REVIEW: Genomics and Biochemistry of Saccharomyces cerevisiae Wine Yeast Strains
M. A. Eldarov1*, S. A. Kishkovskaia2, T. N. Tanaschuk2, and A. V. Mardanov1
1Institute of Bioengineering, Federal Research Center “Fundamentals of Biotechnology”, Russian Academy of Sciences, 119071 Moscow, Russia; E-mail: eldarov@biengi.ac.ru2All-Russian National Research Institute of Viticulture and Winemaking “Magarach”, Russian Academy of Sciences, 298600 Yalta, Republic of Crimea, Russia
* To whom correspondence should be addressed.
Received July 29, 2016; Revision received September 19, 2016
Saccharomyces yeasts have been used for millennia for the production of beer, wine, bread, and other fermented products. Long-term “unconscious” selection and domestication led to the selection of hundreds of strains with desired production traits having significant phenotypic and genetic differences from their wild ancestors. This review summarizes the results of recent research in deciphering the genomes of wine Saccharomyces strains, the use of comparative genomics methods to study the mechanisms of yeast genome evolution under conditions of artificial selection, and the use of genomic and postgenomic approaches to identify the molecular nature of the important characteristics of commercial wine strains of Saccharomyces. Succinctly, data concerning metagenomics of microbial communities of grapes and wine and the dynamics of yeast and bacterial flora in the course of winemaking is provided. A separate section is devoted to an overview of the physiological, genetic, and biochemical features of sherry yeast strains used to produce biologically aged wines. The goal of the review is to convince the reader of the efficacy of new genomic and postgenomic technologies as tools for developing strategies for targeted selection and creation of new strains using “classical” and modern techniques for improving winemaking technology.
KEY WORDS: wine, yeast strains, diversity, artificial selection, stress response, evolution, comparative genomics, metagenomics, microbial interactions, flor yeast, adhesion, aromatic compounds, gene regulation, strain improvementDOI: 10.1134/S0006297916130046
Abbreviations: ARS, autonomously replicating sequence; CNV, copy number variation; GCR, gross chromosomal rearrangement; HGT, horizontal gene transfer; Indel, insertion/deletion polymorphism; LTR, long terminal repeat; MLF, malolactic fermentation; NGS, next-generation sequencing; ORF, open reading frame; QTL, quantitative trait loci; SNP, single-nucleotide polymorphism.
Bread, wine, and beer are some of the oldest products known to man.
Their preparation is impossible without the use of Saccharomyces
yeast. The history of viticulture and winemaking roots back millennia
and is inextricably linked with the history of mankind. The earliest
archaeological evidence of the rise of fermented beverage production
technology based on rice, honey, and fruit was found in China in
antiquity tombs dating back more than 7000 years BC [1]. Chemical analysis data confirming the beginning of
winemaking date back to 5400 BC [2], while data on
the use of yeast in wine fermentation processes in ancient Egypt date
as far back as 3150 BC [3]. From Egypt and
Mesopotamia, fermented beverage production technology spread to Europe
and then further to the New World [4].
The role of yeast in alcoholic fermentation was established by Pasteur in 1860 [5]. In the early 1980s, Emil Christian Hansen of Carlsberg Laboratory isolated the first pure yeast culture, which was then used as an inoculum for the fermentation of wine musts. Surprisingly, this practice began to spread effectively only in the middle of the twentieth century, and now almost all commercial wine production in the world is based on the use of starter cultures. Using carefully selected commercial yeast strains has significantly improved the controllability and reliability of the wine fermentation process, limited variability in wine microbial composition, and made a significant contribution into the improvement of wine quality in recent decades.
The widespread application of yeast in winemaking and other fields of traditional biotechnology had soon pushed these microorganisms to the forefront of genetic, biochemical, and cell biology research and advanced studies in the fields of physiology, genomics, and evolutionary biology. Methods of yeast cultivation and genetic manipulations are simple and convenient. These factors together with other advantages soon converted yeast into one of the most popular model eukaryotes.
Elucidation of the genomic sequence of the laboratory strain of baker’s yeast Saccharomyces cerevisiae in 1996 became a turning point in genomics, for the first time opening the opportunity for the global study of the expression and functioning of the eukaryotic genome [5] and creating opportunities for further studies in comparative, functional, and evolutionary genomics [6]. The rapid progress in the development of next-generation sequencing methods markedly increased the number of sequenced genomes of natural, industrial, and clinical S. cerevisiae isolates and greatly expanded our understanding of the diversity, origin, and natural history of S. cerevisiae and related yeasts.
This review summarizes some recent advances in sequencing of the genomes of wine yeast strains, the use of comparative genomics to study the mechanisms of evolution of the yeast genome under artificial selection, and the application of genomic and post-genomic approaches to identify the molecular nature of important winemaking properties of commercial S. cerevisiae wine strains. A separate section is devoted to the physiological, genetic, and biochemical features of sherry yeast strains used to produce biologically aged wines.
PHYLOGENY AND BIOCHEMISTRY OF S. cerevisiae WINE
YEAST
Origin of wine yeast strains. Saccharomyces cerevisiae wine yeast strains are characterized by a mixed haplo-diploid life cycle and predominantly vegetative proliferation mode. Most natural isolates are diploid with significant degree of heterozygosity [6]. The frequency of heterozygous single-nucleotide polymorphisms (SNPs) is from 1000 to 18,000 SNP per genome [7, 8], reflecting frequent crossing and meiosis events under conditions of artificial selection [9].
Numerous recent studies [10-15] have contributed to in-depth understanding of S. cerevisiae population structure and evolutionary history. It was shown that industrial S. cerevisiae strains emerged for natural ancestors from independent “domestication” events [12, 16]. Notably, the clear majority (95%) of wine strains belong to a single phylogenetic cluster [12, 15], thus assuming their unique origin and subsequent population expansion due to human activity.
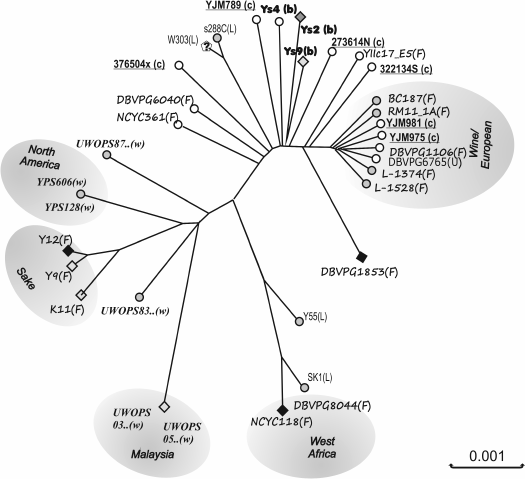
Whole-genome analysis of S. cerevisiae isolates from various geographic areas all over the world has revealed five major phylogenetic groups differing both in place of origin and in technological properties (Fig. 1). Many strains with mosaic genomes were also identified that emerged from crossing between strains belonging to these groups [14]. It is important that stable correlations were found between strain genotypes and experimentally determined phenotypic properties.
Fig. 1. Yeast phylogenomics. Phylogenetic tree of S. cerevisiae isolates from different geographical regions and ecological niches. The “pure” non-mosaic lineages are outlined in gray. Geographical origin: white circles – Europe, gray circles – America, gray diamonds – Asia, black diamonds – Africa. Strain origin: (w) – natural (wild) isolate, (c) – clinical isolate (conditional pathogen), (b) – baking, (L) – laboratory, (F) – fermentative (wine, beer, ethanol). Reproduced with modifications from [14].
The genetic studies of yeast within small populations had shown that their population structure generally reflects adaptation to different ecological niches. Wine yeast strains constitute a separate phylogenetic group having low variability [13, 14].
Genotyping data obtained with microsatellite markers suggests Mesopotamian origin of European wine strains [12, 17] and their subsequent migration to Europe via the Mediterranean Sea or the Danube, together with grape varieties [17].
The center for the origin and diversity of S. cerevisiae might be primeval forests in China, as indicated by the analysis of a large sample of data on isolates from geographically and ecologically different habitats [11].
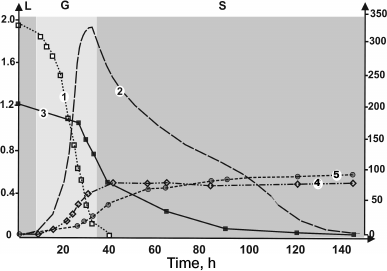
Biochemical peculiarities of wine yeast strains. The process of wine fermentation is accompanied by various types of stress – osmotic stress, resulting from high concentrations of sugar, stress related to acidification of the medium, high sulfite content, anaerobic conditions, insufficient sources of nitrogen, lipids, and vitamins, high ethanol concentrations, and temperature fluctuations. Good wine strains should be resistant to the combined action of these abiotic factors in all the major phases of wine fermentation (Fig. 2) and possess several other important technological characteristics (Table 1).
Fig. 2. Main phases of wine fermentation. Evolution of the main fermentation parameters during winemaking on synthetic medium containing 200 g/liter glucose/fructose and 330 mg/liter assimilable nitrogen, with the commercial wine strain EC1118. Curves: 1) nitrogen (mg/liter); 2) fermentation rate, dCO2/dt (g/liter per h); 3) sugars (g/liter); 4) cell number (106 cells/ml); 5) ethanol (g/liter); L, lag phase; G, growth phase; S, stationary phase. Left ordinate axis is for curve 2, right ordinate axis is for curves 1, 3, 4, 5. Reproduced with modifications from [20].
Table 1. Properties of wine yeast
strains

Extremely important for wine sensory and organoleptic properties is the content of numerous fermentation byproducts, among which are glycerol, carboxylic acids, aldehydes, higher alcohols, esters and sulfides, etc., formed during the degradation of grape sugars, amino acids, fatty acids, terpenes, and thiols, which is also largely dependent on the peculiarities of the strains used [18, 19].
The outcome of fermentation depends on many factors – in particular, the content and the quality of the grape must. Essentially these are sugars (glucose and fructose in equimolar high concentrations of 180-300 g/liter), organic acids (tartaric and malic), inorganic cations (potassium), nitrogen compounds, and lipids (phytosterols). Since fructose metabolism is subject to glucose repression, at farther stages of fermentation the relative fructose content can reach significant values. Wine yeasts must ferment these sugars in conditions of long starvation periods and in the presence of high ethanol concentrations. The ability of wine strains to ferment fructose efficiently is critical to maintaining a high rate of fermentation at advanced stages of alcohol formation.
Nitrogen is another essential nutrient influencing the fermentation rate and volatile matter content. The nitrogen content in the musts is limited, while the lack of nitrogen is the most common cause of slow or arrested fermentations [20]. Ammonium ions, amino acids, oligopeptides, proteins, amides, biogenic amines, and nucleic acids can all be nitrogen sources for yeast. It is widely accepted that fermentation of 200 g/liter of sugar requires 140 mg/liter of nitrogen sources [20].
Many other components of the grape must (lipids, vitamins, various inhibitors) affect the yeast viability and metabolism. However, the major stress factor and “selective property” of wine yeast is their resistance to high ethanol concentrations.
GENOMICS OF S. cerevisiae WINE STRAINS
Already in the “pre-genomic” period, it became clear that natural and industrial yeast strains possess extensive genetic variability leading to significant differences in their properties compared to the reference strain S288C. Evolutionary constraints that were involuntarily imposed on S. cerevisiae genome during domestication led to selection of a huge number of yeast strains with highly specialized phenotypes optimally suited to specific applications [12]. The number of yeast strains with deciphered complete genomes sequenced during the 20 years since the first published sequence of strain S288C have reached several hundreds, and 237 complete sequences of various S. cerevisiae isolates are deposited at GenBank. The progress of these studies is due to the recent revolution in the next-generation sequencing methods. Yeast are well suited for these tasks. Their genomes are relatively small (7-12 Mb) and contain small numbers of repeats. Thus, they are ideal targets for high throughput next-generation sequencing (NGS) (Table 2).
Table 2. Basic properties of S.
cerevisiae genome*
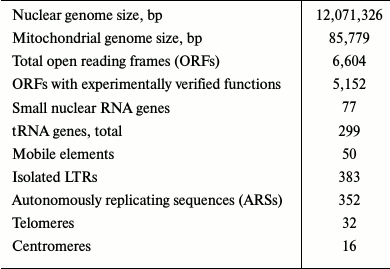
* Reference strain S288C from the Saccharomyces genome database
(July 23, 2016).
