REVIEW: Hevein-Like Antimicrobial Peptides of Plants
A. A. Slavokhotova1,2*, A. A. Shelenkov2, Ya. A. Andreev1,3, and T. I. Odintsova2
1Shemyakin–Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, 117997 Moscow, Russia; E-mail: annslav82@gmail.com2Vavilov Institute of General Genetics, Russian Academy of Sciences, 119991 Moscow, Russia
3Sechenov First Moscow State Medical University, Ministry of Healthcare of the Russian Federation, Institute of Molecular Medicine, 119991 Moscow, Russia
* To whom correspondence should be addressed.
Received January 15, 2017; Revision received May 27, 2017
Plant antimicrobial peptides represent one of the evolutionarily oldest innate immunity components providing the first line of host defense to pathogen attacks. This review is dedicated to a small, currently actively studied family of hevein-like peptides that can be found in various monocot and dicot plants. The review thoroughly describes all known peptides belonging to this family including data on their structures, functions, and antimicrobial activity. The main features allowing to assign these peptides to a separate family are given, and the specific characteristics of each peptide are described. Further, the mode of action for hevein-like peptides, their role in plant immune system, and the applications of these molecules in biotechnology and medicine are considered.
KEY WORDS: hevein-like peptides, antimicrobial peptides, plant immunityDOI: 10.1134/S0006297917130065
Abbreviations: a.a., amino acid residue; AMP, antimicrobial peptide.
Plants are attacked by microorganisms and pests throughout their life
cycle. As a result of the long-term evolutionary process, plants have
developed an arsenal of protective agents being components of their
innate immune system. The most important components of
“chemical” protection are secondary metabolites, as well as
antimicrobial proteins and peptides. Recently, defense (antimicrobial)
peptides are being intensively studied. Antimicrobial peptides (AMP)
include short polypeptides that are expressed in the cells of living
organisms continuously or in response to pathogenic attacks. They occur
in all organisms, not only plants, but also animals and microorganisms.
The AMPs are components of the innate immune system, one of the oldest
mechanisms of resistance providing fast-acting and non-energy-consuming
“first line of defense”. Although AMP structure is highly
varied, we can distinguish several common properties that are
characteristic for all AMPs. They include, first of all, low molecular
mass (up to 10 kDa), antimicrobial activity, positive charge and
amphiphilic structure of the molecule, which together with features
mentioned above enable interaction between the peptides and the
membranes of pathogens. Most of the plant AMPs are cysteine-rich and
contain 2 to 12 cysteine residues forming disulfide bonds, which
contributes to their structure density, as well as resistance to
chemical and proteolytic cleavage. The role of AMPs in resistance is
demonstrated in numerous works showing that transgenic plants
expressing AMP genes become more resistant to phytopathogens, and
plants show the enhanced expression of AMP-encoding genes in response
to biotic and abiotic stresses. It is interesting that AMPs have been
revealed in all plant organs [1], and those
constitutively expressed in plants are usually found in peripheral cell
layers covering the surface of organs. Transcriptomic and genomic data
analysis shows that plants contain hundreds of AMP-like sequences;
however, the expression of their genes and the synthesis of functional
AMPs have been confirmed only for some peptides [2-4].
It should be noted that the increasing interest in plant AMPs is associated with their unique properties and primarily with the mechanism of their action. It is considered that the main target of AMPs is the cytoplasmic membrane of microorganisms, the integrity of which is crippled by plant peptides. Therefore, in contrast to conventional antibiotics causing the formation of resistant forms of pathogens, the development of AMP resistance reduces them to a minimum. In addition, the advantages of AMPs are the broad spectrum of action and the ability to rapidly kill target cells. All of the above makes AMPs highly promising objects for research and practical applications aimed at the development of resistant crops and a new generation of drugs [5-7].
Plant AMPs are molecules of extreme structural heterogeneity, which are usually grouped into families depending on amino acid sequence similarity, spatial structure, the number and arrangement of cysteine residues in molecules (the so-called “cysteine motif”). Plant AMPs are currently grouped into the following families: defensins, thionins, lipid transfer proteins, alpha-harpinins, hevein-like peptides, snakins, knottins, and cyclotides [8]. This review is dedicated to the family of hevein-like peptides, which have been intensively studied for some time past.
GENERAL CHARACTERISTICS OF HEVEIN-LIKE PEPTIDES
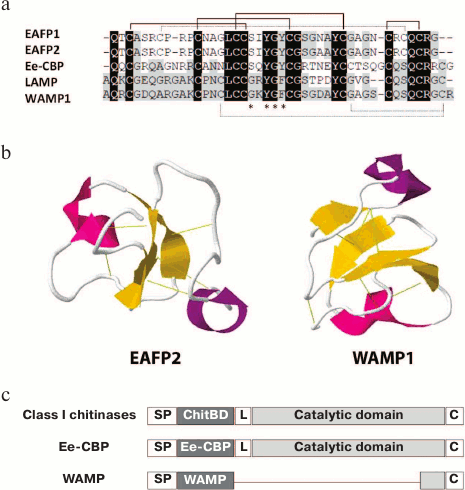
The family of hevein-like AMPs has got its name from hevein isolated from the latex of the rubber tree (Hevea brasiliensis Moll. Arg.). This small family now includes about 20 members isolated from different mono- and dicotyledonous plants (see table). Hevein-like AMPs have several characteristics: first, they are short basic peptides of 29-45 amino acid residues (a.a.) in length enriched in cysteine and glycine residues. The classical hevein-like AMPs, including hevein, contain 8 cysteine residues; however, there are many forms with 6 or 10 cysteines. Six of these residues take rather conservative positions in the peptide molecule and form a cysteine motif that can be expressed by the following formula: C1X4-5C2X4C3C4X5C5X6C6, where C represents cysteine and X is any residue except cysteine. At the same time, disulfide bond formation occurs between the following cysteine residues: C1–C4, C2–C5, and C3–C6.
Antimicrobial activity of hevein-like peptides
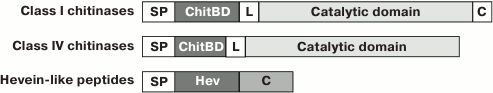
The key feature of all hevein-like peptides is that one of their structural components is a conservative chitin-binding site with the amino acid sequence SXFGY/SXYGY, where X is any amino acid residue. The chitin-binding site is typical not only for hevein-like AMPs, but also for other proteins capable of binding chitin, which is a polymer of N-acetylglucosamine (GlcNAc), as well as the related compounds containing GlcNAc or N-acetylneuraminic acid (NeuNAc) [9]. Chitin is a cell wall component, that is present in numerous plant pathogens (fungi, insects, and nematodes) but is absent in plants [8]. Chitin-binding proteins are believed to be involved in plant protection from various chitin-containing pathogens [1]. The chitin-binding proteins of plants also include lectins, class I and IV chitinases, and a few proteins induced by wounding [9]. All these proteins have at least one chitin-binding domain consisting of 30-43 a.a. and containing the above-mentioned site of conservative amino acid residues. Among the chitin-binding proteins listed above, the proteins with the highest similarity to hevein-like AMPs are class I and IV chitinases, which are monomeric enzymes with molecular weight of 25-35 kDa [1] acting as endochitinases and forming chitooligosaccharides with various polymerization degrees. In addition to the chitin-binding domain, the precursors of these chitinases contain an N-terminal hydrophobic signal peptide, a “hinge” (linker) proline-rich region, and a catalytic domain. The structural characteristics of precursors of hevein-like AMPs and class I and IV chitinases are shown in the Fig. 1. It is interesting that the antifungal and catalytic activities of class I chitinase retain after removal of the chitin-binding domain, but are much lower than in the intact enzyme [10]. Consequently, the chitin-binding domain is not essential for fungal growth suppression but seems to provide efficient interaction between the enzyme and the substrate.
Fig. 1. Characteristic structural features of precursors of hevein-like AMPs and class I and IV chitinases. SP, the N-terminal signal peptide; ChitBD, the chitin-binding domain; Hev, the mature hevein-like peptide; L, the linker, or “hinge” region; C, the C-terminal prodomain.
Thus, the main structural characteristics that can be used to assign the peptide to the family of hevein-like AMPs are its small size (29-45 a.a), positively charged molecule, abundance of glycine and cysteine residues (6, 8, or 10), and the presence of the characteristic cysteine motif and chitin-binding site.
