Enzyme–Substrate Reporters for Evaluation of Substrate Specificity of HIF Prolyl Hydroxylase Isoforms
A. I. Osipyants1, N. A. Smirnova1, A. Yu. Khristichenko1, D. M. Hushpulian1, S. V. Nikulin1, T. A. Chubar2, A. A. Zakhariants3, V. I. Tishkov2,3, I. G. Gazaryan1,2*, and A. A. Poloznikov1
1Rogachev National Medical Research Center for Pediatric Hematology, Oncology and Immunology, Healthcare Ministry of Russia, 117997 Moscow, Russia; E-mail: igazaryan@gmail.com2Lomonosov Moscow State University, Chemistry Faculty, 119991 Moscow, Russia
3Innovations and High Technologies MSU Ltd., 109451 Moscow, Russia
* To whom correspondence should be addressed.
Received August 26, 2017; Revision received September 8, 2017
An organism naturally responds to hypoxia via stabilization of hypoxia-inducible factor (HIF). There are three isoforms of HIFα subunits whose stability is regulated by three isozymes of HIF prolyl hydroxylase (PHD1-3). Despite intense studies on recombinant enzyme isoforms using homogeneous activity assay, there is no consensus on the PHD isoform preference for the HIF isoform as a substrate. This work provides a new approach to the problem of substrate specificity using cell-based reporters expressing the enzyme and luciferase-labeled substrate pair encoded in the same expression vector. The cell is used as a microbioreactor for running the reaction between the overexpressed enzyme and substrate. Using this novel approach, no PHD3 activity toward HIF3 was demonstrated, indirectly pointing to the hydroxylation of the second proline in 564PYIP567 (HIF1) catalyzed by this isozyme. The use of “paired” enzyme–substrate reporters to evaluate the potency of “branched tail” oxyquinoline inhibitors of HIF PHD allows higher precision in revealing the optimal structural motif for each enzyme isoform.
KEY WORDS: activation, luciferase fusion protein, enzyme inhibitors, branched oxyquinolines, ciclopirox, real-time PCRDOI: 10.1134/S0006297917100145
Abbreviations: αKG, alpha-ketoglutarate; DMSO, dimethylsulfoxide; HIF, hypoxia-inducible factor; ODD, oxygen-dependent degradation; PHD, prolyl hydroxylase domain; VHL, von Hippel–Lindau protein.
Hypoxia-inducible factor (HIF) triggers the genetic program of cell
survival under conditions of limited oxygen. HIF consists of two
subunits, where the HIFα subunit is subject to proteasomal
degradation in the presence of oxygen and is stabilized under hypoxia
[1]. HIF prolyl hydroxylase (PHD), an
α-ketoglutarate-dependent nonheme iron dioxygenase, is the major
regulator of HIFα protein stability. HIF PHD hydroxylates the
C-terminal Pro564 and N-terminal Pro402 in HIFα, providing their
recognition by von Hippel–Lindau protein (VHL) in the ubiquitin
ligase complex, labeling HIFα with ubiquitin for subsequent
proteasomal degradation. Currently, three forms of the enzyme and three
isoforms of HIFα [1] are well characterized,
and the existence of another, fourth enzyme isoform has been reported
[2]. In addition to HIFα, the enzymes
hydroxylate prolines in several other proteins, with PHD3 recognizing
the largest pool of protein substrates [3-7]. It has been unequivocally demonstrated that the
C-terminal proline is equally well recognized by all enzyme isoforms,
whereas the N-terminal proline is better recognized by PHD2, less by
PHD1, and is not recognized by PHD3 [8].
