Quantitative Affinity Interaction of Ubiquitinated and Non-ubiquitinated Proteins with Proteasome Subunit Rpn10
O. A. Buneeva1, O. V. Gnedenko1, A. T. Kopylov1, M. V. Medvedeva2, V. G. Zgoda1, A. S. Ivanov1, and A. E. Medvedev1*
1Orekhovich Institute of Biomedical Chemistry, 119121 Moscow, Russia; E-mail: professor57@yandex.ru2Lomonosov Moscow State University, Biological Faculty, 119991 Moscow, Russia
* To whom correspondence should be addressed.
Received May 10, 2017
Recent proteomic profiling of mouse brain preparations using the ubiquitin receptor, Rpn10 proteasome subunit, as an affinity ligand revealed a representative group of proteins bound to this sorbent (Medvedev, A. E., et al. (2017) Biochemistry (Moscow), 82, 330-339). In the present study, we investigated interaction of the Rpn10 subunit of proteasomes with some of these identified proteins: glyceraldehyde-3-phosphate dehydrogenase (GAPDH), pyruvate kinase, and histones H2A and H2B. The study revealed: (i) quantitative affinity interaction of the proteasome subunit immobilized on a Biacore-3000 optical biosensor cuvette with both the GAPDH (Kd = 2.4·10–6 M) and pyruvate kinase (Kd = 2.8·10–5 M); (ii) quantitative high-affinity interaction of immobilized histones H2A and H2B with the Rpn10 subunit (Kd values of 6.5·10–8 and 3.2·10–9 M, respectively). Mass spectrometric analysis revealed the presence of the ubiquitin signature (GG) only in a highly purified preparation of GAPDH. We suggest that binding (especially high-affinity binding) of non-ubiquitinated proteins to the Rpn10 proteasome subunit can both regulate the functioning of this proteasomal ubiquitin receptor (by competing with ubiquitinated substrates) and promote activation of other pathways for proteolytic degradation of proteins destined to the proteasome.
KEY WORDS: Rpn10 proteasome subunit, Rpn10-binding proteins, optical biosensor, ubiquitin signatureDOI: 10.1134/S0006297917090073
Abbreviations: GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Kd, equilibrium dissociation constant.
Ubiquitin is a small protein (8 kDa) widely distributed in all
eukaryotic cells, where it targets proteins for subsequent proteasomal
degradation [1-4]. The
ubiquitination process includes several sequential stages that involve
several enzymes: ubiquitin activating enzyme (E1),
ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3) [1-5]. Ubiquitin molecules are
attached to the target protein as a polyubiquitin chain, which serves
as a signal for its proteolytic degradation in proteasomes. In the
context of proteasomal degradation of ubiquitinated proteins, the 19S
proteasome Rpn10 subunit plays the role of the ubiquitin receptor [6] responsible for delivery of client proteins to the
20S proteolytically active proteasome, where their subsequent
proteolytic degradation occurs [7].
Recent affinity-based proteomic profiling of mouse brain preparations using the Rpn10 proteasome subunit as the ubiquitin receptor revealed a representative group of proteins that could bind to this affinity ligand [8]. The presence of the ubiquitin tag in the identified proteins was not investigated. Taking into consideration that affinity-based isolation can involve not only direct interaction of specific partner proteins with the immobilized ligand [9], the interaction of the identified proteins with the Rpn10 subunit should be confirmed in direct experiments. Previously, for validation of such interactions between an immobilized ligand and proteins from complex biological systems we successfully used biosensor analysis based on surface plasmon resonance using both low molecular weight substances [10] and proteins [11] as affinity ligands.
In this biosensor-based study, we investigated the interaction of the Rpn10 proteasome subunit with highly purified glyceraldehyde-3-phosphate dehydrogenase (GAPDH), pyruvate kinase, and histones H2A and H2B identified as Rpn10-binding proteins [8] and performed mass-spectrometric search for the “ubiquitin signature” in the studied proteins.
MATERIALS AND METHODS
The following proteins were used in this study: electrophoretically homogeneous GAPDH and pyruvate kinase (140 and 260 µmol/min per mg protein, respectively) isolated from rabbit skeletal muscles [12], recombinant human H2A and H2B histones (New England Biolabs, USA; electrophoretic purity ≥ 95%), and the recombinant Rpn10 subunit of 19S proteasomes (Enzo Life Sciences, USA; electrophoretic purity ≥ 95%).
