding of G-quadruplexes by UV and
circular dichroism spectroscopies and calorimetry techniques,
Methods Mol. Biol., 608, 147-158.
108.Karsisiotis, A. I., Hessari, N. M., Novellino,
E., Spada, G. P., Randazzo, A., and Webba da Silva, M. (2011)
Topological characterization of nucleic acid G-quadruplexes by UV
absorption and circular dichroism, Angew. Chem. Int. Ed. Engl.,
50, 10645-10648.
109.Tran, P. L., Mergny, J. L., and Alberti, P.
(2011) Stability of telomeric G-quadruplexes, Nucleic Acids
Res., 39, 3282-3294.
110.Shek, Y. L., Noudeh, G. D., Nazari, M.,
Heerklotz, H., Abu-Ghazalah, R. M., Dubins, D. N., and Chalikian, T. V.
(2014) Folding thermodynamics of the hybrid-1 type intramolecular human
telomeric G-quadruplex, Biopolymers, 101, 216-227.
111.Petraccone, L., Spink, C., Trent, J. O.,
Garbett, N. C., Mekmaysy, C. S., Giancola, C., and Chaires, J. B.
(2011) Structure and stability of higher-order human telomeric
quadruplexes, J. Am. Chem. Soc., 133, 20951-20961.
112.Russo Krauss, I., Pica, A., Merlino, A.,
Mazzarella, L., and Sica, F. (2013) Duplex-quadruplex motifs in a
peculiar structural organization cooperatively contribute to thrombin
binding of a DNA aptamer, Acta Crystallogr. D Biol.
Crystallogr., 69, 2403-2411.
113.Sun, D., and Hurley, L. H. (2009) The
importance of negative superhelicity in inducing the formation of
G-quadruplex and i-motif structures in the c-Myc promoter: implications
for drug targeting and control of gene expression, J. Med.
Chem., 52, 2863-2874.
114.Sun, D., Guo, K., and Shin, Y. J. (2011)
Evidence of the formation of G-quadruplex structures in the promoter
region of the human vascular endothelial growth factor gene, Nucleic
Acids Res., 39, 1256-1265.
115.Mendoza, O., Gueddouda, N. M., Boule, J. B.,
Bourdoncle, A., and Mergny, J. L. (2015) A fluorescence-based helicase
assay: application to the screening of G-quadruplex ligands, Nucleic
Acids Res., 43, e71.
116.Kreig, A., Calvert, J., Sanoica, J., Cullum,
E., Tipanna, R., and Myong, S. (2015) G-quadruplex formation in double
strand DNA probed by NMM and CV fluorescence, Nucleic Acids
Res., 43, 7961-7970.
117.Long, X., Parks, J. W., Bagshaw, C. R., and
Stone, M. D. (2013) Mechanical unfolding of human telomere G-quadruplex
DNA probed by integrated fluorescence and magnetic tweezers
spectroscopy, Nucleic Acids Res., 41, 2746-2755.
118.Ray, S., Bandaria, J. N., Qureshi, M. H.,
Yildiz, A., and Balci, H. (2014) G-quadruplex formation in telomeres
enhances POT1/TPP1 protection against RPA binding, Proc. Natl. Acad.
Sci. USA, 111, 2990-2995.
119.Palacky, J., Vorlickova, M., Kejnovska, I., and
Mojzes, P. (2013) Polymorphism of human telomeric quadruplex structure
controlled by DNA concentration: a Raman study, Nucleic Acids
Res., 41, 1005-1016.
120.Zhang, X., Xu, C. X., Di Felice, R., Sponer,
J., Islam, B., Stadlbauer, P., Ding, Y., Mao, L., Mao, Z. W., and Qin,
P. Z. (2016) Conformations of human telomeric G-quadruplex studied
using a nucleotide-independent nitroxide label, Biochemistry,
55, 360-372.
121.Singh, V., Azarkh, M., Drescher, M., and
Hartig, J. S. (2012) Conformations of individual quadruplex units
studied in the context of extended human telomeric DNA, Chem.
Commun. (Camb.), 48, 8258-8260.
122.Haider, S., and Neidle, S. (2010) Molecular
modeling and simulation of G-quadruplexes and quadruplex–ligand
complexes, Methods Mol. Biol., 608, 17-37.
123.Husby, J., Todd, A. K., Platts, J. A., and
Neidle, S. (2013) Small-molecule G-quadruplex interactions: systematic
exploration of conformational space using multiple molecular dynamics,
Biopolymers, 99, 989-1005.
124.Islam, B., Stadlbauer, P., Neidle, S., Haider,
S., and Sponer, J. (2016) Can we execute reliable MM-PBSA free energy
computations of relative stabilities of different guanine quadruplex
folds? J. Phys. Chem. B, 120, 2899-2912.
125.Collie, G. W., Parkinson, G. N., Neidle, S.,
Rosu, F., De Pauw, E., and Gabelica, V. (2010) Electrospray mass
spectrometry of telomeric RNA (TERRA) reveals the formation of stable
multimeric G-quadruplex structures, J. Am. Chem. Soc.,
132, 9328-9334.
126.Adrian, M., Heddi, B., and Phan, A. T. (2012)
NMR spectroscopy of G-quadruplexes, Methods, 57,
11-24.
127.Lim, K. W., Ng, V. C., Martin-Pintado, N.,
Heddi, B., and Phan, A. T. (2013) Structure of the human telomere in
Na+ solution: an antiparallel (2+2) G-quadruplex scaffold
reveals additional diversity, Nucleic Acids Res., 41,
10556-10562.
128.Lane, A. N., Chaires, J. B., Gray, R. D., and
Trent, J. O. (2008) Stability and kinetics of G-quadruplex structures,
Nucleic Acids Res., 36, 5482-5515.
129.Chaires, J. B. (2010) Human telomeric
G-quadruplex: thermodynamic and kinetic studies of telomeric quadruplex
stability, FEBS J., 277, 1098-1106.
130.Stegle, O., Payet, L., Mergny, J. L., MacKay,
D. J., and Leon, J. H. (2009) Predicting and understanding the
stability of G-quadruplexes, Bioinformatics, 25,
i374-382.
131.Lane, A. N. (2012) The stability of
intramolecular DNA G-quadruplexes compared with other macromolecules,
Biochimie, 94, 277-286.
132.Kankia, B. I. (2011) Self-dissociative primers
for nucleic acid amplification and detection based on DNA quadruplexes
with intrinsic fluorescence, Anal. Biochem., 409,
59-65.
133.Heddi, B., Martin-Pintado, N., Serimbetov, Z.,
Kari, T. M., and Phan, A. T. (2016) G-quadruplexes with (4n–1)
guanines in the G-tetrad core: formation of a G-triad water complex and
implication for small-molecule binding, Nucleic Acids Res.,
44, 910-916.
134.Cerofolini, L., Amato, J., Giachetti, A.,
Limongelli, V., Novellino, E., Parrinello, M., Fragai, M., Randazzo,
A., and Luchinat, C. (2014) G-triplex structure and formation
propensity, Nucleic Acids Res., 42, 13393-13404.
135.Virgilio, A., Petraccone, L., Esposito, V.,
Citarella, G., Giancola, C., and Galeone, A. (2012) The abasic site
lesions in the human telomeric sequence
d[TAG3T2A3G3]: a
thermodynamic point of view, Biochim. Biophys. Acta,
1820, 2037-2043.
136.Harkness, R. W. T., and Mittermaier, A. K.
(2016) G-register exchange dynamics in guanine quadruplexes, Nucleic
Acids Res., 44, 3481-3494.
137.Mathias, J., Okyere, R., Lomidze, L.,
Gvarjaladze, D., Musier-Forsyth, K., and Kankia, B. (2014) Thermal
stability of quadruplex primers for highly versatile isothermal DNA
amplification, Biophys. Chem., 185, 14-18.
138.Hatzakis, E., Okamoto, K., and Yang, D. (2010)
Thermodynamic stability and folding kinetics of the major G-quadruplex
and its loop isomers formed in the nuclease hypersensitive element in
the human c-Myc promoter: effect of loops and flanking segments on the
stability of parallel-stranded intramolecular G-quadruplexes,
Biochemistry, 49, 9152-9160.
139.Piazza, A., Adrian, M., Samazan, F., Heddi, B.,
Hamon, F., Serero, A., Lopes, J., Teulade-Fichou, M. P., Phan, A. T.,
and Nicolas, A. (2015) Short loop length and high thermal stability
determine genomic instability induced by G-quadruplex-forming
minisatellites, EMBO J., 34, 1718-1734.
140.Mathad, R. I., Hatzakis, E., Dai, J., and Yang,
D. (2011) c-MYC promoter G-quadruplex formed at the 5′-end of NHE
III1 element: insights into biological relevance and parallel-stranded
G-quadruplex stability, Nucleic Acids Res., 39,
9023-9033.
141.Selvam, S., Yu, Z. B., and Mao, H. B. (2016)
Exploded view of higher order G-quadruplex structures through
click-chemistry assisted single-molecule mechanical unfolding,
Nucleic Acids Res., 44, 45-55.
142.Kuryavyi, V., Phan, A. T., and Patel, D. J.
(2010) Solution structures of all parallel-stranded monomeric and
dimeric G-quadruplex scaffolds of the human c-kit2 promoter, Nucleic
Acids Res., 38, 6757-6773.
143.Zhou, J., Tateishi-Karimata, H., Mergny, J. L.,
Cheng, M., Feng, Z., Miyoshi, D., Sugimoto, N., and Li, C. (2016)
Reevaluation of the stability of G-quadruplex structures under crowding
conditions, Biochimie, 121, 204-208.
144.Nagatoishi, S., Isono, N., Tsumoto, K., and
Sugimoto, N. (2011) Hydration is required in DNA
G-quadruplex–protein binding, ChemBioChem, 12,
1822-1826.
145.Miyoshi, D., Fujimoto, T., and Sugimoto, N.
(2013) Molecular crowding and hydration regulating of G-quadruplex
formation, Top. Curr. Chem., 330, 87-110.
146.You, H., Zeng, X., Xu, Y., Lim, C. J., Efremov,
A. K., Phan, A. T., and Yan, J. (2014) Dynamics and stability of
polymorphic human telomeric G-quadruplex under tension, Nucleic
Acids Res., 42, 8789-8795.
147.Gray, R. D., Li, J., and Chaires, J. B. (2009)
Energetics and kinetics of a conformational switch in G-quadruplex DNA,
J. Phys. Chem. B, 113, 2676-2683.
148.Buscaglia, R., Gray, R. D., and Chaires, J. B.
(2013) Thermodynamic characterization of human telomere quadruplex
unfolding, Biopolymers, 99, 1006-1018.
149.Boncina, M., Lah, J., Prislan, I., and
Vesnaver, G. (2012) Energetic basis of human telomeric DNA folding into
G-quadruplex structures, J. Am. Chem. Soc., 134,
9657-9663.