Mechanisms of yeast genome variability. The results of genome sequencing of wine and other yeast strains confirmed the “pre-genomic” data concerning their significant genetic heterogeneity, the presence of multiple single-nucleotide polymorphisms (SNPs), insertion/deletion polymorphisms (Indels), gross chromosomal rearrangements, and copy number variations (CNVs) [12, 21] compared to the reference genome, variations in open reading frame (ORF) size and gene sets [22], and presence of multiple strain-specific ORFs.
Comparison of genomic sequences of 100 natural, clinical, and industrial isolates with the genome of strain S288C had shown that mean SNP frequency is 78,140 per genome, Indel frequency is 7840, and 566 ORFs have significant size variations.
The degree of nucleotide sequence variation of wine yeast genomes determined for 236 strains appeared to be significantly lower, indicating that these strains represent a highly inbred population. However, wine yeast genomes contain tall types of chromosomal polymorphisms as well as traces of introgression and horizontal gene transfer (HGT) [23-26] with obvious adaptive significance (see below).
Translocations and amplifications. The main source of gross chromosomal rearrangements (GCRs) in yeast is the ectopic recombination between Ty-elements and other repeats [27]. The most frequent targets for deletions and amplifications are the subtelomeric genes, genes encoding ion, sugar, and metal transporters, transcription and translation factors, and various adhesion proteins. These observations confirm the high plasticity of the subtelomeric regions and their important role as sources of variation for rapid adaptation of wine yeast to changing external conditions [7, 8].
The high GCR frequency in wine strains may be related to mutagenic effects of high ethanol concentrations, but the adaptive significance of these variations is not clear.
A well-documented example of chromosomal rearrangement with an adaptive advantage is the widespread among wine yeast reciprocal translocation between chromosomes VIII and XVI. This translocation generated a dominant allele of the sulfite pump, SSU1-R1, which is expressed at much higher levels than SSU1 [29] and confers a high level of sulfite resistance [29, 30]. Another translocation, between chromosomes XV and XVI, was recently identified in several wine strains, also leading to increased SSU1 gene expression [31]. These translocations have only been observed in wine yeasts and occur with high frequency, conferring selective advantage by shortening the growth lag phase in media containing sulfite. Thus, the long-lasting practice of using sulfites [32] likely caused an evolutionary bottleneck favoring convergent evolutionary rearrangements that confer a growth advantage to strains carrying the SSU1 recombinant forms.
Acquisition of resistance to CuSO4 is another possibly domestication-related trait. Copper sulfate is widely used as a fungicide to protect grape yards from powdery mildew [33]. Copper-resistant European and Sake lineages possess a copy number variation (CNV) of CUP1 encoding a copper-binding metallothionein [33, 34].
Another means of enhancing CUP1 gene expression, observed in the EC1118 strain, is related to CUP1 promoter variation [35].
Recent genome-wide studies have provided insights into structural variation, revealing the existence of many CNVs in wine yeasts that correspond to genes encoding transporters or dehydrogenases or genes involved in drug response perhaps having adaptive roles [8, 33, 36].
Gene loss and inactivation. The aquaporin water transporters (AQYs) are critical for surviving freeze–thaw stress by preventing intracellular shearing due to water crystallization [37]. On the other hand, loss of AQY function is beneficial for utilization of high-sugar substrates by overcoming the effect of high osmolarity. Laboratory and industrial strains as well as several vineyard isolates are known to harbor nonfunctional alleles of AQY2 or AQY1 [38, 39]. Mutational inactivation of these paralogs has occurred many times [40].
Horizontal gene transfer. Comparative genomics methods have unexpectedly revealed significant contribution of HGT to the genome landscape of wine yeasts. The genome of the EC1118 strain was shown to contain three large chromosomal segments – A, B, and C – acquired through independent HGT events from distant yeast species [7]. Region B originated from Zygosaccharomyces bailii, a major contaminant of wine fermentations [7, 41]. Multiple copy insertions and different arrangements of region B have been identified in various wine strains [8, 41]. Recent HGT from Torulaspora microellipsoides yeast was the source of region C [42].
Thirty-nine genes annotated within these three segments encode functions potentially important for winemaking, being related to nitrogen and sugar metabolism [7]. Some of the region C genes were characterized in more detail. The high-affinity fructose/H+ symporter encoded by FSY1 may help to utilize abundant fructose at the end of wine fermentation [43]. The XDH1 gene is necessary for xylose utilization [44]. Tandemly duplicated FOT1-2 genes encode oligopeptide transporters, and their adaptive role was experimentally demonstrated [45]. Strains with increased Fot-protein expression provide for the transport of a larger set of oligopeptides, including those rich in glutamate, leading to the increase in growth, fermentation, and survival rates under winemaking conditions [42]. Moreover, Fot-mediated peptide transport significantly affected central carbon and nitrogen metabolic pathways, amino acid and peptide synthesis, oxidative stress response, and redox metabolism. Altogether, this led to decrease in acetic acid production and increase in ester formation, positively affecting organoleptic balance of wine. The ability to use various di- and tripeptides varies significantly among various strains, and the presence of FOT genes is an important contribution to this phenotypic variability.
In addition to FOT genes, several genes present in new regions acquired by wine yeasts have putative functions associated with nitrogen metabolism, including asparaginase, oxoprolinase, an ammonium transporter, and allantoate transporter transcription factors associated with the biosynthesis enzymes involved in lysine and proline utilization [7, 46].
Introgression. Several regions of introgression from S. paradoxus, S. uvarum, and S. eubayanus strains have been identified in S. cerevisiae wine yeasts. An extended S. paradoxus genome segment transferred to commercial wine yeast strains includes the SUC2 gene for invertase and the glucan-α-1,4-glucosidase gene, which is important for production of white wines [47]. The introgressed region also contains the AWA1 gene, present in S. cerevisiae sake strains but absent from S288C, encoding a putative GPI-anchored protein localized to the cell wall and conferring hydrophobicity to the cell surface.
A recent study was devoted to experimental verification of evolutionary advantages of interspecies hybridization. An experimental S. cerevisiae × S. uvarum hybrid was obtained and subjected to artificial selection under nitrogen starvation conditions. The authors [48] selected a strain with a modified chimeric MEP2 gene encoding a high-affinity ammonium permease. This translocation occurred by introgression of a small fragment of the S. uvarum genome into chromosome XIV of S. cerevisiae [48].
Interspecific hybrids. Interspecific hybrids of Saccharomyces are widely used in both brewing and winemaking. Lager yeasts, S. pastorianus (S. cerevisiae × S. eubayanus), are the most famous example [54]. Some commercial wine strains are also hybrids and polyploids – VIN7 (S. cerevisiae × S. kudriavzevii [57]), S6U (S. cerevisiae × S. uvarum [53, 54]), EP2 (S. cerevisiae × S. kudriavzevii [55]), and NT50 (S. cerevisiae × S. kudriavzevii [51, 55]).
Under wine fermentation conditions, hybrids may have certain advantages, including higher tolerance to various kinds of stress [56]. For example, S. kudriavzevii and S. bayanus are better adapted to growth at lower temperatures, while S. cerevisiae is more resistant to ethanol. Natural hybrids of these strains are better adapted to grow under temperature and ethanol stress conditions, having benefitted from the competitive advantages of each of the parents [57]. Such hybrids can be quite valuable in white wine production, where reduced fermentation temperature is used to prevent the loss of aromatic compounds.
The genetic characteristics of hybrids can also directly affect the organoleptic characteristics of wine. Hybrids of S. cerevisiae × S. kudriavzevii have been described that are characterized by increased production of esters and higher alcohols during fermentation of musts from certain grape varieties [58, 59]. Under wine fermentation conditions, such hybrids produce more thiol-4-mercapto-4-methylpentandione – an aromatic compound with a strong fruity aroma [60]. The frequent occurrence of such hybrids may result from their adaptive advantages, although we cannot exclude that stress conditions themselves stimulate the hybridization processes [61].
Genetic instability of hybrids results in considerable variability in their chromosome sets and other chromosomal rearrangements and modifications [52, 56, 62]. Molecular analysis of 24 natural hybrids of S. cerevisiae × S. kudriavzevii revealed significant variations in their ploidy (from 2n to 4n) and in the S. kudriavzevii genetic material content [63].
As repeatedly cited, the genomes of 212 sequenced wine strains have been analyzed for the presence of genetic material from other strains of the Saccharomyces sensu stricto clade [15].
In general, the contribution of sequences of strains other than S. cerevisiae has been rather insignificant. However, 17 strains possess extensive areas (more than 10% of a single chromosome) that was transferred from S. paradoxus, S. uvarum, S. eubayanus, S. mikitae, or S. kudriavzevii genomes. However, 12 out of these 17 strains contain a practically complete second genome. Five hybrids were obtained under laboratory conditions from rare occurrences of crosses between wine strains and other strains of non-Saccharomyces to create new industrial lines [64, 65]. Other strains were distinguished by highly variable degree of polyploidy. For example, the NT50 strain contained the tetraploid genome of S. cerevisiae and one copy of chromosome 13 of S. kudriavzevii. The S6U and WLP862 strains contain a significant share of the genetic material from three different species.
MICROBIAL ECOLOGY OF WINE
Before the widespread introduction of starter cultures of Saccharomyces, wine production was based on spontaneous fermentation by microorganisms naturally present on the surface of the grapes, harvesting and winemaking equipment, etc. A detailed picture of the microbial community of wine has become available only recently, thanks to the introduction of modern methods of metagenomic analysis. It was found that wine must and wine form a complex ecosystem, mainly represented by yeast, bacteria, and filamentous fungi [66]. Intermicrobial interaction starts on the surface of grape berries and continues in wineries as the process of alcoholic fermentation by yeasts and malolactic fermentation by lactic acid bacteria.
According to literature data, 93 different yeast species have been detected on the surface of 49 grape varieties grown in 22 countries [67].
At least 15 of these yeast species are involved in the winemaking process. Kloeckera and Hanseniaspora are most widely represented on the surface of grapes (up to 50% of the microflora) [68]. Other yeasts inhabiting the surface of the grapes, such as C. stellata, C. pulcherrima, B. intermedius, B. lambicus, B. custeri, Cryptococcus, Kluyveromyces spp., Metschnikowia spp., P. membranaefaciens, and H. anomalae [66] are also involved in the process of winemaking.
The bacterial community of grapes is comprised of more than 50 species belonging mainly to the firmicutes and proteobacteria. The firmicutes are represented by gram-positive families of Lactobacillaceae and Leuconostocaceae related to industrial strains of Bacillaceae and Enterococcaceae lactic acid bacteria [69].
The ratio between the yeast and bacterial microflora and Saccharomyces and non-Saccharomyces yeasts depends on biotic and abiotic factors, and it is important for the chemical and sensory changes that occur in the fermentation process and affect the quality of the produced wine. The presence of filamentous fungi from the genera Aspergillus and Penicillium and grape pathogens (oidium, gray mold) is undesirable as they can produce mycotoxins and compounds with unpleasant odor and taste [70].
At the beginning of the fermentation process, the wine must microflora is represented mainly by the naturally occurring species that are characterized by low resistance to ethanol, such as Hanseniaspora/Kloeckera, Hansenula, Metschnikowia, Candida, and some strains of S. cerevisiae [71]. At the end of the fermentation process, the composition of the wine microflora changes dramatically – the ethanol sensitive yeasts, bacteria, and fungi are almost completely displaced by S. cerevisiae. The key role in this process is played by such adaptive advantages of Saccharomyces as high speed of fermentation, alcohol endurance, anaerobic growth properties, etc. One of the most remarkable characteristics of S. cerevisiae, important from the point of view of understanding their evolution and ecology as well as for practical winemaking, is their ability for alcoholic fermentation under aerobic conditions (Crabtree effect).
For “Crabtree-positive” yeast species, the ability to effectively catabolize the previously accumulated ethanol underlies the adaptive strategy of “make-accumulate-consume” (MAC) [72]. In line with the MAC strategy, the yeasts first rapidly ferment the sugars with the release of ethanol, thus suppressing the growth of other species. Then, after the expulsion of competitors from the given ecological niche, the accumulated ethanol is used by ethanol-resistant yeast as a carbon source [73]. It is believed that the biochemical mechanisms underlying the Crabtree effect emerged in the evolutionary history of Saccharomyces about 125-150 million years ago as a result of ancestral genome-duplication [74]; their formation has paralleled the evolution of angiosperms and the appearance of their fruit that is juicy and rich in easily fermentable sugars (125 million years ago). Duplication resulted in increased copy number of the genes responsible for glucose transport and catabolism, improved the efficiency of anaerobic energy metabolism, and allowed division of ethanol production and consumption functions facilitated by two isoforms of alcohol dehydrogenases [75]. Another important factor for the emergence of the MAC strategy was genome-wide genetic effect, apparently, engaged global change in respiratory metabolism gene transcription regulation mechanisms due to the loss of cis-regulatory elements [74].
