Glutathione-Dependent Formaldehyde Dehydrogenase Homolog from Bacillus subtilis Strain R5 is a Propanol-Preferring Alcohol Dehydrogenase
Raza Ashraf1, Naeem Rashid1*, Saadia Basheer1, Iram Aziz1, and Muhammad Akhtar2
1School of Biological Sciences, University of the Punjab, Quaid-e-Azam Campus, Lahore 54590, Pakistan; E-mail: naeem.ff.sbs@pu.edu.pk, naeemrashid37@hotmail.com2School of Biological Sciences, University of Southampton, Southampton SO16 7PX, UK
* To whom correspondence should be addressed.
Received July 2, 2016; Revision received August 26, 2016
Genome search of Bacillus subtilis revealed the presence of an open reading frame annotated as glutathione-dependent formaldehyde dehydrogenase/alcohol dehydrogenase. The open reading frame consists of 1137 nucleotides corresponding to a polypeptide of 378 amino acids. To examine whether the encoded protein is glutathione-dependent formaldehyde dehydrogenase or alcohol dehydrogenase, we cloned and characterized the gene product. Enzyme activity assays revealed that the enzyme exhibits a metal ion-dependent alcohol dehydrogenase activity but no glutathione-dependent formaldehyde dehydrogenase or aldehyde dismutase activity. Although the protein is of mesophilic origin, optimal temperature for the enzyme activity is 60°C. Thermostability analysis by circular dichroism spectroscopy revealed that the protein is stable up to 60°C. Presence or absence of metal ions in the reaction mixture did not affect the enzyme activity. However, metal ions were necessary at the time of protein production and folding. There was a marked difference in the enzyme activity and CD spectra of the proteins produced in the presence and absence of metal ions. The experimental results obtained in this study demonstrate that the enzyme is a bona-fide alcohol dehydrogenase and not a glutathione-dependent formaldehyde dehydrogenase.
KEY WORDS: Bacillus subtilis, formaldehyde dehydrogenase, alcohol dehydrogenase, metal dependent, protein folding, circular dichroismDOI: 10.1134/S0006297917010023
Abbreviations: ADH, alcohol dehydrogenase; CD, circular dichroism; GSH, glutathione; GSH-FDH, glutathione-dependent formaldehyde dehydrogenase.
Alcohol dehydrogenases (ADH) with lowest enzyme commission number (EC
1.1.1.1) catalyze the interconversion of primary or secondary alcohols
and corresponding aldehydes or ketones, respectively [1]. They play an important role in a broad range of
physiological processes including alcohol metabolism and cell defense
against exogenous alcohol and aldehyde stress [2].
These enzymes are widely distributed in all the three domains of life
[3]. ADHs can be divided into three superfamilies
based on the cofactor specificity. They are: (i) NAD(P)+
dependent; (ii) pyrroloquinoline quinine, heme, or coenzyme F420
dependent; (iii) FAD dependent [4]. The
NAD+ or NADP+ dependent ADHs are further divided
into three classes: (a) short-chain, (b) medium chain, usually
zinc-containing, and (c) iron-activated long-chain alcohol
dehydrogenases [3, 5, 6]. Medium-chain zinc-containing ADHs are further
divided into various classes, and among them class I, classical liver
alcohol dehydrogenase [7], and class III, the
ancestral glutathione-dependent formaldehyde dehydrogenase (GSH-FDH)
[8] are the most studied [9].
GSH-FDH oxidizes formaldehyde to formate, which is further oxidized to
carbon dioxide or incorporated into the one-carbon pool [10]. In contrast to several ADHs, GSH-FDHs do not
exhibit substantial activity towards short aliphatic alcohols [11]. Instead, they catalyze the NAD-dependent
oxidation of long-chain alcohols in addition to their normal
formaldehyde dehydrogenase activity [12]. It is
thought that in the course of evolution tandem gene duplication of
GSH-FDH led to the new form of class I enzymes [13] with an inclination towards ethanol related
activities. Further duplication gave rise to different classes and
different isozymes within these classes [14]. Each
class has specific conserved substrate-binding residues, which are
helpful in the recognition of the particular class [15, 16].
Genome search of Bacillus subtilis strain SZMC 6179J, available at http://www.ncbi.nlm.nih.gov/nucleotide/1012899741?report=genbank&log$=nuclalign&blast_rank=1&RID=NJPWZC4B01R, revealed the presence of an open reading frame (AMS48275) annotated as GSH-FDH/ADH but not yet characterized. In this study, we cloned and characterized GSH-FDH/ADH homolog (GSH-FDH/ADHR5) from B. subtilis strain R5 exhibiting a 100% identity with AMS48275. Biochemical characterization revealed that the enzyme is not a glutathione-dependent formaldehyde dehydrogenase, but a metal ion-activated alcohol dehydrogenase.
MATERIALS AND METHODS
Bacterial strains and growth conditions. Bacillus subtilis strain R5 [17] was cultivated in LB (tryptone 1%, yeast extract 0.5%, NaCl 0.5%, pH 7.0) medium at 30°C. All chemicals used were of analytical grade and purchased from either Sigma-Aldrich Company (USA), Fluka Chemical Corporation (USA), or Thermo Fisher Scientific Inc. (USA). DNA polymerase, restriction enzymes, cloning vectors, and DNA purification kits were purchased from Thermo Fisher Scientific Inc. or Novagen-Merck Millipore (USA). Gene-specific primers were commercially synthesized by Gene Link, Inc. (USA). Escherichia coli DH5α strain was used for cloning and plasmid preparation, and the E. coli CodonPlus(DE3)-RIL (Stratagene, USA) strain was used as a host for the heterologous expression of the gene.