HEVEIN: PROPERTIES, STRUCTURE, AND FUNCTIONS
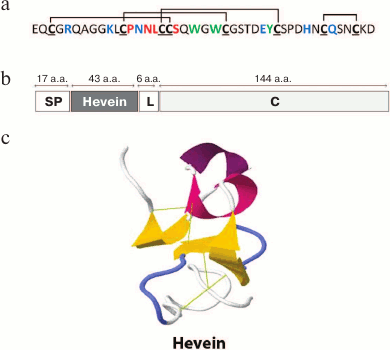
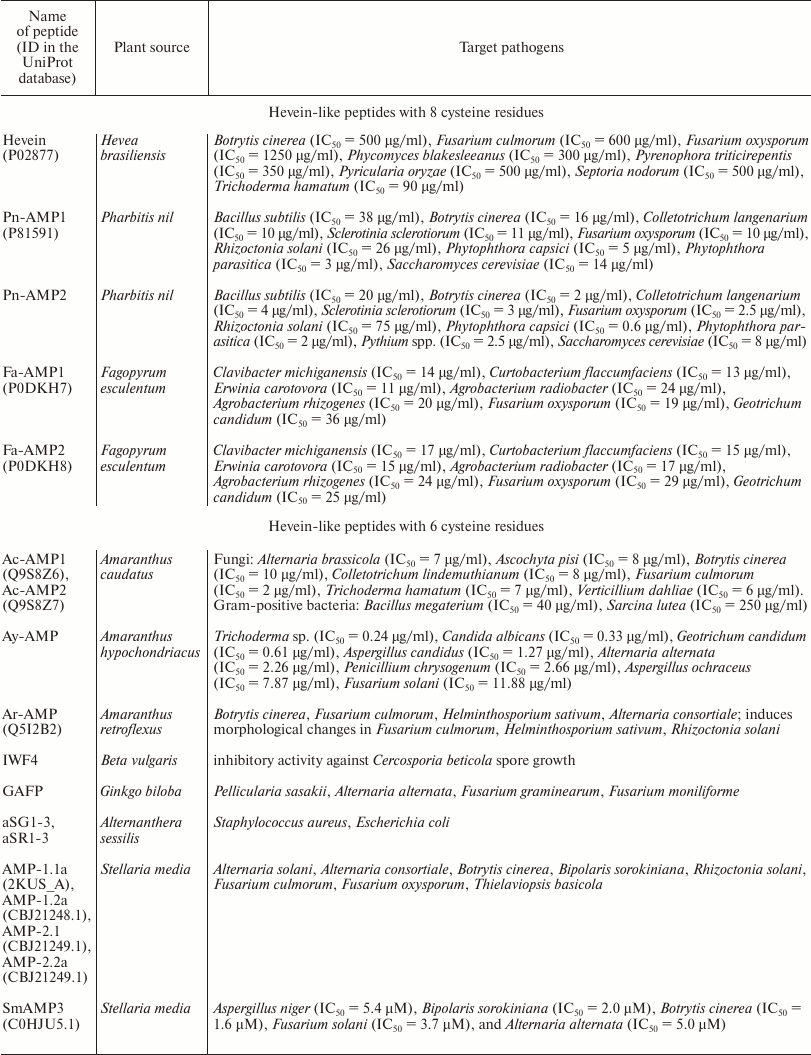
Hevein was first described as early as in 1960 [11]. This short cysteine-rich peptide was assigned to the basic components of vacuolar structures (the so-called “lutoids”) of the latex of rubber tree (Hevea brasiliensis) of the spurge family (Euphorbiaceae). The amino acid sequence of hevein was determined in 1975 [12] and, unexpectedly, proved to be highly similar to the chitin-binding agglutinin (UDA) of great nettle (Urtica dioica L.) [13, 14]. It is known that UDA is accumulated in large amounts in the underground organs of nettle and inhibits in vitro the growth of many chitin-containing fungi [15]. It was shown that hevein had a number of characteristics similar to UDA. Like the chitin-binding domain of UDA, hevein contains 8 cysteine residues forming a typical cysteine motif (Fig. 2a), is able to specifically bind to chitin, and possesses antifungal activity in vitro against several chitin-containing phytopathogenic fungi [16]. The antifungal activity of hevein is retained even after high temperature exposure (90°C for 10 min). However, in contrast to lectins, hevein does not exhibit agglutinating properties [16]. Hevein inhibited hyphal growth, but its activity was low compared to those of other known hevein-like peptides (see table).
Fig. 2. Primary (a) and spatial ((c) 1HEV in the PDB database) structure of hevein and its precursor (b); cysteine residues are shown in bold and underlined; disulfide bonds are shown with black lines; Ser19 and the loop (13-16 a.a.) playing the key role in the binding of hevein with oligosaccharides are red; Arg5, Lys10, Asn14, Glu29, His35, and Gln38 residues involved in the interaction with immunoglobulin E are blue; Trp21, Trp23, Tyr30 residues important for the interaction with both oligosaccharides and immunoglobulin E are green; Hevein is the mature peptide hevein; for other designations see Fig. 1 legend.
The structure of hevein-encoding cDNA was determined in 1990 [17]. The hevein precursor turned out to be a rather long protein (210 a.a.) consisting of a signal peptide (17 a.a.), the hevein itself (43 a.a.), a linker (6 a.a.), and a C-terminal domain (144 a.a.; Fig. 2b). Using the in vitro translation system and the experiments on translocation of hevein domains as hybrid protein components, it was shown that the N-terminal peptide was in fact a signal peptide responsible for pro-hevein transport into the rough endoplasmic reticulum [18]. Then, the signal peptide is cleaved and the remaining precursor undergoes further processing including the cleavage of 6 a.a. between mature hevein and the C-terminal domain [18]. Mass spectromery showed the presence of minor components containing one or two additional residues at the C-termunis of hevein. This indicated that the processing most likely included the cleavage of N-terminal region of C-terminal domain, followed by the cleavage of 4-6 a.a. from the C-terminus of mature hevein [19].
Interestingly, cDNA encoding the C-terminal domain of hevein showed high similarity with cDNA of the potato protein WIN1 and WIN2 genes induced by wounding. Plant wounding and the treatment with phytohormones (ethylene and abscisic acid) induced the accumulation of hevein transcripts in leaves, stems, and latex, but not in roots [17]. Hence, it is necessary to mention the initial function of hevein in hevea plant. It has been shown that hevein specifically interacts with glycoprotein receptors on the surface of rubber particles resulting in particle aggregation and coagulation of latex [20]. Coagulation is necessary for blocking the casual wounds or those created by frequent tapping of rubber trees for latex. As a result, the wound quickly closes and, accordingly, the possibility for pathogenic microorganisms to get into and spread over the plant is eliminated. Thus, hevein, initiating the described cascade of reactions, actively participates in the protection of tissues of rubber tree stem [20].
Five novel genes encoding hevein precursors were discovered in 2005 [21]. These genes were highly conservative in transcribed regions and differed only in promoter sequences. It turned out that, though all of the discovered promoters were functional, only the longest of them provided the high level of expression of heterologous gene in differnet tissues of transgenic rice. This promoter was shown to induce enhanced transgene experssion in case of mechanical damage and fungal infection [21].
The spatial structure of hevein was determined by nuclear magnetic resonance (NMR) and X-ray structure analysis [22, 23]. Both methods have shown that hevein contains a characteristic structural motif including the α-helix–β1–β2–α-helix–β3, the central β-sheet with antiparallel β-chains being surrounded by two small helices stabilized by disulfide bridges (Fig. 2c). Various techniques including NMR, molecular modeling, and isothermal titration calorimetry were used to study the interaction between hevein and saccharides [24-26]. As a result, it was shown that Trp21, Trp23, Tyr30, Ser19, and residues from 13 to 16 play a key role in the binding of hevein to oligosaccharides [24, 27]. Moreover, it was shown that the minimal number of monomeric elements of polymer (GlcNAc)n, with which hevein can bind at a ratio of 1 : 1, is 2. With the number of monomers increasing to 5 or 8, the hevein/oligosaccharide ratio in solution becomes 2 : 1 [24]. It is interesting that, in case of substantial shortening of hevein molecule and substitution of Asp for one of the key residues, Ser19, the ability to bind to oligomers remains, but the binding constant decreases significantly [28].
HEVEIN AS AN ALLERGEN IN LATEX-FRUIT SYNDROME
In the field of hevein research the considerable number of studies is devoted to the ability of this peptide to cause allergic reactions. As mentioned above, hevein is a component of natural latex, which represents the concentrated and stabilized milky sap of the rubber tree. Natural latex is extensively used in the production of natural rubbers, as well as for various latex products. The first cases of allergic reactions to latex were described as early as in 1927; however, beginning from the 1980s, the frequency of occurrence of this type of allergy has dramatically increased, especially for medical personnel using latex gloves [29]. In 2002, the proportion of medical personnel exposed to this allergic reaction reached 36% [30]; in 2008, this index slightly decreased in the developed countries due to using gloves with low content of allergens [31]. However, the risk group still includes patients (children and adults) with various diseases that require frequent repeated surgery, as well as the use of catheters, drainage tubes, and other latex-containing accessories [31]. Interestingly, 30-50% of individuals suffering from latex allergy also demonstrate allergic reactions to some plant products, mainly fresh fruit (banana, avocado, nuts, kiwi fruit, etc.); consequently, this type of allergy is referred to as latex-fruit syndrome [31]. It has been shown that the major allergens of the latex-fruit syndrome are hevein and its precursor protein (prohevein), since they cause allergic reactions in 70% of patients [32, 33]. The cross-reactivity between latex and some fruits is caused by class I endochitinases, in particular their chitin-binding domain, the sequence of which is homologous to the hevein sequence [34]. There are about 12 protein allergens causing the latex-fruit syndrome [35], including endo-β-1,3-glucanase [36], profilin [37], patatin-like proteins [38], nonspecific lipid transfer protein [39], etc.
One of the methods of allergy control is efficient allergen-specific immunotherapy with allergen isoforms or modified proteins having a reduced ability to bind with immunoglobulin E (IgE) [40]. The production of such compounds, in addition, associated with the minimal risk of occurrence of systemic side effects, requires the identification of IgE-related epitopes. The study of the hevein linear epitopes in several independent works has shown that the regions critical for interaction with immunoglobulin E are located at the N- and C-termini of the peptide [41, 42]. In addition, it has been shown that the tertiary structure of the allergen is crucial for the binding to IgE, because recognition of mutant peptide forms by IgE in case of Ser or Ala substitutions for Cys residues in hevein did not occur [43]. The amino acid residues directly involved in the interaction with IgE were identified: Arg5, Lys10, Glu29, Tyr30, His35, Gln38 [44] (Fig. 2a). Based on the data obtained, different mutant forms of hevein with alanine substitutions for all or some of the critical amino acid residues were constructed. This substantially reduced their ability to bind with IgE, as well as the sensitivity of skin scarification tests [44]. The list of amino acids involved in the interaction with IgE was later supplemented with Trp21, Trp23 [45], and Asn14 [46] (Fig. 2a). Further studies of natural hevein isoforms and mutant forms with chemically modified residues of aromatic amino acids demonstrated the existence of several conformations of the hevein epitope; however, the precise mechanism of hevein recognition by IgE is yet unknown [47]. At the same time, it is believed that the derived mutant hevein analogs having a reduced ability to bind to IgE can be used as candidates for immunotherapy; moreover, such compounds should have lower risk of development of systemic or anaphylactic side effects in allergy treatment.