150.Gray, R. D., Trent, J. O., and Chaires, J. B.
(2014) Folding and unfolding pathways of the human telomeric
G-quadruplex, J. Mol. Biol., 426, 1629-1650.
151.David Wilson, W., and Paul, A. (2014) Kinetics
and structures on the molecular path to the quadruplex form of the
human telomere, J. Mol. Biol., 426, 1625-1628.
152.Li, W., Hou, X. M., Wang, P. Y., Xi, X. G., and
Li, M. (2013) Direct measurement of sequential folding pathway and
energy landscape of human telomeric G-quadruplex structures, J. Am.
Chem. Soc., 135, 6423-6426.
153.Koirala, D., Ghimire, C., Bohrer, C., Sannohe,
Y., Sugiyama, H., and Mao, H. (2013) Long-loop G-quadruplexes are
misfolded population minorities with fast transition kinetics in human
telomeric sequences, J. Am. Chem. Soc., 135,
2235-2241.
154.Stadlbauer, P., Trantirek, L., Cheatham, T. E.,
3rd, Koca, J., and Sponer, J. (2014) Triplex intermediates in folding
of human telomeric quadruplexes probed by microsecond-scale molecular
dynamics simulations, Biochimie, 105, 22-35.
155.Gray, R. D., and Chaires, J. B. (2008) Kinetics
and mechanism of K+- and Na+-induced folding of
models of human telomeric DNA into G-quadruplex structures, Nucleic
Acids Res., 36, 4191-4203.
156.Zhang, A. Y., and Balasubramanian, S. (2012)
The kinetics and folding pathways of intramolecular G-quadruplex
nucleic acids, J. Am. Chem. Soc., 134, 19297-19308.
157.You, H., Wu, J., Shao, F., and Yan, J. (2015)
Stability and kinetics of c-MYC promoter G-quadruplexes studied by
single-molecule manipulation, J. Am. Chem. Soc., 137,
2424-2427.
158.Stadlbauer, P., Kuhrova, P., Banas, P., Koca,
J., Bussi, G., Trantirek, L., Otyepka, M., and Sponer, J. (2015)
Hairpins participating in folding of human telomeric sequence
quadruplexes studied by standard and T-REMD simulations, Nucleic
Acids Res., 43, 9626-9644.
159.Bessi, I., Jonker, H. R., Richter, C., and
Schwalbe, H. (2015) Involvement of long-lived intermediate states in
the complex folding pathway of the human telomeric G-quadruplex,
Angew. Chem. Int. Ed. Engl., 54, 8444-8448.
160.Prislan, I., Lah, J., Milanic, M., and
Vesnaver, G. (2011) Kinetically governed polymorphism of
d(G4T4G3) quadruplexes in
K+ solutions, Nucleic Acids Res., 39,
1933-1942.
161.Yu, Z., Gaerig, V., Cui, Y., Kang, H., Gokhale,
V., Zhao, Y., Hurley, L. H., and Mao, H. (2012) Tertiary DNA structure
in the single-stranded hTERT promoter fragment unfolds and refolds by
parallel pathways via cooperative or sequential events, J. Am. Chem.
Soc., 134, 5157-5164.
162.Abraham Punnoose, J., Cui, Y., Koirala, D.,
Yangyuoru, P. M., Ghimire, C., Shrestha, P., and Mao, H. (2014)
Interaction of G-quadruplexes in the full-length 3′ human
telomeric overhang, J. Am. Chem. Soc., 136,
18062-18069.
163.Dhakal, S., Yu, Z., Konik, R., Cui, Y.,
Koirala, D., and Mao, H. (2012) G-quadruplex and i-motif are mutually
exclusive in ILPR double-stranded DNA, Biophys. J., 102,
2575-2584.
164.Dhakal, S., Cui, Y., Koirala, D., Ghimire, C.,
Kushwaha, S., Yu, Z., Yangyuoru, P. M., and Mao, H. (2013) Structural
and mechanical properties of individual human telomeric G-quadruplexes
in molecularly crowded solutions, Nucleic Acids Res., 41,
3915-3923.
165.Bergues-Pupo, A. E., Arias-Gonzalez, J. R.,
Moron, M. C., Fiasconaro, A., and Falo, F. (2015) Role of the central
cations in the mechanical unfolding of DNA and RNA G-quadruplexes,
Nucleic Acids Res., 43, 7638-7647.
166.De Messieres, M., Chang, J. C., Brawn-Cinani,
B., and La Porta, A. (2012) Single-molecule study of G-quadruplex
disruption using dynamic force spectroscopy, Phys. Rev. Lett.,
109, 058101.
167.Ghimire, C., Park, S., Iida, K., Yangyuoru, P.,
Otomo, H., Yu, Z., Nagasawa, K., Sugiyama, H., and Mao, H. (2014)
Direct quantification of loop interaction and pi-pi stacking for
G-quadruplex stability at the submolecular level, J. Am. Chem.
Soc., 136, 15537-15544.
168.Long, X., and Stone, M. D. (2013) Kinetic
partitioning modulates human telomere DNA G-quadruplex structural
polymorphism, PLoS One, 8, e83420.
169.Arora, A., Nair, D. R., and Maiti, S. (2009)
Effect of flanking bases on quadruplex stability and Watson–Crick
duplex competition, FEBS J., 276, 3628-3640.
170.Mendoza, O., Elezgaray, J., and Mergny, J. L.
(2015) Kinetics of quadruplex to duplex conversion, Biochimie,
118, 225-233.
171.Kuo, M. H., Wang, Z. F., Tseng, T. Y., Li, M.
H., Hsu, S. T., Lin, J. J., and Chang, T. C. (2015) Conformational
transition of a hairpin structure to G-quadruplex within the
WNT1 gene promoter, J. Am. Chem. Soc., 137,
210-218.
172.Gupta, A., Lee, L. L., Roy, S., Tanious, F. A.,
Wilson, W. D., Ly, D. H., and Armitage, B. A. (2013) Strand invasion of
DNA quadruplexes by PNA: comparison of homologous and complementary
hybridization, ChemBioChem, 14, 1476-1484.
173.Lv, B., Li, D., Zhang, H., Lee, J. Y., and Li,
T. (2013) DNA gyrase-driven generation of a G-quadruplex from plasmid
DNA, Chem. Commun. (Camb.), 49, 8317-8319.
174.Zhabinskaya, D., and Benham, C. J. (2012)
Theoretical analysis of competing conformational transitions in
superhelical DNA, PLoS Comput. Biol., 8, e1002484.
175.Selvam, S., Koirala, D., Yu, Z., and Mao, H.
(2014) Quantification of topological coupling between DNA superhelicity
and G-quadruplex formation, J. Am. Chem. Soc., 136,
13967-13970.
176.Brazier, J. A., Shah, A., and Brown, G. D.
(2012) I-motif formation in gene promoters: unusually stable formation
in sequences complementary to known G-quadruplexes, Chem. Commun.
(Camb.), 48, 10739-10741.
177.Kendrick, S., Kang, H. J., Alam, M. P.,
Madathil, M. M., Agrawal, P., Gokhale, V., Yang, D., Hecht, S. M., and
Hurley, L. H. (2014) The dynamic character of the BCL2 promoter i-motif
provides a mechanism for modulation of gene expression by compounds
that bind selectively to the alternative DNA hairpin structure, J.
Am. Chem. Soc., 136, 4161-4171.
178.Bhavsar-Jog, Y. P., Van Dornshuld, E., Brooks,
T. A., Tschumper, G. S., and Wadkins, R. M. (2014) Epigenetic
modification, dehydration, and molecular crowding effects on the
thermodynamics of i-motif structure formation from C-rich DNA,
Biochemistry, 53, 1586-1594.
179.Reilly, S. M., Morgan, R. K., Brooks, T. A.,
and Wadkins, R. M. (2015) Effect of interior loop length on the thermal
stability and pKa of i-motif DNA,
Biochemistry, 54, 1364-1370.
180.Konig, S. L., Huppert, J. L., Sigel, R. K., and
Evans, A. C. (2013) Distance-dependent duplex DNA destabilization
proximal to G-quadruplex/i-motif sequences, Nucleic Acids Res.,
41, 7453-7461.
181.Rajendran, A., Nakano, S., and Sugimoto, N.
(2010) Molecular crowding of the cosolutes induces an intramolecular
i-motif structure of triplet repeat DNA oligomers at neutral pH,
Chem. Commun. (Camb.), 46, 1299-1301.
182.Cui, J., Waltman, P., Le, V. H., and Lewis, E.
A. (2013) The effect of molecular crowding on the stability of human
c-MYC promoter sequence i-motif at neutral pH, Molecules,
18, 12751-12767.
183.Cui, Y., Kong, D., Ghimire, C., Xu, C., and
Mao, H. (2016) Mutually exclusive formation of G-quadruplex and i-motif
is a general phenomenon governed by steric hindrance in duplex DNA,
Biochemistry, 55, 2291-2299.
184.Kang, H. J., Kendrick, S., Hecht, S. M., and
Hurley, L. H. (2014) The transcriptional complex between the BCL2
i-motif and hnRNP LL is a molecular switch for control of gene
expression that can be modulated by small molecules, J. Am. Chem.
Soc., 136, 4172-4185.
185.Lim, K. W., and Phan, A. T. (2013) Structural
basis of DNA quadruplex-duplex junction formation, Angew. Chem. Int.
Ed. Engl., 52, 8566-8569.
186.Lim, K. W., Khong, Z. J., and Phan, A. T.
(2014) Thermal stability of DNA quadruplex-duplex hybrids,
Biochemistry, 53, 247-257.
187.Lim, K. W., Nguyen, T. Q., and Phan, A. T.
(2014) Joining of multiple duplex stems at a single quadruplex loop,
J. Am. Chem. Soc., 136, 17969-17973.
188.Lim, K. W., Jenjaroenpun, P., Low, Z. J.,
Khong, Z. J., Ng, Y. S., Kuznetsov, V. A., and Phan, A. T. (2015)
Duplex stem-loop-containing quadruplex motifs in the human genome: a
combined genomic and structural study, Nucleic Acids Res.,
43, 5630-5646.
189.Spiridonova, V. A., Barinova, K. V., Glinkina,
K. A., Melnichuk, A. V., Gainutdynov, A. A., Safenkova, I. V., and
Dzantiev, B. B. (2015) A family of DNA aptamers with varied duplex
region length that forms complexes with thrombin and prothrombin,
FEBS Lett., 589, 2043-2049.
190.Dolinnaya, N. G., Yuminova, A. V., Spiridonova,
V. A., Arutyunyan, A. M., and Kopylov, A. M. (2012) Coexistence of
G-quadruplex and duplex domains within the secondary structure of
31-mer DNA thrombin-binding aptamer, J. Biomol. Struct. Dyn.,
30, 524-531.