Some yeast strains can secrete protein toxins characterized by lethal effect on sensitive strains [76]. These killer strains, or “killers”, are widespread not only among S. cerevisiae, but also among Pichia, Kluyveromyces, Candida, and other yeast genera. There are also neutral strains unable to secrete their own toxins, but resistant to the toxins of other strains. The “killer” phenomenon has a double effect on winemaking. The presence of a killer factor in the starter cultures can contribute to the suppression of endogenous yeast flora to ensure a more stable fermentation process and prevent contamination with “spoiler” yeast species (e.g. Brettanomyces). On the other hand, “killers” of foreign yeast species may prevent full fermentation by killing or inhibiting the starter or endogenous cultures. There are occasional reports on negative impact of killer strains on the fermentation process, but in general, this impact is rather insignificant. The impact might result from relatively high incidence of “neutral” strains among the endogenous and the starter winemaking microflora, and inactivation of the killer factor in winemaking conditions under the influence of tannins, ethanol, sulfite, etc. [23].
The molecular nature of the “killer” and “neutrality” phenomena in yeasts is diverse and not fully understood. Several useful reviews can be recommended for more detailed acquaintance with the results of the latest research in this field [77, 78].
MECHANISMS CONTROLLING WINE FERMENTATION
In addition to comparative genomics methods, various “omics” approaches (transcriptomics, proteomics, metabolomics, functional genomics, etc.) may be used efficiently for identification of biochemical mechanisms controlling important winemaking properties – ethanol production, nitrogen uptake, residual sugar content, volatile acids content, ability to produce aroma compounds, etc. The “top-down” approach is exemplified by quantitative trait loci (QTL) mapping. The “bottom-up” strategy is related to high-throughput screening of yeast strain knockout collections. Such collections are not developed for wine yeast yet, but some parameters of wine fermentation can be modeled using laboratory strains. The degree and rate of wine fermentation can be controlled by epigenetic mechanisms that are modulated by bacterial chemical signalization. More detailed description of applied approaches and their results is provided below.
Quantitative trait loci (QTL). Winemaking properties of yeasts are complex and quantitative in nature, as convincingly shown recently during comparison of 72 strains of varying origin under experimental winemaking conditions [79]. QTL mapping is performed by crossing of phenotypically distant species and searching for statistically significant associations between phenotypes and genetic markers in hybrid progeny [80]. Knowledge of genomic sequence then allows narrowing QTL regions to separate genes and separate polymorphisms of these genes. Several yeast QTL, for instance degree of heat shock resistance, telomere length, and efficiency of DNA repair were mapped using this approach [33, 81].
To search for QTL specific for winemaking, special tester strains and many hybrids were created that, together with the development of methods for high-throughput screening, revealed more than 80 loci important for winemaking [82].
A recent study [83] applied of this approach to identifying the polymorphism of separate genes controlling important winemaking properties – production of malic, acetic, and succinic acids and characteristics of ethanol, glycerol, residual sugar content, and nitrogen assimilation. The genetic analysis enabled identification of 18 QTL, some of which had pleiotropic effects. Further analysis revealed specific polymorphic sites in genes responsible for transport of sugars, mitochondrial respiration, and nitrogen metabolism that were key to determining observed phenotypic variation in winemaking properties.
A polymorphic variant of the ALD6 gene promoter provided five-fold increase in the level of aldehyde dehydrogenase expression and acetic acid production. Alterations in a conserved region of Hap4 transcription factor and linked Mbr1 ORF reduced the level of respiratory gene expression and increased the concentration of residual sugars, and more complex epistatic interrelations were observed between the FLX1 and MDH2 genes controlling succinic acid levels.
Screening of a knockout gene collection. Recently, for identification of genes important for efficient wine fermentation, an approach based on high-throughput screening of a yeast deletion mutant collection was described [84]. The authors used a collection of 5100 knockout mutants of the laboratory strain S. cerevisiae BY4741 developed during the “functional yeast genome analysis project”. Cells were grown under conditions of microfermentation in 96-well plates on media that mimicked wine must in nitrogen and glucose content. Strains with defects in fermentation rate were selected (slow or “arrested” fermentation) and subjected to additional tests under conditions of experimental winemaking. Even though laboratory strain BY4741 is phenotypically distant from wine strains, the developed strategy identified a group of 93 genes forming a “fermentome”, i.e. coding proteins important for fast and complete fermentation.
Analysis of “fermentome” gene function in gene ontology terms showed that their products are involved in adaptation and response to different stress conditions, regulation of autophagy, signaling pathways to carbon source availability, cation transport, etc. Nine mutants with complete fermentation arrest were inactivated in diverse genes involved in trehalose synthesis, ion and pH homeostasis, and other functions (Table 3). However, all these genes were united by the earlier proven role in combined resistance to different types of stresses – ethanol, osmotic, cold and heat-shock, desiccation, strong media acidification, etc.
Table 3. Key “fermentome”
genes
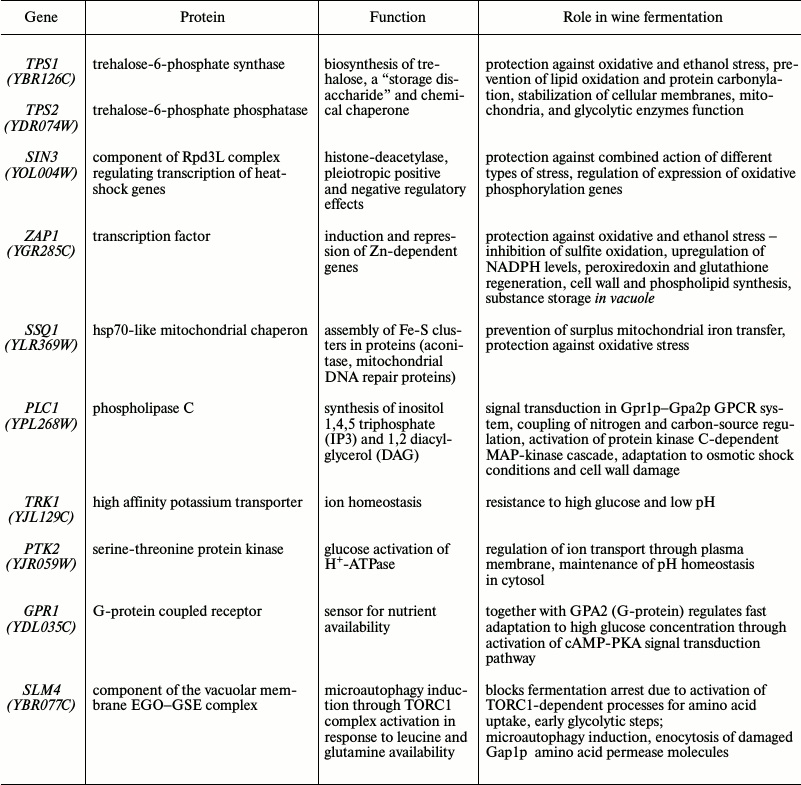
Eighteen “fermentome” genes were related to vacuolar function (components of vacuolar H+-ATPase complex and other proteins). Vacuoles are important for many cellular processes, such as proteolysis and protein recycling, calcium and phosphorus homeostasis, detoxification of heavy metal ions, osmoregulation, amino acid storage, and membrane protein transport. It is not surprising that inactivation of these functions, especially of the V-ATPase as the main ion transporter between the vacuole and cytoplasm, calamitously affects the resistance of yeast strains to stressful winemaking conditions.
Epigenetic control of fermentation efficiency. Sluggish or arrested fermentations are widespread winemaking problems, leading to wine spoilage, oxidation, and contamination with extraneous microflora. With this outcome, yeasts go into “latent state”, fermentation retardation results in reduction of ethanol concentration, and increase in residual sugar forms conditions for microbial spoilage. The main reasons for arrested fermentations include nitrogen shortage, inappropriate temperature regimen, and low-quality raw materials with excess sugar content [85].
However, recently yet another mechanism leading to arrested fermentation was identified; it relates to epigenetic regulation of glucose repression in yeast and its modulation under the control of a specific chemical signaling system between yeast and bacterial cells [86, 87]. The basis of this effect is the ability to overcome glucose repression of genes responsible for utilization of non-fermented carbon sources under the control of an unusual epigenetic mechanism. The authors explain this mechanism by the existence of a nonamyloid form of a yeast prion named as [GAR+] (resistant to Glucose-Associated Repression) [86]. Formation of this prion is mediated by interaction between the plasma membrane ATPase Pma1 with Std1 transcription factor and Hsp70 heat shock protein. An important consequence of [GAR+] induction is a 40-fold decrease in transcription of high capacity sugar transporter HXT3, leading to reduction in ethanol fermentation [86]. The adaptive role of [GAR+] as a mechanism enabling yeast to simultaneously utilize a broad range of carbon sources was proved by its widespread occurrence in natural and wine S. cerevisiae strains, as well as phylogenetically distant representatives of other yeast clades [88]. For winemaking practice, another aspect of [GAR+] formation is important, the mediating symbiotic relations between yeast and bacteria inhabiting a particular ecological niche. It was shown that bacteria from phylogenetically diverse species can generate a yet unidentified chemical signal that significantly enhances the frequency of [GAR+] formation in neighboring yeast cells [87].
Of course, [GAR+] strains cannot use the “MAC” strategy, but due to enlargement of their metabolic repertoire they can survive longer during growth on mixed carbon substrates. The benefits for bacteria are obvious – the concentration of deleterious ethanol is reduced and the concentration of easily accessible sugars is increased.
How should one tackle these effects in winemaking practice? One way to reduce [GAR+] induction frequency by microbes is just to add sulfite to inhibit bacterial growth [89]. Other ways to control bacterial contamination of wine must to detect strains capable of [GAR+] induction is use of starter strains with low frequency of [GAR+] induction. Such strains may be created through transgenesis establishing a “species” barrier for [GAR+] induction through expression of heterologous Pma1 and Std1 from related yeast species [86].
Transcriptome analysis. Advances in yeast genomics have created the premises for intensive use of microchips and RNAseq technology for analysis of gene expression in yeast.
In one of the first studies, significant alterations in the expression of more than 2000 genes were detected during adaption of wine yeast strain EC118 to changing physiological and biochemical conditions of wine fermentation. Genes involved in many metabolic and signaling pathways underwent strictly coordinated regulation, including genes associated with wine fermentation, genes involved in nitrogen catabolism, and others [90, 91]. Under conditions of low-temperature fermentation, genes important to improve wine quality (cold shock genes) involved in cell cycle progression and cell proliferation are upregulated. These and other transcriptional changes correlated with increased cell viability, improved ethanol tolerance, and increased production of short chain fatty acids and esters [92]. Significant interstrain differences in the expression patterns of about 30% of genes responsible for aroma formation were detected in three yeast strains during wine fermentation at normal (28°C) and low (12°C) temperatures. Notably, altered expression was observed for many genes important for winemaking, those encoding proteins of amino acid transport and metabolism, acyl acetyltransferases, decarboxylases, aldehyde dehydrogenases, and alcohol dehydrogenases [93]. This and many other studies, in particular for the analysis of gene expression in wine yeast strains depending on the availability of nitrogen sources [94, 95], clearly show that variations in gene expression are closely associated with phenotypic variation of wine strains.
The most significant changes in wine yeast transcriptome occur during the transition to the stationary phase [96]. Thus, 223 genes dramatically induced at different stages of fermentation have been allocated to a specific group of “fermentation stress response (FSR)” genes [91], and more than 60% of these genes are novel, with previously undescribed functions.
Thus, in-depth understanding of the global dynamics of gene expression in wine yeast strains seems to be crucial for establishing the functions of previously uncharacterized yeast genes, for improving the technology of wine fermentation, and for finding target genes for subsequent strain improvement programs.