Construction of recombinant plasmids. The gene encoding GSH-FDH/ADHR5 was amplified by polymerase chain reaction using a set of forward (5′-AGATATACATATGCACCACCACCACCACCACAAGGCAGTAACGTATCAAGGC-3′) and reverse (5′-AAAGCTGAGCTCAGTATGCCCTCCTGTAAATCTC-3′) primers and genomic DNA of B. subtilis R5 as template. A His-tag (shown in bold) and NdeI site (underlined sequence) were added in the forward primer. The reverse primer contained a Bpu1102I site (underlined). The PCR amplified gene product was cloned in pTZ57R/T cloning vector. The resulting plasmid was named pTZ-FDH/ADH. The GSH-FDH/ADHR5 gene was excised from pTZ-FDH/ADH plasmid using NdeI and Bpu1102I and ligated at the corresponding position in GroEL-pETDuet (prepared in our laboratory for another study). The resulting plasmid was named GroEL-pETDuet-FDH/ADH.
Production of recombinant GSH-FDH/ADHR5 in E. coli. Plasmid GroEL-pETDuet-FDH/ADH was used to transform E. coli BL21-CodonPlus(DE3)-RIL cells. Gene expression was induced by the addition of isopropyl-β-D-1-thiogalactopyranoside (IPTG) at a final concentration of 0.07 mM. After induction, the cells were grown overnight at 17°C and harvested by centrifugation. Cells, almost 1.5 g wet weight from 1 liter of culture, were suspended in 50 ml of 50 mM Tris-HCl buffer, pH 8.0, and disrupted by sonication. Soluble and insoluble fractions were separated by centrifugation at 12,000g for 30 min. Proteins were analyzed by denaturing polyacrylamide gel electrophoresis (SDS-PAGE). Supernatant, containing GSH-FDH/ADHR5 in the soluble form, was supplemented with 0.5 M NaCl and 5 mM imidazole and applied to a column containing high capacity nickel chelate affinity matrix (Sigma). The bound proteins were eluted with 50 mM Tris-HCl containing 20, 50, 100, 150, 200, and 300 mM imidazole. The fractions containing recombinant GSH-FDH/ADHR5 were pooled and applied to a Superdex 200 10/300 GL (GE Healthcare, USA) gel-filtration chromatography column. The molecular mass of recombinant GSH-FDH/ADHR5 was determined by plotting a standard curve with thyroglobulin (669 kDa), ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), and ovalbumin (44 kDa). Solutions of the standard and sample proteins were prepared in 20 mM Tris-HCl (pH 8.0) containing 150 mM NaCl. Protein concentration was determined spectrophotometrically at every step of purification using Bradford reagent.
Enzyme assays. Enzyme activity was examined spectrophotometrically using a Shimadzu UV-1601 spectrophotometer equipped with a thermoelectric cell. To examine the glutathione-dependent formaldehyde dehydrogenase activity, the assay mixture consisted of either 50 mM Tris-HCl, pH 8.0, or 50 mM glycine NaOH, pH 9.5, 1-50 mM GSH, 2 mM NAD+, 1-20 mM formaldehyde, and 25 µg of purified enzyme in a total volume of 1 ml. The reduction of NAD+ was examined at 37°C by measuring the increase in absorbance at 340 nm.
To examine the alcohol dehydrogenase activity, the reaction mixture consisted of 50 mM glycine-NaOH buffer (pH 9.5), 50 mM of various alcohols, and 0.2 mM NAD+, and 25 µg of purified protein. The reduction reaction mixture consisted of 50 mM sodium phosphate buffer (pH 6.0), 20 mM aldehyde or ketone, 0.2 mM NADH, and 25 µg of purified protein. The routine enzyme assays were conducted at 37°C. One unit of ADH was defined as the amount of enzyme that catalyzes the formation of 1 µmol of aldehyde or ketone per minute.
Formaldehyde dismutase activity was examined at 37°C in a 10 ml reaction mixture containing 20 mM formaldehyde, 100 mM KCl, 50 mM Tris-HCl, pH 7.0, 25 µg of the enzyme under N2 gas, and formation of formate was monitored by pH-stat titration with 5 mM NaOH [18].
Effect of temperature and pH was determined by measuring the enzyme activity at various temperatures (30-70°C) while keeping the pH constant or varying the pH values (4-11) and keeping the temperature unchanged by using ethanol as substrate.
Effect of metal ions on the enzyme activity was analyzed either by the addition of 25 µM of metal ions in the reaction mixture or growth medium during protein production in E. coli. Chloride salts of Cu2+, Fe2+, and Zn2+ were used in the growth medium prepared in deionized water. Control protein was produced without addition of any metal ions in the growth medium prepared in either tap water or deionized water. ADH activity was measured as described above using ethanol as substrate.
For zymography, 50 µg of GSH-FDH/ADHR5 was electrophoresed on native polyacrylamide gel, in duplicate, at room temperature. The gel was cut in two halves. For ADH activity, one half of the gel was incubated, after washing with deionized water, in 50 mM glycine-NaOH buffer (pH 9.5) containing 100 mM ethanol, 1.5 mM NAD+, 0.24 mM nitroblue tetrazolium, and 0.063 mM phenazine methosulfate [19]. For glutathione-dependent formaldehyde dehydrogenase activity, the other half of the gel was incubated with 50 mM Tris-HCl (pH 8.0) containing 500 mM KCl, 10 mM formaldehyde, 1.5 mM NAD+, 0.24 mM nitroblue tetrazolium, and 0.063 mM phenazine methosulfate [12]. Both halves of the gel were incubated on a shaker at room temperature for 30 min.