191.Yu, H., Gu, X., Nakano, S., Miyoshi, D., and
Sugimoto, N. (2012) Beads-on-a-string structure of long telomeric DNAs
under molecular crowding conditions, J. Am. Chem. Soc.,
134, 20060-20069.
192.Martadinata, H., Heddi, B., Lim, K. W., and
Phan, A. T. (2011) Structure of long human telomeric RNA (TERRA):
G-quadruplexes formed by four and eight UUAGGG repeats are stable
building blocks, Biochemistry, 50, 6455-6461.
193.Bauer, L., Tluckova, K., Tohova, P., and
Viglasky, V. (2011) G-quadruplex motifs arranged in tandem occurring in
telomeric repeats and the insulin-linked polymorphic region,
Biochemistry, 50, 7484-7492.
194.Hansel, R., Lohr, F., Trantirek, L., and
Dotsch, V. (2013) High-resolution insight into G-overhang architecture,
J. Am. Chem. Soc., 135, 2816-2824.
195.Selvam, S., Yu, Z., and Mao, H. (2016) Exploded
view of higher order G-quadruplex structures through click-chemistry
assisted single-molecule mechanical unfolding, Nucleic Acids
Res., 44, 45-55.
196.Kankia, B., Gvarjaladze, D., Rabe, A., Lomidze,
L., Metreveli, N., and Musier-Forsyth, K. (2016) Stable domain assembly
of a monomolecular DNA quadruplex: implications for DNA-based
nanoswitches, Biophys. J., 110, 2169-2175.
197.Wang, H., Nora, G. J., Ghodke, H., and Opresko,
P. L. (2011) Single molecule studies of physiologically relevant
telomeric tails reveal POT1 mechanism for promoting G-quadruplex
unfolding, J. Biol. Chem., 286, 7479-7489.
198.Rajendran, A., Endo, M., Hidaka, K., and
Sugiyama, H. (2014) Direct and single-molecule visualization of the
solution-state structures of G-hairpin and G-triplex intermediates,
Angew. Chem. Int. Ed. Engl., 53, 4107-4112.
199.Payet, L., and Huppert, J. L. (2012) Stability
and structure of long intramolecular G-quadruplexes,
Biochemistry, 51, 3154-3161.
200.Chen, Y., Agrawal, P., Brown, R. V., Hatzakis,
E., Hurley, L., and Yang, D. (2012) The major G-quadruplex formed in
the human platelet-derived growth factor receptor beta promoter adopts
a novel broken-strand structure in K+ solution, J. Am.
Chem. Soc., 134, 13220-13223.
201.Bugaut, A., and Alberti, P. (2015)
Understanding the stability of DNA G-quadruplex units in long human
telomeric strands, Biochimie, 113, 125-133.
202.Vy Thi Le, T., Han, S., Chae, J., and Park, H.
J. (2012) G-quadruplex binding ligands: from naturally occurring to
rationally designed molecules, Curr. Pharm. Des., 18,
1948-1972.
203.Rodriguez, R., Miller, K. M., Forment, J. V.,
Bradshaw, C. R., Nikan, M., Britton, S., Oelschlaegel, T., Xhemalce,
B., Balasubramanian, S., and Jackson, S. P. (2012)
Small-molecule-induced DNA damage identifies alternative DNA structures
in human genes, Nat. Chem. Biol., 8, 301-310.
204.Rizzo, A., Salvati, E., and Biroccio, A. (2012)
Methods of studying telomere damage induced by quadruplex–ligand
complexes, Methods, 57, 93-99.
205.Muller, S., Kumari, S., Rodriguez, R., and
Balasubramanian, S. (2010) Small-molecule-mediated G-quadruplex
isolation from human cells, Nat. Chem., 2, 1095-1098.
206.Kwok, C. K., and Balasubramanian, S. (2015)
Targeted detection of G-quadruplexes in cellular RNAs, Angew. Chem.
Int. Ed. Engl., 54, 6751-6754.
207.Largy, E., Granzhan, A., Hamon, F., Verga, D.,
and Teulade-Fichou, M. P. (2013) Visualizing the quadruplex: from
fluorescent ligands to light-up probes, Top. Curr. Chem.,
330, 111-177.
208.Monchaud, D., and Teulade-Fichou, M. P. (2008)
A hitchhiker’s guide to G-quadruplex ligands, Org. Biomol.
Chem., 6, 627-636.
209.Di Leva, F. S., Novellino, E., Cavalli, A.,
Parrinello, M., and Limongelli, V. (2014) Mechanistic insight into
ligand binding to G-quadruplex DNA, Nucleic Acids Res.,
42, 5447-5455.
210.Ou, T. M., Lu, Y. J., Tan, J. H., Huang, Z. S.,
Wong, K. Y., and Gu, L. Q. (2008) G-quadruplexes: targets in anticancer
drug design, ChemMedChem, 3, 690-713.
211.Il’inskii, N. S., Varizhuk, A. M.,
Beniaminov, A. D., Puzanov, M. A., Shchelkina, A. K., and Kaliuzhnyi,
D. N. (2014) G-quadruplex ligands: mechanisms of anticancer action and
target binding, Mol. Biol. (Moscow), 48, 891-907.
212.Douarre, C., Mergui, X., Sidibe, A., Gomez, D.,
Alberti, P., Mailliet, P., Trentesaux, C., and Riou, J. F. (2013) DNA
damage signaling induced by the G-quadruplex ligand 12459 is modulated
by PPM1D/WIP1 phosphatase, Nucleic Acids Res., 41,
3588-3599.
213.Piazza, A., Boule, J. B., Lopes, J., Mingo, K.,
Largy, E., Teulade-Fichou, M. P., and Nicolas, A. (2010) Genetic
instability triggered by G-quadruplex interacting Phen-DC compounds in
Saccharomyces cerevisiae, Nucleic Acids Res., 38,
4337-4348.
214.Chung, W. J., Heddi, B., Tera, M., Iida, K.,
Nagasawa, K., and Phan, A. T. (2013) Solution structure of an
intramolecular (3 + 1) human telomeric G-quadruplex bound to a
telomestatin derivative, J. Am. Chem. Soc., 135,
13495-13501.
215.Bazzicalupi, C., Ferraroni, M., Bilia, A. R.,
Scheggi, F., and Gratteri, P. (2013) The crystal structure of human
telomeric DNA complexed with berberine: an interesting case of stacked
ligand to G-tetrad ratio higher than 1 : 1, Nucleic Acids Res.,
41, 632-638.
216.Clark, G. R., Pytel, P. D., and Squire, C. J.
(2012) The high-resolution crystal structure of a parallel
intermolecular DNA G-4 quadruplex/drug complex employing syn
glycosyl linkages, Nucleic Acids Res., 40, 5731-5738.
217.Lavrado, J., Borralho, P. M., Ohnmacht, S. A.,
Castro, R. E., Rodrigues, C. M., Moreira, R., dos Santos, D. J.,
Neidle, S., and Paulo, A. (2013) Synthesis, G-quadruplex stabilization,
docking studies, and effect on cancer cells of indolo[3,2-b]quinolines
with one, two, or three basic side chains, ChemMedChem,
8, 1648-1661.
218.Koirala, D., Dhakal, S., Ashbridge, B.,
Sannohe, Y., Rodriguez, R., Sugiyama, H., Balasubramanian, S., and Mao,
H. (2011) A single-molecule platform for investigation of interactions
between G-quadruplexes and small-molecule ligands, Nat. Chem.,
3, 782-787.
219.Jain, A. K., and Bhattacharya, S. (2011)
Interaction of G-quadruplexes with nonintercalating duplex-DNA minor
groove binding ligands, Bioconjug. Chem., 22,
2355-2368.
220.Hamon, F., Largy, E., Guedin-Beaurepaire, A.,
Rouchon-Dagois, M., Sidibe, A., Monchaud, D., Mergny, J. L., Riou, J.
F., Nguyen, C. H., and Teulade-Fichou, M. P. (2011) An acyclic
oligoheteroaryle that discriminates strongly between diverse
G-quadruplex topologies, Angew. Chem. Int. Ed. Engl., 50,
8745-8749.
221.Sabharwal, N. C., Savikhin, V., Turek-Herman,
J. R., Nicoludis, J. M., Szalai, V. A., and Yatsunyk, L. A. (2014)
N-methylmesoporphyrin IX fluorescence as a reporter of strand
orientation in guanine quadruplexes, FEBS J., 281,
1726-1737.
222.Dhamodharan, V., Harikrishna, S., Bhasikuttan,
A. C., and Pradeepkumar, P. I. (2015) Topology specific stabilization
of promoter over telomeric G-quadruplex DNAs by bisbenzimidazole
carboxamide derivatives, ACS Chem. Biol., 10,
821-833.
223.Wang, J., Chen, Y., Ren, J., Zhao, C., and Qu,
X. (2014) G-Quadruplex binding enantiomers show chiral selective
interactions with human telomere, Nucleic Acids Res., 42,
3792-3802.
224.Yu, Q., Liu, Y., Wang, C., Sun, D., Yang, X.,
Liu, Y., and Liu, J. (2012) Chiral ruthenium(II) polypyridyl complexes:
stabilization of G-quadruplex DNA, inhibition of telomerase
activity and cellular uptake, PLoS One, 7, e50902.
225.Zhao, A., Zhao, C., Ren, J., and Qu, X. (2015)
Enantioselective targeting left-handed Z-G-quadruplex, Chem. Commun.
(Camb.), 52, 1365-1368.
226.Boncina, M., Podlipnik, C., Piantanida, I.,
Eilmes, J., Teulade-Fichou, M. P., Vesnaver, G., and Lah, J. (2015)
Thermodynamic fingerprints of ligand binding to human telomeric
G-quadruplexes, Nucleic Acids Res., 43, 10376-10386.
227.Waller, Z. A., Howell, L. A., Macdonald, C. J.,
O’Connell, M. A., and Searcey, M. (2014) Identification and
characterisation of a G-quadruplex forming sequence in the promoter
region of nuclear factor (erythroid-derived 2)-like 2 (Nrf2),
Biochem. Biophys. Res. Commun., 447, 128-132.
228.Wang, Z. F., Li, M. H., Chen, W. W., Hsu, S.
T., and Chang, T. C. (2016) A novel transition pathway of
ligand-induced topological conversion from hybrid forms to parallel
forms of human telomeric G-quadruplexes, Nucleic Acids Res.,
44, 3958-3968.
229.McLuckie, K. I., Waller, Z. A., Sanders, D. A.,
Alves, D., Rodriguez, R., Dash, J., McKenzie, G. J., Venkitaraman, A.