The question of the specificity of individual enzyme isoforms toward the individual HIF isoform is still open, since the traditional enzyme assay of PHD isoform activity generates contradictory data. Namely, PHD1 Michaelis constants for HIF1 and HIF2 oxygen-dependent degradation domains (ODD) are in the range of 10-20 nM, whereas their values for the pairs PHD2-HIF1 and PHD3-HIF2 range from 100 to 140 nM; for the PHD2-HIF2 and PHD3-HIF1 pairs, the constants show intermediate values of ca. 60-70 nM [9]. The maximum rate values are equal within experimental error. Hence, based on these results, PHD1 is the most active isoform with respect to HIF1 and HIF2, exhibiting 5-6-fold better values of Michaelis constants (Km) compared to the other two enzyme isoforms. Using an approach independent of catalytic activity – a yeast two-hybrid system expressing an individual enzyme isoform and HIF1 to monitor protein–protein recognition – no interaction between PHD1 and HIF1 ODD was found [10], in contrast to data reported for the enzymes expressed in the baculovirus system [9]. For the corresponding 35-mer HIF peptides, Michaelis constants in the enzyme assay show micromolar values [9], and they are useless for making any conclusion concerning the enzyme isoform specificity. Works performed on cell cultures indicate the dominant role of PHD2 in HIF1α hydroxylation under normoxia [11], and an important role of PHD3 in controlling HIF stability under hypoxia and re-oxygenation [12]. In accord with the most recent review summarizing existing knowledge in this field, HIF1 is the best substrate for PHD2, whereas HIF2 is the best for PHD1 and PHD3 [1].
The use of HIF1α ODD-luciferase fusion reporter stably expressed in the neuroblastoma SH-SY5Y cell line for enzyme inhibitor screening has demonstrated the benefits of this approach. In particular, one of the identified inhibitors [13], without additional structural optimization, is specific only for PHD among other αKG-dependent nonheme iron dioxygenases and is neuroprotective in two in vivo models of hemorrhagic stroke [14]. The principle of the HIF1α ODD-luc reporter performance is equivalent to intracellular recognition of HIFα and its subsequent degradation, with the PHD-catalyzed step being rate limiting in the reporter response [13]. The major problem of cellular studies on PHD specificity towards HIF isoforms is the presence of all three enzyme isoforms in the cells: PHD1 and PHD2 in the cell are comparable, whereas the hypoxia-induced PHD3 is negligible under normoxia [13]. To solve the problem, we constructed vectors expressing paired combinations of individual HIF and PHD isoforms. This approach guarantees the dominant presence of an overexpressed enzyme isoform, with the overexpressed substrate determined by the luciferase fused HIF isoform. In other words, the cell is used as a microbioreactor to carry on the reaction between the desired pairs of PHD and HIF isoforms. As demonstrated below, the reporter baseline and its response to enzyme inhibitors reveals novel answers to the problem of PHD isoform substrate specificity as well as specifying the optimal structure of inhibitors recognizing an individual enzyme isoform.
MATERIALS AND METHODS
Reagents. Oxyquinoline compounds 8 (Adaptaquin) (cat. No. 7706-0075) and 7 (cat. No. 3460-0065) were purchased from ChemDiv Research Institute (Skolkovo, Moscow, Russia). PHD pan-inhibitor FG-4592 (Roxadustat) was purchased from Cayman Chemical Co. (USA). Stock solutions (10 mM) were made in DMSO and stored at –20°C. Immediately before the addition, 1 mM DMSO stocks were prepared, and then, by their sequential dilution, the desired range of 50× concentrations was prepared. Cell media and reagents were purchased from Gibco (USA). Luciferase reagent was from Promega (USA). All other reagents were from Sigma-Aldrich (USA). All reagents were used without additional purification.
Enzyme–luciferase-labeled HIF substrate “paired” vector construction. DNA fragments coding for the C-terminal portion of HIF2α and HIF3α ODD (including the full N-terminal transactivation domain; see Table 1) were made by PCR using the corresponding primers (Table 2) and inserted into HIND3 and NAR1 sites in pGL3-control plasmid (Promega). The HindIII-XbaI fragments in the selected plasmids encoded HIFα ODD fusion proteins with luciferase. The cDNA for PHD1 and PHD3 was obtained by PCR with the corresponding primers (Table 2) and cloned under the strong EF-1α promoter by inserting into KPN1 and XHO1 sites of pBudCE4.1 plasmid (Invitrogen, USA). To clone PHD2 cDNA, the KPN1 and BGL2 restriction sites were used. These intermediate constructs were used to generate stable cell lines expressing individual enzyme isoforms.