Steady state enzyme kinetics for oxidation reaction was measured in triplicates in 50 mM glycine-NaOH buffer (pH 9.5) by varying the concentration of the substrate (0.5-30 mM 1-propanol). Steady state enzyme kinetics for reduction reaction was measured in triplicates in 50 mM sodium phosphate buffer (pH 6.0). The kinetic parameters Km, Vmax, kcat, and kcat/Km were calculated using the Michaelis–Menten equation.
Metal ion content and structural stability. The metal ion contents of the recombinant protein were analyzed by inductively coupled plasma-atomic absorption mass spectrometry (ICP-MS) using an Elemental Solaar M6 AA atomic absorption spectrophotometer (Thermo Scientific, USA). The blank solution, 20 mM Tris-HCl buffer (pH 8.0), was used for dilution of standards and samples. Calibration curves were obtained by analysis of the calculated external standards. All solutions were prepared in deionized double distilled water.
Structural stability of the protein was analyzed by circular dichroism (CD) spectroscopy using a Chirascan-plus CD Spectrometer (Applied Photophysics, UK). Recombinant protein was produced in E. coli in the presence or absence of different metal ions. The CD spectra of the protein samples were recorded in 10 mM Tris-HCl, pH 8.0, in the far UV-range of 200-260 nm. Solvent spectra were subtracted from those of the experimental values. Equal amount (0.3 mg/ml) of protein was used in each case, and the CD spectra were measured at 30°C.
Structure prediction and homology modeling. For structure prediction and homology modeling, the amino acid sequence of GSH-FDH/ADHR5 was submitted to NCBI Blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to get the most similar protein whose crystal structure had been determined. Among these proteins, ADH from Thermus ATN1 alcohol dehydrogenase (PDB code: 4CPD) [20] was found most similar to GSH-FDH/ADHR5 with identity of 34%, and therefore it was used as a template for structure prediction and homology modeling studies. For construction of the predicted model, the sequence was submitted to SWISS-MODEL ExPASy (https://swissmodel.expasy.org/). Analysis and comparison of the predicted model were carried out by using PyMOL provided at https://www.pymol.org/.
Sequence accession number. The sequence of GSH-FDH/ADHR5 is available under accession number LC159199 in the GenBank/EMBL/DDBJ databases.
RESULTS AND DISCUSSION
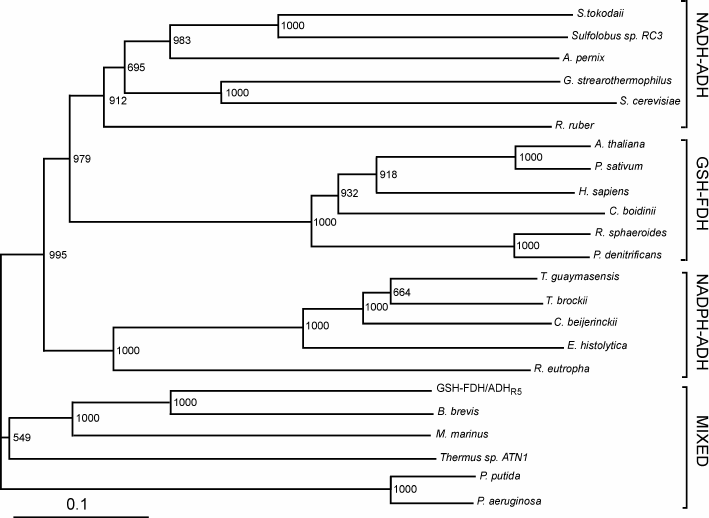
Sequence comparison. The GSH-FDH/ADHR5 gene from B. subtilis strain R5 consisted of 1137 nucleotides encoding a protein of 378 amino acids with theoretical molecular mass of 41,208 Da and isoelectric point of 5.72. Amino acid sequence comparison of the protein displayed the highest identity of 100% with an uncharacterized glutathione-dependent formaldehyde dehydrogenase from B. subtilis strain SZMC 6179J. In an unrooted phylogenetic tree of characterized medium-chain formaldehyde dehydrogenases and alcohol dehydrogenase, constructed by the Neighbor-Joining method, GSH-FDH/ADHR5 clustered neither with NAD(P)-ADH nor GSH-FDH. Rather, it made a mixed group comprising bacterial formaldehyde dehydrogenases and an alcohol dehydrogenase from Thermus sp. ATN1 (Tadh) (Fig. 1). Among these characterized enzymes highest identity, 61%, was found with formaldehyde dehydrogenase from Brevibacillus brevis [21], followed by 43% with glutathione-independent formaldehyde dehydrogenase from Pseudomonas aeruginosa [22], 34% with alcohol dehydrogenase from Thermus sp. ATN1 [20], 31% with formaldehyde dismutase from Pseudomonas putida [18], and 24% with glutathione-dependent formaldehyde dehydrogenase from Arabidopsis thaliana [23]. A glycine rich conserved motif “GXGXXG”, reported to be involved in dinucleotide binding [24], was found (186GCGPVG191) in GSH-FDH/ADHR5. A bifurcation of NAD+ or NADP+ dependence is marked by the presence or absence of Asp223 (human numbering), respectively [25]. Alignment with the human enzyme demonstrated that this Asp is conserved in GSH-FDH/ADHR5 at position 210. Four cysteine residues – Cys97, Cys100, Cys103, and Cys111 – in human GSH-FDH are involved in structural Zn2+ binding. These residues were also conserved in GSH-FDH/ADHR5 as Cys90, Cys93, Cys96, and Cys104. Similarly, Cys46, His67, and Cys174 are involved in catalytic Zn2+ binding in human GSH-FDH. Comparison demonstrated that the first two residues – Cys44 and His66 – are conserved in all the Zn2+-dependent medium-chain dehydrogenases. However, the third residue may be Cys or Asp. In GSH-FDH/ADHR5, these residues were Cys38, His60, and Asp162. Although GSH-FDH/ADHR5 exhibited highest homology with the uncharacterized GSH-FDH from B. subtilis, the four amino acid residues (Thr48, Asp57, Glu59, and Arg115, human numbering) reported to be involved in glutathione binding [25] and conserved in all the characterized GSH-FDHs were missing in GSH-FDH/ADHR5 (Fig. 2). The absence of these amino acid residues created ambiguity whether GDH-FDH/ADHR5 is a bona-fide GSH-FDH or not. A glycyl-aspartyl dipeptide that is involved in maintaining the structural integrity of the N-terminal segment of medium-chain alcohol dehydrogenases [26] was also found, 79GD (Fig. 2).