R., and Balasubramanian, S. (2011) G-quadruplex-binding
benzo[a]phenoxazines down-regulate c-KIT expression in human gastric
carcinoma cells, J. Am. Chem. Soc., 133, 2658-2663.
230.Alzeer, J., and Luedtke, N. W. (2010)
pH-mediated fluorescence and G-quadruplex binding of amido
phthalocyanines, Biochemistry, 49, 4339-4348.
231.Lin, S., He, B., Yang, C., Leung, C. H.,
Mergny, J. L., and Ma, D. L. (2015) Luminescence switch-on assay of
interferon-gamma using a G-quadruplex-selective iridium(III) complex,
Chem. Commun. (Camb.), 51, 16033-16036.
232.Guo, Y., Zhou, L., Xu, L., Zhou, X., Hu, J.,
and Pei, R. (2014) Multiple types of logic gates based on a single
G-quadruplex DNA strand, Sci. Rep., 4, 7315.
233.Gabelica, V., Maeda, R., Fujimoto, T., Yaku,
H., Murashima, T., Sugimoto, N., and Miyoshi, D. (2013) Multiple and
cooperative binding of fluorescence light-up probe thioflavin T with
human telomere DNA G-quadruplex, Biochemistry, 52,
5620-5628.
234.Doria, F., Nadai, M., Folini, M., Di Antonio,
M., Germani, L., Percivalle, C., Sissi, C., Zaffaroni, N., Alcaro, S.,
Artese, A., Richter, S. N., and Freccero, M. (2012) Hybrid
ligand-alkylating agents targeting telomeric G-quadruplex structures,
Org. Biomol. Chem., 10, 2798-2806.
235.Di Antonio, M., Biffi, G., Mariani, A., Raiber,
E. A., Rodriguez, R., and Balasubramanian, S. (2012) Selective RNA
versus DNA G-quadruplex targeting by in situ click chemistry,
Angew. Chem. Int. Ed. Engl., 51, 11073-11078.
236.Morris, M. J., Wingate, K. L., Silwal, J.,
Leeper, T. C., and Basu, S. (2012) The porphyrin TmPyP4 unfolds the
extremely stable G-quadruplex in MT3-MMP mRNA and alleviates its
repressive effect to enhance translation in eukaryotic cells,
Nucleic Acids Res., 40, 4137-4145.
237.Faudale, M., Cogoi, S., and Xodo, L. E. (2012)
Photoactivated cationic alkyl-substituted porphyrin binding to g4-RNA
in the 5′-UTR of KRAS oncogene represses translation, Chem.
Commun. (Camb.), 48, 874-876.
238.Alcaro, S., Costa, G., Distinto, S., Moraca,
F., Ortuso, F., Parrotta, L., and Artese, A. (2012) The polymorphisms
of DNA G-quadruplex investigated by docking experiments with
telomestatin enantiomers, Curr. Pharm. Des., 18,
1873-1879.
239.Li, Q., Xiang, J. F., Yang, Q. F., Sun, H. X.,
Guan, A. J., and Tang, Y. L. (2013) G4LDB: a database for discovering
and studying G-quadruplex ligands, Nucleic Acids Res.,
41, D1115-1123.
240.Le, D. D., Di Antonio, M., Chan, L. K., and
Balasubramanian, S. (2015) G-quadruplex ligands exhibit differential
G-tetrad selectivity, Chem. Commun. (Camb.), 51,
8048-8050.
241.Kaluzhny, D. N., Mamaeva, O. K., Beniaminov, A.
D., Shchyolkina, A. K., and Livshits, M. A. (2016) The thermodynamics
of binding of low-molecular-weight ligands at extreme tetrads of
telomeric G-quadruplexes, Biophysics, 61, 28-33.
242.Gai, W., Yang, Q., Xiang, J., Jiang, W., Li,
Q., Sun, H., Guan, A., Shang, Q., Zhang, H., and Tang, Y. (2013) A
dual-site simultaneous binding mode in the interaction between
parallel-stranded G-quadruplex [d(TGGGGT)]4 and cyanine dye
2,2′-diethyl-9-methyl-selenacarbocyanine bromide, Nucleic
Acids Res., 41, 2709-2722.
243.Li, Q., Xiang, J., Li, X., Chen, L., Xu, X.,
Tang, Y., Zhou, Q., Li, L., Zhang, H., Sun, H., Guan, A., Yang, Q.,
Yang, S., and Xu, G. (2009) Stabilizing parallel G-quadruplex DNA by a
new class of ligands: two non-planar alkaloids through interaction in
lateral grooves, Biochimie, 91, 811-819.
244.Giancola, C., and Pagano, B. (2013) Energetics
of ligand binding to G-quadruplexes, Top. Curr. Chem.,
330, 211-242.
245.Kaluzhny, D., Ilyinsky, N., Shchekotikhin, A.,
Sinkevich, Y., Tsvetkov, P. O., Tsvetkov, V., Veselovsky, A., Livshits,
M., Borisova, O., Shtil, A., and Shchyolkina, A. (2011) Disordering of
human telomeric G-quadruplex with novel antiproliferative
anthrathiophenedione, PLoS One, 6, e27151.
246.McLuckie, K. I., Di Antonio, M., Zecchini, H.,
Xian, J., Caldas, C., Krippendorff, B. F., Tannahill, D., Lowe, C., and
Balasubramanian, S. (2013) G-quadruplex DNA as a molecular target for
induced synthetic lethality in cancer cells, J. Am. Chem. Soc.,
35, 9640-9643.
247.Campbell, N. H., Patel, M., Tofa, A. B., Ghosh,
R., Parkinson, G. N., and Neidle, S. (2009) Selectivity in ligand
recognition of G-quadruplex loops, Biochemistry, 48,
1675-1680.
248.Luo, D., and Mu, Y. (2015) All-atomic
simulations on human telomeric G-quadruplex DNA binding with thioflavin
T, J. Phys. Chem. B, 119, 4955-4967.
249.Cummaro, A., Fotticchia, I., Franceschin, M.,
Giancola, C., and Petraccone, L. (2011) Binding properties of human
telomeric quadruplex multimers: a new route for drug design,
Biochimie, 93, 1392-1400.
250.Petraccone, L. (2013) Higher-order quadruplex
structures, Top. Curr. Chem., 330, 23-46.
251.Zhu, L. N., Wu, B., and Kong, D. M. (2013)
Specific recognition and stabilization of monomeric and multimeric
G-quadruplexes by cationic porphyrin TMPipEOPP under molecular crowding
conditions, Nucleic Acids Res., 41, 4324-4335.
252.Zhao, C., Wu, L., Ren, J., Xu, Y., and Qu, X.
(2013) Targeting human telomeric higher-order DNA: dimeric G-quadruplex
units serve as preferred binding site, J. Am. Chem. Soc.,
135, 18786-18789.
253.Georgiades, S. N., Abd Karim, N. H.,
Suntharalingam, K., and Vilar, R. (2010) Interaction of metal complexes
with G-quadruplex DNA, Angew. Chem. Int. Ed. Engl., 49,
4020-4034.
254.Ralph, S. F. (2011) Quadruplex DNA: a promising
drug target for the medicinal inorganic chemist, Curr. Top. Med.
Chem., 11, 572-590.
255.Zheng, K. W., Zhang, D., Zhang, L. X., Hao, Y.
H., Zhou, X., and Tan, Z. (2011) Dissecting the strand folding
orientation and formation of G-quadruplexes in single- and
double-stranded nucleic acids by ligand-induced photocleavage
footprinting, J. Am. Chem. Soc., 133, 1475-1483.
256.Di Antonio, M., Rodriguez, R., and
Balasubramanian, S. (2012) Experimental approaches to identify cellular
G-quadruplex structures and functions, Methods, 57,
84-92.
257.Di Antonio, M., Doria, F., Richter, S. N.,
Bertipaglia, C., Mella, M., Sissi, C., Palumbo, M., and Freccero, M.
(2009) Quinone methides tethered to naphthalene diimides as selective
G-quadruplex alkylating agents, J. Am. Chem. Soc., 131,
13132-13141.
258.Hampel, S. M., Sidibe, A., Gunaratnam, M.,
Riou, J. F., and Neidle, S. (2010) Tetrasubstituted naphthalene diimide
ligands with selectivity for telomeric G-quadruplexes and cancer cells,
Bioorg. Med. Chem. Lett., 20, 6459-6463.
259.Tseng, T. Y., Wang, Z. F., Chien, C. H., and
Chang, T. C. (2013) In-cell optical imaging of exogenous G-quadruplex
DNA by fluorogenic ligands, Nucleic Acids Res., 41,
10605-10618.
260.Haudecoeur, R., Stefan, L., Denat, F., and
Monchaud, D. (2013) A model of smart G-quadruplex ligand, J. Am.
Chem. Soc., 135, 550-553.
261.Bare, G. A., Liu, B., and Sherman, J. C. (2013)
Synthesis of a single G-quartet platform in water, J. Am. Chem.
Soc., 135, 11985-11989.
262.Laguerre, A., Stefan, L., Larrouy, M., Genest,
D., Novotna, J., Pirrotta, M., and Monchaud, D. (2014) A twice-as-smart
synthetic G-quartet: PyroTASQ is both a smart quadruplex ligand and a
smart fluorescent probe, J. Am. Chem. Soc., 136,
12406-12414.
263.Laguerre, A., Hukezalie, K., Winckler, P.,
Katranji, F., Chanteloup, G., Pirrotta, M., Perrier-Cornet, J. M.,
Wong, J. M., and Monchaud, D. (2015) Visualization of RNA-quadruplexes
in live cells, J. Am. Chem. Soc., 137, 8521-8525.
264.Sissi, C., Gatto, B., and Palumbo, M. (2011)
The evolving world of protein-G-quadruplex recognition: a medicinal
chemist’s perspective, Biochimie, 93,
1219-1230.
265.De Cian, A., Lacroix, L., Douarre, C.,
Temime-Smaali, N., Trentesaux, C., Riou, J. F., and Mergny, J. L.
(2008) Targeting telomeres and telomerase, Biochimie, 90,
131-155.
266.Bochman, M. L., Sabouri, N., and Zakian, V. A.
(2010) Unwinding the functions of the Pif1 family helicases, DNA
Repair (Amst.), 9, 237-249.
267.Biffi, G., Tannahill, D., and Balasubramanian,
S. (2012) An intramolecular G-quadruplex structure is required for
binding of telomeric repeat-containing RNA to the telomeric protein
TRF2, J. Am. Chem. Soc., 134, 11974-11976.
268.Chakraborty, P., and Grosse, F. (2011) Human
DHX9 helicase preferentially unwinds RNA-containing displacement loops
(R-loops) and G-quadruplexes, DNA Repair (Amst.), 10,
654-665.
269.Gonzalez, V., Guo, K., Hurley, L., and Sun, D.