Table 1. Cloned HIFα cDNA
fragments
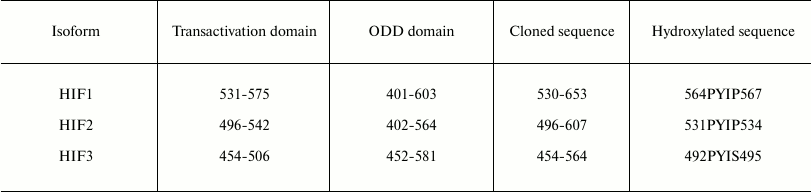
Table 2. Primers for PHD/HIF ODD-luc paired
reporter constructs

Stable cell lines overexpressing the corresponding enzyme isoform were validated to confirm the overexpression of the PHD isoform. The selected plasmids with individual PHD isoforms were used to clone under pCMV promoter the HindIII-XbaI fragments coding for luciferase-labeled ODD of HIFα isoforms (as described above). The resulting plasmid provided the simultaneous expression of one individual PHD isoform and the luciferase labeled ODD of an individual HIFα isoform. Altogether, nine combinations were generated.
Transient expression. Neuroblastoma SH-SY5Y cell line was cultivated in 6-well plates to 70% confluence, and then the plasmids were added in accord with the Lipofectamine 2000 transformation protocol (Invitrogen). The cells were cultivated with 250 µg/ml zeocin. Cell lines expressing either paired reporter construct were produced simultaneously, and all experiments were performed simultaneously on nine cell lines. After overnight incubation, the cells were collected and plated at 25,000 cells per well density in 96-well white sterile microplates, in 100 µl DMEM/F12+GlutaMAX media, and grown at 37°C and 5% CO2.
Luciferase assay. After 20 h cultivation, 2 µl of DMSO solutions of compounds 7, 8, or FG-4592 were added to each well (with the final concentrations varied in the 1-10 µM range) and incubated for 3 h under the same conditions. Then cell media was carefully removed using a multichannel pipette (to keep the cell monolayer untouched), 20 µl of lysis buffer (Promega) was added to each well, and the cells incubated for 7 min on a microplate shaker at room temperature. After a 4 µl aliquot was taken from each well for protein assay, 80 µl of luciferase reagent was added to each well, mixed, and luminescence recorded on a SpectraMax M5e spectrophotometer (Molecular Devices, USA). Ciclopirox (1-25 µM) was used as a positive control. Luminescent signal in the presence of PHD inhibitors was normalized to the baseline (DMSO aliquot without drugs). To compare the baselines of the different reporters, the corresponding background luminescent signals in the absence of any added drugs were normalized to the protein content.
Real-time PCR for individual enzyme isoforms. Expression was studied quantitatively with primers selected using the Clone Manager program: PHD1_f: CATCCGTGGGGACCAGATTGC; PHD1_rev: TACCCGAGCCCGTTGCCTG; PHD3_f: GGTCCTCTACTGCGGGAGCC; PHD3_rev: CAAGCCACCATTGCCTTAGACCT; PHD2_f: GACCTGATACGCCACTGTAAC; PHD2_rev: CGGATAACAAGCAACCATG. GAPDH and β-actin (ACTB) genes were used as control. Each cell line (1,700,000 cells) was lysed with QIAzol reagent (Qiagen, USA). RNA was purified with an RNeasy Mini Kit (Qiagen), and the concentrations were determined on a NanoDrop 1000 spectrophotometer. Reverse transcription of 500 ng RNA was performed with a MMLV RT Kit (Evrogen, Russia). All measurements were done in triplicate, and the results were normalized to control genes.