The HBS buffer (150 mM NaCl, 3 mM EDTA, 0.005% P20 detergent, 10 mM HEPES, pH 7.4), 10 mM acetate buffer (pH 4.0), 10 mM acetate buffer (pH 4.5), and reagents for covalent immobilization of proteins for primary amino groups (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide-HCl (EDC), N-hydroxysuccinimide (NHS), 1 M ethanolamine-HCl (pH 8.5)) were obtained from GE Healthcare, USA.
Interaction of investigated proteins with their immobilized partners (Rpn10 proteasome subunit, histone H2A or H2B) was studied using optical SPR biosensors Biacore 3000 and Biacore T200 and special Biacore Control software (GE Healthcare). All measurements were performed at 25°C using standard CM5 (GE Healthcare) optical chips coated with a carboxymethylated dextran layer. Molecular interactions were recorded as sensograms of the time-dependent biosensor signal (in resonant units, RU).
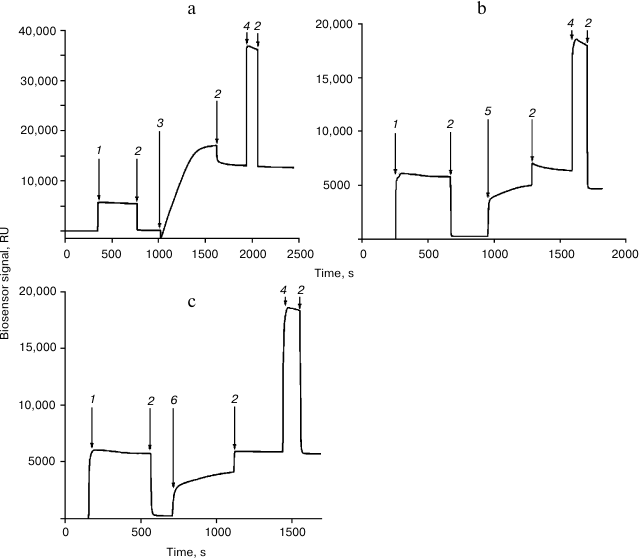
Immobilization of the Rpn10 subunit and histone H2A or H2B was performed in accordance with a recently developed protocol [11] (Fig. 1). Interaction of proteins with immobilized Rpn10 subunit and histone H2A or H2B was examined for 5 min at flow rate 10 µl/min. Before subsequent use, the chip surface was regenerated by washing with 1 M NaCl in 50 mM phosphate buffer (pH 7.4) for 0.5 min at flow rate 50 µl/min.
Fig. 1. Immobilization of the proteasome Rpn10 subunit (a), histone H2A (b), and histone H2B (c) on the surface of a CM5 optical chip. The arrows show injections: 1) EDC/NHS; 2) HBS; 3) Rpn10 subunit (50 µg/ml in 10 mM acetate buffer, pH 4.0); 4) 1 M ethanolamine-HCl; 5) histone H2A (50 µg/ml in 10 mM acetate buffer, pH 4.5); 6) histone H2B (50 µg/ml in 10 mM acetate buffer, pH 4.5).
The resulting sensorgrams were analyzed using BIAevaluation version 4.1 software (GE Healthcare).
Mass spectrometry analysis was carried out on an Orbitrap Fusion instrument (Thermo Scientific, USA) equipped with an NSI ion source. The instrument was operated in positive ionization mode; the drying gas temperature (nitrogen) was 280°C and the emitter voltage was set to 2.1 kV. Precursor ions were surveyed in the range 400-1200 m/z, tandem scanning of fragment ions was performed with fixed first mass of 110 m/z (upper limit of m/z was defined by charge state of the fragmented precursor ion, but no more than 2100 m/z). Precursor ions with charge states from z = 2+ to z = 6+ were triggered for tandem scanning. The tandem scanning was conducted in two parallelized ways: conventional MS2 scanning and MS3 scanning accompanied with synchronous selection of fragment ions conditioned by monoisotopic mass difference of either ΔM = 114.0429 u (with asymmetric selection tolerance –3 ppm/+7 ppm), corresponding to the ubiquitin tag GG, or ΔM = 383.2281 u (mass tolerance –3 ppm/+7 ppm), corresponding to the ubiquitin tag LRGG. The maximum number of ions for simultaneous synchronous selection in MS3 mode was n = 2 in case of mass difference between ions in a pair. The maximum integration time for precursor ions was set to 80 ms, for fragment ions in MS2 110 ms, and for fragment ions in MS3 250 ms.