Fig. 1. Phylogenetic tree of GSH-FDH/ADHR5. The unrooted tree with branch length was constructed using the Neighbor-Joining method. Bootstrap values and segments corresponding to evolutionary distance of 0.1 are shown. Following are the sources of sequences with accession numbers used for the alignment: NADP-dependent alcohol dehydrogenases (NADP-ADH) from Thermoanaerobacter brockii (ADV78851), Thermococcus guaymasensis (ADV18977.1), Clostridium beijerinckii (P25984.2), Entamoeba histolytica (BAN39903), and Ralstonia eutropha (P14940); NAD-dependent alcohol dehydrogenases (NAD-ADH) from Sulfolobus tokodaii (WP_010977990.1), Sulfolobus sp. RC3 (P50381), Aeropyrum pernix (BAA81251), Geobacillus stearothermophilus (P42327.1), Saccharomyces cerevisiae (EWG88693.1), and Rhodococcus ruber (WP_010594839); glutathione-dependent formaldehyde dehydrogenases (GSH-FDH) from Arabidopsis thaliana (CAA57973.1), Pisum sativum (P80572.1), Candida boidinii (BAC16635.1), Homo sapiens (NP_000662), Rhodobacter sphaeroides (L47326.1), and Pseudomonas denitrificans (AAC44551); and mixed group formed by NADH-ADH from Thermus sp. ATN1 (ACD50896.1) and glutathione-independent FDHs from Brevibacillus brevis (AEM59539), Methylobacter marinus (P47734), Pseudomonas putida (P46154.3), and Pseudomonas aeruginosa (WP_061181139).
Fig. 2. Comparison of amino acid sequence of GSH-FDH/ADHR5 with characterized NADH-ADH (S. tokodaii and A. pernix), NADPH-ADH (T. brockii), GSH-FDH (H. sapiens and A. thaliana), and mixed (glutathione-independent FDH from P. putida and NADH-ADH from Thermus sp. ATN1). Comparison was made using the ClustalW program. The accession numbers are given in the legend to Fig. 1. The residues shown with gray background are involved in the glutathione binding. Residues highlighted in black background are involved in catalytic zinc binding. Amino acids shown with black filled circles at the top are the structural zinc binding residues. Residues enclosed in a box are involved in maintaining the structural integrity of the N-terminal segment. The sequence segment in braces corresponds to the dinucleotide-binding region. The aspartate shown with an unfilled circle is expected to be involved in preference of NAD+ over NADP+. Asterisks designate the residues, which are the same in all eight aligned sequences.
Gene expression in E. coli and purification of recombinant GSH-FDH/ADHR5. Production of recombinant GSH-FDH/ADHR5 in E. coli cells harboring GroEL-pETDuet-FDH/ADHR5 was analyzed by SDS-PAGE, and it was found that a high level of recombinant protein was produced in the cells induced with IPTG. The recombinant protein, having a His-tag at the N-terminal, was purified to homogeneity using nickel affinity chromatography. About 5 mg of purified GSH-FDH/ADHR5 was obtained from 1.5 g of E. coli cells (1 liter of culture).
The molecular mass and subunit number of the recombinant protein were determined by gel-filtration chromatography. GSH-FDH/ADHR5 eluted at retention volume of 14 ml, which corresponded to an approximate molecular mass of 164 kDa on the standard curve. Keeping in view the subunit mass of 42 kDa, this result indicated that the recombinant GSH-FDH/ADHR5 existed as a tetramer.
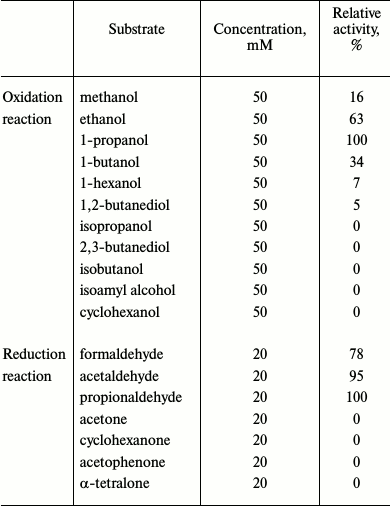
Biochemical characterization. When glutathione-dependent or -independent formaldehyde dehydrogenase activity was examined, we could not detect any activity even when the glutathione and formaldehyde concentrations were varied from 1 to 50 mM and from 1 to 20 mM, respectively. Similarly, no aldehyde dismutase activity could be detected. However, GSH-FDH/ADHR5 exhibited significant alcohol dehydrogenase activity (450 nmol/min per mg) when ethanol was used as the substrate. Zymogram analysis also confirmed the presence of alcohol dehydrogenase activity in GSH-FDH/ADHR5 and no formaldehyde dehydrogenase activity (data not shown). GSH-FDH/ADHR5 exhibited a clear preference for aliphatic primary alcohols and corresponding aldehydes as substrates. In the oxidative direction, highest activity was found when 1-propanol was used as the substrate (Table 1).