(2009) Identification and characterization of nucleolin as a c-myc
G-quadruplex-binding protein, J. Biol. Chem., 284,
23622-23635.
270.Fan, G., Lin, Y. X., Yang, L., Gao, F. P.,
Zhao, Y. X., Qiao, Z. Y., Zhao, Q., Fan, Y. S., Chen, Z., and Wang, H.
(2015) Co-self-assembled nanoaggregates of BODIPY amphiphiles for dual
colour imaging of live cells, Chem. Commun. (Camb.), 51,
12447-12450.
271.Von Hacht, A., Seifert, O., Menger, M.,
Schutze, T., Arora, A., Konthur, Z., Neubauer, P., Wagner, A., Weise,
C., and Kurreck, J. (2014) Identification and characterization of RNA
guanine-quadruplex binding proteins, Nucleic Acids Res.,
42, 6630-6644.
272.Chiarella, S., De Cola, A., Scaglione, G. L.,
Carletti, E., Graziano, V., Barcaroli, D., Lo Sterzo, C., Di Matteo,
A., Di Ilio, C., Falini, B., Arcovito, A., De Laurenzi, V., and
Federici, L. (2013) Nucleophosmin mutations alter its nucleolar
localization by impairing G-quadruplex binding at ribosomal DNA,
Nucleic Acids Res., 41, 3228-3239.
273.Cogoi, S., Shchekotikhin, A. E., and Xodo, L.
E. (2014) HRAS is silenced by two neighboring G-quadruplexes and
activated by MAZ, a zinc-finger transcription factor with DNA unfolding
property, Nucleic Acids Res., 42, 8379-8388.
274.Raiber, E. A., Kranaster, R., Lam, E., Nikan,
M., and Balasubramanian, S. (2012) A non-canonical DNA structure is a
binding motif for the transcription factor SP1 in vitro,
Nucleic Acids Res., 40, 1499-1508.
275.Wolfe, A. L., Singh, K., Zhong, Y., Drewe, P.,
Rajasekhar, V. K., Sanghvi, V. R., Mavrakis, K. J., Jiang, M.,
Roderick, J. E., Van der Meulen, J., Schatz, J. H., Rodrigo, C. M.,
Zhao, C., Rondou, P., De Stanchina, E., Teruya-Feldstein, J., Kelliher,
M. A., Speleman, F., Porco, J. A., Jr., Pelletier, J., Ratsch, G., and
Wendel, H. G. (2014) RNA G-quadruplexes cause eIF4A-dependent oncogene
translation in cancer, Nature, 513, 65-70.
276.Tosoni, E., Frasson, I., Scalabrin, M.,
Perrone, R., Butovskaya, E., Nadai, M., Palu, G., Fabris, D., and
Richter, S. N. (2015) Nucleolin stabilizes G-quadruplex structures
folded by the LTR promoter and silences HIV-1 viral transcription,
Nucleic Acids Res., 43, 8884-8897.
277.Brosh, R. M., Jr. (2013) DNA helicases involved
in DNA repair and their roles in cancer, Nat. Rev. Cancer,
13, 542-558.
278.Wickramasinghe, C. M., Arzouk, H., Frey, A.,
Maiter, A., and Sale, J. E. (2015) Contributions of the specialised DNA
polymerases to replication of structured DNA, DNA Repair
(Amst.), 29, 83-90.
279.Eddy, S., Maddukuri, L., Ketkar, A., Zafar, M.
K., Henninger, E. E., Pursell, Z. F., and Eoff, R. L. (2015) Evidence
for the kinetic partitioning of polymerase activity on G-quadruplex
DNA, Biochemistry, 54, 3218-3230.
280.Li, Q. J., Tong, X. J., Duan, Y. M., and Zhou,
J. Q. (2013) Characterization of the intramolecular G-quadruplex
promoting activity of Est1, FEBS Lett., 587, 659-665.
281.Paeschke, K., Juranek, S., Rhodes, D., and
Lipps, H. J. (2008) Cell cycle-dependent regulation of telomere
tethering in the nucleus, Chromosome Res., 16,
721-728.
282.Paeschke, K., Bochman, M. L., Garcia, P. D.,
Cejka, P., Friedman, K. L., Kowalczykowski, S. C., and Zakian, V. A.
(2013) Pif1 family helicases suppress genome instability at
G-quadruplex motifs, Nature, 497, 458-462.
283.Gray, L. T., Vallur, A. C., Eddy, J., and
Maizels, N. (2014) G quadruplexes are genomewide targets of
transcriptional helicases XPB and XPD, Nat. Chem. Biol.,
10, 313-318.
284.Kang, H. J., Le, T. V., Kim, K., Hur, J., Kim,
K. K., and Park, H. J. (2014) Novel interaction of the Z-DNA binding
domain of human ADAR1 with the oncogenic c-Myc promoter G-quadruplex,
J. Mol. Biol., 426, 2594-2604.
285.Phan, A. T., Kuryavyi, V., Darnell, J. C.,
Serganov, A., Majumdar, A., Ilin, S., Raslin, T., Polonskaia, A., Chen,
C., Clain, D., Darnell, R. B., and Patel, D. J. (2011)
Structure-function studies of FMRP RGG peptide recognition of an RNA
duplex-quadruplex junction, Nat. Struct. Mol. Biol., 18,
796-804.
286.Gallo, A., Lo Sterzo, C., Mori, M., Di Matteo,
A., Bertini, I., Banci, L., Brunori, M., and Federici, L. (2012)
Structure of nucleophosmin DNA-binding domain and analysis of its
complex with a G-quadruplex sequence from the c-MYC promoter, J.
Biol. Chem., 287, 26539-26548.
287.Hayashi, T., Oshima, H., Mashima, T., Nagata,
T., Katahira, M., and Kinoshita, M. (2014) Binding of an RNA aptamer
and a partial peptide of a prion protein: crucial importance of water
entropy in molecular recognition, Nucleic Acids Res., 42,
6861-6875.
288.Heddi, B., Cheong, V. V., Martadinata, H., and
Phan, A. T. (2015) Insights into G-quadruplex specific recognition by
the DEAH-box helicase RHAU: solution structure of a
peptide–quadruplex complex, Proc. Natl. Acad. Sci. USA,
112, 9608-9613.
289.Vasilyev, N., Polonskaia, A., Darnell, J. C.,
Darnell, R. B., Patel, D. J., and Serganov, A. (2015) Crystal structure
reveals specific recognition of a G-quadruplex RNA by a beta-turn in
the RGG motif of FMRP, Proc. Natl. Acad. Sci. USA, 112,
E5391-5400.
290.Takahama, K., and Oyoshi, T. (2013) Specific
binding of modified RGG domain in TLS/FUS to G-quadruplex RNA:
tyrosines in RGG domain recognize 2′-OH of the riboses of loops
in G-quadruplex, J. Am. Chem. Soc., 135, 18016-18019.
291.Boyer, A. S., Grgurevic, S., Cazaux, C., and
Hoffmann, J. S. (2013) The human specialized DNA polymerases and non-B
DNA: vital relationships to preserve genome integrity, J. Mol.
Biol., 425, 4767-4781.
292.Haeusler, A. R., Donnelly, C. J., Periz, G.,
Simko, E. A., Shaw, P. G., Kim, M. S., Maragakis, N. J., Troncoso, J.
C., Pandey, A., Sattler, R., Rothstein, J. D., and Wang, J. (2014)
C9orf72 nucleotide repeat structures initiate molecular cascades of
disease, Nature, 507, 195-200.
293.Ivanov, P., O'Day, E., Emara, M. M., Wagner,
G., Lieberman, J., and Anderson, P. (2014) G-quadruplex structures
contribute to the neuroprotective effects of angiogenin-induced tRNA
fragments, Proc. Natl. Acad. Sci. USA, 111,
18201-18206.
294.Paeschke, K., Capra, J. A., and Zakian, V. A.
(2011) DNA replication through G-quadruplex motifs is promoted by the
Saccharomyces cerevisiae Pif1 DNA helicase, Cell,
145, 678-691.
295.Castillo Bosch, P., Segura-Bayona, S., Koole,
W., van Heteren, J. T., Dewar, J. M., Tijsterman, M., and Knipscheer,
P. (2014) FANCJ promotes DNA synthesis through G-quadruplex structures,
EMBO J., 33, 2521-2533.
296.Aguilera, A., and Garcia-Muse, T. (2013) Causes
of genome instability, Annu. Rev. Genet., 47, 1-32.
297.Lopes, J., Piazza, A., Bermejo, R., Kriegsman,
B., Colosio, A., Teulade-Fichou, M. P., Foiani, M., and Nicolas, A.
(2011) G-quadruplex-induced instability during leading-strand
replication, EMBO J., 30, 4033-4046.
298.Ribeyre, C., Lopes, J., Boule, J. B., Piazza,
A., Guedin, A., Zakian, V. A., Mergny, J. L., and Nicolas, A. (2009)
The yeast Pif1 helicase prevents genomic instability caused by
G-quadruplex-forming CEB1 sequences in vivo, PLoS Genet.,
5, e1000475.
299.Lormand, J. D., Buncher, N., Murphy, C. T.,
Kaur, P., Lee, M. Y., Burgers, P., Wang, H., Kunkel, T. A., and
Opresko, P. L. (2013) DNA polymerase delta stalls on telomeric lagging
strand templates independently from G-quadruplex formation, Nucleic
Acids Res., 41, 10323-10333.
300.Sarkies, P., Reams, C., Simpson, L. J., and
Sale, J. E. (2010) Epigenetic instability due to defective replication
of structured DNA, Mol. Cell, 40, 703-713.
301.Byrd, A. K., and Raney, K. D. (2015) A parallel
quadruplex DNA is bound tightly but unfolded slowly by pif1 helicase,
J. Biol. Chem., 290, 6482-6494.
302.Duan, X. L., Liu, N. N., Yang, Y. T., Li, H.
H., Li, M., Dou, S. X., and Xi, X. G. (2015) G-quadruplexes
significantly stimulate Pif1 helicase-catalyzed duplex DNA unwinding,
J. Biol. Chem., 290, 7722-7735.
303.Chen, M. C., Murat, P., Abecassis, K.,
Ferre-D’Amare, A. R., and Balasubramanian, S. (2015) Insights
into the mechanism of a G-quadruplex-unwinding DEAH-box helicase,
Nucleic Acids Res., 43, 2223-2231.
304.Qin, W., Lam, J. W., Yang, Z., Chen, S., Liang,
G., Zhao, W., Kwok, H. S., and Tang, B. Z. (2015) Red emissive AIE
luminogens with high hole-transporting properties for efficient
non-doped OLEDs, Chem. Commun. (Camb.), 51,
7321-7324.
305.Wu, W. Q., Hou, X. M., Li, M., Dou, S. X., and
Xi, X. G. (2015) BLM unfolds G-quadruplexes in different structural
environments through different mechanisms, Nucleic Acids Res.,
43, 4614-4626.