OTHER HEVEIN-LIKE PEPTIDES WITH EIGHT CYSTEINE RESIDUES
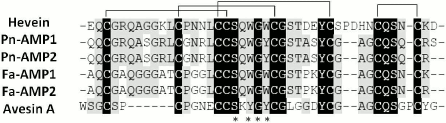
In addition to hevein, hevein-like peptides with eight cysteine residues (8C-Hev peptides) include the molecules isolated from different plants of the Poaceae, Convolvulaceae, and Polygonaceae families. First, we should mention the two short (about 40 a.a.) antifungal peptides isolated from the seeds of morning glory (Pharbitis nil) belonging to Convolvulaceae family [48]. The Pn-AMP1 and Pn-AMP2 peptides have identical primary structures except for the presence of an additional serine residue at the C-terminus of the Pn-AMP1 molecule (Fig. 3). Both peptides possess a remarkable antifungal activity against fungi either containing or not containing chitin in their cell walls. It was shown that these peptides penetrated very rapidly into fungal hyphae and concentrated near the septum and at the hyphal tips, which caused actin depolarization and burst of hyphal tips, followed by membrane disruption and the crippling of cytoplasm integrity [48]. The precursors of ipomoea peptides contain N-terminal signal peptide, a mature protein, and C-terminal domain. The transcripts encoding Pn-AMP1 and Pn-AMP2 have been found in seeds only.
Fig. 3. Alignment of amino acid sequences of hevein (number in UniProt database: P02877), Pn-AMP2, Pn-AMP1 (P81591), Fa-AMP1 (P0DKH7), Fa-AMP2 (P0DKH8), and Avesin A [50]; identical amino acids are shown as white letters on black background; conservative amino acids are shown as black letters on gray background; amino acids comprising the chitin-binding site are asterisked.
In addition to the peptides from ipomoea, 8C-Hev peptides include peptides from buckwheat (Fa-AMP1, Fa-AMP2) [49] and a peptide from oats (Avesin A) [50]. It is interesting that Avesin A was the first hevein-like peptide isolated from cereals by 2003 [50]. All of the peptides given above are short (up to 40 a.a.) and contain, in addition to 8 cysteines, 10 glycine residues and a chitin-binding site, similar to all hevein-like peptides (Fig. 3). It should be noted that buckwheat peptides were identical to each other except for the terminal residue: Fa-AMP2 peptide had a Lys40 → Arg40 substitution (Fig. 3). Both peptides inhibited the growth of phytopathogenic fungi and were active against Gram-positive and Gram-negative bacteria in micromolar concentrations (see table) [49].
HEVEIN-LIKE PEPTIDES WITH SIX CYSTEINE RESIDUES
The shortest molecules demonstrating antifungal activity and ability to bind to chitin are 6-cysteine hevein-like peptides (6C-Hev peptides) with three disulfide bonds and a total length of 29-30 a.a. Most of the 6C-Hev peptides were isolated from plants of the Amaranthaceae family; however, some representatives were obtained from plants of the Caryophyllaceae family. The molecules of these peptides have a deletion of the C-terminal region as compared to the 8C-Hev peptides. The size of deletion is about 10 a.a. including two cysteine residues, which form the fourth disulfide bond in the 8C-Hev peptides.
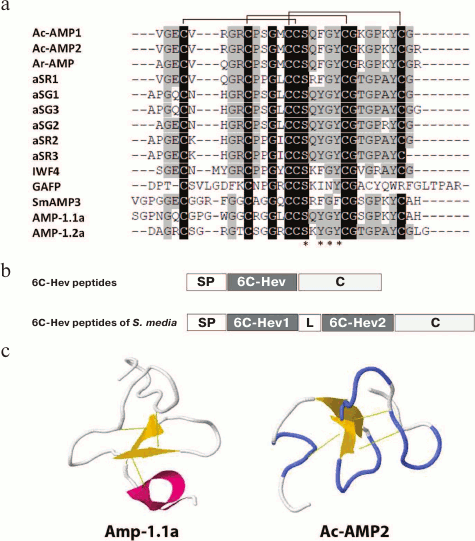
The first 6C-Hev peptides (Ac-AMP1, Ac-AMP2) were isolated in 1992 from the seeds of love-lies-bleeding (Amaranthus caudatus L.) [51]. Both peptides had identical primary structures and differed only in the presence of additional arginine residue at the C-terminus of Ac-AMP2 (Fig. 4a). It has been shown that these peptides are reversibly bound to chitin by means of the chitin-binding site. It should be noted that Ac-AMP1 and Ac-AMP2 inhibited the growth of phytopathogenic fungi at much lower effective concentrations (see table) than many antifungal chitin-binding proteins known by that time. These peptides inhibited the growth of Gram-positive bacteria, but were inactive against Gram-negative bacteria (see table), while their antimicrobial activity decreased considerably in the presence of cations [51].
Fig. 4. Alignment of amino acid sequences (a) of 6C-hevein-like plant peptides Ac-AMP1 (Q9S8Z6), Ac-AMP2 (Q9S8Z7), Ar-AMP (Q5I2B2), IWF4 [56], aSG1-3, aSR1-3 [54], AMP-1.1a (CBJ21248.1) and AMP-1.2a (CBJ21248.1), the typical structure of their precursors and its comparison with the precursor of 6C-Hev peptides of Stellaria media (b) and the spatial structure (c) of Ac-AMP2 (1MMC) and AMP1.1a (2KUS). 6C-Hev, the mature 6C-Hev peptide; 6C-Hev1, the first mature 6C-Hev peptide of S. media (AMP-1.1a); 6C-Hev2, the second mature 6C-Hev peptide of S. media (AMP-1.2a).
In 2007, a peptide identical to Ac-AMP2 was isolated from Prince-of-Wales feather (A. hypochondriacus) and designated as Ay-AMP [52]. It was suggested, on the basis of the hypothesis of monophyletic origin of amaranths, that the identical sequences of Ay-AMP and Ac-AMP2 result from domestication of green amaranth (A. hybridus), which is probably the common ancestor of A. caudatus and A. hypochondriacus. This hypothesis is confirmed by the fact that the sequence of nonspecific lipid transfer peptide isolated from A. hypochondriacus is identical to the sequence of the A. caudatus peptide [52]. Ay-AMP possesses antifungal properties identical to those of Ac-AMP2 (see table) and can bind to chitin [52].
Another 6C-Hev peptide, Ar-AMP isolated from red-root amaranth (A. retroflexus), differs from Ac-AMP2 only in three amino acid substitutions (Fig. 4a) [53]. It is interesting that this peptide was obtained from seeds that had lost their germination ability, because they were collected almost 20 years before the beginning of the experiment. Nevertheless, it retained its antimicrobial activity against phytopathogenic fungi and Gram-positive bacteria (see table). In particular, it inhibited, in micromolar concentrations, the development of helminthosporium leaf blight caused by Helminthosporium sativum on barley seedlings (see table) [53].
Six highly homologous 6C-Hev peptides were isolated from a perennial herbaceous plant, sessile joyweed (Alternanthera sessilis L.), which also belongs to the Amaranthaceae family [54]. The comparison with Ac-AMP1 showed that these peptides had similar structures of the precursors and several substitutions (Fig. 4a). The spatial structure of the aSG1 peptide was determined by NMR, and it was found that its aromatic amino acid residues comprising the chitin-binding site directly interacted with the chitin of pathogens, similarly to other hevein-like peptides [54].
The structures of precursors of all described 6C-Hev peptides were typical for hevein-like peptides (Fig. 4b): N-terminal signal peptide (about 25 a.a.), the mature peptide (about 30 a.a.), and C-terminal prodomain (about 34 a.a.). The genes of 6C peptides from different amaranth species were highly similar in their nucleotide sequences. At the same time, the genes encoding the Ac-AMP1 and Ac-AMP2 peptides were also found, in addition to A. caudatus, in A. retroflexus [53]. Interestingly, the Ac-AMP transcripts were revealed in developing seeds but did not present in roots, leaves, and leaves exposed to stress [55].
The intercellular washing fluid from leaves of sugar beet (Beta vulgaris L.), also belonging to the Amaranthaceae family, was shown to contain another 6C-Hev peptide designated as IWF4 [56]. This peptide had 10 a.a. substitutions compared to Ac-AMP (Fig. 4a), exhibited higher affinity to chitin compared to chitin-binding chitinases, and efficiently inhibited spore germination in the phytopathogenic fungus Cercospora beticola (see table). The IWF4 precursor had the same structure as the above 6C peptides (Fig. 4b) and was expressed in the aerial parts of plants only [56].
Another 6C-Hev peptide designated as GAFP was isolated from the leaves of the medical plant Ginkgo biloba of the family Ginkgoaceae [57]. It contained 38 a.a., and its primary structure was substantially different from the structure of peptides from the Amaranthaceae family (Fig. 4a). GAFP was active against such fungi as Pellicularia sasakii, Alternaria alternata, Fusarium graminearum, Fusarium moniliforme; it increased the permeability of hyphal membranes, causing a rapid alkalization of the medium.
Finally, the peptide designated as Sm-AMP3 was isolated from the leaves of the common chickweed (Stellaria media L.) from the Caryophyllaceae family and was characterized [58]. This 6C-Hev peptide was 35-a.a. long, its amino acid sequence was rather different from that of Ac-AMP1 (Fig. 4a), and it showed high antifungal activity against several phytopathogenic fungi, but not against bacteria (see table). It was shown that SmAMP3 could bind chitin in vitro. Interestingly, this peptide was constitutively expressed in all above-ground organs and seeds of chickweed, which may be evidence of its important role in plant immunity. It should be noted that the precursors of 6C-Hev peptides isolated also from the common chickweed seeds proved to be homologous to the SmAMP3 peptide and had a modular structure, in contrast to all the precursors described above. Thus, besides the signal and mature peptides and the C-terminal prodomain typical to this family, they contained an additional domain, which represented another 6C-Hev peptide, as well as a linker binding it to the first mature peptide (GenBank: CBJ21248.1; CBJ21249.1; Fig. 4b) [1]. The two mature 6C-Hev peptides formed during the processing of such prepropeptide, AMP-1.1a and AMP-1.2a (Fig. 4a), were shown to have high antifungal activities, but against different pathogens [59]. This, probably, enabled the plant to be resistant to a wide range of phytopathogens (see table) [60].