Table 1. Substrate specificity of
GSH-FDH/ADHR5
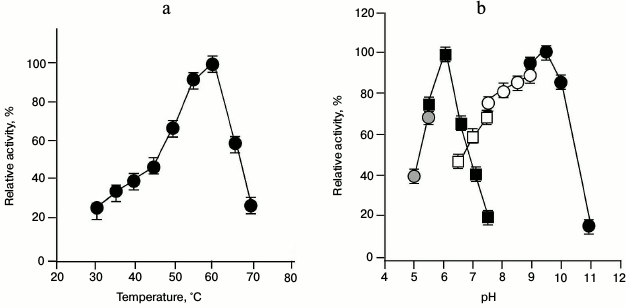
When the enzyme activity of GSH-FDH/ADHR5 was examined at various temperatures at constant pH, the activity increased with increasing temperature until it reached 60°C. The enzyme activity decreased drastically above 60°C (Fig. 3a). Similarly, when the enzyme activity was examined at 60°C in different buffers of various pH, the highest activity for the oxidation reaction, using ethanol as substrate, was found at pH 9.5 in glycine sodium hydroxide buffer, whereas the highest activity for the reduction reaction, using acetaldehyde as substrate, was observed at pH 6.0 in sodium phosphate buffer (Fig. 3b).
Fig. 3. Effect of temperature and pH on the enzyme activity of GSH-FDH/ADHR5 produced in cells grown in the presence of 25 µM Cu2+. a) Effect of temperature on the enzyme activity. Activity assays were performed in 50 mM glycine-NaOH buffer at 30-70°C using ethanol as the substrate. b) Optimal pH for the enzyme activity of GSH-FDH/ADHR5. The activity assays were conducted at 60°C using sodium acetate buffer (pH 5.0-5.5; gray filled circles) and sodium phosphate buffer (pH 5.5-7.5; black filled squares) for reduction reaction using acetaldehyde as the substrate. Oxidation reactions were conducted using sodium phosphate buffer (pH 6.5-7.5; unfilled squares); Tris-HCl buffer (pH 7.5-9.0; unfilled circles), and glycine NaOH buffer (pH 9-11; black filled circles), and ethanol as the substrate.
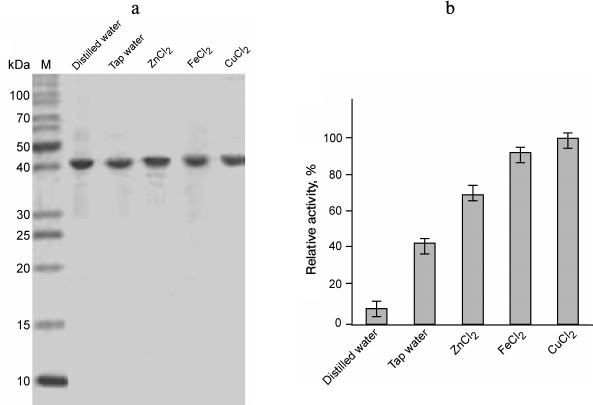
The presence or absence of metal ions in the reaction mixture did not significantly affect the enzyme activity of GSH-FDH/ADHR5. Instead of increase, there was a slight decrease in enzyme activity when various metal ions were added in the reaction mixture at final concentration of 100 µM. This observation indicated that the enzyme activity of GSH-FDH/ADHR5 is independent of added metal ions. However, the presence of EDTA in the reaction mixture at final concentration of 10 mM significantly decreased the enzyme activity. This is in contrast to the first observation. We hypothesized that metal ions might be required in the structural stability during production and folding of GSH-FDH/ADHR5. Therefore, the GSH-FDH/ADHR5 gene was expressed in the presence of various metal ions including Cu2+, Fe2+, and Zn2+ at final concentration of 25 µM. Two control experiments, expression in double distilled water and in tap water, were included. These proteins were purified to apparent homogeneity (Fig. 4a), and the enzyme activities were compared (Fig. 4b). There was very low activity, 20 nmol/min per mg, in the protein produced in double distilled water, which increased 5-fold in the protein produced in tap water. The highest activity, 22-fold increase, was found in the protein produced in the presence of 25 µM Cu2+, followed by Fe2+ (20-fold). Although all the amino acid residues reported for structural Zn2+ binding [25] were conserved in GSH-FDH/ADHR5, the protein produced in the presence of 25 µM Zn2+ exhibited lower activity compared to the protein produced in the presence of same concentration of Fe2+ or Cu2+ (Fig. 4b).
Fig. 4. Purification and enzyme activity of GSH-FDH/ADHR5 produced in cells grown in the presence and absence of metal ions. a) Coomassie brilliant blue stained 14% SDS-PAGE showing the purified GSH-FDH/ADHR5. b) Comparison of enzyme activity of the same proteins as shown in (a). The activity was measured at 60°C and pH 9.5.