RESULTS AND DISCUSSION
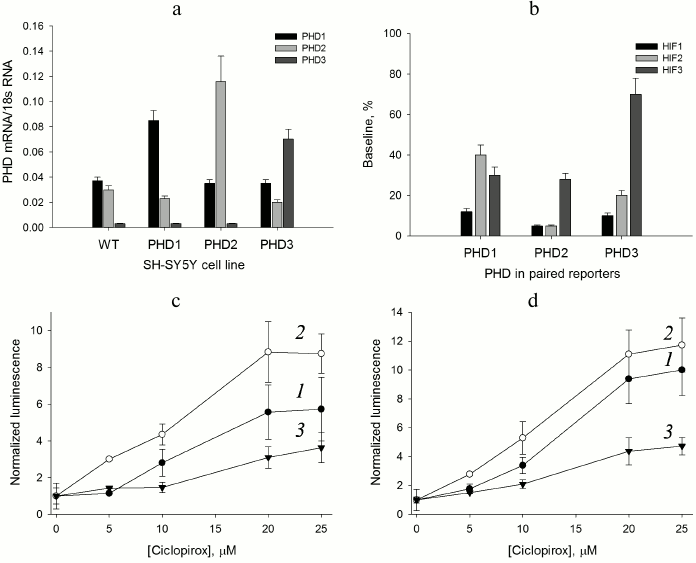
HIF prolyl hydroxylase isoform expression. The use of cDNA fragments listed in Table 1 for the construction of luciferase fusions with the C-terminal portion of HIF2 and HIF3 ODD yields reporters recognized by PHD and the ubiquitinylation system, subject to subsequent proteasomal degradation. The preliminary validation of HIF2 ODD-luc and HIF3 ODD-luc single reporters was successful, and hence these fragments were used to design “paired” enzyme–substrate reporters. With respect to enzyme isoforms, the level of their overexpression in the paired constructs should be preliminarily evaluated, and for this purpose the intermediate constructs in pBudCE4.1 were used to generate stable cell lines expressing individual enzyme isoforms. To evaluate the expression of each isoform, RT-PCR was performed for each stable cell line (Fig. 1a).
Fig. 1. Validation of paired PHD1-3 and HIF1-3 ODD reporters. a) RT-PCR estimate of PHD isoform expression level in the PHD-overexpressing cell lines compared to the original SH-SY5Y cell line (WT). b) Baseline signal for “paired” reporters as a characteristic of HIF ODD isoform recognition by the enzyme isoforms. c) PHD1 “paired” reporter activation by ciclopirox. Curves: 1) PHD1HIF1; 2) PHD1HIF2; 3) PHD1HIF3. d) PHD3 “paired” reporter activation by ciclopirox. Curves: 1) PHD3HIF1; 2) PHD3HIF2; 3) PHD3HIF3.
As seen from the data for the original neuroblastoma cell line (Fig. 1a), the PHD1 and PHD2 expression levels can be considered equal within the experimental error, whereas the PHD3 level is negligibly small. Of note, PHD1 and PHD3 overexpression results in some decrease in PHD2 mRNA level (Fig. 1a). The level of PHD1 mRNA doubles upon overexpression and is 3-fold over the level of PHD2 mRNA. Hence, PHD1 in this line can be considered as a dominant isoform. The level of PHD3 mRNA is increased more than 15-fold upon overexpression and is 2- and 3-fold over the levels of PHD1 and PHD2 mRNA, respectively. The overexpression of PHD3 brings it to the 50% level of the total PHD expressed, whereas for the other two PHD isoforms the overexpression is at the 70% level of the total PHD expressed. Therefore, for the reporter lines overexpressing PHD1 and PHD2, one may assume the dominant character of the overexpressed enzyme isoform, whereas in the case of PHD3 overexpression its contribution is significantly increased, but cannot be considered dominant. Since contribution of PHD1 and PHD2 into the PHD3 overexpressed reporter line cannot be ignored completely, the question on PHD3 substrate specificity can be answered only in a simultaneous comparative study of all “paired” reporters.
“Paired” reporter validation. The background signal of a reporter cell line corresponds to the equilibrium concentration of HIF ODD-luc fusion protein, i.e. the concentration that provides equal rates of fusion protein synthesis (Ko) and degradation. The validation of the HIF1 ODD-luc reporter demonstrated the PHD enzyme-catalyzed step to be rate limiting in the degradation cascade of the fusion substrate [13]. An estimate of the steady-state (baseline) concentration of luciferase fusion protein was reported earlier in [13] as ca. 4 nM, which is much below all reported Km values for HIF1 and, thus, corresponds to substrate non-saturation conditions. In this case, the background luminescent signal is proportional to the equilibrium concentration [HIF–ODD] = KoKm/Vmax, where Ko is the rate of fusion protein synthesis, Km and Vmax are Michaelis constant and maximum velocity for the enzymatic step, respectively, and the change in the reporter background signal indirectly indicates the change in the enzyme activity with respect to a particular HIF ODD isoform fusion. For the “paired” reporters, the increase in total mRNA for all PHD isoforms is about 2-fold (Fig. 1a), which may be interpreted as a 2-fold increase in the total PHD concentration, definitely not sufficient to switch the rate limiting step of the reporter performance.