The spatial structures of 6C-Hev peptides such as Ac-AMP2, AMP-1.1a, and aSG1 (IDs 1MMC, 2KUS in the database RSCB PDB) were determined by the NMR technique [54, 61] (Fig. 4c). The cysteine motif typical to all hevein-like peptides forms a knot in the spatial structure, in which a ring of two disulfide bonds and atoms of the main chain is “threaded” by the third disulfide. At the same time, the main structural element of the Ac-AMP2 peptide is an anti-parallel β-sheet (Met13-Lys23) including two β-strands, N-terminal region with unordered structure, and C-terminal β-turn (Gly24-Cys28) [61]. It has been shown that the N-terminal region is linked with the two main β-strands by two disulfide bonds, while the C-terminal region containing a helical turn is linked with the first β-chain by the third disulfide bond [8].
HEVEIN-LIKE PEPTIDES WITH TEN CYSTEINE RESIDUES
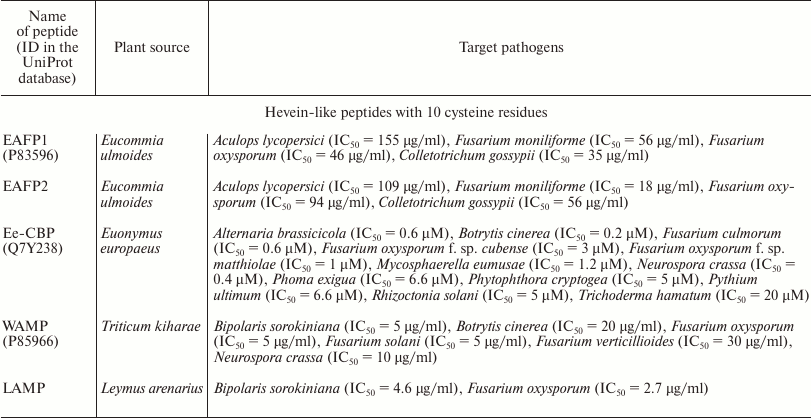
Along with truncated 6C-Hev peptides having the fourth disulfide bond deleted, some peptides with 10 cysteine residues and, accordingly, five disulfide bonds, have been isolated. The first 10-cysteine hevein-like peptides (10C-Hev peptides) were isolated in 2002 from the bark of elm-leaved pineapple-flower (Eucommia ulmoides) belonging to the family Eucommiaceae, and were designated as EAFP1 and EAFP2 [62]. They contained 41 a.a., had a pyroglutamic acid residue at the N-terminus, and differed only in one amino acid substitution (Asn27 → Ala). It should be noted that eight basic cysteine residues were located at conservative positions, like in hevein, while the additional fifth disulfide bond was formed between the second and ninth cysteine residues in its molecule Cys7–Cys37 (Fig. 5a). The presence of five disulfide bonds in peptide molecules from Eucommia made them extremely stable. Specifically, they maintained their antifungal activity even after 30-min boiling and formed crystals easily. The peptides had a broad spectrum of antifungal activity: they were active against eight major pathogenic fungi attacking cotton, wheat, potato, tomato and tobacco. At the same time, the inhibitory activity was observed against fungi either containing or not containing chitin. The effective concentration varied within a range of IC50 = 18-155 µg/ml, while the antifungal effect decreased dramatically in the presence of calcium ions [62] (see table).
Fig. 5. The alignment of amino acid sequences (a) of such 10C-Hev plant peptides as EAFP1 (P83596), EAFP2 (P83597), Ee-CBP (Q7Y238), LAMP (P86521), and WAMP-1 (H6S4F1), the comparison of structures of EeCBP and WAMP precursors with class I chitinase precursors (c), and the spatial structures (b) of EAFP2 (1P9Z) and WAMP1 (2LB7); the formation of the additional fifth disulfide bond is shown by dashed lines; Ee-CBP, WAMP represent the mature Ee-CBP and WAMP peptides.
The methods of X-ray structure analysis and NMR spectroscopy were used to determine the spatial structure of the EAFP2 peptide in crystal and in solution [63, 64] (Fig. 5b). These two methods showed similar results, namely, that the tertiary structure of EAFP2 was a compact globule consisting of a 310 helix (Cys3-Arg6), α-helix (Gly26-Cys30), and three regions of anti-parallel β-strands (Cys16-Ser18; Tyr22-Gly24; Arg36-Cys37). The chitin-binding site of EAFP2 had a conformation similar to the analogous domains of other hevein-like peptides; its characteristic feature was that the amino acids surrounding the site formed a hydrophobic surface. Interestingly, the fifth disulfide bond connected the first ten N-terminal residues with C-terminal residues at positions 35-41, formed a cluster of positively charged amino acid residues Arg6, Arg9, Arg36, and Arg40 on the surface of the molecule [63, 64].
Another 10C-Hev peptide (Ee-CBP) was isolated from the bark and leaves of European spindle-tree (Euonymus europaeus) belonging to the family Celastraceae. The fifth disulfide bond of this peptide was located in C-terminal part of the molecule, connecting the seventh and tenth cysteine residues (Cys32–Cys44) [65] (Fig. 5a). Like the Eucommia peptides, this bond provided higher stability for Ee-CBP. It retained antifungal activity after 10-min boiling, long-term storage, and in a broad pH range (from 2 to 11). Ee-CBP had a high antifungal activity, which exceeded the activity of the 6C-peptide Ac-AMP2 from amaranth, one of the most active hevein-like antimicrobial peptides (see table). In particular, Ee-CBP inhibited the growth of the fungus B. cinerea at a concentration lower than 1 µg/ml. In addition, the peptide possessed antibacterial activity against Gram-positive bacteria [65].
Interestingly, class I chitinase (Ee-chitinase) was isolated from the leaves and bark of European spindle-tree simultaneously with the Ee-CBP peptide [66]. It was shown that the sequences of Ee-CBP and Ee-chitinase precursors had a high similarity (about 70%) but, in contrast to Ee-CBP, the chitin-binding domain of the chitinase contained only eight cysteine residues forming four disulfide bonds. The 10C-Hev peptide and chitinase precursors had almost the same size, and the difference between them was mainly located in the linker between the hevein-like and C-terminal domains. It should be noted that, compared to other peptides of the family, Ee-CBP had the longest precursor including the C-terminal prodomain of the same length as in class I/IV chitinases (Fig. 5c). This prodomain consisted of a catalytic chitinase domain and a linker, by which the proteolysis took place. In chitinase, only short C-terminal propeptide was truncated as a result of processing; in Ee-CBP, the entire C-terminal domain underwent processing. The study of antimicrobial activity showed that the Ee-CBP peptide had a higher activity than chitinase. It has been found for the first time that the joint effects of Ee-CBP and Ee-chitinase resulted in synergism, and the antimicrobial activity increased many times. To establish whether this effect could be observed in planta, the plant was treated with a phytohormone (methyl jasmonate), whereupon the Ee-CBP and Ee-chitinase transcripts were found in the bark and leaves of European spindle-tree; at the same time, the level of expression of both transcripts increased substantially. The analysis of leaf and bark extracts confirmed the presence of 10C-Hev peptide and chitinase in the organs being investigated, which was also an evidence of the joint effect of Ee-CBP and Ee-chitinase [66].
Other peptides containing 10 cysteine residues are WAMP and LAMP isolated from the seeds of highly resistant wheat species Triticum kiharae L. and wild lyme grass (Leymus arenarius L.) [67, 68]. The positions of four disulfides in the WAMP peptide structure precisely matched their localization in the chitin-binding domain of class I chitinases isolated from cereals. At the same time, the fifth disulfide bond in the peptides was located between the third and tenth cysteine residues (Cys16–Cys44; enumerated by the WAMP peptide), and this bond brought together the N- and C-terminal regions of the polypeptide chain, enriched in basic amino acids (Fig. 5a). Thus, a cluster of positively charged amino acid residues was formed. On the other side of the molecule, there was a hydrophobic cluster containing the chitin-binding site. It should be noted that the WAMP and LAMP peptides had unique chitin-binding sites, the characteristic of which was that that the conservative serine residue at position 20 was replaced by glycine (Ser20 → Gly20); at the same time, three other conservative residues of aromatic amino acids (Tyr22, Phe24, Tyr31) remained unchanged (Fig. 5a). It is interesting that this substitution had no effect on the ability of WAMP and LAMP to bind to polymeric chitin in vitro; however, the WAMP peptide lost its ability to bind penta-N-acetylchitopentose [1, 69]. A method of heterologous expression in E. coli cells was developed to obtain the recombinant analogs of WAMP and LAMP in the amounts necessary for testing their antimicrobial activities and determining their spatial structures. It was shown that these peptides, at their micromolar concentrations, were active against a wide range of microorganisms and demonstrated antimicrobial activity against fungi either containing or not containing chitin in the cell wall (see table). In addition, the WAMP peptide was active against Gram-positive and Gram-negative bacteria [67]. The structural studies by NMR spectroscopy showed that WAMP had a 3D structure typical to all hevein-like peptides [69]. The two disulfide bonds (between the first and fourth, second and fifth cysteine residues) were located actually at right angle to each other and formed a nottin-like core, while the whole spatial structure of the molecule included four anti-parallel β-strands (2-3, 18-20, 24-26, and 37-39 a.a.) and two short helical regions (helix 310 – residues 6-8 and α-helix 29-32 a.a.) [69] (Fig. 5b).