Kinetic parameters of GSH-FDH/ADHR5 produced in the presence of Cu2+ and Zn2+ were determined at 60°C and pH 9.5 by plotting the initial velocities for various concentrations of 1-propanol. The Km and Vmax values of GSH-FDH/ADHR5 produced in the presence of Cu2+ were 1.12 mM and 885 nmol/min per mg, respectively. The kinetic parameters of GSH-FDH/ADHR5 produced in cells grown in the presence of Cu2+ or Zn2+ are compared in Table 2.
Table 2. Kinetic parameters of
GSH-FDH/ADHR5 produced in the presence of metal ions
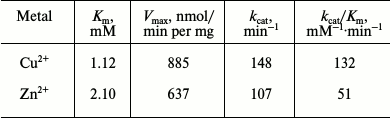
The specific activity of recombinant GSH-FDH/ADHR5 was found to be much lower compared to the alcohol dehydrogenase from Thermus sp. ATN1 (30.6 µmol/min per mg) [20], horse liver alcohol dehydrogenase (3.2 µmol/min per mg) [27], and human liver alcohol dehydrogenase (3.3 µmol/min per mg) [28]. Low activity of GSH-FDH/ADHR5 may be attributed to incorrectly or misfolded recombinant protein produced in E. coli.
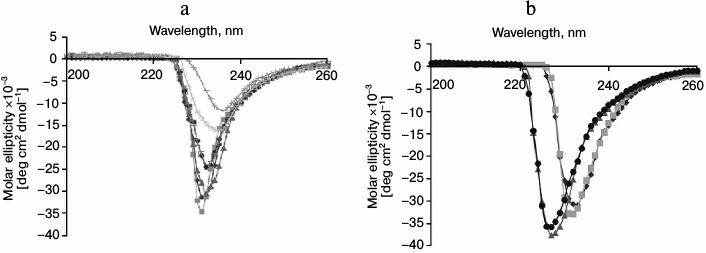
Structural stability. The structural stability of GSH-FDH/ADHR5 was analyzed at different temperatures (20-80°C). The CD spectra showed that there was no significant change in the spectrum up to 60°C (Fig. 5a), indicating that the enzyme maintains its secondary structure up to 60°C, which is in good agreement with the optimal temperature for the enzyme activity. The structure of GSH-FDH/ADHR5 produced in the presence or absence of divalent metal ions was analyzed by CD spectroscopy at 30°C. A significant change in the CD spectra of the protein produced in the presence, particularly Fe2+ or Cu2+, and absence of metal ions was observed, indicating that binding of Cu2+ or Fe2+ may have resulted in structural changes during folding of the protein that contributed to higher activity of the enzyme as compared to Zn2+-bound enzyme (Fig. 5b).
The metal ion contents of the proteins produced in the presence or absence of metal ions (25 µM Zn2+ or Cu2+) were determined, and it was found that there was 1.5 mol of Zn2+ (in the protein produced in the presence of 25 µM Zn2+) and 1.3 mol of Cu2+ (in the protein produced in the presence of 25 µM Cu2+) per mol of the protein. However, no metal ion could be detected when the protein was produced in the absence of any metal ion, though it exhibited weak enzyme activity, possibly due to the low sensitivity of detection.
Fig. 5. Circular dichroism studies on GSH-FDH/ADHR5. a) Far-UV spectrum of GSH-FDH/ADHR5 was analyzed by examining the circular dichroism spectra from 200-260 nm at 20 (closed diamond), 30 (closed squares), 40 (closed triangles), 50 (closed circles), 60 (open squares), 70 (open circles), and 80°C (crosses). b) Comparison of circular dichroism spectra of GSH-FDH/ADHR5 produced in E. coli cells grown in the medium prepared either in double distilled deionized water (diamonds) or containing 25 µM of either Zn2+ (squares), or Fe2+ (circles), or Cu2+ (triangles).
Protein 3D structure comparison. Among the proteins whose crystal structures have been determined, GSH-FDH/ADHR5 displayed the highest identity of 34% with Tadh from Thermus sp. ATN1 (4CPD), followed by 31% with formaldehyde dehydrogenase from P. putida (1KOL), and 24% with human alcohol dehydrogenase (1TEH). A theoretical model of GSH-FDH/ADHR5 was constructed using the crystal structure of 4CPD as a template. There was 24% β-strand and 28% α-helix in the theoretical structure of GSH-FDH/ADHR5. The Ramachandran plot assessment of the predicted model by RAMPAGE showed that a high number of the amino acids (91.8%) were present in the favored region.
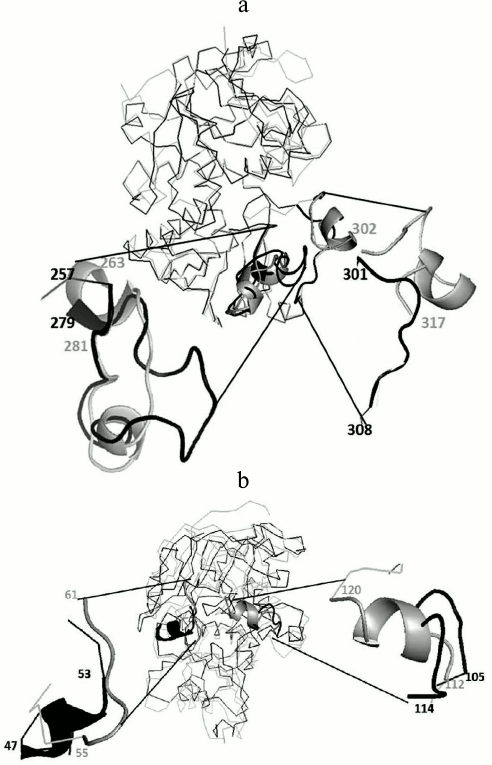
To analyze the tertiary structural differences in GSH-FDH/ADHR5, the predicted model was superimposed on the crystal structure of either 1KOL or 1TEH or 4CPD. The crystal structure of 1KOL shows the presence of two insertion loops comprising 263-281 and 302-317 a.a. [29]. The former loop makes a compact shield and hydrogen bonding with the adenine part of NAD+, thus restricting the release of NAD+ from the enzyme [30]. The predicted structure of GSH-FDH/ADHR5 also shows the presence of an insertion loop (257-279 a.a.), but this loop is not as compact as in 1KOL. Therefore, the release of NAD+ may not be restricted and might be exchanged with the solvent. The second loop in 1KOL (301-308 a.a.) was absent in GSH-FDH/ADHR5 (Fig. 6a).