Metabolic engineering of wine yeast. Saccharomyces yeasts are easily subjected to various genetic manipulations, and many genetic engineering techniques developed for laboratory yeast strains can be used to modify the genome of wine strains to improve their winemaking characteristics.
These characteristics include genetic stability, high efficiency of fermentation, sulfite and ethanol tolerance, high osmotolerance, resistance to drying, high level of glycerol production, low level of ethyl carbamate production, etc. [23].
Examples of successful application of genetic engineering approaches to create superior wine yeast strains are numerous.
Thus, through constitutive overexpression of the GPD1 and FPS1 genes encoding glycerol-3-phosphate dehydrogenase and glycerol channel, respectively, it was possible to increase 2-4-fold the production level of extracellular glycerol in the recombinant wine yeast strains [97]. Increasing the glycerol to ethanol ratio is important for the production of low alcohol wines and to improve the “body” of the wine [98].
The speed of fermentation of Palomino grape by wine yeast strains can be substantially improved through the expression of the FSY1 gene encoding fructose transporter, which leads to a significant decrease in the amount of residual sugar in the must [99].
The transgenic VIN13 wine yeast strain that improves the flavor of Sauvignon wines was constructed by constitutive expression of the gene encoding E. coli carbon sulfurylase [100]. Two transgenic yeast strains are approved for use in USA, Canada, and Moldova. The ML01 strain is capable of malolactic fermentation (MLF), which usually takes place under the action of the lactic acid bacterium Oenococcus oeni. These bacteria are quite whimsical, and therefore the effectiveness of MLF is quite variable, thus affecting wine quality. The ML01 strain contains two foreign genes – the mae1 gene encoding malate transporter from Schizosaccharomyces pombe and the O. oeni mleA gene encoding the MLF enzyme [101].
Strain 522 was created to reduce the risk of ethylcarbamate formation, which is a potential carcinogen. The strain contains the DUR1,2 genes for carbamate metabolic enzyme under the control of the strong constitutive PGK1 gene promoter [102], does not contain foreign DNA (only yeast genes), and thus is not subject to regulations and restrictions typical for transgenic strains (it is considered a “cis-genic” strain).
Despite the obvious benefits of genetically modified (GM) yeast strains in winemaking, they are not widespread and are generally rejected by the international community [110]. This is largely due to the peculiarities of existing legislation on the use of genetically modified organisms (GMOs) in food in different countries.
BIOLOGY AND BIOTECHNOLOGY OF SHERRY YEAST
Sherry is a wine produced under a film of sherry yeasts cultivated on the surface of dry wine, fortified to a strength of 16.5% vol. in vessels filled not to their full capacity, or by the bottom fermentation method [105]. Due to the prolonged biological aging, this wine acquires unique bouquet and taste. Techniques for biological aging of sherry wine are traditional for several European countries (Hungary, France, Spain, and Italy) and are used to produce world-famous brands like yellow wines from the Jura Region (France), Vernaccia di Oristano (Sardinia, Italy), dry Tokay (Hungary), different types of Jerez (Spain and, in particular the “Sherry triangle”) [106]. Methods of producing biologically aged wines differ from country to country. In Sardinia, at the end of fermentation the wine is placed into oak and chestnut butts completed up to 9/10 of the volume, where biological aging takes place under a layer of sherry yeasts, followed by oxidative aging. In France (Jura Region), the wine fermentation is first carried out in separate tanks, after which the wine undergoes biological aging that involves yeast starter cultures. In Spain, a different technology is used, the one that combines the “sobretablas” and “soleras” systems. Sobretablas is the first phase that comprises must fermentation in 500-600-liter oak barrels to produce a young wine, which is then fortified to 15-16% alcohol. In the second phase the oak butts are used, which are stacked on top of each other to form what is known as a scale and contain wine at different fermentation stages aged under a film of sherry yeast. The bottom row is called the “solera” and contains the oldest wine. Above the “solera” row the 1st, 2nd, and 3rd criaderas are placed containing successively younger wines. The wine for sale is taken from the “solera” row, and it is replaced with an equal volume of wine from the row above (1/3 of the total volume). This occurs successively up through the rows until the turn comes to the young wine at the top row [106].
Thus, wines are mixed to guarantee a final product with consistent organoleptic properties within a certain row. Reserve sherry wines are some of the most precious, and the cost of one bottle can amount to tens of thousands of dollars.
In Russia and then in the USSR, work on microbiology of sherry wines was started in 1909 by A. M. Frolov-Bagreev and was continued by N. F. Saenko at the Magarach enochemical laboratory. The sherry yeasts were selected based on their tolerance to alcohol and intensity of aldehyde production, which produced strains that quickly form a film on the wine at 16.5% vol. alcohol. In his experiments, N. F. Saenko also identified the optimum concentrations of ethanol, sulfite, iron, residual sugar, and temperature and pH values that are important for sherry fermentation and formation of a stable flor film. By combining selected races of yeasts with optimal winemaking techniques, an accelerated technology of producing biologically aged sherry wines was developed at the “Magarach” Research Institute of Viticulture and Winemaking (Yalta). From this point on, local races of sherry yeasts characterized by high tolerance to alcohol and high biochemical activity have been isolated in various wine regions of the former Soviet Union (Armenia, Uzbekistan, and Dagestan) and used in the production of local sherry-like wines.
Technological peculiarities of biological aging. Wine fermentation and biological aging are two fundamentally different processes that occur in a different biochemical environment using strains with different biochemical and physiological characteristics.
First, the location of the sherry and wine yeast cells in the vessel with wine material is different. In the process of wine fermentation, the yeasts are spread evenly throughout the whole volume, as a suspension, while during biological aging of the wine the yeast cells form multicellular aggregates that rise to the surface of the wine to form a biofilm (vellum, flor). This biofilm can form both immediately after the end of the fermentation and at subsequent stages.
To grow, the yeasts require carbon and nitrogen sources, microelements and vitamins, in which the wine must is rich, containing high concentrations of glucose and fructose (from 150 to 260 g/liter) and sufficient concentrations of nitrogenous substances (from 50 up to 400 mg/liter).
During biological aging, glucose and fructose are almost completely absent and are replaced by ethanol and byproducts – glycerol, organic acids, higher alcohols, and their acetates. Ethanol concentrations reach 16%, and those of glycerol come up to 6-10 g/liter.
Deficient glucose and fructose levels in the presence of oxygen cause switching of sherry yeast metabolism from fermentative to oxidative (diauxic growth) using nonfermentable carbon sources [107].
Sufficient content of nitrogen sources is important not only to support the growth and metabolism of the yeast, but also as a factor determining the completeness and the speed of the fermentation [108]. During biological aging, proline becomes the main source of nitrogen [124].
Aerobic growth on available carbon and nitrogen sources results in significant changes in the overall composition of the wine and its organoleptic properties. One of the main reactions of biological aging of wine is oxidation of ethanol to acetaldehyde occurring under the influence of alcohol dehydrogenase.
For the development of characteristic sherry flavor, the acetaldehyde content should be at the level of 200-350 mg/dm3. During biological aging, its content may amount to 1000 mg/dm3, while the amount of acetyls, diethylacetal in particular, can come up to 50-60 mg/dm3. The influence of acetaldehyde is mainly manifested in the appearance of fruity notes in the aroma of the wine materials.
During aging, the content of glycerol, volatile acids (especially acetate), malic acid, and amino acids in sherry wines goes significantly down (from 9-7 to 3-2 g/dm3). At the same time, under the sherry flor film some organic acids (oxalic, fumaric, glycolic, glutaric, etc.) are produced, which were not present in the original wine material.
In the process of sherry yeast oxidative metabolism, the major flavor-building substances are produced including: aliphatic acetals (1,1-diethoxyethane); cyclic acetals (dioxanes and dioxolanes), which are formed from glycerol and acetaldehyde; ketones (diacetyl and acetoin) that enrich the sherry aroma with milky cheese shades [111].
Special attention is paid to research on ketones of the furan series, sotolon, which is responsible for the manifestation of nutty range in the aroma and flavor of the wine. Research data [112] showed that sotolon is formed by aldol condensation between acetaldehyde and α-oxobutyric acid by threonine degradation. It is thought that this process can take place only under the influence of sherry yeasts. Lactones are other components specific for sherry obtained under a film of yeast.
Genetic diversity and origin of sherry yeast. The yeast used to produce biologically aged wines (wines aged under a film of yeast known as flor) should be alcohol-tolerant and able to proliferate quickly to form a film on the surface of wine with an alcohol content of 16.5% vol. They should also be able to actively oxidize ethanol to aldehydes and acetals under conditions of low pH and high sulfite content. Unlike other film yeasts (non-Saccharomyces), these yeasts cannot spoil wine by deep oxidation of alcohols to water and carbon dioxide. The high alcohol content in wine ensures the purity of fermentation, inhibiting the development of acetic acid bacteria and foreign yeast species.
Thus, stringent conditions of biological aging are an important factor for the genetic selection of the sherry yeast that forms them into different groups.
The film yeasts isolated from the flor on the surface of sherry wines have long been considered specific S. cerevisiae races due to their unique physiology; however, until recently we had no idea about the extent of their relationship with each other.
The first attempt to classify sherry yeasts based on their ability to ferment various sugars (galactose, dextrose, lactose, maltose, melibiose, raffinose, sucrose) led to the isolation of four races: Saccharomyces cerevisiae var. beticus, Saccharomyces cerevisiae var. cheresiensis, Saccharomyces cerevisiae var. montuliensis, and Saccharomyces cerevisiae var. rouxii [113]. These strains were also found among the flor yeast from the Jura Region [114]. Subsequent studies revealed that, despite the high degree of mtDNA polymorphism, all Spanish sherry yeast are characterized by a 24-bp deletion in the ITS1 region, suggesting their close phylogenetic kinship [115]. French yeasts have a different ITS1 allele [114].
The variability of the sherry yeast mtDNA pattern and significant aneuploidy of certain strain groups is believed to be a consequence of the mutagenic effect of high ethanol and acetaldehyde concentrations [116, 117].
Genotyping of sherry yeasts from different geographical regions using microsatellite markers has revealed that, oddly enough, most of their representatives from Spain, Italy, France, and Hungary belong to the same genetic group [118], which breaks easily into subclusters specific for strains of each country. Some of the strains of the Jura group differ in the size of the coding sequence of the FLO11 gene and the presence of a 111-bp deletion in the promoter. The strains from the Jura Region with this deletion form a thicker, more solid film; the film produced by strains with an extended FLO11 ORF and promoter of the “wild-type” is thinner [118]. Interestingly, some of the Hungarian strains are heterozygous for this deletion. Suggesting a common origin of the sherry strains and their migration within Europe, the phylogeny data thus illustrate their evolutionary success in adaptation to an ecological niche mediated by the presence of certain genetic characteristics (FLO11 polymorphism, etc.).
Mechanisms of film formation and physiology of sherry yeast. The ability to float on the surface of the wine and form a velum is a key property of flor yeast, tightly linked to a high cell surface hydrophobicity and, in turn, with specific cell wall mannoprotein content and lipid content modification [134]. A classical genetic analysis concluded that the FLO11 gene is key for velum formation [121], and its role in yeast biofilm formation has since been confirmed by other studies [122-124]. FLO11 encodes one of the yeast surface adhesins and is a GPI-anchored protein with extended central part rich in Ser and Thr residues. Deletion of FLO11 prevents velum formation, but not all S. cerevisiae strains that possess the gene can sherry wine production. Comparisons of the FLO11 gene sequence have revealed numerous differences in promoter and coding regions [118, 124, 125].
Some flor yeast strains contain a specific 111-bp deletion in the FLO11 gene promoter [124], as well as other deletions and rearrangements. The 111-bp deletion was shown later to be responsible for inactivation of ICR1 ncRNA coded by the (–) strand of the FLO11 gene promoter, stimulating in this way Flo11p synthesis [126]. Variation in the FLO11 coding sequence size (from 3 to 6 kb) is determined mainly by the number of repeats within the central domain. Initially, it was shown that flor strains encode extended Flo11p variants with enhanced hydrophobicity. However, subsequent studies showed that the central domain of Flo11 is unstable under nonselective conditions, and that other repeats and direct FLO11 expression level are also important for film formation [127].