306.Qureshi, M. H., Ray, S., Sewell, A. L., Basu,
S., and Balci, H. (2012) Replication protein A unfolds G-quadruplex
structures with varying degrees of efficiency, J. Phys. Chem. B,
116, 5588-5594.
307.Ray, S., Qureshi, M. H., Malcolm, D. W.,
Budhathoki, J. B., Celik, U., and Balci, H. (2013) RPA-mediated
unfolding of systematically varying G-quadruplex structures,
Biophys. J., 104, 2235-2245.
308.Hwang, H., Kreig, A., Calvert, J., Lormand, J.,
Kwon, Y., Daley, J. M., Sung, P., Opresko, P. L., and Myong, S. (2014)
Telomeric overhang length determines structural dynamics and
accessibility to telomerase and ALT-associated proteins,
Structure, 22, 842-853.
309.Zhuang, X. Y., and Yao, Y. G. (2013)
Mitochondrial dysfunction and nuclear-mitochondrial shuttling of TERT
are involved in cell proliferation arrest induced by G-quadruplex
ligands, FEBS Lett., 587, 1656-1662.
310.Shuai, L., Deng, M., Zhang, D., Zhou, Y., and
Zhou, X. (2010) Quadruplex-duplex motifs as new topoisomerase I
inhibitors, Nucleosides Nucleotides Nucleic Acids, 29,
841-853.
311.Ogloblina, A. M., Bannikova, V. A., Khristich,
A. N., Oretskaya, T. S., Yakubovskaya, M. G., and Dolinnaya, N. G.
(2015) Parallel G-quadruplexes formed by guanine-rich microsatellite
repeats inhibit human topoisomerase I, Biochemistry (Moscow),
80, 1026-1038.
312.Gao, J., Zybailov, B. L., Byrd, A. K., Griffin,
W. C., Chib, S., Mackintosh, S. G., Tackett, A. J., and Raney, K. D.
(2015) Yeast transcription co-activator Sub1 and its human homolog PC4
preferentially bind to G-quadruplex DNA, Chem. Commun. (Camb.),
51, 7242-7244.
313.Metifiot, M., Amrane, S., Litvak, S., and
Andreola, M. L. (2014) G-quadruplexes in viruses: function and
potential therapeutic applications, Nucleic Acids Res.,
42, 12352-12366.
314.Tan, J., Vonrhein, C., Smart, O. S., Bricogne,
G., Bollati, M., Kusov, Y., Hansen, G., Mesters, J. R., Schmidt, C. L.,
and Hilgenfeld, R. (2009) The SARS-unique domain (SUD) of SARS
coronavirus contains two macrodomains that bind G-quadruplexes, PLoS
Pathog., 5, e1000428.
315.Mukundan, V. T., Do, N. Q., and Phan, A. T.
(2011) HIV-1 integrase inhibitor T30177 forms a stacked dimeric
G-quadruplex structure containing bulges, Nucleic Acids Res.,
39, 8984-8991.
316.Metifiot, M., Amrane, S., Mergny, J. L., and
Andreola, M. L. (2015) Anticancer molecule AS1411 exhibits low
nanomolar antiviral activity against HIV-1, Biochimie,
118, 173-175.
317.Fernando, H., Rodriguez, R., and
Balasubramanian, S. (2008) Selective recognition of a DNA G-quadruplex
by an engineered antibody, Biochemistry, 47,
9365-9371.
318.Biffi, G., Tannahill, D., McCafferty, J., and
Balasubramanian, S. (2013) Quantitative visualization of DNA
G-quadruplex structures in human cells, Nat. Chem., 5,
182-186.
319.Biffi, G., Di Antonio, M., Tannahill, D., and
Balasubramanian, S. (2014) Visualization and selective chemical
targeting of RNA G-quadruplex structures in the cytoplasm of human
cells, Nat. Chem., 6, 75-80.
320.Lam, E. Y., Beraldi, D., Tannahill, D., and
Balasubramanian, S. (2013) G-quadruplex structures are stable and
detectable in human genomic DNA, Nat. Commun., 4,
1796.
321.Fernando, H., Sewitz, S., Darot, J., Tavare,
S., Huppert, J. L., and Balasubramanian, S. (2009) Genome-wide analysis
of a G-quadruplex-specific single-chain antibody that regulates gene
expression, Nucleic Acids Res., 37, 6716-6722.
322.Bryan, T. M., and Baumann, P. (2011)
G-quadruplexes: from guanine gels to chemotherapeutics, Mol.
Biotechnol., 49, 198-208.
323.Yang, D., and Okamoto, K. (2010) Structural
insights into G-quadruplexes: towards new anticancer drugs, Future
Med. Chem., 2, 619-646.
324.Sekaran, V., Soares, J., and Jarstfer, M. B.
(2014) Telomere maintenance as a target for drug discovery, J. Med.
Chem., 57, 521-538.
325.Gunes, C., and Rudolph, K. L. (2013) The role
of telomeres in stem cells and cancer, Cell, 152,
390-393.
326.Smith, J. S., Chen, Q., Yatsunyk, L. A.,
Nicoludis, J. M., Garcia, M. S., Kranaster, R., Balasubramanian, S.,
Monchaud, D., Teulade-Fichou, M. P., Abramowitz, L., Schultz, D. C.,
and Johnson, F. B. (2011) Rudimentary G-quadruplex-based telomere
capping in Saccharomyces cerevisiae, Nat. Struct. Mol.
Biol., 18, 478-485.
327.Hasegawa, D., Okabe, S., Okamoto, K., Nakano,
I., Shin-Ya, K., and Seimiya, H. (2016) G-quadruplex ligand-induced DNA
damage response coupled with telomere dysfunction and replication
stress in glioma stem cells, Biochem. Biophys. Res. Commun.,
471, 75-81.
328.Chen, Z., Zheng, K. W., Hao, Y. H., and Tan, Z.
(2009) Reduced or diminished stabilization of the telomere G-quadruplex
and inhibition of telomerase by small chemical ligands under molecular
crowding condition, J. Am. Chem. Soc., 131,
10430-10438.
329.Heddi, B., and Phan, A. T. (2011) Structure of
human telomeric DNA in crowded solution, J. Am. Chem. Soc.,
133, 9824-9833.
330.Wang, Z. F., and Chang, T. C. (2012) Molecular
engineering of G-quadruplex ligands based on solvent effect of
polyethylene glycol, Nucleic Acids Res., 40,
8711-8720.
331.Buscaglia, R., Miller, M. C., Dean, W. L.,
Gray, R. D., Lane, A. N., Trent, J. O., and Chaires, J. B. (2013)
Polyethylene glycol binding alters human telomere G-quadruplex
structure by conformational selection, Nucleic Acids Res.,
41, 7934-7946.
332.Miller, M. C., Buscaglia, R., Chaires, J. B.,
Lane, A. N., and Trent, J. O. (2010) Hydration is a major determinant
of the G-quadruplex stability and conformation of the human telomere
3′-sequence of d(AG3(TTAG3)3), J. Am. Chem. Soc.,
132, 17105-17107.
333.Petraccone, L., Malafronte, A., Amato, J., and
Giancola, C. (2012) G-quadruplexes from human telomeric DNA: how many
conformations in PEG containing solutions? J. Phys. Chem. B,
116, 2294-2305.
334.Kejnovska, I., Vorlickova, M., Brazdova, M.,
and Sagi, J. (2014) Stability of human telomere quadruplexes at high
DNA concentrations, Biopolymers, 101, 428-438.
335.Lannan, F. M., Mamajanov, I., and Hud, N. V.
(2012) Human telomere sequence DNA in water-free and high-viscosity
solvents: G-quadruplex folding governed by Kramers rate theory, J.
Am. Chem. Soc., 134, 15324-15330.
336.Noer, S. L., Preus, S., Gudnason, D.,
Aznauryan, M., Mergny, J. L., and Birkedal, V. (2016) Folding dynamics
and conformational heterogeneity of human telomeric G-quadruplex
structures in Na+ solutions by single molecule FRET
microscopy, Nucleic Acids Res., 44, 464-471.
337.Petraccone, L., Garbett, N. C., Chaires, J. B.,
and Trent, J. O. (2010) An integrated molecular dynamics (MD) and
experimental study of higher order human telomeric quadruplexes,
Biopolymers, 93, 533-548.
338.Wang, Q., Liu, J. Q., Chen, Z., Zheng, K. W.,
Chen, C. Y., Hao, Y. H., and Tan, Z. (2011) G-quadruplex formation at
the 3′-end of telomere DNA inhibits its extension by telomerase,
polymerase and unwinding by helicase, Nucleic Acids Res.,
39, 6229-6237.
339.Marusic, M., Veedu, R. N., Wengel, J., and
Plavec, J. (2013) G-rich VEGF aptamer with locked and unlocked nucleic
acid modifications exhibits a unique G-quadruplex fold, Nucleic
Acids Res., 41, 9524-9536.
340.Martin-Pintado, N., Yahyaee-Anzahaee, M.,
Deleavey, G. F., Portella, G., Orozco, M., Damha, M. J., and Gonzalez,
C. (2013) Dramatic effect of furanose C2′ substitution on
structure and stability: directing the folding of the human telomeric
quadruplex with a single fluorine atom, J. Am. Chem. Soc.,
135, 5344-5347.
341.Sagi, J. (2014) G-quadruplexes incorporating
modified constituents: a review, J. Biomol. Struct. Dyn.,
32, 477-511.
342.Hellman, L. M., Spear, T. J., Koontz, C. J.,
Melikishvili, M., and Fried, M. G. (2014) Repair of O6-methylguanine
adducts in human telomeric G-quadruplex DNA by O6-alkylguanine-DNA
alkyltransferase, Nucleic Acids Res., 42, 9781-9791.
343.Virgilio, A., Esposito, V., Mayol, L.,
Giancola, C., Petraccone, L., and Galeone, A. (2015) The oxidative
damage to the human telomere: effects of
5-hydroxymethyl-2′-deoxyuridine on telomeric G-quadruplex
structures, Org. Biomol. Chem., 13, 7421-7429.
344.Konvalinova, H., Dvorakova, Z., Renciuk, D.,
Bednarova, K., Kejnovska, I., Trantirek, L., Vorlickova, M., and Sagi,
J. (2015) Diverse effects of naturally occurring base lesions on the
structure and stability of the human telomere DNA quadruplex,
Biochimie, 118, 15-25.
345.Cheong, V. V., Heddi, B., Lech, C. J., and
Phan, A. T. (2015) Xanthine and 8-oxoguanine in G-quadruplexes:
formation of a G. G. X. O. tetrad, Nucleic Acids Res.,
43, 10506-10514.
346.Babinsky, M., Fiala, R., Kejnovska, I.,
Bednarova, K., Marek, R., Sagi, J., Sklenar, V., and Vorlickova, M.