Peptides were separated using a UPLC chromatographic system Ultimate 3000 RSLCnano (Thermo Scientific). Samples were loaded for 3.7 min onto an enrichment Acclaim µ-Precolumn (0.5 mm × 3 mm, 5 µm) (Thermo Scientific) in isocratic mode of mobile phase C (2% acetonitrile, 0.1% formic acid, 0.02% acetic acid, pH 2.63) at flow rate 20 µl/min. Separation was carried out on an Acclaim Pepmap® C18 analytical column (75 µm × 150 mm, 2 µm) (Thermo Scientific) in a gradient of mobile phase A (water with 0.1% formic acid and 0.02% acetic acid, pH 2.66) and mobile phase B (acetonitrile with 0.1% formic acid and 0.02% acetic acid) at flow rate 0.3 µl/min. Analytical separation was performed in the following gradient: 2-37% of mobile phase B for 45 min following column washing in 90% of mobile phase B for 8 min and equilibrating the column in initial gradient conditions (2% of mobile phase B) for 15 min before starting the next run.
RESULTS AND DISCUSSION
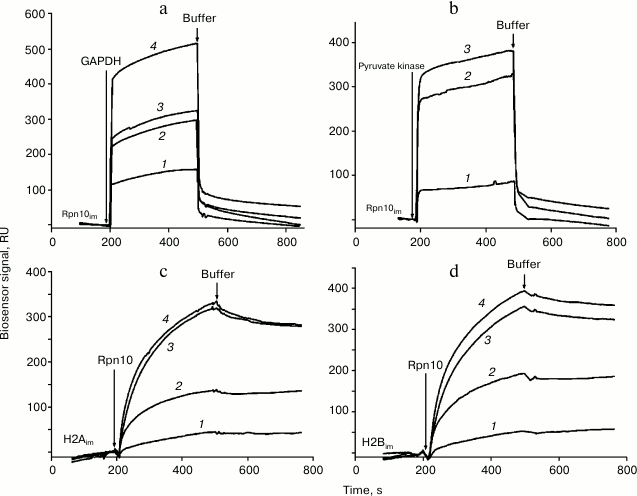
Injection of solutions of GAPDH, pyruvate kinase, and histones H2A and H2B through the optical biosensor channel with immobilized 19S proteasome Rpn10 resulted in appearance of a clear biosensor signal that depended on concentrations of the added protein (Fig. 2). Because of the high nonspecific binding of both histones to the chip surface, correct calculation of Kd values was possible only for GAPDH and pyruvate kinase (table). These results demonstrate significantly higher affinity of GAPDH for the immobilized Rpn10 than that of pyruvate kinase.
Fig. 2. Typical sensorgrams of binding of GAPDH (a) and pyruvate kinase (b) to immobilized Rpn10 subunit of the proteasome, as well as binding of the Rpn10 subunit of proteasomes with immobilized histone H2A (c) and histone H2B (d). Concentrations of protein solutions were as follows. a: 1) 2; 2) 3; 3) 5; 4) 10 µM. b: 1) 1; 2) 5; 3) 7 µM. c: 1) 182; 2) 214; 3) 257; 4) 300 nM. d: 1) 182; 2) 214; 3) 257; 4) 300 nM.
Kinetic parameters of protein binding to immobilized ligands
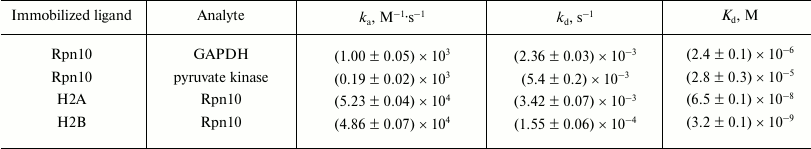
Note: ka, association rate constant;
kd, dissociation rate constant.
Specific interaction of Rpn10 with histones was confirmed under conditions when each histone was immobilized on the optical biosensor cuvette as a ligand and Rpn10 was used as an analyte. Earlier, this approach (immobilization of histones on the surface of an optical biosensor chip) was used by Wang et al. studying specificity of histone binding to various ligands [13]. The Kd values calculated from resulting sensorgrams (table) were two or three orders of magnitude lower than for the interaction of GAPDH and pyruvate kinase with the immobilized Rpn10 (table).