Comparison of baseline luminescence for “paired” reporters (Fig. 1b) leads to some conclusions. Based on the observed level of background luminescence for reporter lines with overexpressed PHD1, this particular isoform is mainly active towards HIF1 and to a much lesser extent towards HIF3 and HIF2. PHD2 hydroxylates HIF1 and HIF2 equally well, but HIF3 much less than the other two. PHD3 is only 2-fold less active toward HIF2 than HIF1, and almost inactive against HIF3, since the baseline for PHD3/HIF3 ODD-luc reporter line is ca. 70% of the luminescence threshold achieved upon complete inhibition of the total PHD (compare to a 40% level of the threshold for PHD1/HIF3 ODD-luc and PHD2/HIF3 ODD-luc reporter lines in Fig. 1b). The activity profile with respect to HIF ODD isoform differs for individual PHD isoforms (Fig. 1b). There is some similarity in the PHD2 and PHD3 activity profiles, whereas the PHD1 activity profile is strikingly different: HIF2 is the poorest substrate for this isoform. This conclusion is in agreement with the effect of PHD pan inhibitor of chelator nature – ciclopirox – on activation of the corresponding PHD1- and PHD3-based reporters: the activation of PHD1/HIF2 ODD-luc “paired” reporter (Fig. 1c) is less pronounced (5-fold maximum) compared to that for PHD3/HIF2 ODD-luc (10-fold maximum) (Fig. 1d). An interesting observation is an increase in the half-activation concentration to 12 µM, whereas for the original HIF1 ODD-luc reporter this number was 4 µM, despite the fact that in both cases the maximum activation (of 8-10-fold) was the same [13]. A 3-fold increase in the half-activation concentration is in good agreement with the assumption of a 2-fold increase in the total PHD concentration as follows. The concentration dependence for the reporter activation by a competitive PHD inhibitor can be described by the equation [HIF–ODD] = Ko(1 + [I]/Ki)Km/Vmax, where I is an inhibitor, and Ki is an inhibition constant; then, the half-activation concentration is the concentration of an inhibitor equal to the inhibition constant: [HIF–ODD]1/2 = 2KoKm/Vmax. Upon increase in the total enzyme expression by 2-fold, which may be interpreted as a 2-fold increase in the total concentration of the enzyme, Vmax will increase 2-fold, and to compensate for this increase the concentration of an inhibitor has to be increased 3-fold in accord with the definition for the half-activation concentration: [HIF–ODD]1/2 = Ko(1 + 3[I]/Ki)Km/2Vmax = 4KoKm/2Vmax = 2KoKm/Vmax.
Thus, we conclude that the performance of the novel “paired” reporters does obey basic equations of enzyme kinetics, and such reporters can be used both for substrate specificity studies on the individual enzyme isoforms, as well as for enzyme isoform-specific inhibitor screening as we show below.
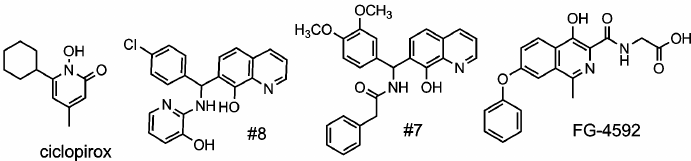
Distinct response of paired reporters on “branched tail” oxyquinoline inhibitors. The use of HIF1 ODD-luc reporter for high throughput screening of HIF PHD potential inhibitors resulted in the identification of potent oxyquinoline inhibitors with a “branched tail” in the 7th position of the oxyquinoline ring [13]. Two hits – compounds 7 and 8 – were selected as the best representatives of two structural subgroups differing in the presence of a carbonyl group (see compound 7 structure versus compound 8 in the scheme).
Structure of studied PHD pan inhibitors
Scheme
The two compounds are indistinguishable in terms of the activation parameters when studied with the original HIF1 ODD-luc reporter (7-fold maximum activation, 2.2 µM half-activation concentration [13]). In the previous work on homogeneous preparations of PHD isoforms used for inhibitor testing [15], it was demonstrated that construction of inhibitors discriminating PHD2 from the two other isoforms with a 10-fold efficiency is feasible. However, it has been stated that inhibitors discriminating between PHD1 and PHD3 cannot be constructed because both the active center and an entry to the active center in these two PHD isoforms are barely different. Therefore, we decided to compare PHD1 and PHD3 in the form of “paired” reporters with respect to compounds 7 and 8 we identified before.