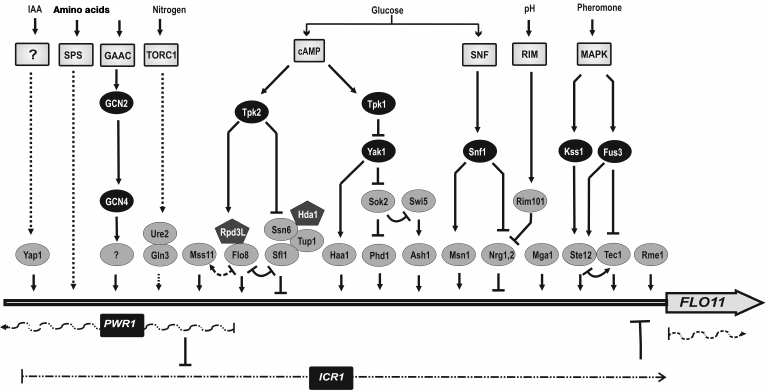
Mechanisms of FLO11 gene regulation are extremely complex. The FLO11 promoter is one of the longest in the yeast genome, containing cis-elements for four known positive regulators and nine negative ones [128]. The majority of these regulatory elements are targets for the MAPK-dependent kinase pathway, the cAMP dependent cascade, and the Gcn4p-dependent pathway [129]. Regulation of FLO11 is also controlled by pH-dependent pathways and chromatin remodeling complexes [130]. The scheme presented in Fig. 3 of course is not intended to clarify how all these numerous factors interact (detailed description is beyond current review), but to show the complex network and interplay of different pathways and mechanisms controlled by more than 50 genes involved in FLO11 regulation.
Another important yet insufficiently studied factor affecting sherry wine aging is the degree of Flo11p glycosylation. Velum formation is lost in mutants with Flo11p glycosylation defects [131].
Other genes besides FLO11, e.g. HSP12 and NRG1 [132, 133] also contribute to velum formation. HSP12 deletion mutants loose film-forming capability, while mutants deleted in the Nrg1 C-terminal domain activate this process [133]. Film formation is indirectly affected by Btn2 protein that encodes one of the components of intracellular protein sorting [134].
Fig. 3. Levels of FLO11 promoter regulation. Complex character of FLO11 regulation under the control of molecular genetic and epigenetic mechanisms. Arrows indicate positive regulation and inhibition is shown by bars. Different stimuli and corresponding signaling pathways targeting FLO11 are indicated at the top (IAA – indole acetic acid; ? – unknown pathway). Light-gray squares – signal transduction pathway, black ovals – kinases involved in regulatory cascades, black pentagons – chromatin-remodeling complexes. Transcription factors targeting the FLO11 promoter are shown in gray ovals. The input of the different transcription factors is only shown schematically and does not correspond to the positions of known binding sites. Regulatory ncRNA and mRNA affecting FLO11 expression are shown as dashed and waved lines. Reproduced with modifications from [128].
A distinct role in film formation was ascribed to Ccw7/Pir2/Hsp150 cell wall glycoprotein. The size of this protein varies between sherry (87 kDa) and lab (117 kDa) strains [135] due to deletion of distinct repeat-containing motifs. Besides flotation capability, ethanol and acetaldehyde resistance are other important properties of flor yeast. Resistance to ethanol and acetaldehyde stress is mediated to a large extent by heat-shock proteins – Hsp12, Hsp82, Hsp104, Hsp26 [136]. It is important to note differential expression of aldehyde dehydrogenases (ALD2/3, ALD4, ALD6) in wine and flor yeast [137]. In flor yeast, levels of these enzymes are higher, providing their ability to use ethanol as the main carbon source. Resistance to enhanced aldehyde levels is determined by general mechanisms mediated by the Msn2/4 and Hsf1 transcription factors [137].
Aerobic ethanol oxidation leads to increased ROS generation in mitochondria, potentially leading to enhanced mtDNA damage [116]. To protect from this stress, flor yeasts have elaborated an efficient antioxidant system based also on elevated expression of SOD1 superoxide dismutase [138]. Forced overexpression of antioxidant genes (HSP12, SOD1, SOD2) in sherry yeast leads to several positive effects – it accelerates film formation, elevates intracellular glutathione concentration, reduces peroxy lipid concentration, enhances survival, etc., clearly showing the importance of antioxidant defense for physiological and winemaking properties [139].
Genomics, proteomics, metabolomics of sherry yeast. Comparative genome hybridization (CGH) was the first instrument applied for understanding the mechanisms of adaptation of wine and sherry yeast to specific winemaking environments [131]. CGH is very precise in CNV detection. Initial karyotype data obtained with pulsed-field electrophoresis assumed a high level of aneuploidy for flor yeasts. However, subsequent studies showed that they are true diploids. The CGH data for six strains confirmed this observation. Extended amplified or deleted regions compared to reference S288C strain were not found [118]. Moderate gene loss was detected in subtelomeric regions, and only three genes – FRE2, MCH2, and YKL222C – were subjected to amplification [118]. MCH2 codes for monocarboxylic acid transporter and is important for yeast survival at the second stage of ethanol fermentation [141]. The role of another two genes is not clear. It is obvious that in-depth analysis of genetic features of sherry yeast requires application of novel methods of whole-genome sequencing, and postgenomic methods (transcriptomics, proteomics, etc.) should be fruitful.
In recent years, proteomics and metabolomics have been applied to the study of flor yeast metabolism and their responses to environmental conditions. Proteome analysis of mitochondrion-localized proteins of flor yeast strains [142] has revealed groups of differentially expressed proteins involved in carbohydrate oxidative metabolism, biofilm formation, apoptosis, and responses to stresses typical of biological aging, e.g. ethanol, acetaldehyde, and reactive oxygen species. In addition, proteins associated with nonfermentable carbon uptake, glyoxylate, and the TCA cycle, cellular respiration, and inositol metabolism are more expressed in yeast growing under biofilms than under fermentative conditions [143]. For example, the level of Ino1p, which participates in inositol biosynthesis, was five-fold elevated under biofilm conditions. The presence of proteins involved in cell wall biosynthesis and protein glycosylation, which are important for cell–cell adhesion and hence for biofilm formation, was also reported in this study.
Through the combination of proteomics and innovative metabolomics techniques that were aimed at quantifying minor volatile compounds under exhaustively controlled biofilm conditions, 33 proteins were shown to be directly involved in the metabolism of glycerol, ethanol, and 17 aroma compounds [144].
Production of specific aroma compounds has an important impact on organoleptic properties of sherry wines. Biological wine aging is accompanied by acetaldehyde accumulation and reduction of volatile acid and glycerol content [145-148]. Among 35 aroma compounds quantified [147], acetaldehyde, 1,1-diethoxyethane, 2,3-butanediol, isoamyl alcohols, ethyl and isoamyl acetates, butanoic acid, 2,3-methylbutanoic acids, and 4-butyrolactone were defined as the most active odorant compounds. The concentrations of all these compounds showed significant variation after biological aging compared to control non-aerated wine material [146, 149].
These data were confirmed and extended in a study devoted to effects of periodic aeration on the content of acetaldehyde and its derivatives, higher alcohols, their acetic acid esters [147]. In contrast, the acids of 4, 5, and 6 carbon atoms showed lower concentrations in aged wines, and levels close to zero were obtained for 2-butanol, various lactones, 3-ethoxy-1-propanol, and neral.
The link between intracellular proteins and metabolites excreted by yeast that are strongly related to sensorial properties constitutes new and useful information for development of future fermentative winemaking technologies.
Dynamics of wine microflora in the production of sherry. The microflora dynamics during biological aging of wine is determined by the peculiarities of its different stages. The indigenous microflora present on grapes is reduced to just a few yeast species present in the composition of the sherry film by increasing the alcohol content, reducing the pH level, and adding sulfur. However, the indigenous microflora has a significant impact on the final organoleptic characteristics of wine. The effect of this flora is ambiguous – on one hand, the presence of Pichia yeast strains is undesirable because they produce ethyl acetate, and strains from the genera Hanseniaspora and Kloeckera have been found to produce high, undesirable levels of acetate, acetaldehyde, and acetoin. On the other hand, these strains can synthesize various enzymes – proteases, lipases, esterases, and pectinases having favorable effect on wine aroma. Other species from the genera Kluyveromyces, Torulaspora, and Saccharomyces have a significant influence on the final aroma of wine due to their ability to convert monoterpene alcohols during fermentation [52].
The ratio between different races of sherry yeasts along with lactic acid bacteria in the composition of the flor films on the surface of sherry musts produced in Spain, France, Italy, and Hungary is quite different and largely determines the difference in wine organoleptic characteristics. In-depth understanding of the impact of the presence of certain yeast species on the final organoleptic characteristics of the wine will help create mixed starter cultures of yeasts of a certain composition that would yield wine with desired characteristics of bouquet and aroma [46].
The rapid progress in yeast genomics have provided deep insights into the population structure and evolutionary history of Saccharomyces. The provided evidence of yeast strain adaptation showed that wine yeast use various genetic mechanisms to adapt to winemaking environments. Expansion of the dataset of sequenced strains form wine niches and other anthropic environments provides better understanding of the evolutionary history of Saccharomyces and relative contribution of different mechanisms of genome plasticity. The availability of a higher number of genome sequences might facilitate the identification of allelic variants and other divergent regions responsible for winemaking properties. For example, flor and wine strains belonging to closely related groups with contrasting lifestyles might constitute a relevant model to identify divergent regions that might explain the adaptation to these niches. New important insights concerning the origin of cellular functions at the basis of yeast strain response and adaptation to winemaking environments were obtained using genetic, proteomic, transcriptomic approaches.
Thus, the use of modern genomic and postgenomic instruments has significantly enhanced our knowledge about the nature of molecular differences underpinning the phenotypic diversity of wine strains, interrelation between molecular genetic data, and specific industrial characteristics. Without doubt, these achievements will promote the development of novel strategies for targeted selection and creation of novel strains using classical and modern approaches for improvement of winemaking conditions.
Acknowledgements
We thank Nikolay Ravin for valuable advice and comments during the preparation of this article.
This work was supported by the Russian Science Foundation (project No. 16-16-00109).
REFERENCES
1.McGovern, P. E., Zhang, J., Tang, J., Zhang, Z.,
Hall, G. R., Moreau, R. A., Nunez, A., Butrym, E. D., Richards, M. P.,
Wang, C.-S., Cheng, G., Zhao, Z., and Wang, C. (2004) Fermented
beverages of pre- and proto-historic China, Proc. Natl. Acad. Sci.
USA, 101, 17593-17598.
2.McGovern, P. E., Glusker, D. L., Exner, L. J., and
Voigt, M. M. (1996) Neolithic resinated wine, Nature,
381, 480-481.
3.Cavalieri, D., McGovern, P. E., Hartl, D. L.,
Mortimer, R., and Polsinelli, M. (2003) Evidence for S.
cerevisiae fermentation in ancient wine, J. Mol. Evol.,
57, 226-232.
4.Griffith, M. P. (2004) Ancient wine: the search for
the origins of viniculture, Econ. Bot., 58, 488-488.
5.Pasteur, L. (1858) Nouveaux faits concernant
l’histoire de la fermentation alcoolique, Comptes Rendus
Chim., 47, 1011-1013.
6.Cubillos, F. A., Vasquez, C., Faugeron, S., Ganga,
A., and Martinez, C. (2009) Self-fertilization is the main sexual
reproduction mechanism in native wine yeast populations, FEMS
Microbiol. Ecol., 67, 162-170.
7.Novo, M., Bigey, F., Beyne, E., Galeote, V.,
Gavory, F., Mallet, S., Cambon, B., Legras, J.-L., Wincker, P.,
Casaregola, S., and Dequin, S. (2009) Eukaryote-to-eukaryote gene
transfer events revealed by the genome sequence of the wine yeast
Saccharomyces cerevisiae EC1118, Proc. Natl. Acad. Sci.
USA, 106, 16333-16338.
8.Borneman, A. R., Desany, B. A., Riches, D.,
Affourtit, J. P., Forgan, A. H., Pretorius, I. S., Egholm, M., and
Chambers, P. J. (2011) Whole-genome comparison reveals novel genetic
elements that characterize the genome of industrial strains of
Saccharomyces cerevisiae, PLoS Genet., 7,
e1001287.
9.Magwene, P. M. (2014) Revisiting Mortimer’s
genome-renewal hypothesis: heterozygosity, homothallism, and the
potential for adaptation in yeast, Adv. Exp. Med. Biol.,
781, 37-48.