It was shown that the WAMP peptide had a precursor protein typical to hevein-like AMPs, which consisted of the signal and mature peptides and C-terminal prodomain (Fig. 5c) [70]. The comparion of the WAMP precursor with chitinase and with the 10C-Hev peptide precursor Ee-CBP showed that the C-terminal domain of the WAMP precursor was strongly reduced. In spite of a vast deletion (about 700 bp), a minor part of the chitinase domain showing high similarity to the analogous domains of class I chitinases was retained in the structure of the WAMP prepropeptide. In addition to this domain, high sequence similarity was observed between the hevein domain of chitinase and the mature region of the WAMP peptide (Fig. 5c). Taking into account this fact, as well as the similarity between spatial structures of the WAMP peptide and class I/IV chitinases, it was supposed that the WAMP gene could evolutionarily descend from the chitinase genes. The selection of these genes during evolution was probably a result of pronounced antimicrobial activity of WAMP peptides, which enhanced the resistance of plants to phytopathogens [70].
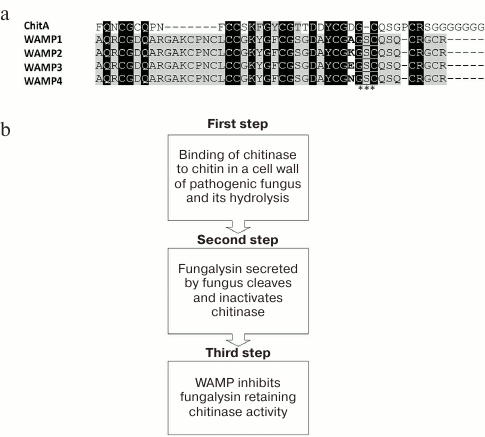
The wheat and goat grass species used for obtaining T. kiharae were investigated. The formula of the T. kiharae genome is AbAbGGDD. It has been shown that the species T. monococcum and T. urartu, being the donors of genome A, contain no wamp gene homologs. At the same time, the species Ae. tauschii (the donor of genome D) and T. timopheevii (the donor of genome G) contain homologous genes, which encode the precursors different from the wamp gene-encoded precursors in single amino acid substitutions or small deletions concentrated mainly in the region of the signal peptide and the C-terminal prodomain. The most conservative domain of such homologous genes was the mature peptide region having, however, one non-conservative position (34th a.a.) where the amino acid residues Ala, Lys, Glu, Asn were observed depending on the species being investigated (Fig. 6a).
Fig. 6. Mechanism of action of WAMP peptides. a) Comparison of the chitin-binding domain of chitinase ChitA (D0EM57) and peptides WAMP-1 (H6S4F1), WAMP-2 (H6S4F4), WAMP-3 (H6S4F5), and WAMP-4 (H6S4F6). Non-conservative position 34 in WAMP peptides is shown in bold; the site of proteolysis of the enzyme fungalysin is asterisked. b) Model of plant–pathogen interaction (see explanations in text).
It was shown that wamp gene expression was activated in response to salt stress, while no changes in its expression were observed in response to temperature stress (temperature increase/decrease). It turned out that wamp transcripts accumulated in response to infection with the phytopathogenic fungus F. oxysporum, while the reprogramming of transcription and alternative polyadenylation were switched on in response to infection with A. niger [70].
MECHANISM OF ACTION OF HEVEIN-LIKE PEPTIDES
Until recently, it was considered that the main mechanism of action of hevein-like peptides is associated with their ability to interact with the cell wall chitin of pathogens. Using Ac-AMP, as well as truncated hevein mutant (Hev32), it has been shown that the interaction with hydrophobic C-H groups of the fungal saccharide occurred through the π-electrons of aromatic amino acid residues of the chitin-binding domain [26, 28, 71, 72]. It was shown that the 8C-Hev peptide Pn-AMP1 quickly penetrated into fungal hypha and induced actin depolarization in the cytoskeleton, membrane disrupture, and leakage of cytoplasmic materials; as a result, hyphal growth of the fungi (Saccharomyces cerevisiae and Candida albicans) was stopped [73]. However, it should be noted that mannans, rather than chitin, are responsible for this process in the fungal cell wall [73]. It was supposed that hevein-like AMPs and chitinases could work together, exerting a targeted impact on the fungi, since they were structurally similar and had a common target of action. This was shown for 10C-Hev peptide Ee-CBP [66]. However, the mechanism of action was described in detail and the molecular target in the pathogenic organism, to which the major activity of hevein-like AMP is directed, was identified as a result of investigating the 10C-Hev peptide WAMP.
It should be noted that the class IV chitinases ChitA and ChitB were isolated previously from the seeds of maize (Zea mays L.) and proved to have high antimicrobial activity against Fusarium and Bipolaris fungal species [74]. It was also shown that some phytopathogenic fungi, in turn, secreted proteins that targeted these chitinases and cleaved the peptide bond between the chitin-binding and chitinase domains; as a result, the chitinases were inactivated and, accordingly, the protection of plants from pathogens was weakened (Fig. 6a) [75]. One of the chitinase-modifying proteins was the enzyme secreted by the fungus F. verticillioides and designated as Fv-cmp. After primary structure identification, this enzyme was assigned to the class of fungalysins, the family of M36 metalloproteases [75]. It was shown that Fv-cmp cleaved class IV chitinases of not only maize (ChitA and ChitB) but also of A. thaliana (AtchitIV3, AtchitIV5). It cleaved the conservative peptide bond between glycine and serine residues located in the region between two chitinase domains. As a result, the chitinases lost their activity and could not affect pathogens (Fig. 6a) [75].
WAMP was also supposed to be a target of fungalysin because of high structural similarity between the WAMP peptide and plant chitinases. However, it turned out that the proteolysis did not occur during the joint incubation of this peptide and the purified protease Fv-cmp in vitro. The absence of proteolysis by Fv-cmp was also observed for the peptides homologous to WAMP having Lys (WAMP2), Glu (WAMP3) or Asn residues (WAMP4) in place of Ala at non-conservative position 34 (Fig. 6a), even under conditions of prolonged incubation. It is supposed that the observed stability is associated with the presence of the additional Ser36 residue in the region that should undergo proteolysis; however, this amino acid residue is absent in the structure of both chitinase and other hevein-like peptides. Moreover, ChitA chitinase remained intact when added to fungalysin and WAMP. No changes in the properties of ChitA were observed in case of WAMP replacement by its homolog WAMP34K; however, the chitinase underwent partial or complete proteolysis in the presence of other homologs (WAMP34E, WAMP34N). Thus, it turned out that the amino acid residue at position 34 is one of the key residues for the binding with fungalysin, and the inhibition of the enzyme could be either observed or not depending on its nature.
A model of plant–pathogen interaction has been proposed (Fig. 6b). In response to the attack of a pathogenic fungus, a plant expresses chitinase that can bind and cleave hyphal chitin. In its turn, the fungus produces fungalysin that specifically cleaves and inactivates chitinase, suppressing its function. Then the plant begins to express hevein-like peptides (WAMP) that bind to fungalysin and inhibit its activity. At the same time, fungalysin cannot cleave chitinase so that chitinase remains intact and can cleave the fungal chitin and suppress the effect of the pathogen.
Since both WAMP and chitinases are expressed in wheat seedlings in response to the infection with Fusarium fungal species [70, 76], we can suppose that the synergism between WAMP and chitinases demonstrated in vitro is also implemented in planta. It is still unclear whether the described mechanism of action is specific only for WAMP peptides or represents a characteristic of other hevein-like AMPs as well [77].
BIOTECHNOLOGICAL AND MEDICAL APPLICATIONS OF HEVEIN-LIKE
PEPTIDES
Due to the pronounced antimicrobial activity against phytopathogens, hevein-like peptides are used primarily for obtaining transgenic plants with enhanced resistance to pathogenic microorganisms. A vector construction based on hevein-encoding cDNA was made to obtain transgenic mustard (Brassica juncea) resistant to the fungus A. brassicae [78]. The biological testing in greenhouses showed that the obtained trangenic plants possessed all characteristics of resistance to alternariosis: longer incubation and latent periods, smaller size of necrosis, and smaller degree of disease development [78]. The hevein precursor expressed in transgenic tomatos was used in addition to the mature hevein. Interestingly, there was no processing, and no mature hevein was formed. However, the obtained plants acquired resistance to the phytopathogen Trichoderma hamatum, which was demonstrated by fungal growth inhibition [79].
Transgenic plants were also obtained using the promoter region of the hevein-encoding gene. The hevein promoter (HEV2.1) was used to obtain constructions allowing the expression of reporter protein β-glucuronidase (GUS) in transgenic Hevea. Interestingly, the expression of GUS in the roots and stems of transgenic plants was observed only in laticifer cells (the cells producing latex), while its expression in leaves was observed in all cells [80]. Currently there are attempts to use the hevein promoter (pHevP) for obtaining transgenic Hevea plants producing higher amounts of recombinant proteins used in pharmaceutics, namely, human protamine 1 and atrial natriuretic peptide [81, 82]. It is supposed that greater amounts of heterologous proteins can be obtained since latex is produced in Hevea in large amounts. Furthermore, it will not be necessary to use the whole plant for obtaining target proteins, but only to make an incision in the bark of the tree, without damaging the cambium, and to collect the dripping sap with the target protein [80-82].
In addition to hevein, transgenic plants were also obtained by using other 8C-Hev peptides [83]. In particular, the Pn-AMP2-encoding cDNA was successfully expressed in tobacco plants under the control of 35S promoter, resulting in their enhanced resistance to the oomycete Phytophthora parasitica causing late blight disease [84]. Transgenic tomato plants expressing the Pn-AMP2 peptide and possessing higher resistance to the phytopathogenic fungi P. capsici and F. oxysporum were obtained [85]. Since chitin was present in the cell walls of F. oxysporum but absent in the same cells of P. capsici, it was concluded that the presence of chitin was not a necessary condition for the exhibition of antifungal activity of the Pn-AMP2 peptide [85].