Fig. 6. Superimposition of predicted structure of GSH-FDH/ADHR5 (shown in black) and 1KOL or 1TEH (shown in gray). a) Presence of relaxed loop in GSH-FDH/ADHR5 at a.a. position 257 to 279 reported to allow the release of NAD+ and absence of another loop at position 301 to 308, which helps to make a compact structure, which may result in the structural stability of GSH-FDH/ADHR5 at higher temperature. b) Replacement of loop in 1TEH by a helix in GSH-FDH/ADHR5 at a.a. position 47 to 53 contrary to replacement of helix in 1TEH by a loop in GSH-FDH/ADHR5 at position 105 to 114. This replacement is expected to hinder the entrance of large molecules into the catalytic cavity.
The superimposition on 1TEH revealed a distinct orientation of amino acids around catalytically important conserved residues. In 1TEH, amino acids 55-61 form a loop and 112-120 form a helix, which results in an open entrance for large substrate molecules [25]. In GSH-FDH/ADHR5, instead of the helix, a loop comprising 105-114 a.a. is formed that covers the entrance and may restrict the entry of large molecules (Fig. 6b).
In 4CPD, the above-described loops are absent, which results in increased compact nature and thermostability [31]. Two of these loops are also absent in GSH-FDH/ADHR5. The absence of these loops may increase the thermostability, similar to 4CPD, which is evident from the optimal temperature (60°C) for the enzyme activity. The CD spectrum of GSH-FDH/ADHR5 also showed that there was no significant change in the secondary structure of the protein up to 60°C.
The experimental data obtained in this study demonstrate that metal ions are necessary for the proper folding and enzyme activity of GSH-FDH/ADHR5. Furthermore, it exhibits only alcohol dehydrogenase activity and no glutathione-dependent formaldehyde dehydrogenase or aldehyde dismutase activity. These results indicate that the enzyme is not a glutathione-dependent formaldehyde dehydrogenase but a bona-fide metal ion-dependent alcohol dehydrogenase.
REFERENCES
1.De Pouplana, L. R., Atrian, S., Gonzalex-Duarte,
R., Fothergill-Gilmore, L. A., Kelly, S. M., and Price, N. C. (1991)
Structural properties of long-and short-chain alcohol dehydrogenases.
Contribution of NAD+ to stability, Biochem. J.,
276, 433-438.
2.Burdette, D. S., Jung, S. H., Shen, G. J.,
Hollingsworth, R. I., and Zeikus, J. G. (2002) Physiological function
of alcohol dehydrogenases and long-chain (C30) fatty acids in alcohol
tolerance of Thermoanaerobacter ethanolicus, Appl. Environ.
Microbiol., 68, 1914-1918.
3.Reid, M. F., and Fewson, C. A. (1994) Molecular
characterization of microbial alcohol dehydrogenases, Crit. Rev.
Microbiol., 20, 13-56.
4.Machielsen, R., Uria, R. A., Kengen, W. M., and
Oost, J. (2006) Production and characterization of a thermostable
alcohol dehydrogenase that belongs to the aldo-keto reductase
superfamily, Appl. Environ. Microbiol., 72, 233-238.
5.Kallberg, Y., Oppermann, U., Jornvall, H., and
Persson, B. (2002) Short‐chain dehydrogenases/reductases (SDRs),
Eur. J. Biochem., 269, 4409-4417.
6.Persson, B., Hedlund, J., and Jornvall, H. (2008)
Medium- and short-chain dehydrogenase/reductase gene and protein
families, Cell. Mol. Life Sci., 65, 3879-3894.
7.Danielsson, O., and Jornvall, H. (1992)
“Enzymogenesis”: classical liver alcohol dehydrogenase
origin from the glutathione-dependent formaldehyde dehydrogenase line,
Proc. Natl. Acad. Sci. USA, 89, 9247-9251.
8.Koivusalo, M., Baumann, M., and Uotila, L. (1989)
Evidence for the identity of glutathione-dependent formaldehyde
dehydrogenase and class III alcohol dehydrogenase, FEBS Lett.,
257, 105-109.
9.Danielsson, O., Atrian, S., Luque, T., Hjelmqvist,
L., Gonzalez-Duarte, R., and Jornvall, H. (1994) Fundamental molecular
differences between alcohol dehydrogenase classes, Proc. Natl. Acad.
Sci. USA, 91, 4980-4984.
10.Mashford, P. M., and Jones, A. R. (1982)
Formaldehyde metabolism by the rat: a re-appraisal, Xenobiotica,
12, 119-124.
11.Wagner, F. W., Pares, X., Holmquist, B., and
Vallee, B. L. (1984) Physical and enzymatic properties of a class III
isozyme of human liver alcohol dehydrogenase: chi-ADH,
Biochemistry, 23, 2193-2199.