10.Stefanini, I., Dapporto, L., Legras, J.-L.,
Calabretta, A., Di Paola, M., De Filippo, C., Viola, R., Capretti, P.,
Polsinelli, M., Turillazzi, S., and Cavalieri, D. (2012) Role of social
wasps in Saccharomyces cerevisiae ecology and evolution,
Proc. Natl. Acad. Sci. USA, 109, 13398-13403.
11.Wang, Q. M., Liu, W. Q., Liti, G., Wang, S. A.,
and Bai, F. Y. (2012) Surprisingly diverged populations of
Saccharomyces cerevisiae in natural environments remote from
human activity, Mol. Ecol., 21, 5404-5417.
12.Legras, J. L., Merdinoglu, D., Cornuet, J.-M.,
and Karst, F. (2007) Bread, beer and wine: Saccharomyces
cerevisiae diversity reflects human history, Mol. Ecol.,
16, 2091-2102.
13.Schacherer, J., Shapiro, J. A., Ruderfer, D. M.,
and Kruglyak, L. (2009) Comprehensive polymorphism survey elucidates
population structure of Saccharomyces cerevisiae, Nature,
458, 342-345.
14.Liti, G., Carter, D. M., Moses, A. M., Warringer,
J., Parts, L., James, S. A., Davey, R. P., Roberts, I. N., Burt, A.,
Koufopanou, V., Tsai, I. J., Bergman, C. M., Bensasson, D.,
O’Kelly, M. J., Van Oudenaarden, A., Barton, D. B., Bailes, E.,
Nguyen, A. N., Jones, M., Quail, M. A., Goodhead, I., Sims, S., Smith,
F., Blomberg, A., Durbin, R., and Louis, E. J. (2009) Population
genomics of domestic and wild yeasts, Nature, 458,
337-341.
15.Borneman, A. R., Forgan, A. H., Kolouchova, R.,
Fraser, J. A., and Schmidt, S. A. (2016) Whole genome comparison
reveals high levels of inbreeding and strain redundancy across the
spectrum of commercial wine strains of Saccharomyces cerevisiae,
G3 Genes Genom. Genet., 6, 957-971.
16.Fay, J. C., and Benavides, J. A. (2005) Evidence
for domesticated and wild populations of Saccharomyces
cerevisiae, PLoS Genet., 1, e5.
17.Sicard, D., and Legras, J. L. (2011) Bread, beer
and wine: yeast domestication in the Saccharomyces sensu stricto
complex, Compt. Rendus Biol., 334, 229-236.
18.Cordente, A. G., Curtin, C. D., Varela, C., and
Pretorius, I. S. (2012) Flavor-active wine yeasts, Appl. Microbiol.
Biotechnol., 96, 601-618.
19.Styger, G., Prior, B., and Bauer, F. F. (2011)
Wine flavor and aroma, J. Industr. Microbiol. Biotechnol.,
38, 1145-1159.
20.Marsit, S., and Dequin, S. (2015) Diversity and
adaptive evolution of Saccharomyces wine yeast: a review,
FEMS Yeast Res., 15, No. 7; pii: fov067.
21.Sipiczki, M. (2010) Diversity, variability and
fast adaptive evolution of the wine yeast (Saccharomyces
cerevisiae) genome – a review, Ann. Microbiol.,
61, 85-93.
22.Strope, P. K., Skelly, D. A., Kozmin, S. G.,
Mahadevan, G., Stone, E. A., Magwene, P. M., Dietrich, F. S., and
McCusker, J.-H. (2015) The 100-genomes strains, an S. cerevisiae
resource that illuminates its natural phenotypic and genotypic
variation and emergence as an opportunistic pathogen, Gen. Res.,
25, 762-774.
23.Pretorius, I. S. (2000) Tailoring wine yeast for
the new millennium: novel approaches to the ancient art of winemaking,
Yeast, 16, 675-729.
24.Barrio, E., Gonzalez, S. S., Arias, A., Belloch,
C., and Querol, A. (2006) Molecular mechanisms involved in the adaptive
evolution of industrial yeasts, in Yeasts in Food and Beverages,
Springer, Berlin-Heidelberg, pp. 153-174.
25.Blondin, B., Dequin, S., Querol, A., and Legras,
J.-L. (2009) Genome of Saccharomyces cerevisiae and related
yeasts, in Biology of Microorganisms on Grapes, in Must and
in Wine, Springer, Berlin-Heidelberg, pp. 361-378.
26.Dequin, S., and Casaregola, S. (2011) The genomes
of fermentative Saccharomyces, Comptes Rendus Biol.,
334, 687-693.
27.Louis, E. J., Fischer, G., James, S. A., Roberts,
I. N., and Oliver, S. G. (2000) Chromosomal evolution in
Saccharomyces, Nature, 405, 451-454.
28.Borneman, A. R., Forgan, A. H., Pretorius, I. S.,
and Chambers, P. J. (2008) Comparative genome analysis of a
Saccharomyces cerevisiae wine strain, FEMS Yeast Res.,
8, 1185-1195.
29.Goto-Yamamoto, N., Kitano, K., Shiki, K.,
Yoshida, Y., Suzuki, T., Iwata, T., Yamane, Y., and Hara, S. (1998)
SSU1-R, a sulfite resistance gene of wine yeast, is an allele of SSU1
with a different upstream sequence, J. Ferment. Bioeng.,
86, 427-433.
30.Yuasa, N., Nakagawa, Y., Hayakawa, M., and
Iimura, Y. (2004) Distribution of the sulfite resistance gene SSU1-R
and the variation in its promoter region in wine yeasts, J. Biosci.
Bioeng., 98, 394-397.
31.Zimmer, A., Durand, C., Loira, N., Durrens, P.,
Sherman, D. J., and Marullo, P. (2014) QTL dissection of lag phase in
wine fermentation reveals a new translocation responsible for
Saccharomyces cerevisiae adaptation to sulfite, PLoS One,
9, e86298.
32.Perez-Ortin, J. E., Querol, A., Puig, S., and
Barrio, E. (2002) Molecular characterization of a chromosomal
rearrangement involved in the adaptive evolution of yeast strains,
Gen. Res., 12, 1533-1539.
33.Warringer, J., Zorgo, E., Cubillos, F. A., Zia,
A., Gjuvsland, A., Simpson, J. T., Forsmark, A., Durbin, R., Omholt, S.
W., Louis, E. J., Liti, G., Moses, A., and Blomberg, A. (2011) Trait
variation in yeast is defined by population history, PLoS
Genet., 7, e1002111.
34.Almeida, P., Barbosa, R., Zalar, P., Imanishi,
Y., Shimizu, K., Turchetti, B., Legras, J. L., Serra, M., Dequin, S.,
Couloux, A., Guy, J., Bensasson, D., Goncalves, P., and Sampaio, J. P.
(2015) A population genomics insight into the Mediterranean origins of
wine yeast domestication, Mol. Ecol., 24, 5412-5427.
35.Liu, J., Martin-Yken, H., Bigey, F., Dequin, S.,
Francois, J. M., and Capp, J. P. (2015) Natural yeast promoter variants
reveal epistasis in the generation of transcriptional-mediated noise
and its potential benefit in stressful conditions, Gen. Biol.
Evol., 7, 969-984.
36.Carreto, L., Eiriz, M. F., Gomes, A. C., Pereira,
P. M., Schuller, D., and Santos, M. A. S. (2008) Comparative genomics
of wild type yeast strains unveils important genome diversity, BMC
Genom., 9, 524.
37.Tanghe, A., Van Dijck, P., Dumortier, F.,
Teunissen, A., Hohmann, S., and Thevelein, J. M. (2002) Aquaporin
expression correlates with freeze tolerance in baker’s yeast, and
overexpression improves freeze tolerance in industrial strains,
Appl. Environ. Microbiol., 68, 5981-5989.
38.Bonhivers, M., Carbrey, J. M., Gould, S. J., and
Agre, P. (1998) Aquaporins in Saccharomyces: genetic and
functional distinctions between laboratory and wild-type strains, J.
Biol. Chem., 273, 27565-27572.
39.Laize, V., Tacnet, F., Ripoche, P., and Hohmann,
S. (2000) Polymorphism of Saccharomyces cerevisiae aquaporins,
Yeast, 16, 897-903.
40.Will, J. L., Kim, H. S., Clarke, J., Painter, J.
C., Fay, J. C., and Gasch, A. P. (2010) Incipient balancing selection
through adaptive loss of aquaporins in natural Saccharomyces
cerevisiae populations, PLoS Genet., 6, e1000893.
41.Galeote, V., Bigey, F., Beyne, E., Novo, M.,
Legras, J. L., Casaregola, S., and Dequin, S. (2011) Amplification of a
Zygosaccharomyces bailii DNA segment in wine yeast genomes by
extrachromosomal circular DNA formation, PLoS One, 6,
e17872.
42.Marsit, S., Mena, A., Bigey, F., Sauvage, F. X.,
Couloux, A., Guy, J., Legras, J. L., Barrio, E., Dequin, S., and
Galeote, V. (2015) Evolutionary advantage conferred by an
eukaryote-to-eukaryote gene transfer event in wine yeasts, Mol.
Biol. Evol., 32, 1695-1707.
43.Galeote, V., Novo, M., Salema-Oom, M., Brion, C.,
Valerio, C., Goncalves, P., and Dequin, S. (2010) FSY1, a horizontally
transferred gene in the Saccharomyces cerevisiae EC1118 wine
yeast strain, encodes a high-affinity fructose/H+ symporter,
Microbiology, 156, 3754-3761.
44.Wenger, J. W., Schwartz, K., and Sherlock, G.
(2010) Bulk segregant analysis by high-throughput sequencing reveals a
novel xylose utilization gene from Saccharomyces cerevisiae,
PLoS Genet., 6, e1000942.
45.Damon, C., Vallon, L., Zimmermann, S., Haider, M.
Z., Galeote, V., Dequin, S., Luis, P., Fraissinet-Tachet, L., and
Marmeisse, R. (2011) A novel fungal family of oligopeptide transporters
identified by functional metatranscriptomics of soil eukaryotes,
ISME J., 5, 1871-1880.
46.Ibstedt, S., Stenberg, S., Bages, S., Gjuvsland,
A. B., Salinas, F., Kourtchenko, O., Samy, J. K. A., Blomberg, A.,
Omholt, S. W., Liti, G., Beltran, G., and Warringer, J. (2015)
Concerted evolution of life stage performances signals recent selection
on yeast nitrogen use, Mol. Biol. Evol., 32, 153-161.
47.Brown, S. L., Stockdale, V. J., Pettolino, F.,
Pocock, K. F., De Barros Lopes, M., Williams, P. J., Bacic, A.,
Fincher, G. B., Hoj, P. B., and Waters, E. J. (2007) Reducing haziness
in white wine by overexpression of Saccharomyces cerevisiae
genes YOL155c and YDR055w, Appl. Microbiol. Biotechnol.,
73, 1363-1376.
48.Dunn, B., Paulish, T., Stanbery, A., Piotrowski,
J., Koniges, G., Kroll, E., Louis, E.-J., Liti, G., Sherlock, G., and
Rosenzweig, F. (2013) Recurrent rearrangement during adaptive evolution
in an interspecific yeast hybrid suggests a model for rapid
introgression, PLoS Genet., 9, e1003366.
49.Dunn, B., and Sherlock, G. (2008) Reconstruction
of the genome origins and evolution of the hybrid lager yeast
Saccharomyces pastorianus, Gen. Res., 18,
1610-1623.
50.Libkind, D., Hittinger, C. T., Valerio, E.,
Goncalves, C., Dover, J., Johnston, M., Goncalves, P., and Sampaio, J.
P. (2011) Microbe domestication and the identification of the wild
genetic stock of lager-brewing yeast, Proc. Natl. Acad. Sci.
USA, 108, 14539-14544.
51.Bradbury, J. E., Richards, K. D., Niederer, H.
A., Lee, S. A., Rod Dunbar, P., and Gardner, R. C. (2006) A homozygous
diploid subset of commercial wine yeast strains, Antonie Van
Leeuwenhoek, 89, 27-38.
52.Borneman, A. R., Desany, B. A., Riches, D.,
Affourtit, J. P., Forgan, A. H., Pretorius, I. S., Egholm, M., and
Chambers, P. J. (2012) The genome sequence of the wine yeast VIN7
reveals an allotriploid hybrid genome with Saccharomyces
cerevisiae and Saccharomyces kudriavzevii origins, FEMS
Yeast Res., 12, 88-96.