6C-Hev peptide Ac-AMP2 was used to obtain transgenic plants of tobacco [86] and tomato [87]. Transgenic tobacco plants were obtained using the following two types of constructions: one with the wild-type gene encoding Ac-AMP2 and the other with the mutant gene carrying no nucleotide sequence encoding the C-terminal propeptide. The transgenic plants obtained from both constructions contained, instead of the original peptide Ac-AMP2, its isoform with deleted Arg residue at the C-terminus, which was present only in intracellular fluid. This peptide isoform was isolated from transgenic tobacco plants, and its antimicrobial activity was determined in vitro. It was shown to possess an antifungal activity similar to that of Ac-AMP2. At the same time, the transgenic plants did not possess the enhanced resistance either to B. cinerea or to A. longipes, against which the isolated isopeptide was active [86].
The genes encoding 6C-Hev peptides of chickweed (SmAMP1, SmAMP2) were used for expression in tobacco plants to provide enhanced resistance to fungal infection [60]. It should be noted that the precursors of these hevein-like AMPs have a modular structure and contain two mature peptides in one cassette. It was shown that SmAMP precursors underwent processing in the obtained trangenic plants, resulting in the formation of SmAMP1 and SmAMP2 peptides. The obtained tobacco plants showed the highest resistance to the phytopathogenic fungi Bipolaris sorokiniana and Thielaviopsis basicola.
Hevein-like peptides are a comparatively small AMP family, the interest in which has increased recently. Due to the high number of cysteine residues, which form a motif typical to these AMPs, hevein-like peptides represent highly stable compounds resistant to heating and protease action. The chitin-binding site provides these peptides with the ability to bind to chitin, the major cell wall component of pathogens, which is absent in plants. Hevein-like AMPs have been found in mono- and dicotyledonous plants from different families. Almost all hevein-like peptides are active, at their micromolar concentrations, against the wide range of phytopathogens, often including bacteria besides fungi. These and other characteristic features of the peptides from this family make them important components of plant innate immunity. Based on two hevein-like peptides it has been shown that they act together with protective proteins, chitinases. It has been established that the WAMP peptide inhibited the secreted metalloprotease of the pathogenic fungus, which protected the plant chitinase from degradation by the enzyme of the pathogen. However, it is still unclear whether this mechanism is universal for all peptides from this family or is typical only for some of them. Hevein-like AMPs were used in biotechnology for obtaining transgenic plants with enhanced resistance to phytopathogens. A number of such plants expressing different hevein-like peptides have been obtained. It is attempted to use the promoter regions of the genes of these peptides for producing large amounts of drugs. The data presented above show that hevein-like AMPs represent a distinctive family, the investigation of which seems promising in both fundamental and practical respect.
Acknowledgments
This work was supported by the Russian Science Foundation (project No. 16-15-00167).
T. I. Odintsova thanks the Russian Foundation for Basic Research for support (grant No. 15-29-02480).
REFERENCES
1.Egorov, Ts. A., and Odintsova, T. I. (2012) Defense
peptides of plant immune system, Bioorg. Khim., 38,
7-17.
2.Silverstein, K. A., Moskal, W. A., Jr., Wu, H. C.,
Underwood, B. A., Graham, M. A., Town, C. D., and VandenBosch, K. A.
(2007) Small cysteine-rich peptides resembling antimicrobial peptides
have been under-predicted in plants, Plant J., 51,
262-280.
3.Slavokhotova, A. A., Shelenkov, A. A., and
Odintsova, T. I. (2015) Prediction of Leymus arenarius (L.)
antimicrobial peptides based on de novo transcriptome assembly,
Plant Mol. Biol., 89, 203-214.
4.Slavokhotova, A. A., Shelenkov, A. A., Korostyleva,
T. V., Rogozhin, E. A., Melnikova, N. V., Kudryavtseva, A. V., and
Odintsova, T. I. (2017) Defense peptide repertoire of Stellaria
media predicted by high throughput next generation sequencing,
Biochimie, 135, 15-27.
5.Costa, F. T., Neto, S. M., Bloch, C., Jr., and
Franco, O. L. (2007) Susceptibility of human pathogenic bacteria to
antimicrobial peptides from sesame kernels, Curr. Microbiol.,
55, 162-166.
6.Oard, S. V., and Enright, F. M. (2006) Expression
of the antimicrobial peptides in plants to control phytopathogenic
bacteria and fungi, Plant Cell Rep., 25, 561-572.
7.Loeza-Angeles, H., Sagrero-Cisneros, E.,
Lara-Zarate, L., Villagomez-Gomez, E., Lopez-Meza, J. E., and
Ochoa-Zarzosa, A. (2008) Thionin Thi2.1 from Arabidopsis
thaliana expressed in endothelial cells shows antibacterial,
antifungal and cytotoxic activity, Biotechnol. Lett., 30,
1713-1719.
8.Tam, J. P., Wang, S., Wong, K. H., and Tan, W. L.
(2015) Antimicrobial peptides from plants, Pharmaceuticals
(Basel), 8, 711-757.
9.Beintema, J. J. (1994) Structural features of plant
chitinases and chitin-binding proteins, FEBS Lett., 350,
159-163.
10.Iseli, B., Boller, T., and Neuhaus, J. M. (1993)
The N-terminal cysteine-rich domain of tobacco class I chitinase is
essential for chitin binding but not for catalytic or antifungal
activity, Plant Physiol., 103, 221-226.
11.Archer, B. L. (1960) The proteins of Hevea
brasiliensis latex. 4. Isolation and characterization of
crystalline hevein, Biochem. J., 75, 236-240.
12.Walujono, K., Scholma, R. A., Beintema, J. J.,
Mariono, A., and Hahn, A. M. (1975) Amino acid sequence of hevein,
Proc. Int. Rubber Conference, Kuala Lumpur, Vol. 2, pp.
518-531.
13.Chapot, M. P., Peumans, W. J., and Strosberg, A.
D. (1986) Extensive homologies between lectins from non-leguminous
plants, FEBS Lett., 195, 231-234.
14.Peumans, W. J., De Ley, M., and Broekaert, W. F.
(1983) An unusual lectin from stinging nettle (Urtica dioica)
rhizomes, FEBS Lett., 177, 99-103.
15.Broekaert, W. F., Vanparijs, J., Leyns, F., Joos,
H., and Peumans, W. J. (1989) A chitin-binding lectin from stinging
nettle rhizomes with antifungal properties, Science, 245,
1100-1102.
16.Van Parijs, J., Broekaert, W. F., Goldstein, I.
J., and Peumans, W. J. (1991) Hevein – an antifungal protein from
rubber-tree (Hevea brasiliensis) latex, Planta,
183, 258-264.
17.Broekaert, I., Lee, H. I., Kush, A., Chua, N. H.,
and Raikhel, N. (1990) Wound-induced accumulation of mRNA containing a
hevein sequence in laticifers of rubber tree (Hevea
brasiliensis), Proc. Natl. Acad. Sci. USA, 87,
7633-7637.
18.Lee, H. I., Broekaert, W. F., and Raikhel, N. V.
(1991) Co- and post-translational processing of the hevein
preproprotein of latex of the rubber tree (Hevea brasiliensis),
J. Biol. Chem., 266, 15944-15948.
19.Soedjanaatmadja, U. M., Hofsteenge, J.,
Jeronimus-Stratingh, C. M., Bruins, A. P., and Beintema, J. J. (1994)
Demonstration by mass spectrometry that pseudo-hevein and hevein have
ragged C-terminal sequences, Biochim. Biophys. Acta,
1209, 144-148.
20.Gidrol, X., Chrestin, H., Tan, H. L., and Kush,
A. (1994) Hevein, a lectin-like protein from Hevea brasiliensis
(rubber tree) is involved in the coagulation of latex, J. Biol.
Chem., 269, 9278-9283.
21.Pujade-Renaud, V., Sanier, C., Cambillau, L.,
Pappusamy, A., Jones, H., Ruengsri, N., Tharreau, D., Chrestin, H.,
Montoro, P., and Narangajavana, J. (2005) Molecular characterization of
new members of the Hevea brasiliensis hevein multigene family
and analysis of their promoter region in rice, Biochim. Biophys.
Acta, 1727, 151-161.
22.Andersen, N. H., Cao, B., Rodriguezromero, A.,
and Arreguin, B. (1993) Hevein-NMR assignment and assessment of
solution-state folding for the agglutinin-toxin motif,
Biochemistry, 32, 1407-1422.
23.Rodriguez, A., Tablero, M., Barragan, B., Lara,
P., Rangel, M., Arreguin, B., Possani, L., and Sorianogarcia, M. (1986)
Crystallization of hevein – a protein from latex of Hevea
brasiliensis (rubber tree), J. Crystal Growth, 76,
710-714.
24.Asensio, J. L., Canada, F. J., Siebert, H. C.,
Laynez, J., Poveda, A., Nieto, P. M., Soedjanaamadja, U. M., Gabius, H.
J., and Jimenez-Barbero, J. (2000) Structural basis for chitin
recognition by defense proteins: GlcNAc residues are bound in a
multivalent fashion by extended binding sites in hevein domains,
Chem. Biol., 7, 529-543.
25.Colombo, G., Meli, M., Canada, J., Asensio, J.
L., and Jimenez-Barbero, J. (2004) Toward the understanding of the
structure and dynamics of protein–carbohydrate interactions:
molecular dynamics studies of the complexes between hevein and
oligosaccharidic ligands, Carbohydr. Res., 339,
985-994.
26.Aboitiz, N., Vila-Perello, M., Groves, P.,
Asensio, J. L., Andreu, D., Canada, F. J., and Jimenez-Barbero, J.