(2014) Loss of loop adenines alters human telomere
d[AG3(TTAG3)3] quadruplex folding,
Nucleic Acids Res., 42, 14031-14041.
347.Hsu, S. T., Varnai, P., Bugaut, A., Reszka, A.
P., Neidle, S., and Balasubramanian, S. (2009) A G-rich sequence within
the c-kit oncogene promoter forms a parallel G-quadruplex having
asymmetric G-tetrad dynamics, J. Am. Chem. Soc., 131,
13399-13409.
348.Islam, B., Stadlbauer, P., Krepl, M., Koca, J.,
Neidle, S., Haider, S., and Sponer, J. (2015) Extended molecular
dynamics of a c-kit promoter quadruplex, Nucleic Acids Res.,
43, 8673-8693.
349.Zidanloo, S. G., Hosseinzadeh Colagar, A.,
Ayatollahi, H., and Raoof, J. B. (2016) Downregulation of the WT1 gene
expression via TMPyP4 stabilization of promoter G-quadruplexes in
leukemia cells, Tumour Biol., 37, 9967-9977.
350.Agrawal, P., Hatzakis, E., Guo, K., Carver, M.,
and Yang, D. (2013) Solution structure of the major G-quadruplex formed
in the human VEGF promoter in K+: insights into loop
interactions of the parallel G-quadruplexes, Nucleic Acids Res.,
41, 10584-10592.
351.Salvati, E., Zizza, P., Rizzo, A., Iachettini,
S., Cingolani, C., D'Angelo, C., Porru, M., Randazzo, A., Pagano, B.,
Novellino, E., Pisanu, M. E., Stoppacciaro, A., Spinella, F., Bagnato,
A., Gilson, E., Leonetti, C., and Biroccio, A. (2014) Evidence for
G-quadruplex in the promoter of vegfr-2 and its targeting to inhibit
tumor angiogenesis, Nucleic Acids Res., 42,
2945-2957.
352.Tong, X., Lan, W., Zhang, X., Wu, H., Liu, M.,
and Cao, C. (2011) Solution structure of all parallel G-quadruplex
formed by the oncogene RET promoter sequence, Nucleic Acids
Res., 39, 6753-6763.
353.Viglasky, V. (2009) Platination of telomeric
sequences and nuclease hypersensitive elements of human c-myc and
PDGF-A promoters and their ability to form G-quadruplexes, FEBS
J., 276, 401-409.
354.Qin, Y., Fortin, J. S., Tye, D.,
Gleason-Guzman, M., Brooks, T. A., and Hurley, L. H. (2010) Molecular
cloning of the human platelet-derived growth factor receptor beta
(PDGFR-beta) promoter and drug targeting of the G-quadruplex-forming
region to repress PDGFR-beta expression, Biochemistry,
49, 4208-4219.
355.Shen, W., Gorelick, R. J., and Bambara, R. A.
(2011) HIV-1 nucleocapsid protein increases strand transfer
recombination by promoting dimeric G-quartet formation, J. Biol.
Chem., 286, 29838-29847.
356.Farhath, M. M., Thompson, M., Ray, S., Sewell,
A., Balci, H., and Basu, S. (2015) G-quadruplex-enabling sequence
within the human tyrosine hydroxylase promoter differentially regulates
transcription, Biochemistry, 54, 5533-5545.
357.Lim, K. W., Lacroix, L., Yue, D. J., Lim, J.
K., Lim, J. M., and Phan, A. T. (2010) Coexistence of two distinct
G-quadruplex conformations in the hTERT promoter, J. Am. Chem.
Soc., 132, 12331-12342.
358.Chaires, J. B., Trent, J. O., Gray, R. D.,
Dean, W. L., Buscaglia, R., Thomas, S. D., and Miller, D. M. (2014) An
improved model for the hTERT promoter quadruplex, PLoS One,
9, e115580.
359.Schonhoft, J. D., Bajracharya, R., Dhakal, S.,
Yu, Z., Mao, H., and Basu, S. (2009) Direct experimental evidence for
quadruplex–quadruplex interaction within the human ILPR,
Nucleic Acids Res., 37, 3310-3320.
360.Chen, Y., and Yang, D. (2012) Sequence,
stability, and structure of G-quadruplexes and their interactions with
drugs, Curr. Protoc. Nucleic Acid Chem., Chap. 17, Unit
17.5.
361.Wei, D., Parkinson, G. N., Reszka, A. P., and
Neidle, S. (2012) Crystal structure of a c-kit promoter quadruplex
reveals the structural role of metal ions and water molecules in
maintaining loop conformation, Nucleic Acids Res., 40,
4691-4700.
362.Sun, H., Xiang, J., Shi, Y., Yang, Q., Guan,
A., Li, Q., Yu, L., Shang, Q., Zhang, H., Tang, Y., and Xu, G. (2014) A
newly identified G-quadruplex as a potential target regulating Bcl-2
expression, Biochim. Biophys. Acta, 1840, 3052-3057.
363.Sheu, S. Y., Huang, C. H., Zhou, J. K., and
Yang, D. Y. (2014) Relative stability of G-quadruplex structures:
interactions between the human Bcl2 promoter region and derivatives of
carbazole and diphenylamine, Biopolymers, 101,
1038-1050.
364.Onel, B., Carver, M., Wu, G., Timonina, D.,
Kalarn, S., Larriva, M., and Yang, D. (2016) A new G-quadruplex with
hairpin loop immediately upstream of the human BCL2 P1 promoter
modulates transcription, J. Am. Chem. Soc., 138,
2563-2570.
365.Selvam, S., Yu, Z., and Mao, H. (2016) Exploded
view of higher order G-quadruplex structures through click-chemistry
assisted single-molecule mechanical unfolding, Nucleic Acids
Res., 44, 45-55.
366.Podbevsek, P., and Plavec, J. (2016) KRAS
promoter oligonucleotide with decoy activity dimerizes into a unique
topology consisting of two G-quadruplex units, Nucleic Acids
Res., 44, 917-925.
367.Cogoi, S., Zorzet, S., Rapozzi, V., Geci, I.,
Pedersen, E. B., and Xodo, L. E. (2013) MAZ-binding G4-decoy with
locked nucleic acid and twisted intercalating nucleic acid
modifications suppresses KRAS in pancreatic cancer cells and delays
tumor growth in mice, Nucleic Acids Res., 41,
4049-4064.
368.Agarwal, T., Roy, S., Kumar, S., Chakraborty,
T. K., and Maiti, S. (2014) In the sense of transcription regulation by
G-quadruplexes: asymmetric effects in sense and antisense strands,
Biochemistry, 53, 3711-3718.
369.Holder, I. T., and Hartig, J. S. (2014) A
matter of location: influence of G-quadruplexes on Escherichia
coli gene expression, Chem. Biol., 21, 1511-1521.
370.Cui, Y., Koirala, D., Kang, H., Dhakal, S.,
Yangyuoru, P., Hurley, L. H., and Mao, H. (2014) Molecular population
dynamics of DNA structures in a bcl-2 promoter sequence is regulated by
small molecules and the transcription factor hnRNP LL, Nucleic Acids
Res., 42, 5755-5764.
371.Baral, A., Kumar, P., Halder, R., Mani, P.,
Yadav, V. K., Singh, A., Das, S. K., and Chowdhury, S. (2012)
Quadruplex-single nucleotide polymorphisms (Quad-SNP) influence gene
expression difference among individuals, Nucleic Acids Res.,
40, 3800-3811.
372.Ji, X., Sun, H., Zhou, H., Xiang, J., Tang, Y.,
and Zhao, C. (2011) Research progress of RNA quadruplex, Nucleic
Acid Ther., 21, 185-200.
373.Mullen, M. A., Assmann, S. M., and Bevilacqua,
P. C. (2012) Toward a digital gene response: RNA G-quadruplexes with
fewer quartets fold with higher cooperativity, J. Am. Chem.
Soc., 134, 812-815.
374.Jodoin, R., Bauer, L., Garant, J. M., Mahdi
Laaref, A., Phaneuf, F., and Perreault, J. P. (2014) The folding of
5′-UTR human G-quadruplexes possessing a long central loop,
RNA, 20, 1129-1141.
375.Arora, A., and Suess, B. (2011) An RNA
G-quadruplex in the 3′-UTR of the proto-oncogene PIM1 represses
translation, RNA Biol., 8, 802-805.
376.Endoh, T., Kawasaki, Y., and Sugimoto, N.
(2013) Suppression of gene expression by G-quadruplexes in open reading
frames depends on G-quadruplex stability, Angew. Chem. Int. Ed.
Engl., 52, 5522-5526.
377.Xu, S., Li, Q., Xiang, J., Yang, Q., Sun, H.,
Guan, A., Wang, L., Liu, Y., Yu, L., Shi, Y., Chen, H., and Tang, Y.
(2015) Directly lighting up RNA G-quadruplexes from test tubes to
living human cells, Nucleic Acids Res., 43,
9575-9586.
378.Luke, B., and Lingner, J. (2009) TERRA:
telomeric repeat-containing RNA, EMBO J., 28,
2503-2510.
379.Collie, G. W., Sparapani, S., Parkinson, G. N.,
and Neidle, S. (2011) Structural basis of telomeric RNA
quadruplex–acridine ligand recognition, J. Am. Chem. Soc.,
133, 2721-2728.
380.Zhang, D. H., Fujimoto, T., Saxena, S., Yu, H.
Q., Miyoshi, D., and Sugimoto, N. (2010) Monomorphic RNA G-quadruplex
and polymorphic DNA G-quadruplex structures responding to cellular
environmental factors, Biochemistry, 49, 4554-4563.
381.Collie, G. W., Haider, S. M., Neidle, S., and
Parkinson, G. N. (2010) A crystallographic and modelling study of a
human telomeric RNA (TERRA) quadruplex, Nucleic Acids Res.,
38, 5569-5580.
382.Tang, C. F., and Shafer, R. H. (2006)
Engineering the quadruplex fold: nucleoside conformation determines
both folding topology and molecularity in guanine quadruplexes, J.
Am. Chem. Soc., 128, 5966-5973.
383.Joachimi, A., Benz, A., and Hartig, J. S.
(2009) A comparison of DNA and RNA quadruplex structures and
stabilities, Bioorg. Med. Chem., 17, 6811-6815.
384.Zhang, A. Y., Bugaut, A., and Balasubramanian,
S. (2011) A sequence-independent analysis of the loop length dependence
of intramolecular RNA G-quadruplex stability and topology,
Biochemistry, 50, 7251-7258.
385.Reddy, K., Zamiri, B., Stanley, S. Y.,
Macgregor, R. B., Jr., and Pearson, C. E. (2013) The disease-associated
r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent
uni- and multimolecular RNA G-quadruplex structures, J. Biol.