53.Naumova, E. S., Naumov, G. I., Masneuf-Pomarede,
I., Aigle, M., and Dubourdieu, D. (2005) Molecular genetic study of
introgression between Saccharomyces bayanus and S.
cerevisiae, Yeast (Chichester, England), 22,
1099-1115.
54.Almeida, P., Goncalves, C., Teixeira, S.,
Libkind, D., Bontrager, M., Masneuf-Pomarede, I., Albertin, W.,
Durrens, P., Sherman, D. J., Marullo, P., Hittinger, C. T., Goncalves,
P., and Sampaio, J. P. (2014) A Gondwanan imprint on global diversity
and domestication of wine and cider yeast Saccharomyces uvarum,
Nat. Commun., 5, 4044.
55.Dunn, B., Richter, C., Kvitek, D. J., Pugh, T.,
and Sherlock, G. (2012) Analysis of the Saccharomyces cerevisiae
pan-genome reveals a pool of copy number variants distributed in
diverse yeast strains from differing industrial environments, Gen.
Res., 22, 908-924.
56.Morales, L., and Dujon, B. (2012) Evolutionary
role of interspecies hybridization and genetic exchanges in yeasts,
Microbiol. Mol. Biol. Rev., 76, 721-739.
57.Belloch, C., Perez-Torrado, R., Gonzalez, S. S.,
Perez-Ortin, J. E., Garcia-Martinez, J., Querol, A., and Barrio, E.
(2009) Chimeric genomes of natural hybrids of Saccharomyces
cerevisiae and Saccharomyces kudriavzevii, Appl. Environ.
Microbiol., 75, 2534-2544.
58.Gonzalez, S. S., Gallo, L., Climent, M. D.,
Barrio, E., and Querol, A. (2007) Enological characterization of
natural hybrids from Saccharomyces cerevisiae and S.
kudriavzevii, Int. J. Food Microbiol., 116,
11-18.
59.Gangl, H., Batusic, M., Tscheik, G.,
Tiefenbrunner, W., Hack, C., and Lopandic, K. (2009) Exceptional
fermentation characteristics of natural hybrids from Saccharomyces
cerevisiae and S. kudriavzevii, New Biotechnol.,
25, 244-251.
60.Swiegers, J. H., Kievit, R. L., Siebert, T.,
Lattey, K. A., Bramley, B. R., Francis, I. L., King, E. S., and
Pretorius, I. S. (2009) The influence of yeast on the aroma of
Sauvignon Blanc wine, Food Microbiol., 26, 204-211.
61.Replansky, T., Koufopanou, V., Greig, D., and
Bell, G. (2008) Saccharomyces sensu stricto as a model system
for evolution and ecology, Trends Ecol. Evol., 23,
494-501.
62.Querol, A., and Bond, U. (2009) The complex and
dynamic genomes of industrial yeasts, FEMS Microbiol. Lett.,
293, 1-10.
63.Erny, C., Raoult, P., Alais, A., Butterlin, G.,
Delobel, P., Matei-Radoi, F., Casaregola, S., and Legras, J. L. (2012)
Ecological success of a GROUP of Saccharomyces
cerevisiae/Saccharomyces kudriavzevii hybrids in the Northern
European wine-making environment, Appl. Environ. Microbiol.,
78, 3256-3265.
64.Bellon, J. R., Eglinton, J. M., Siebert, T. E.,
Pollnitz, A. P., Rose, L., De Barros Lopes, M., and Chambers, P. J.
(2011) Newly generated interspecific wine yeast hybrids introduce
flavor and aroma diversity to wines, Appl. Microbiol.
Biotechnol., 91, 603-612.
65.Bellon, J. R., Yang, F., Day, M. P., Inglis, D.
L., and Chambers, P. J. (2015) Designing and creating
Saccharomyces interspecific hybrids for improved, industry
relevant, phenotypes, Appl. Microbiol. Biotechnol., 99,
8597-8609.
66.Konig, H., Unden, G., and Frohlich, J. (2009)
Biology of microorganisms on grapes, in Must and in Wine,
Springer Science and Business Media.
67.Barata, A., Malfeito-Ferreira, M., and Loureiro,
V. (2012) The microbial ecology of wine grape berries, Int. J. Food
Microbiol., 153, 243-259.
68.Fleet, G. H. (2003) Yeast interactions and wine
flavor, Int. J. Food Microbiol., 86, 11-22.
69.Barata, A., Malfeito-Ferreira, M., and Loureiro,
V. (2012) Changes in sour rotten grape berry microbiota during ripening
and wine fermentation, Int. J. Food Microbiol., 154,
152-161.
70.Steel, C. C., Blackman, J. W., and Schmidtke, L.
M. (2013) Grapevine bunch rots: impacts on wine composition, quality,
and potential procedures for the removal of wine faults, J.
Agricult. Food Chem., 61, 5189-5206.
71.Fleet, G. H. (2007) Yeasts in foods and
beverages: impact on product quality and safety, Curr. Opin.
Biotechnol., 18, 170-175.
72.Hagman, A., Sall, T., Compagno, C., and Piskur,
J. (2013) Yeast “make-accumulate-consume” life strategy
evolved as a multi-step process that predates the whole genome
duplication, PLoS One, 8, e68734.
73.Dashko, S., Zhou, N., Compagno, C., and Piskur,
J. (2014) Why, when, and how did yeast evolve alcoholic fermentation?
FEMS Yeast Res., 14, 826-832.
74.Hagman, A., and Piskur, J. (2015) A study on the
fundamental mechanism and the evolutionary driving forces behind
aerobic fermentation in yeast, PLoS One, 10,
e0116942.
75.Thomson, J. M., Gaucher, E. A., Burgan, M. F., De
Kee, D. W., Li, T., Aris, J. P., and Benner, S. A. (2005) Resurrecting
ancestral alcohol dehydrogenases from yeast, Nat. Genet.,
37, 630-635.
76.Schmitt, M. J., and Breinig, F. (2006) Yeast
viral killer toxins: lethality and self-protection, Nat. Rev.
Microbiol., 4, 212-221.
77.Schmitt, M. J., Sao-Jose, C., and Santos, M. A.
(2009) Phages of yeast and bacteria, in Biology of Microorganisms on
Grapes, in Must and in Wine, Springer, Berlin-Heidelberg,
pp. 89-109.
78.Liu, G.-L., Chi, Z., Wang, G.-Y., Wang, Z.-P.,
Li, Y., and Chi, Z.-M. (2015) Yeast killer toxins, molecular mechanisms
of their action and their applications, Crit. Rev. Biotechnol.,
35, 222-234.
79.Hyma, K. E., Saerens, S. M., Verstrepen, K. J.,
and Fay, J. C. (2011) Divergence in wine characteristics produced by
wild and domesticated strains of Saccharomyces cerevisiae,
FEMS Yeast Res., 11, 540-551.
80.Mackay, T. F. C., Stone, E. A., and Ayroles, J.
F. (2009) The genetics of quantitative traits: challenges and
prospects, Nat. Rev. Genet., 10, 565-577.
81.Steinmetz, L. M., Sinha, H., Richards, D. R.,
Spiegelman, J. I., Oefner, P. J., McCusker, J. H., and Davis, R. W.
(2002) Dissecting the architecture of a quantitative trait locus in
yeast, Nature, 416, 326-330.
82.Cubillos, F. A., Billi, E., Zorgo, E., Parts, L.,
Fargier, P., Omholt, S., Blomberg, A., Warringer, J., Louis, E. J., and
Liti, G. (2011) Assessing the complex architecture of polygenic traits
in diverged yeast populations, Mol. Ecol., 20,
1401-1413.
83.Salinas, F., Cubillos, F. A., Soto, D., Garcia,
V., Bergstrom, A., Warringer, J., Ganga, M. A., Louis, E. J., Liti, G.,
and Martinez, C. (2012) The genetic basis of natural variation in
oenological traits in Saccharomyces cerevisiae, PLoS One,
7, e49640.
84.Walker, M. E., Nguyen, T. D., Liccioli, T.,
Schmid, F., Kalatzis, N., Sundstrom, J. F., Gardner, J. M., and
Jiranek, V. (2014) Genome-wide identification of the fermentome; genes
required for successful and timely completion of wine-like fermentation
by Saccharomyces cerevisiae, BMC Genom., 15,
552.
85.Alexandre, H., and Charpentier, C. (1998)
Biochemical aspects of stuck and sluggish fermentation in grape must,
J. Industr. Microbiol. Biotechnol., 20, 20-27.
86.Brown, J. C. S., and Lindquist, S. (2009) A
heritable switch in carbon source utilization driven by an unusual
yeast prion, Genes Dev., 23, 2320-2332.
87.Jarosz, D. F., Brown, J. C. S., Walker, G. A.,
Datta, M. S., Ung, W. L., Lancaster, A. K., Rotem, A., Chang, A.,
Newby, G. A., Weitz, D. A., Bisson, L. F., and Lindquist, S. (2014)
Cross-kingdom chemical communication drives a heritable, mutually
beneficial prion-based transformation of metabolism, Cell,
158, 1083-1093.
88.Jarosz, D. F., Lancaster, A. K., Brown, J. C. S.,
and Lindquist, S. (2014) An evolutionarily conserved prion-like element
converts wild fungi from metabolic specialists to generalists,
Cell, 158, 1072-1082.
89.Walker, G. A., Hjelmeland, A., Bokulich, N. A.,
Mills, D. A., Ebeler, S. E., and Bisson, L. F. (2016) Impact of the
[GAR+] prion on fermentation and bacterial community
composition with Saccharomyces cerevisiae UCD932, Am. J.
Enol. Viticult., 67, 296-307.
90.Rossignol, T., Dulau, L., Julien, A., and
Blondin, B. (2003) Genome-wide monitoring of wine yeast gene expression
during alcoholic fermentation, Yeast (Chichester, England),
20, 1369-1385.
91.Gasch, A. P., Spellman, P. T., Kao, C. M.,
Carmel-Harel, O., Eisen, M. B., Storz, G., Botstein, D., and Brown, P.
O. (2000) Genomic expression programs in the response of yeast cells to
environmental changes, Mol. Biol. Cell, 11,
4241-4257.
92.Beltran, G., Novo, M., Leberre, V., Sokol, S.,
Labourdette, D., Guillamon, J.-M., Mas, A., Francois, J., and Rozes, N.
(2006) Integration of transcriptomic and metabolic analyses for
understanding the global responses of low-temperature winemaking
fermentations, FEMS Yeast Res., 6, 1167-1183.
93.Gamero, A., Belloch, C., Ibanez, C., and Querol,
A. (2014) Molecular analysis of the genes involved in aroma synthesis
in the species S. cerevisiae, S. kudriavzevii and S.
bayanus var. uvarum in winemaking conditions, PLoS One,
9, e97626.
94.Backhus, L. E., DeRisi, J., and Bisson, L. F.
(2001) Functional genomic analysis of a commercial wine strain of
Saccharomyces cerevisiae under differing nitrogen conditions,
FEMS Yeast Res., 1, 111-125.
95.Barbosa, C., Garcia-Martinez, J., Perez-Ortin, J.
E., and Mendes-Ferreira, A. (2015) Comparative transcriptomic analysis
reveals similarities and dissimilarities in Saccharomyces
cerevisiae wine strains response to nitrogen availability, PLoS
One, 10, e0122709.
96.Marks, V. D., Ho Sui, S. J., Erasmus, D., Van Der
Merwe, G. K., Brumm, J., Wasserman, W. W., Bryan, J., and Van Vuuren,
H. J. J. (2008) Dynamics of the yeast transcriptome during wine
fermentation reveals a novel fermentation stress response, FEMS
Yeast Res., 8, 35-52.
97.Kutyna, D. R., Varela, C., Henschke, P. A.,
Chambers, P. J., and Stanley, G. A. (2010) Microbiological approaches
to lowering ethanol concentration in wine, Trends Food Sci.
Technol., 21, 293-302.
98.Remize, F., Cambon, B., Barnavon, L., and Dequin,
S. (2003) Glycerol formation during wine fermentation is mainly linked
to Gpd1p and is only partially controlled by the HOG pathway, Yeast
(Chichester, England), 20, 1243-1253.
99.Rodrigues de Sousa, H., Spencer-Martins, I., and
Goncalves, P. (2004) Differential regulation by glucose and fructose of
a gene encoding a specific fructose/H+ symporter in
Saccharomyces sensu stricto yeasts, Yeast (Chichester,
England), 21, 519-530.