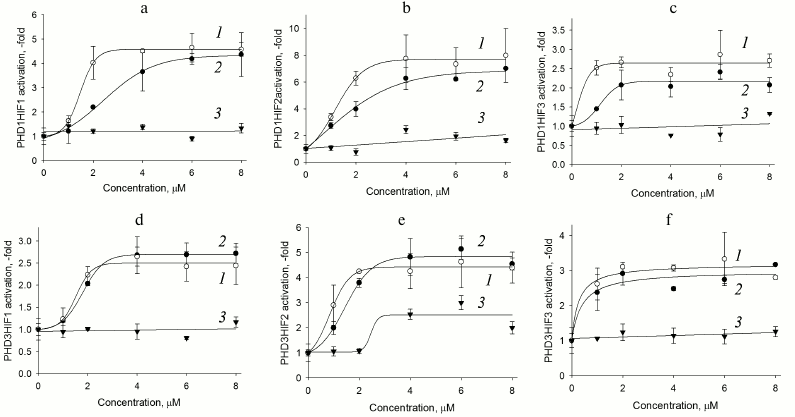
Fig. 2. PHD1HIF1-3 (a-c) and PHD3HIF1-3 (d-f) “paired” reporter activation by PHD pan inhibitors. Curves: 1) compound 8; 2) compound 7; 3) FG-4592 (Fibrogen inhibitor currently undergoing phase III clinical trials for treatment of anemia).
As seen in Fig. 2, there is a clear preference for compound 8 as an inhibitor for the reporters based on PHD1 (Fig. 2, a-c), whereas the two inhibitors are indistinguishable for the reporters based on PHD3 (Fig. 2, d-f). We conclude that further optimization of compound 7 structure will definitely yield an inhibitor preferentially recognizing PHD3, whereas optimization of compound 8 could yield an inhibitor preferentially recognizing PHD1. Of note, PHD1 is the isoform whose deletion or inhibition is prosurvival under conditions of oxidative stress [16]. Comparison of maximum activation induced by the two inhibitors show that HIF3 is a poor substrate for both enzyme isoforms (Fig. 2, c and f). Taking into account a decrease in the apparent half-activation concentration to the level of 0.5 µM, we assume some decrease in the concentration of an enzyme isoform responsible for HIF3 recognition upon PHD1 or PHD3 overexpression. Only PHD2 can be considered for this role, since its mRNA does show a decrease upon overexpression of the other enzyme isoforms (Fig. 1a). HIF3 differs from the other two HIF isoforms by the C-terminal hydroxylated proline sequence (see Table 1), namely, it has Ser in place of Pro567 (numbered in accord with the HIF1 sequence). With this respect, it is of interest to discuss here the results of studying PHD enzyme specificity by the other principally different approaches. The first approach – a yeast two-hybrid system to study PHD3 specificity for the hydroxylated proline Pro564 flanking sequences –demonstrated that the Pro567Ala replacement had no effect on recognition by PHD2, but it results in the loss of recognition by PHD3 [10]. A similar conclusion was made in an independent study on recombinant HIF PHD3 expressed in the active form in E. coli: the authors demonstrated PHD3-catalyzed hydroxylation of Pro567 in HIF1 [17]. Later, by means of computer modeling, VHL was shown to better recognize hydroxylated Pro567, and worse hydroxylated Pro564 in HIF peptide [18]. Considering our results on the very poor activity of PHD3-based reporter towards HIF3 ODD fusion obtained with the novel approach of “paired” cell-based reporters, we conclude that the PHD3 isoform does hydroxylate the second proline in the PYIP sequence, and hence the PYIS sequence in HIF3 ODD is not recognized as a hydroxylation substrate.