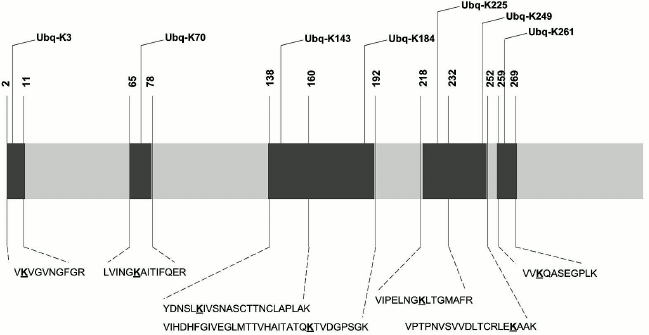
Results of biosensor analysis confirmed the fact of direct interaction of the tested proteins with Rpn10; earlier, these proteins were identified during proteomic profiling of mouse brain preparations with Rpn10 as an affinity ligand [8]. Since the presence of the ubiquitin tag in Rpn10-binding proteins was not investigated during that proteomic profiling, we performed mass-spectrometric search for the protein signature in the purified proteins. This search resulted in identification of the ubiquitin signature only in GAPDH (Fig. 3). Earlier, this glycolytic enzyme was already identified as a substrate for ubiquitination [14, 15]. Although mass-spectrometric analysis used in this study cannot evaluate the structure of ubiquitin chains, the ubiquitin signature has been found in seven regions of the GAPDH polypeptide chain (Fig. 3). All modified peptides of GAPDH contained only the GG type of the ubiquitin signature. However, despite the common modifying group (GG) and conservatism of the amino acid sequence, identified peptides are characterized by some structural heterogeneity. For example, the presence of the ubiquitin signature in the peptide VIHDHFGIVEGLMTTVHAITATQK is always accompanied by the oxidation of the methionine residue (M173), whereas in the peptide VIPELNGKLTGMAFR, the methionine residue always remains intact. The amino acid sequence homology of human, rabbit, and rat GAPDH exceeds 90%, and peptides containing the ubiquitin signature (Fig. 3) are highly conservative and with complete identity for these three species. In this context, the data obtained for the rabbit enzyme can be extrapolated to human and rat GAPDH.
Fig. 3. Scheme of the polypeptide chain of GAPDH with identified ubiquitination sites. The lysine residues carrying the ubiquitin signature are highlighted in bold and underlined.
Although existence of several ubiquitination sites does explain higher affinity of GAPDH to immobilized Rpn10 than that of pyruvate kinase, it should be noted that affinity of Rpn10 to immobilized histones H2A and H2B is at least two orders of magnitude higher than in the ubiquitinated protein.
Thus, the results of this study demonstrate the possibility in principal of interaction of some non-ubiquitinated proteins with the ubiquitin receptor, the 19S proteasome subunit Rpn10, and the affinity of these non-ubiquitinated proteins is significantly higher than in the ubiquitinated proteins.
Known mechanisms of delivery of ubiquitinated proteins to the proteasome include their interaction with ubiquitin receptors, Rpn10 or Rpn13 [16], and with adapter proteins like Rad23 [16, 17]. In the context of delivery of client proteins to the proteasome, Rad23 and Rpn10 are considered as alternative receptors [17]. The results obtained in this study suggest that high-affinity binding of non-ubiquitinated proteins with Rpn10 may thus regulate functioning of this ubiquitin receptor by competing with ubiquitinated substrates. On the other hand, blockade of this ubiquitin receptor may promote activation of other pathways responsible for delivery of client proteins to the proteasome for their proteolytic degradation.
Acknowledgments
This work was performed within the framework of the Program for Basic Research of State Academies of Sciences for 2013-2020 and was partially supported by the Russian Foundation for Basic Research (project No. 16-04-00173a). LC-MS/MS analysis of proteins was carried out in the Center for Collective Use “Human Proteome” (Institute of Biomedical Chemistry).
REFERENCES
1.Hershko, A., and Ciechanover, A. (1998) The
ubiquitin system, Annu. Rev. Biochem., 67, 425-479.
2.Hershko, A., Ciechanover, A., and Varshavsky, A.
(2000) The ubiquitin system, Nature Med., 6,
1073-1081.
3.Schwartz, A. L., and Ciechanover, A. (2009)
Targeting proteins for destruction by the ubiquitin system:
implications for human pathobiology, Annu. Rev. Pharmacol.