Chem., 288, 9860-9866.
386.Dethoff, E. A., Chugh, J., Mustoe, A. M., and
Al-Hashimi, H. M. (2012) Functional complexity and regulation through
RNA dynamics, Nature, 482, 322-330.
387.Bugaut, A., Murat, P., and Balasubramanian, S.
(2012) An RNA hairpin to G-quadruplex conformational transition, J.
Am. Chem. Soc., 134, 19953-19956.
388.Halder, K., Largy, E., Benzler, M.,
Teulade-Fichou, M. P., and Hartig, J. S. (2011) Efficient suppression
of gene expression by targeting 5′-UTR-based RNA quadruplexes
with bisquinolinium compounds, ChemBioChem, 12,
1663-1668.
389.Nagata, T., Sakurai, Y., Hara, Y., Mashima, T.,
Kodaki, T., and Katahira, M. (2012) 'Intelligent' ribozyme whose
activity is altered in response to K+ as a result of
quadruplex formation, FEBS J., 279, 1456-1463.
390.Hu, Y., Wang, F., Lu, C. H., Girsh, J., Golub,
E., and Willner, I. (2014) Switchable enzyme/DNAzyme cascades by the
reconfiguration of DNA nanostructures, Chemistry, 20,
16203-16209.
391.Yamaoki, Y., Nagata, T., Mashima, T., and
Katahira, M. (2015) K+-responsive off-to-on switching of
hammerhead ribozyme through dual G-quadruplex formation requiring no
heating and cooling treatment, Biochem. Biophys. Res. Commun.,
468, 27-31.
392.Martadinata, H., and Phan, A. T. (2014)
Formation of a stacked dimeric G-quadruplex containing bulges by the
5′-terminal region of human telomerase RNA (hTERC),
Biochemistry, 53, 1595-1600.
393.Bugaut, A., and Balasubramanian, S. (2012)
5′-UTR RNA G-quadruplexes: translation regulation and targeting,
Nucleic Acids Res., 40, 4727-4741.
394.Millevoi, S., Moine, H., and Vagner, S. (2012)
G-quadruplexes in RNA biology, Wiley Interdiscip. Rev. RNA,
3, 495-507.
395.Shahid, R., Bugaut, A., and Balasubramanian, S.
(2010) The BCL-2 5′ untranslated region contains an RNA
G-quadruplex-forming motif that modulates protein expression,
Biochemistry, 49, 8300-8306.
396.Yu, C. H., Teulade-Fichou, M. P., and
Olsthoorn, R. C. (2014) Stimulation of ribosomal frameshifting by RNA
G-quadruplex structures, Nucleic Acids Res., 42,
1887-1892.
397.Bhattacharyya, D., Nguyen, K., and Basu, S.
(2014) Rationally induced RNA:DNA G-quadruplex structures elicit an
anticancer effect by inhibiting endogenous eIF-4E expression,
Biochemistry, 53, 5461-5470.
398.Oyaghire, S. N., Cherubim, C. J., Telmer, C.
A., Martinez, J. A., Bruchez, M. P., and Armitage, B. A. (2016) RNA
G-quadruplex invasion and translation inhibition by antisense
gamma-peptide nucleic acid oligomers, Biochemistry, 55,
1977-1988.
399.Panyutin, I. G., Onyshchenko, M. I., Englund,
E. A., Appella, D. H., and Neumann, R. D. (2012) Targeting DNA
G-quadruplex structures with peptide nucleic acids, Curr. Pharm.
Des., 18, 1984-1991.
400.Rouleau, S. G., Beaudoin, J. D., Bisaillon, M.,
and Perreault, J. P. (2015) Small antisense oligonucleotides against
G-quadruplexes: specific mRNA translational switches, Nucleic Acids
Res., 43, 595-606.
401.Agarwala, P., Pandey, S., Mapa, K., and Maiti,
S. (2013) The G-quadruplex augments translation in the
5′-untranslated region of transforming growth factor beta2,
Biochemistry, 52, 1528-1538.
402.Agarwala, P., Pandey, S., and Maiti, S. (2014)
Role of G-quadruplex located at 5′-end of mRNAs, Biochim.
Biophys. Acta, 1840, 3503-3510.
403.Morris, M. J., Negishi, Y., Pazsint, C.,
Schonhoft, J. D., and Basu, S. (2010) An RNA G-quadruplex is essential
for cap-independent translation initiation in human VEGF IRES, J.
Am. Chem. Soc., 132, 17831-17839.
404.Bhattacharyya, D., Diamond, P., and Basu, S.
(2015) An independently folding RNA G-quadruplex domain directly
recruits the 40S ribosomal subunit, Biochemistry, 54,
1879-1885.
405.Arnoult, N., Van Beneden, A., and Decottignies,
A. (2012) Telomere length regulates TERRA levels through increased
trimethylation of telomeric H3K9 and HP1alpha, Nat. Struct. Mol.
Biol., 19, 948-956.
406.Hirashima, K., and Seimiya, H. (2015) Telomeric
repeat-containing RNA/G-quadruplex-forming sequences cause genome-wide
alteration of gene expression in human cancer cells in vivo,
Nucleic Acids Res., 43, 2022-2032.
407.Redon, S., Zemp, I., and Lingner, J. (2013) A
three-state model for the regulation of telomerase by TERRA and
hnRNPA1, Nucleic Acids Res., 41, 9117-9128.
408.Cusanelli, E., and Chartrand, P. (2014)
Telomeric noncoding RNA: telomeric repeat-containing RNA in telomere
biology, Wiley Interdiscip. Rev. RNA, 5, 407-419.
409.Huang, H., Suslov, N. B., Li, N. S., Shelke, S.
A., Evans, M. E., Koldobskaya, Y., Rice, P. A., and Piccirilli, J. A.
(2014) A G-quadruplex-containing RNA activates fluorescence in a
GFP-like fluorophore, Nat. Chem. Biol., 10, 686-691.
410.Tanpure, A. A., and Srivatsan, S. G. (2015)
Conformation-sensitive nucleoside analogues as topology-specific
fluorescence turn-on probes for DNA and RNA G-quadruplexes, Nucleic
Acids Res., 43, e149.
411.Xu, Y., Ishizuka, T., Yang, J., Ito, K.,
Katada, H., Komiyama, M., and Hayashi, T. (2012) Oligonucleotide models
of telomeric DNA and RNA form a hybrid G-quadruplex structure as a
potential component of telomeres, J. Biol. Chem., 287,
41787-41796.
412.Zheng, K. W., Xiao, S., Liu, J. Q., Zhang, J.
Y., Hao, Y. H., and Tan, Z. (2013) Co-transcriptional formation of
DNA:RNA hybrid G-quadruplex and potential function as constitutional
cis element for transcription control, Nucleic Acids Res.,
41, 5533-5541.
413.Xiao, S., Zhang, J. Y., Zheng, K. W., Hao, Y.
H., and Tan, Z. (2013) Bioinformatic analysis reveals an evolutional
selection for DNA:RNA hybrid G-quadruplex structures as putative
transcription regulatory elements in warm-blooded animals, Nucleic
Acids Res., 41, 10379-10390.
414.Zhang, J. Y., Zheng, K. W., Xiao, S., Hao, Y.
H., and Tan, Z. (2014) Mechanism and manipulation of DNA:RNA hybrid
G-quadruplex formation in transcription of G-rich DNA, J. Am. Chem.
Soc., 136, 1381-1390.
415.Wanrooij, P. H., Uhler, J. P., Shi, Y.,
Westerlund, F., Falkenberg, M., and Gustafsson, C. M. (2012) A hybrid
G-quadruplex structure formed between RNA and DNA explains the
extraordinary stability of the mitochondrial R-loop, Nucleic Acids
Res., 40, 10334-10344.
416.Wang, J., Haeusler, A. R., and Simko, E. A.
(2015) Emerging role of RNA–DNA hybrids in C9orf72-linked
neurodegeneration, Cell Cycle, 14, 526-532.
417.Groh, M., Lufino, M. M., Wade-Martins, R., and
Gromak, N. (2014) R-loops associated with triplet repeat expansions
promote gene silencing in Friedreich ataxia and fragile X syndrome,
PLoS Genet., 10, e1004318.
418.Eddy, S., Ketkar, A., Zafar, M. K., Maddukuri,
L., Choi, J. Y., and Eoff, R. L. (2014) Human Rev1 polymerase disrupts
G-quadruplex DNA, Nucleic Acids Res., 42, 3272-3285.
419.Li, D., Lv, B., Zhang, H., Lee, J. Y., and Li,
T. (2014) Positive supercoiling affiliated with nucleosome formation
repairs non-B DNA structures, Chem. Commun. (Camb.), 50,
10641-10644.
420.Mergny, J. L. (2012) Alternative DNA
structures: G4 DNA in cells: itae missa est? Nat. Chem. Biol.,
8, 225-226.
421.Henderson, A., Wu, Y., Huang, Y. C., Chavez, E.
A., Platt, J., Johnson, F. B., Brosh, R. M., Jr., Sen, D., and
Lansdorp, P. M. (2014) Detection of G-quadruplex DNA in mammalian
cells, Nucleic Acids Res., 42, 860-869.
422.Huang, W. C., Tseng, T. Y., Chen, Y. T., Chang,
C. C., Wang, Z. F., Wang, C. L., Hsu, T. N., Li, P. T., Chen, C. T.,
Lin, J. J., Lou, P. J., and Chang, T. C. (2015) Direct evidence of
mitochondrial G-quadruplex DNA by using fluorescent anti-cancer agents,
Nucleic Acids Res., 43, 10102-10113.
423.Biffi, G., Tannahill, D., Miller, J., Howat, W.
J., and Balasubramanian, S. (2014) Elevated levels of G-quadruplex
formation in human stomach and liver cancer tissues, PLoS One,
9, e102711.
424.Yang, T. L., Lin, L., Lou, P. J., Chang, T. C.,
and Young, T. H. (2014) Detection of cell carcinogenic transformation
by a quadruplex DNA binding fluorescent probe, PLoS One,
9, e86143.
425.Hoffmann, R. F., Moshkin, Y. M., Mouton, S.,
Grzeschik, N. A., Kalicharan, R. D., Kuipers, J., Wolters, A. H.,
Nishida, K., Romashchenko, A. V., Postberg, J., Lipps, H., Berezikov,
E., Sibon, O. C., Giepmans, B. N., and Lansdorp, P. M. (2016) Guanine
quadruplex structures localize to heterochromatin, Nucleic Acids
Res., 44, 152-163.
426.David, A. P., Margarit, E., Domizi, P.,
Banchio, C., Armas, P., and Calcaterra, N. B. (2016) G-quadruplexes as
novel cis-elements controlling transcription during embryonic
development, Nucleic Acids Res., 44, 4163-4173.