Comparison of response of HIF1 ODD-luc paired reporters (Fig. 2, a and d) to enzyme inhibitors demonstrates that PHD3 is less active toward HIF1 if compared to PHD1, since the maximum activation of the corresponding reporters is 2.5- and 5-fold, respectively. The highest maximum activation is observed for reporters based on HIF2 ODD-luc: 8-fold in the paired reporter with PHD1 and 5-fold if paired with PHD3. The preliminary validation of HIF2 ODD-luc reporter also demonstrated its higher sensitivity to the branched oxyquinoline inhibitors. The fact that HIF2 ODD is poor but still a substrate for these two enzyme isoforms results in the apparent increase in the efficiency of specific inhibitors, and in this way HIF2-based reporters will permit fine discrimination between the inhibitors. These reporters can be used for future work on inhibitor optimization.
The newly developed approach to evaluate the substrate specificity of HIF prolyl hydroxylase based on the paired enzyme–substrate reporters allows additional data on the enzyme isoform properties to be generated. Transient expression of these paired reporters results in the elucidation of HIF ODD substrate preference patterns for each PHD isoform. Namely, HIF1 is the best substrate for PHD1 if compared to HIF2, whereas PHD2 is equally and highly active towards both HIF isoforms; PHD3 does not recognize HIF3. The study of paired reporters with branched oxyquinoline inhibitors 7 and 8 demonstrates that PHD1 is better inhibited by compound 8 than by compound 7, whereas PHD3 is inhibited by both compounds equally well. PHD1/HIF1-luc, PHD1/HIF2-luc, and PHD3/HIF2-luc reporter performance observed for ciclopirox and branched oxyquinolines results in high activation maximum, and thus exhibit the highest sensitivity toward PHD inhibitors. Therefore, these reporters are of high interest for future work on optimization of isoform-specific enzyme inhibitors.
Acknowledgments
This work was supported by the Russian Science Foundation (project No. 16-14-10226).
REFERENCES
1.Ivan, M., and Kaelin, W. G., Jr. (2017) The
EGLN-HIF O2-sensing system: multiple inputs and feedbacks, Mol.
Cell, 66, 772-779.
2.Koivunen, P., Tiainen, P., Hyvarinen, J., Williams,
K. E., Sormunen, R., Klaus, S. J., Kivirikko, K. I., and Myllyharju, J.
(2007) An endoplasmic reticulum transmembrane prolyl 4-hydroxylase is
induced by hypoxia and acts on hypoxia-inducible factor alpha, J.
Biol. Chem., 282, 30544-30552.
3.Xie, L., Xiao, K., Whalen, E. J., Forrester, M. T.,
Freeman, R. S., Fong, G., Gygi, S. P., Lefkowitz, R. J., and Stamler,
J. S. (2009) Oxygen-regulated beta(2)-adrenergic receptor hydroxylation
by EGLN3 and ubiquitylation by pVHL, Sci. Signal., 2,
ra33.
4.Xie, L., Pi, X., Mishra, A., Fong, G., Peng, J.,
and Patterson, C. (2009) PHD3-dependent hydroxylation of HCLK2 promotes
the DNA damage response, J. Clin. Invest., 122,
2827-2836.
5.Luo, W., Lin, B., Wang, Y., Zhong, J.,
O’Meally, R., Cole, R. N., Pandey, A., Levchenko, A., and
Semenza, G. L. (2014) PHD3-mediated prolyl hydroxylation of nonmuscle
actin impairs polymerization and cell motility, Mol. Biol. Cell,
25, 2788-2796.
6.Heir, P., Srikumar, T., Bikopoulos, G., Bunda, S.,
Poon, B. P., Lee, J. E., Raught, B., and Ohh, M. (2016)
Oxygen-dependent regulation of erythropoietin receptor turnover and
signaling, J. Biol. Chem., 291, 7357-7372.
7.German, N. J., Yoon, H., Yusuf, R. Z., Murphy, J.
P., Finley, L. W., Laurent, G., Haas, W., Satterstrom, F. K.,
Guarnerio, J., Zaganjor, E., Santos, D., Pandolfi, P. P., Beck, A. H.,
Gygi, S. P., Scadden, D. T., Kaelin, W. G., Jr., and Haigis, M. C.
(2016) PHD3 loss in cancer enables metabolic reliance on fatty acid
oxidation via deactivation of ACC2, Mol. Cell, 63,
1006-1020.