Toxicol., 49, 73-96.
4.Kravtsova-Ivantsiv, Y., and Ciechanover, A. (2012)
Non-canonical ubiquitin-based signals for proteasomal degradation,
J. Cell Sci., 125, 539-548.
5.Buneeva, O. A., and Medvedev, A. E. (2016) The role
of atypical ubiquitination in cell regulation, Biomed. Khim.,
62, 496-509.
6.Deveraux, Q., Ustrell, V., Pickart, C., and
Rechsteiner, M. (1994) A 26S protease subunit that binds ubiquitin
conjugates, J. Biol. Chem., 269, 7059-7061.
7.Hamazaki, J., Sasaki, K., Kawahara, H., Hisanaga,
S., Tanaka, K., and Murata, S. (2007) Rpn10-mediated degradation of
ubiquitinated proteins is essential for mouse development, Mol.
Cell. Biol., 19, 6629-6638.
8.Medvedev, A. E., Buneeva, O. A., Kopylov, A. T.,
Tikhonova, O. V., Medvedeva, M. V., Nerobkova, L. N., Kapitsa, I. G.,
and Zgoda, V. G. (2017) Brain mitochondrial subproteome of
Rpn10-binding proteins and its changes induced by the neurotoxin MPTP
and the neuroprotector isatin, Biochemistry (Moscow), 82, 330-339.
9.Ivanov, A., Medvedev, A., Ershov, P., Molnar, A.,
Mezentsev, Y., Yablokov, E., Kaluzhsky, L., Gnedenko, O., Buneeva, O.,
Haidukevich, I., Sergeev, G., Lushchyk, A., Yantsevich, A., Medvedeva,
M., Kozin, S., Popov, I., Novikova, S., Zgoda, V., Gilep, A., Usanov,
S., Lisitsa, A., and Archakov, A. (2014) Protein interactomics based on
direct molecular fishing on paramagnetic particles: practical
realization and further SPR validation, Proteomics, 14,
2261-2274.
10.Buneeva, O., Gnedenko, O., Zgoda, V., Kopylov,
A., Glover, V., Ivanov, A., Medvedev, A., and Archakov, A. (2010)
Isatin binding proteins of rat and mouse brain: proteomic
identification and optical biosensor validation, Proteomics,
10, 23-37.
11.Medvedev, A. E., Buneeva, O. A., Kopylov, A. T.,
Gnedenko, O. V., Medvedeva, M. V., Kozin, S. A., Ivanov, A. S., Zgoda,
V. G., and Makarov, A. A. (2015) The effects of an endogenous
non-peptide molecule isatin and hydrogen peroxide on proteomic
profiling of rat brain amyloid-beta binding proteins: relevance to
Alzheimer’s disease? Int. J. Mol. Sci., 16,
476-495.
12.Scopes, R. K., and Stoter, A. (1982) Purification
of all glycolytic enzymes from one muscle extract, Methods
Enzymol., 90 (Pt. E), 479-490.
13.Wang, H., Walsh, S. T. R., and Parthun, M. R.
(2008) Expanded binding specificity of the human histone chaperone
NASP, Nucleic Acids Res., 36, 5763-5772.
14.Blumenfeld, N., Gonen, H., Mayer, A., Smith, C.
E., Siegel, N. R., Schwartz, A. L., and Ciechanover, A. (1994)
Purification and characterization of a novel species of
ubiquitin-carrier protein, E2, that is involved in degradation
of non-“N-end rule” protein substrates, J. Biol.
Chem., 269, 9574-9581.
15.Jeon, H. B., Choi, E. S., Yoon, J. H., Hwang, J.
H., Chang, J. W., Lee, E. K., Choi, H. W., Park, Z. Y., and Yoo, Y. J.
(2007) A proteomics approach to identify the ubiquitinated proteins in
mouse heart, Biochem. Biophys. Res. Commun., 357,
731-736.
16.Schrader, E. K., Harstad, K. G., and Matouschek,
A. (2009) Targeting proteins for degradation, Nat. Chem. Biol.,
10, 815-822.
17.Elsasser, S., Chandler-Militello, D., Muller, B.,
Hanna, J., and Finley, D. (2004) Rad23 and Rpn10 serve as alternative
ubiquitin receptors for the proteasome, J. Biol. Chem.,
279, 26817-26822.