12.Barber, R. D., Rott, M. A., and Donohue, T. J.
(1996) Characterization of a glutathione-dependent formaldehyde
dehydrogenase from Rhodobacter sphaeroides, J.
Bacteriol., 178, 1386-1393.
13.Canestro, C., Albalat, R., Hjelmqvist, L., Godoy,
L., Jornvall, H., and Gonzalez-Duarte, R. (2002) Ascidian and amphioxus
Adh genes correlate functional and molecular features of the ADH family
expansion during vertebrate evolution, J. Mol. Evol., 54,
81-89.
14.Cederlund, E., Hedlund, J., Hjelmqvist, L.,
Jonsson, A., Shafqat, J., Norin, A., Keung, W. M., Persson, B., and
Jornvall, H. (2011) Characterization of new medium-chain alcohol
dehydrogenases adds resolution to duplications of the class I/III and
the sub-class I genes, Chem. Biol. Interact., 191,
8-13.
15.Sanghani, P. C., Robinson, H., Bosron, W. F., and
Hurley, T. D. (2002) Human glutathione-dependent formaldehyde
dehydrogenase, structures of apo, binary, and inhibitory ternary
complexes, Biochemistry, 41, 10778-10786.
16.Eklund, H., Plapp, B. V., Samama, J. P., and
Branden, C. I. (1982) Binding of substrate in a ternary complex of
horse liver alcohol dehydrogenase, J. Biol. Chem., 257,
14349-14358.
17.Jalal, A., Rashid, N., Rasool, N., and Akhtar, M.
(2009) Gene cloning and characterization of a xylanase from a newly
isolated Bacillus subtilis strain R5, J. Biosci. Bioeng.,
107, 360-365.
18.Kato, N., Yamagami, T., Shimo, M., and Sakazawa,
C. (1986) Formaldehyde dismutase, a novel NAD‐binding
oxidoreductase from Pseudomonas putida F61, Eur. J.
Biochem., 156, 59-64.
19.Yamasue, Y., Tanisaka, T., and Kusanagi, T.
(1990) Alcohol dehydrogenase zymogram, its inheritance and anaerobic
germinability of seeds of Echinochloa weeds, Japan J.
Breed., 40, 53-61.
20.Hollrigl, V., Hollmann, F., Kleeb, A. C.,
Buehler, K., and Schmid, A. (2008) TADH, the thermostable alcohol
dehydrogenase from Thermus sp. ATN1: a versatile new biocatalyst
for organic synthesis, Appl. Microbiol. Biotechnol., 81,
263-273.
21.Nian, H., Meng, Q., Zhang, W., and Chen, L.
(2013) Overexpression of the formaldehyde dehydrogenase gene from
Brevibacillus brevis to enhance formaldehyde tolerance and
detoxification of tobacco, Appl. Biochem. Biotechnol.,
169, 170-180.
22.Liao, Y., Chen, S., Wang, D., Zhang, W., Wang,
S., Ding, J., and Zhu, H. (2013) Structure of formaldehyde
dehydrogenase from Pseudomonas aeruginosa: the binary complex
with the cofactor NAD+, Acta Crystallogr. Sect. F Struct.
Biol. Cryst. Commun., 69, 967-972.
23.Sakamoto, A., Ueda, M., and Morikawa, H. (2002)
Arabidopsis glutathione-dependent formaldehyde dehydrogenase is an
S-nitrosoglutathione reductase, FEBS Lett., 515,
20-24.
24.Bottoms, C. A., Smith, P. E., and Tanner, J. J.
(2002) A structurally conserved water molecule in Rossmann
dinucleotide‐binding domains, Protein Sci., 11,
2125-2137.
25.Yang, Z. N., Bosron, W. F., and Hurley, T. D.
(1997) Structure of human chi chi alcohol dehydrogenase: a
glutathione-dependent formaldehyde dehydrogenase, J. Mol. Biol.,
265, 330-343.
26.Vitale, A., Rosso, F., Barbarisi, A., Labella,
T., and D’Auria, S. (2010) Properties and evolution of an alcohol
dehydrogenase from the crenarchaeota Pyrobaculum aerophilum,
Gene, 46, 26-31.
27.Ryzewski, C. N., and Pietruszko, R. (1977) Horse
liver alcohol dehydrogenase SS: purification and characterization of
the homogeneous isoenzyme, Arch. Biochem. Biophys., 183,
73-82.
28.Woronick, C. L. (1975) Alcohol dehydrogenase from
human liver, Methods Enzymol., 41, 369-374.
29.Tanaka, N., Kusakabe, Y., Ito, K., Yoshimoto, T.,
and Nakamura, K. T. (2003) Crystal structure of glutathione-independent
formaldehyde dehydrogenase, Chem. Biol. Interact., 143,
211-218.
30.Tanaka, N., Kusakabe, Y., Ito, K., Yoshimoto, T.,
and Nakamura, K. T. (2002) Crystal structure of formaldehyde
dehydrogenase from Pseudomonas putida: the structural origin of
the tightly bound cofactor in nicotinoprotein dehydrogenases, J.
Mol. Biol., 324, 519-533.
31.Man, H., Gargiulo, S., Frank, A., Hollmann, F.,
and Grogan, G. (2014) Structure of the NADH-dependent thermostable
alcohol dehydrogenase TADH from Thermus sp. ATN1 provides a
platform for engineering specificity and improved compatibility with
inorganic cofactor-regeneration catalysts, J. Mol. Catal. B
Enzymol., 105, 1-6.