Influence of a Single Point Mutation in the Constant Domain of the Bence-Jones Protein bif on Its Aggregation Properties
M. A. Timchenko1* and A. A. Timchenko2
1Institute of Theoretical and Experimental Biophysics, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; E-mail: maria_timchenko@mail.ru2Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia
* To whom correspondence should be addressed.
Received May 1, 2017; Revision received October 10, 2017
Multiple myeloma nephropathy occurs due to the aggregate formation by monoclonal immunoglobulin light chains (Bence-Jones proteins) in kidneys of patients with multiple myeloma. The mechanism of amyloid deposit formation is still unclear. Earlier, the key role in the fibril formation has been assigned to the variable domains that acquired amyloidogenic properties as a result of somatic mutations. However, fibril formation by the Bence-Jones protein BIF was found to be the function of its constant domain. The substitution of Ser177 by Asn in the constant domain of the BIF protein is most likely an inherited than a somatic mutation. To study the role of this mutation in amyloidogenesis, the recombinant Bence-Jones protein BIF and its mutant with the N177S substitution typical for the known immunoglobulin Cκ allotypes Km1, Km1,2, and Km3 were isolated. The morphology of aggregates formed by the recombinant proteins under conditions similar to those occurring during the protein transport in bloodstream and its filtration into the renal glomerulus, in the distal tubules, and in the proximal renal tubules was analyzed by atomic force microscopy. The nature of the aggregates formed by BIF and its N177S mutant during incubation for 14 days at 37°C strongly differed and depended on both pH and the presence of a reducing agent. BIF formed fibrils at pH 7.2, 6.5, and 10.1, while the N177S mutant formed fibrils only at alkaline pH 10.1. The refolding of both proteins in the presence of 5 mM dithiothreitol resulted in the formation of branched structures.
KEY WORDS: Bence-Jones proteins, myeloma, aggregation, atomic force microscopyDOI: 10.1134/S0006297918020037
Abbreviations: AFM, atomic force microscopy; BJ myeloma, Bence-Jones myeloma; BJPs, Bence-Jones proteins (Ig light chains); CL, constant domain of Ig light chain; MM, multiple myeloma; VL, variable domain of Ig light chain.
Myeloma nephropathy is a major clinical complication of multiple myeloma
(MM), the cancer of B cells. It is related to the overproduction of
abnormal proteins – paraproteins of immunoglobulin (Ig) group
– by malignant clonal plasma cells [1, 2]. Myeloma proteins have an abnormal structure due to
the mutations in immunoglobulin genes. Myeloma cells are able to
produce either only Ig light chains (Bence-Jones myeloma, 10% of
patients) or only heavy chains, as well as molecular fragments of Ig.
In 80-90% cases, Bence-Jones (BJ) myeloma is accompanied by amyloidosis
of light chains that primarily results in renal failure. It is related
to the fact that Bence-Jones proteins (BJPs, Ig light chains) produced
by the myeloma cells enter the bloodstream, reach the kidneys, easily
penetrate intact glomerulus filter, and form large amounts of amyloid
deposits in renal tubules, which results in the obturation of distal
tubules. The amyloid precursor molecules are mostly λ light
chains and less frequently – κ light chains. The mechanisms
of amyloid nephropathy progression have not been completely understood.
It is believed that amyloidogenicity of BJPs is due to the changes in
their primary structure determined by genetic code or acquired
throughout lifetime. The analysis of amyloid deposits in BJ myeloma
revealed that they mainly represent the variable domain of the Ig light
chain (VL) or VL with a fragment of the constant
domain (CL) (up to 60 amino acid residues) whose
amyloidogenic properties are the result of somatic mutations. However,
Solomon et al. [3] found in one of the BJ myeloma
patients amyloid fibrils that were formed entirely by amino acids
109-207 of the CL domain of the BJP κ light chain (BJP
BIF). Its constant domain contained the Ser to Asn substitution at
position 177, which, in author’s opinion, was an inherited
mutation.
In this work, we obtained the recombinant BJP protein BIF and its mutant with the N177S substitution typical for the known Km1, Km1,2, and Km3 allotypes of κ Ig chains and compared their aggregation properties by the method of atomic force microscopy (AFM) under conditions reflecting the microenvironments at various parts of the renal nephron.
MATERIALS AND METHODS
Construction of the BIF-expressing plasmid pETBIF. Amino acid sequence of the BIF protein [3] was translated according to the standard genetic code into the coding nucleotide sequence with codon optimization for the expression in E. coli. The DNA fragment encoding this sequence was synthesized by polymerization reaction with overlapping oligonucleotides using Velocity DNA polymerase (Bioline, Germany) and then amplified by polymerase chain reaction (PCR). The obtained PCR fragment was cloned into NdeI and XhoI restriction sites into the pET23b expression vector (Novagen, USA) under control of the T7 RNA polymerase promotor. The presence of the insert was confirmed by restriction analysis and sequencing. The construct contained (His)6-tag at the C-terminus for further protein purification by affinity chromatography. Recombinant analogs of native BJPs have been studied in a number of earlier works and were found to reliably reflect the structural properties of the native proteins [4, 5].
Site-directed mutagenesis. The N177S mutant of the BIF protein (plasmid pETN177S) was obtained by site-directed mutagenesis. To substitute Asn N (AAC)177 with Ser S (AGC), the following primers were used: BJP(N-S)F: 5′-CCTATAGCCTGAGCAGCACCCTGACCCTGA-3′; BJP(N-S)R: 5′-TCAGGGTCAGGGTGCTGCTCAGGCTATAGG-3′. The accuracy of mutagenesis was confirmed by sequencing.