(2004) NMR and modeling studies of protein–carbohydrate
interactions: synthesis, three-dimensional structure, and recognition
properties of a minimum hevein domain with binding affinity for
chitooligosaccharides, Chembiochem, 5, 1245-1255.
27.Mareska, V., Tvaroska, I., Kralova, B., and
Spiwok, V. (2015) Molecular simulations of hevein/(GlcNAc)(3) complex
with weakened OH/O and CH/pi hydrogen bonds: implications for their
role in complex stabilization, Carbohydr. Res., 408,
1-7.
28.Chavez, M. I., Vila-Perello, M., Canada, F. J.,
Andreu, D., and Jimenez-Barbero, J. (2010) Effect of a
serine-to-aspartate replacement on the recognition of chitin
oligosaccharides by truncated hevein. A 3D view by using NMR,
Carbohydr. Res., 345, 1461-1468.
29.Ownby, D. R. (2002) A history of latex allergy,
J. Allergy Clin. Immunol., 110, S27-32.
30.Bousquet, J., Flahault, A., Vandenplas, O.,
Ameille, J., Duron, J. J., Pecquet, C., Chevrie, K., and
Annesi-Maesano, I. (2006) Natural rubber latex allergy among health
care workers: a systematic review of the evidence, J. Allergy Clin.
Immunol., 118, 447-454.
31.Radauer, C., Adhami, F., Furtler, I., Wagner, S.,
Allwardt, D., Scala, E., Ebner, C., Hafner, C., Hemmer, W., Mari, A.,
and Breiteneder, H. (2011) Latex-allergic patients sensitized to the
major allergen hevein and hevein-like domains of class I chitinases
show no increased frequency of latex-associated plant food allergy,
Mol. Immunol., 48, 600-609.
32.Chen, Z., Duser, M., Flagge, A., Maryska, S.,
Sander, I., Raulf-Heimsoth, M., and Baur, X. (2000) Identification and
characterization of cross-reactive natural rubber latex and Ficus
benjamina allergens, Int. Arch. Allergy Immunol.,
123, 291-298.
33.Alenius, H., Kalkkinen, N., Lukka, M., Reunala,
T., Turjanmaa, K., Makinen-Kiljunen, S., Yip, E., and Palosuo, T.
(1995) Prohevein from the rubber tree (Hevea brasiliensis) is a
major latex allergen, Clin. Exp. Allergy, 25,
659-665.
34.Blanco, C. (2003) Latex-fruit syndrome, Curr.
Allergy Asthma Rep., 3, 47-53.
35.Wagner, S., and Breiteneder, H. (2002) The
latex-fruit syndrome, Biochem. Soc. Trans., 30,
935-940.
36.Barre, A., Culerrier, R., Granier, C., Selman,
L., Peumans, W. J., Van Damme, E. J., Bienvenu, F., Bienvenu, J., and
Rouge, P. (2009) Mapping of IgE-binding epitopes on the major latex
allergen Hev b2 and the cross-reacting 1,3-beta-glucanase fruit
allergens as a molecular basis for the latex-fruit syndrome, Mol.
Immunol., 46, 1595-1604.
37.Ganglberger, E., Radauer, C., Wagner, S.,
Riordain, G., Beezhold, D. H., Brehler, R., Niggemann, B., Scheiner,
O., Jensen-Jarolim, E., and Breiteneder, H. (2001) Hev b8, the Hevea
brasiliensis latex profilin, is a cross-reactive allergen of latex,
plant foods and pollen, Int. Arch. Allergy. Immunol.,
125, 216-227.
38.Schmidt, M. H., Raulf-Heimsoth, M., and Posch, A.
(2002) Evaluation of patatin as a major cross-reactive allergen in
latex-induced potato allergy, Ann. Allergy Asthma Immunol.,
89, 613-618.
39.Beezhold, D. H., Hickey, V. L., Kostyal, D. A.,
Puhl, H., Zuidmeer, L., Van Ree, R., and Sussman, G. L. (2003) Lipid
transfer protein from Hevea brasiliensis (Hev b12), a
cross-reactive latex protein, Ann. Allergy Asthma Immunol.,
90, 439-445.
40.Ferreira, F., Wallner, M., Breiteneder, H.,
Hartl, A., Thalhamer, J., and Ebner, C. (2002) Genetic engineering of
allergens: future therapeutic products, Int. Arch. Allergy
Immunol., 128, 171-178.
41.Banerjee, B., Wang, X., Kelly, K. J., Fink, J.
N., Sussman, G. L., and Kurup, V. P. (1997) IgE from latex-allergic
patients binds to cloned and expressed B cell epitopes of prohevein,
J. Immunol., 159, 5724-5732.
42.Beezhold, D. H., Kostyal, D. A., and Sussman, G.
L. (1997) IgE epitope analysis of the hevein preprotein; a major latex
allergen, Clin. Exp. Immunol., 108, 114-121.
43.Drew, A. C., Eusebius, N. P., Kenins, L., De
Silva, H. D., Suphioglu, C., Rolland, J. M., and O’Hehir, R. E.
(2004) Hypoallergenic variants of the major latex allergen Hev b6.01
retaining human T lymphocyte reactivity, J. Immunol.,
173, 5872-5879.
44.Karisola, P., Mikkola, J., Kalkkinen, N.,
Airenne, K. J., Laitinen, O. H., Repo, S., Pentikainen, O. T., Reunala,
T., Turjanmaa, K., Johnson, M. S., Palosuo, T., Kulomaa, M. S., and
Alenius, H. (2004) Construction of hevein (Hev b6.02) with reduced
allergenicity for immunotherapy of latex allergy by comutation of six
amino acid residues on the conformational IgE epitopes, J.
Immunol., 172, 2621-2628.
45.Reyes-Lopez, C. A., Hernandez-Santoyo, A.,
Pedraza-Escalona, M., Mendoza, G., Hernandez-Arana, A., and
Rodriguez-Romero, A. (2004) Insights into a conformational epitope of
Hev b6.02 (hevein), Biochem. Biophys. Res. Commun., 314,
123-130.
46.Reyes-Lopez, C. A., Pedraza-Escalona, M.,
Mendoza, G., Hernandez-Santoyo, A., and Rodriguez-Romero, A. (2006) A
single amino acid substitution on the surface of a natural hevein
isoform (Hev b6.0202), confers different IgE recognition, FEBS
Lett., 580, 2483-2487.
47.Pedraza-Escalona, M., Becerril-Lujan, B.,
Agundis, C., Dominguez-Ramirez, L., Pereyra, A., Riano-Umbarila, L.,
and Rodriguez-Romero, A. (2009) Analysis of B-cell epitopes from the
allergen Hev b6.02 revealed by using blocking antibodies, Mol.
Immunol., 46, 668-676.
48.Koo, J. C., Lee, S. Y., Chun, H. J., Cheong, Y.
H., Choi, J. S., Kawabata, S., Miyagi, M., Tsunasawa, S., Ha, K. S.,
Bae, D. W., Han, C. D., Lee, B. L., and Cho, M. J. (1998) Two hevein
homologs isolated from the seed of Pharbitis nil L. exhibit
potent antifungal activity, Biochim. Biophys. Acta, 1382,
80-90.
49.Fujimura, M., Minami, Y., Watanabe, K., and
Tadera, K. (2003) Purification, characterization, and sequencing of a
novel type of antimicrobial peptides, Fa-AMP1 and Fa-AMP2, from seeds
of buckwheat (Fagopyrum esculentum Moench.), Biosci.
Biotechnol. Biochem., 67, 1636-1642.
50.Li, S. S., and Claeson, P. (2003) Cys/Gly-rich
proteins with a putative single chitin-binding domain from oat
(Avena sativa) seeds, Phytochemistry, 63,
249-255.
51.Broekaert, W. F., Marien, W., Terras, F. R., De
Bolle, M. F., Proost, P., Van Damme, J., Dillen, L., Claeys, M., Rees,
S. B., Vanderleyden, J., et al. (1992) Antimicrobial peptides from
Amaranthus caudatus seeds with sequence homology to the
cysteine/glycine-rich domain of chitin-binding proteins,
Biochemistry, 31, 4308-4314.
52.Rivillas-Acevedo, L. A., and Soriano-Garcia, M.
(2007) Isolation and biochemical characterization of an antifungal
peptide from Amaranthus hypochondriacus seeds, J. Agric. Food
Chem., 55, 10156-10161.
53.Lipkin, A., Anisimova, V., Nikonorova, A.,
Babakov, A., Krause, E., Bienert, M., Grishin, E., and Egorov, T.
(2005) An antimicrobial peptide Ar-AMP from amaranth (Amaranthus
retroflexus L.) seeds, Phytochemistry, 66,
2426-2431.
54.Kini, S. G., Nguyen, P. Q., Weissbach, S.,
Mallagaray, A., Shin, J., Yoon, H. S., and Tam, J. P. (2015) Studies on
the chitin binding property of novel cysteine-rich peptides from
Alternanthera sessilis, Biochemistry, 54,
6639-6649.
55.De Bolle, M. F., David, K. M., Rees, S. B.,
Vanderleyden, J., Cammue, B. P., and Broekaert, W. F. (1993) Cloning
and characterization of a cDNA encoding an antimicrobial chitin-binding
protein from amaranth, Amaranthus caudatus, Plant. Mol.
Biol., 22, 1187-1190.
56.Nielsen, K. K., Nielsen, J. E., Madrid, S. M.,
and Mikkelsen, J. D. (1997) Characterization of a new antifungal
chitin-binding peptide from sugar beet leaves, Plant Physiol.,
113, 83-91.
57.Huang, X., Xie, W., and Gong, Z. (2000)
Characteristics and antifungal activity of a chitin binding protein
from Ginkgo biloba, FEBS Lett., 478, 123-126.
58.Rogozhin, E. A., Slezina, M. P., Slavokhotova, A.
A., Istomina, E. A., Korostyleva, T. V., Smirnov, A. N., Grishin, E.