100.Swiegers, J. H., Capone, D. L., Pardon, K. H.,
Elsey, G. M., Sefton, M. A., Francis, I. L., and Pretorius, I. S.
(2007) Engineering volatile thiol release in Saccharomyces
cerevisiae for improved wine aroma, Yeast
(Chichester, England), 24, 561-574.
101.Husnik, J. I., Volschenk, H., Bauer, J.,
Colavizza, D., Luo, Z., and Van Vuuren, H. J. J. (2006) Metabolic
engineering of malolactic wine yeast, Metab. Eng., 8,
315-323.
102.Coulon, J., Husnik, J. I., Inglis, D. L., van
der Merwe, G. K., Lonvaud, A., Erasmus, D. J., and van Vuuren, H. J. J.
(2006) Metabolic engineering of Saccharomyces cerevisiae to
minimize the production of ethyl carbamate in wine, Am. J. Enol.
Viticult., 57, 113-124.
103.Borneman, A. R., Schmidt, S. A., and Pretorius,
I. S. (2013) At the cutting-edge of grape and wine biotechnology,
Trends Genet., 29, 263-271.
104.Cebollero, E., Gonzalez-Ramos, D., Tabera, L.,
and Gonzalez, R. (2007) Transgenic wine yeast technology comes of age:
is it time for transgenic wine? Biotechnol. Lett., 29,
191-200.
105.Valuiko, G. G., and Kosura, V. T. (2000)
Handbook on Winemaking [in Russian], 2nd Edn., Tavrida,
Simferopol.
106.Stevenson, T. (2011) The Sotheby’s
Wine Encyclopedia, 5th Edn., Dorling Kindersley, London-New
York.
107.Zara, S., Gross, M. K., Zara, G., Budroni, M.,
and Bakalinsky, A. T. (2010) Ethanol-independent biofilm formation by a
flor wine yeast strain of Saccharomyces cerevisiae, Appl.
Environ. Microbiol., 76, 4089-4091.
108.Mauricio, J. C., Ortega, J. M., and Salmon, J.
M. (1995) Sugar uptake by three strains of Saccharomyces
cerevisiae during alcoholic fermentation at different initial
ammoniacal nitrogen concentrations, Acta Horticult., 388,
197-202.
109.Botella, M. A., Perez-Rodriguez, L., Domecq,
B., and Valpuesta, V. (1990) Amino acid content of fino and oloroso
sherry wines, Am. J. Enol. Viticult., 41, 12-15.
110.Mauricio, J. C., and Ortega, J. M. (1997)
Nitrogen compounds in wine during its biological aging by two flor film
yeasts: an approach to accelerated biological aging of dry sherry-type
wines, Biotechnol. Bioeng., 53, 159-167.
111.Bakker, J., and Clarke, R. J. (2011) Wine
Flavor Chemistry, 2nd Edn., Wiley-Blackwell.
112.Thuy, P. T., Elisabeth, G., Pascal, S., and
Claudine, C. (1995) Optimal conditions for the formation of sotolon
from alpha-ketobutyric acid in the French “Vin Jaune”,
J. Agricult. Food Chem., 43, 2616-2619.
113.Martinez, P., Perez Rodriguez, L., and Benitez,
T. (1997) Evolution of flor yeast population during the biological
aging of fino sherry wine, Am. J. Enol. Viticult., 48,
160-168.
114.Charpentier, C., Colin, A., Alais, A., and
Legras, J. L. (2009) French Jura flor yeasts: genotype and
technological diversity, Antonie Van Leeuwenhoek, 95,
263-273.
115.Esteve-Zarzoso, B., Peris-Toran, M. J.,
Garcia-Maiquez, E., Uruburu, F., and Querol, A. (2001) Yeast population
dynamics during the fermentation and biological aging of sherry wines,
Appl. Environ. Microbiol., 67, 2056-2061.
116.Castrejon, F., Codon, A. C., Cubero, B.,
Benitez, T., Castrejon, F., Codon, A. C., and Benitez, T. (2002)
Acetaldehyde and ethanol are responsible for mitochondrial DNA (mtDNA)
restriction fragment length polymorphism (RFLP) in flor yeasts,
System. Appl. Microbiol., 25, 462-467.
117.Infante, J. J., Dombek, K. M., Rebordinos, L.,
Cantoral, J. M., and Young, E. T. (2003) Genome-wide ampli cations
caused by chromosomal rearrangements play a major role in the adaptive
evolution of natural yeast, Genetics, 165, 1745-1759.
118.Legras, J.-L., Erny, C., and Charpentier, C.
(2014) Population structure and comparative genome hybridization of
European flor yeast reveal a unique group of Saccharomyces
cerevisiae strains with few gene duplications in their genome,
PLoS One, 9, e108089.
119.Martinez, P., Perez Rodriguez, L., and Benitez,
T. (1997) Velum formation by flor yeasts isolated from sherry wine,
Am. J. Enol. Viticult., 48, 55-62.
120.Alexandre, H., Blanchet, S., and Charpentier,
C. (2000) Identification of a 49-kDa hydrophobic cell wall mannoprotein
present in velum yeast which may be implicated in velum formation,
FEMS Microbiol. Lett., 185, 147-150.
121.Reynolds, T. B., and Fink, G. R. (2001)
Bakers’ yeast, a model for fungal biofilm formation,
Science, 291, 878-881.
122.Zara, S., Bakalinsky, A. T., Zara, G., Pirino,
G., Demontis, M. A., and Budroni, M. (2005) FLO11-based model
for air-liquid interfacial biofilm formation by Saccharomyces
cerevisiae, Appl. Environ. Microbiol., 71,
2934-2939.
123.Ishigami, M., Nakagawa, Y., Hayakama, M., and
Iimura, Y. (2006) FLO11 is the primary factor in flor formation caused
by cell surface hydrophobicity in wild-type flor yeast, Biosci.
Biotechnol. Biochem., 70, 660-666.
124.Fidalgo, M., Barrales, R. R., Ibeas, J. I., and
Jimenez, J. (2006) Adaptive evolution by mutations in the FLO11
gene, Proc. Natl. Acad. Sci. USA, 103, 11228-11233.
125.Fidalgo, M., Barrales, R. R., and Jimenez, J.
(2008) Coding repeat instability in the FLO11 gene of
Saccharomyces yeasts, Yeast, 25, 879-889.
126.Bumgarner, S. L., Dowell, R. D., Grisafi, P.,
Gifford, D. K., and Fink, G. R. (2009) Toggle involving
cis-interfering noncoding RNAs controls variegated gene
expression in yeast, Proc. Natl. Acad. Sci. USA, 106,
18321-18326.
127.Zara, G., Zara, S., Pinna, C., Marceddu, S.,
and Budroni, M. (2009) FLO11 gene length and transcriptional
level affect biofilm-forming ability of wild flor strains of
Saccharomyces cerevisiae, Microbiology, 155,
3838-3846.
128.Bruckner, S., and Mosch, H.-U. (2012) Choosing
the right lifestyle: adhesion and development in Saccharomyces
cerevisiae, FEMS Microbiol. Rev., 36, 25-58.
129.Barrales, R. R., Jimenez, J., and Ibeas, J. I.
(2008) Identification of novel activation mechanisms for FLO11
regulation in Saccharomyces cerevisiae, Genetics,
178, 145-156.
130.Barrales, R. R., Korber, P., Jimenez, J., and
Ibeas, J. I. (2012) Chromatin modulation at the FLO11 promoter of
Saccharomyces cerevisiae by HDAC and Swi/Snf complexes,
Genetics, 191, 791-803.
131.Meem, M. H., and Cullen, P. J. (2012) The
impact of protein glycosylation on Flo11-dependent adherence in
Saccharomyces cerevisiae, FEMS Yeast Res., 12,
809-818.
132.Ishigami, M., Nakagawa, Y., Hayakawa, M., and
Iimura, Y. (2004) FLO11 is essential for flor formation caused
by the C-terminal deletion of Nrg1 in Saccharomyces cerevisiae,
FEMS Microbiol. Lett., 237, 425-430.
133.Zara, S., Antonio Farris, G., Budroni, M., and
Bakalinsky, A. T. (2002) HSP12 is essential for biofilm formation by a
Sardinian wine strain of S. cerevisiae, Yeast, 19,
269-276.
134.Espinazo-Romeu, M., Cantoral, J. M., Matallana,
E., and Aranda, A. (2008) Btn2p is involved in ethanol tolerance and
biofilm formation in flor yeast, FEMS Yeast Res., 8,
1127-1136.
135.Kovacs, M., Stuparevic, I., Mrsa, V., and
Maraz, A. (2008) Characterization of Ccw7p cell wall proteins and the
encoding genes of Saccharomyces cerevisiae wine yeast strains:
relevance for flor formation, FEMS Yeast Res., 8,
1115-2116.
136.Aranda, A., Querol, A., and Del Olmo, M. l.
(2002) Correlation between acetaldehyde and ethanol resistance and
expression of HSP genes in yeast strains isolated during the
biological aging of sherry wines, Arch. Microbiol., 177,
304-312.
137.Aranda, A., and Del Olmo, M. L. (2003) Response
to acetaldehyde stress in the yeast Saccharomyces cerevisiae
involves a strain-dependent regulation of several ALD genes and is
mediated by the general stress response pathway, Yeast
(Chichester, England), 20, 747-759.
138.Alexandre, H. (2013) Flor yeasts of
Saccharomyces cerevisiae – their ecology, genetics and
metabolism, Int. J. Food Microbiol., 167, 269-275.
139.Fierro-Risco, J., Rincon, A. M., Benitez, T.,
and Codon, A. C. (2013) Overexpression of stress-related genes enhances
cell viability and velum formation in Sherry wine yeasts, Appl.
Microbiol. Biotechnol., 97, 6867-6881.
140.Dunn, B., Levine, R. P., and Sherlock, G.
(2005) Microarray karyotyping of commercial wine yeast strains reveals
shared, as well as unique, genomic signatures, BMC Genom.,
6, 53.
141.Novo, M., Mangado, A., Quiros, M., Morales, P.,
Salvado, Z., and Gonzalez, R. (2013) Genome-wide study of the
adaptation of Saccharomyces cerevisiae to the early stages of
wine fermentation, PLoS One, 8, e74086.
142.Moreno-Garcia, J., Garcia-Martinez, T., Moreno,
J., Millan, M. C., and Mauricio, J. C. (2014) A proteomic and
metabolomic approach for understanding the role of the flor yeast
mitochondria in the velum formation, Int. J. Food Microbiol.,
172, 21-29.
143.Moreno-Garcia, J., Garcia-Martinez, T., Moreno,
J., and Mauricio, J. C. (2015) Proteins involved in flor yeast carbon
metabolism under biofilm formation conditions, Food Microbiol.,
46, 25-33.
144.Moreno-Garcia, J., Garcia-Martinez, T., Millan,
M. C., Mauricio, J. C., and Moreno, J. (2015) Proteins involved in wine
aroma compounds metabolism by a Saccharomyces cerevisiae
flor-velum yeast strain grown in two conditions, Food
Microbiol., 51, 1-9.
145.Mauricio, J. C., Valero, E., Millan, C., and
Ortega, J. M. (2001) Changes in nitrogen compounds in must and wine
during fermentation and biological aging by flor yeasts, J.
Agricult. Food Chem., 49, 3310-3315.
146.Begona Cortes, M., Moreno, J. J., Zea, L.,
Moyano, L., and Medina, M. (1999) Response of the aroma fraction in
Sherry wines subjected to accelerated biological aging, J. Agricult.
Food Chem., 47, 3297-3302.
147.Munoz, D., Peinado, R. A., Medina, M., and
Moreno, J. (2007) Biological aging of sherry wines under periodic and
controlled microaerations with Saccharomyces cerevisiae var.
capensis: effect on odorant series, Food Chem.,
100, 1188-1195.
148.Moreno-Garcia, J., Raposo, R. M., and Moreno,
J. (2013) Biological aging status characterization of Sherry type wines
using statistical and oenological criteria, Food Res. Int.,
54, 285-292.
149.Zea, L., Moyano, L., Moreno, J., Cortes, B.,
and Medina, M. (2001) Discrimination of the aroma fraction of Sherry
wines obtained by oxidative and biological ageing, Food Chem.,
75, 79-84.