8.Pappalardi, M. B., McNulty, D. E., Martin, J. D.,
Fisher, K. E., Jiang, Y., Burns, M. C., Zhao, H., Ho, T., Sweitzer, S.,
Schwartz, B., Annan, R. S., Copeland, R. A., Tummino, P. J., and Luo,
L. (2011) Biochemical characterization of human HIF hydroxylases using
HIF protein substrates that contain all three hydroxylation sites,
Biochem. J., 436, 363-369.
9.Koivunen, P., Hirsila, M., Kivirikko, K. I., and
Myllyharju, J. (2006) The length of peptide substrates has a marked
effect on hydroxylation by the hypoxia-inducible factor prolyl
4-hydroxylases, J. Biol. Chem., 281, 28712-28720.
10.Landazuri, M. O., Vara-Vega, A., Viton, M.,
Cuevas, Y., and Del Peso, L. (2006) Analysis of HIF-prolyl hydroxylases
binding to substrates, Biochem. Biophys. Res. Commun.,
351, 313-320.
11.Berra, E., Benizri, E., Ginouves, A., Volmat, V.,
Roux, D., and Pouyssegur, J. (2003) HIF prolyl-hydroxylase 2 is the key
oxygen sensor setting low steady-state levels of HIF-1alpha in
normoxia, EMBO J., 22, 4082-4090.
12.Appelhoff, R. J., Tian, Y. M., Raval, R. R.,
Turley, H., Harris, A. L., Pugh, C. W., Ratcliffe, P. J., and Gleadle,
J. M. (2004) Differential function of the prolyl hydroxylases PHD1,
PHD2, and PHD3 in the regulation of hypoxia-inducible factor, J.
Biol. Chem., 279, 38458-38465.
13.Smirnova, N. A., Rakhman, I., Moroz, N., Basso,
M., Payappilly, J., Kazakov, S., Hernandez-Guzman, F., Gaisina, I. N.,
Kozikowski, A. P., Ratan, R. R., and Gazaryan, I. G. (2010) Utilization
of an in vivo reporter for high throughput identification of
branched small molecule regulators of hypoxic adaptation, Chem.
Biol., 17, 380-391.
14.Karuppagounder, S. S., Alim, I., Khim, S. J.,
Bourassa, M. W., Sleiman, S. F., John, R., Thinnes, C. C., Yeh, T. L.,
Demetriades, M., Neitemeier, S., Cruz, D., Gazaryan, I., Killilea, D.
W., Morgenstern, L., Xi, G., Keep, R. F., Schallert, T., Tappero, R.
V., Zhong, J., Cho, S., Maxfield, F. R., Holman, T. R., Culmsee, C.,
Fong, G. H., Su, Y., Ming, G. L., Song, H., Cave, J. W., Schofield, C.
J., Colbourne, F., Coppola, G., and Ratan, R. R. (2016) Therapeutic
targeting of oxygen-sensing prolyl hydroxylases abrogates
ATF4-dependent neuronal death and improves outcomes after brain
hemorrhage in several rodent models, Sci. Transl. Med.,
8, 328ra29.
15.Murray, J. K., Balan, C., Allgeier, A. M.,
Kasparian, A., Viswanadhan, V., Wilde, C., Allen, J. R., Yoder, S. C.,
Biddlecome, G., Hungate, R. W., and Miranda, L. P. (2010)
Dipeptidyl-quinolone derivatives inhibit hypoxia inducible
factor-1α prolyl hydroxylases-1, -2, and -3 with altered
selectivity, J. Combin. Chem., 12, 676-686.
16.Siddiq, A., Aminova, L. R., Troy, C. M., Suh, K.,
Messer, Z., Semenza, G. L., and Ratan, R. R. (2009) Selective
inhibition of hypoxia-inducible factor prolyl-hydroxylase 1 mediates
neuroprotection against normoxic oxidative death via HIF and CREB
independent pathways, J. Neurosci., 29, 8828-8838.
17.Fedulova, N., Hanrieder, J., Bergquist, J., and
Emren, L. O. (2007) Expression and purification of catalytically active
human PHD3 in Escherichia coli, Protein Express. Purif.,
54, 1-10.
18.Minervini, G., Masiero, A., Moro, S., and
Tosatto, S. C. (2013) In silico investigation of PHD-3 specific
HIF1-α proline 567 hydroxylation: a new player in the
VHL/HIF-1α interaction pathway? FEBS Lett., 587,
2996-3001.