V., Egorov, T. A., and Odintsova, T. I. (2015) A novel antifungal
peptide from leaves of the weed Stellaria media L.,
Biochimie, 116, 125-132.
59.Slavokhotova, A. A., Shelenkov, A. A.,
Korostyleva, T. V., Rogozhin, E. A., Melnikova, N. V., Kudryavtseva, A.
V., and Odintsova, T. I. (2017) Defense peptide repertoire of
Stellaria media predicted by high throughput next generation
sequencing, Biochimie, 135, 15-27.
60.Shukurov, R. R., Voblikova, V. D., Nikonorova, A.
K., Komakhin, R. A., Komakhina, V. V., Egorov, T. A., Grishin, E. V.,
and Babakov, A. V. (2011) Transformation of tobacco and
Arabidopsis plants with Stellaria media genes encoding
novel hevein-like peptides increases their resistance to fungal
pathogens, Transgenic Res., 21, 313-325.
61.Martins, J. C., Maes, D., Loris, R., Pepermans,
H. A., Wyns, L., Willem, R., and Verheyden, P. (1996) H-NMR study of
the solution structure of Ac-AMP2, a sugar binding antimicrobial
protein isolated from Amaranthus caudatus, J. Mol. Biol.,
258, 322-333.
62.Huang, R. H., Xiang, Y., Liu, X. Z., Zhang, Y.,
Hu, Z., and Wang, D. C. (2002) Two novel antifungal peptides distinct
with a five-disulfide motif from the bark of Eucommia ulmoides
Oliv., FEBS Lett., 521, 87-90.
63.Huang, R. H., Xiang, Y., Tu, G. Z., Zhang, Y.,
and Wang, D. C. (2004) Solution structure of Eucommia antifungal
peptide: a novel structural model distinct with a five-disulfide motif,
Biochemistry, 43, 6005-6012.
64.Xiang, Y., Huang, R. H., Liu, X. Z., Zhang, Y.,
and Wang, D. C. (2004) Crystal structure of a novel antifungal protein
distinct with five disulfide bridges from Eucommia ulmoides
Oliver at an atomic resolution, J. Struct. Biol., 148,
86-97.
65.Van den Bergh, K. P., Proost, P., Van Damme, J.,
Coosemans, J., Van Damme, E. J., and Peumans, W. J. (2002) Five
disulfide bridges stabilize a hevein-type antimicrobial peptide from
the bark of spindle tree (Euonymus europaeus L.), FEBS
Lett., 530, 181-185.
66.Van den Bergh, K. P., Rouge, P., Proost, P.,
Coosemans, J., Krouglova, T., Engelborghs, Y., Peumans, W. J., and Van
Damme, E. J. (2004) Synergistic antifungal activity of two
chitin-binding proteins from spindle tree (Euonymus europaeus
L.), Planta, 219, 221-232.
67.Odintsova, T. I., Vassilevski, A. A.,
Slavokhotova, A. A., Musolyamov, A. K., Finkina, E. I., Khadeeva, N.
V., Rogozhin, E. A., Korostyleva, T. V., Pukhalsky, V. A., Grishin, E.
V., and Egorov, T. A. (2009) A novel antifungal hevein-type peptide
from Triticum kiharae seeds with a unique 10-cysteine motif,
FEBS J., 276, 4266-4275.
68.Utkina, L. L., Zhabon, E. O., Slavokhotova, A.
A., Rogozhin, E. A., Shiian, A. N., Grishin, E. V., Egorov, Ts. A.,
Odintsova, T. I., and Pukhal’skii, V. A. (2010) Heterologous
expression of a synthetic gene encoding a novel hevein-type
antimicrobial peptide of Leymus arenarius in Escherichia
coli cells, Genetika, 46, 1645-1651.
69.Dubovskii, P. V., Vassilevski, A. A.,
Slavokhotova, A. A., Odintsova, T. I., Grishin, E. V., Egorov, T. A.,
and Arseniev, A. S. (2011) Solution structure of a defense peptide from
wheat with a 10-cysteine motif, Biochem. Biophys. Res. Commun.,
411, 14-18.
70.Andreev, Y. A., Korostyleva, T. V., Slavokhotova,
A. A., Rogozhin, E. A., Utkina, L. L., Vassilevski, A. A., Grishin, E.
V., Egorov, T. A., and Odintsova, T. I. (2012) Genes encoding
hevein-like defense peptides in wheat: distribution, evolution, and
role in stress response, Biochimie, 94, 1009-1016.
71.Muraki, M. (2002) The importance of CH/pi
interactions to the function of carbohydrate binding proteins,
Protein Pept. Lett., 9, 195-209.
72.Chavez, M. I., Andreu, C., Vidal, P., Aboitiz,
N., Freire, F., Groves, P., Asensio, J. L., Asensio, G., Muraki, M.,
Canada, F. J., and Jimenez-Barbero, J. (2005) On the importance of
carbohydrate–aromatic interactions for the molecular recognition
of oligosaccharides by proteins: NMR studies of the structure and
binding affinity of AcAMP2-like peptides with non-natural naphthyl and
fluoroaromatic residues, Chemistry, 11, 7060-7074.
73.Koo, J. C., Lee, B., Young, M. E., Koo, S. C.,
Cooper, J. A., Baek, D., Lim, C. O., Lee, S. Y., Yun, D. J., and Cho,
M. J. (2004) Pn-AMP1, a plant defense protein, induces actin
depolarization in yeasts, Plant Cell Physiol., 45,
1669-1680.
74.Naumann, T. A., Wicklow, D. T., and Price, N. P.
(2011) Identification of a chitinase-modifying protein from Fusarium
verticillioides: truncation of a host resistance protein by a
fungalysin metalloprotease, J. Biol. Chem., 286,
35358-35366.
75.Naumann, T. A., and Price, N. P. (2012)
Truncation of class IV chitinases from Arabidopsis by secreted
fungal proteases, Mol. Plant Pathol., 13, 1135-1139.
76.Caruso, C., Chilosi, G., Caporale, C., Leonardi,
L., Bertini, L., Magro, P., and Buonocore, V. (1990) Induction of
pathogenesis-related proteins in germinating wheat seeds infected with
Fusarium culmorum, Plant Sci., 140, 87-97.
77.Slavokhotova, A. A., Naumann, T. A., Price, N.
P., Rogozhin, E. A., Andreev, Y. A., Vassilevski, A. A., and Odintsova,
T. I. (2014) Novel mode of action of plant defense peptides –
hevein-like antimicrobial peptides from wheat inhibit fungal
metalloproteases, FEBS J., 281, 4754-4764.
78.Kanrar, S., Venkateswari, J. C., Kirti, P. B.,
and Chopra, V. L. (2002) Transgenic expression of hevein, the rubber
tree lectin, in Indian mustard confers protection against Alternaria
brassicae, Plant Sci., 162, 441-448.
79.Lee, H. I., and Raikhel, N. V. (1995) Prohevein
is poorly processed but shows enhanced resistance to a chitin-binding
fungus in transgenic tomato plants, Braz. J. Med. Biol. Res.,
28, 743-750.
80.Montoro, P., Lagier, S., Baptiste, C., Marteaux,
B., Pujade-Renaud, V., Leclercq, J., and Alemanno, L. (2008) Expression
of the HEV2.1 gene promoter in transgenic Hevea brasiliensis,
Plant Cell Tissue Organ Cult., 94, 55-63.
81.Sunderasan, E. B., Badaruddin, B. E., Azharuddin,
A., and Arokiaraj, P. (2012) Genetic transformation of Hevea
brasiliensis with human atrial natriuretic factor, J. Rubber
Res., 15, 255-264.
82.Sunderasan, E. S., Shuib, S. S., Badaruddin, B.
E., Azharuddin, A., and Arokiaraj, P. (2010) Hevea genetic
transformation for enhanced recombinant pharmaceutical production by
the use of hevein promoter, in National Biotechnology Seminar,
Kuala Lumpur, Malaysia.
83.Berthelot, K., Peruch, F., and Lecomte, S. (2016)
Highlights on Hevea brasiliensis (pro)hevein proteins,
Biochimie, 127, 258-270.
84.Koo, J. C., Chun, H. J., Park, H. C., Kim, M. C.,
Koo, Y. D., Koo, S. C., Ok, H. M., Park, S. J., Lee, S. H., Yun, D. J.,
Lim, C. O., Bahk, J. D., Lee, S. Y., and Cho, M. J. (2002)
Over-expression of a seed specific hevein-like antimicrobial peptide
from Pharbitis nil enhances resistance to a fungal pathogen in
transgenic tobacco plants, Plant Mol. Biol., 50,
441-452.
85.Lee, O. S., Lee, B., Park, N., Koo, J. C., Kim,
Y. H., Prasad, D. T., Karigar, C., Chun, H. J., Jeong, B. R., Kim, D.
H., Nam, J., Yun, J. G., Kwak, S. S., Cho, M. J., and Yun, D. J. (2003)
Pn-AMPs, the hevein-like proteins from Pharbitis nil confers
disease resistance against phytopathogenic fungi in tomato,
Lycopersicum esculentum, Phytochemistry, 62,
1073-1079.
86.De Bolle, M. F., Osborn, R. W., Goderis, I. J.,
Noe, L., Acland, D., Hart, C. A., Torrekens, S., Van Leuven, F., and
Broekaert, W. F. (1996) Antimicrobial peptides from Mirabilis
jalapa and Amaranthus caudatus: expression, processing,
localization and biological activity in transgenic tobacco, Plant
Mol. Biol., 31, 993-1008.
87.Liapkova, N. S., Loskutova, N. A., Maisurian, A.
N., Mazin, V. V., Korableva, N. P., Platonova, T. A., Ladyzhenskaia, E.
P., and Evsiunina, A. S. (2001) Isolation of genetically modified
potato plant containing the gene of defensive peptide from
Amaranthus, Prikl. Biokhim. Mikrobiol., 37,
349-354.