Expression and purification of the recombinant BIF protein and its mutant analog. Escherichia coli BL21(DE3) cells were transformed with the pETBIF and pETN177S plasmids by a standard method [6]. Expression was performed in Erlenmeyer flasks on a shaker in 1 liter of LB medium [6] containing 0.1 g/liter carbenicillin (Bioline). The cells were grown at 37°C, and the overnight culture was added to the flasks at a 1 : 100 dilution. When the optical density of the culture reached 0.5 (λ = 550 nm), expression of the recombinant proteins was induced by addition of isopropyl-β-D-thiogalactopyranoside (IPTG) (Bioline) to a final concentration of 1 mM. After 6 h of incubation at 37°C, the cells were pelleted by centrifugation at 3000g for 15 min at 4°C. The obtained cell pellet was frozen and stored at –20°C before use.
Cells collected from 1 liter of culture were resuspended in 50 ml of NPI-10 buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0), and lysozyme (Amresco, USA) was added to a final concentration of 1 mg/ml. The cells were vortexed on ice for 30 min and then disintegrated by sonication. The obtained lysate was centrifuged for 30 min at 10,000g at 4°C. The supernatant was then discarded, and the pellet containing inclusion bodies was resuspended in 20 ml of NPI-10 for washing and centrifuged for 30 min at 10,000g at 4°C. After discarding the supernatant, the pellet was divided into two portions. One of them was resuspended in 10 ml of NPI-10 buffer containing 6 M guanidine hydrochloride (Amresco) and 1 mM PMSF (phenylmethylsulfonyl fluoride) (Amresco) and sonicated. After sonication, Tween-20 (Amresco) was added to a final concentration of 2%, and the suspension was vortexed on ice for 1 h. The resulting lysate was centrifuged for 30 min at 10,000g at 20°C to remove the remaining insoluble fraction and then transferred into clean microtubes. The lysate was dialyzed against NPI-10 buffer containing 6 M urea (Amresco) and loaded on a Ni-NTA-Sepharose column (Novagen, USA) equilibrated with NPI-10 buffer containing 6 M urea. The column was washed with 10 volumes of NPI-20 buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0) containing 6 M urea, and the bound proteins were eluted with one volume of NPI-250 buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0) containing 6 M urea. Proteins from the second portion of the pellet were isolated using the same procedure except all used buffers contained 5 mM dithiothreitol (DTT) (Serva, Germany). The presence of protein was confirmed by Laemmli gel electrophoresis [7].
Refolding of the recombinant BIF protein and its mutant analog. The refolding of proteins was performed according to the procedure [8]. Protein-containing fractions after affinity chromatography were dialyzed for 12 h against NP phosphate buffer (50 mM NaH2PO4, 300 mM NaCl, pH 8.0) sequentially containing 6, 3, and 1 M urea. Insoluble protein aggregates precipitated and were removed by centrifugation after each stage of dialysis. Subsequent refolding was performed by controlled dialysis. NP buffer containing 1 M urea was replaced with 50 mM sodium phosphate buffer, pH 7.2, without urea for 48 h (10% buffer volume per hour). After centrifugation, the supernatant was divided into four fractions that were dialyzed for 12 h against (i) 50 mM sodium phosphate buffer, pH 7.2, containing 0.1 M NaCl; (ii) 50 mM sodium phosphate buffer, pH 6.5, containing 0.4 M NaCl and 0.4 M urea; (iii) 30 mM sodium acetate buffer, pH 4.5, containing 0.245 M NaCl; (iv) 25 mM bicarbonate buffer (NaHCO3-NaOH, pH 10.1) to analyze the effect of alkalinization on the process of amyloid formation.
When protein fractions were obtained under reducing conditions, refolding was performed in the presence of 5 mM DTT, and the fractions were dialyzed for 12 h against 50 mM sodium phosphate buffer, pH 7.2, containing 0.1 M NaCl and 5 mM DTT.
The obtained samples were filtered through a 0.22-µm filter (Millipore, USA). The protein yield was 0.1-0.3 mg/ml. The presence of protein and its purity was confirmed by Laemmli gel electrophoresis [7].
Formation of aggregates by BIF and its mutant analog in solutions. The filtered protein samples in an appropriate buffer were incubated at 37°C for 14 days. Every 24 h, 50-µl aliquot was collected to analyze self-association of proteins by AFM. The first aliquot (day 1) was taken immediately after dialysis and filtration.
Thioflavin T (ThT) assay. A 10-µl sample of protein solution (0.1 mg/ml) was added to 1 ml of 10 mM Na2HPO4, 150 mM NaCl, pH 7.0, containing ThT (0.016 mg/ml; Sigma-Aldrich, USA). The intensity of solution fluorescence was measured with a Cary Eclipse spectrofluorimeter (Varian, Australia) at 482 nm (excitation at 440 nm) [9], and the fluorescence intensity of ThT (0.016 mg/ml) in the absence of protein was subtracted from the obtained value.
Congo Red spectroscopic assay. A 10-µl sample of protein solution (0.1 mg/ml) was added to 1 ml of 5 mM Na2HPO4, 150 mM NaCl, pH 7.4, containing 34.8 µg/ml Congo Red (Sigma-Aldrich). After incubation for 30 min at room temperature, the protein spectrum was recorded in the 400-700 nm range (quartz cell, 1 cm) using a UV-2401 spectrophotometer (Shimadzu, Japan). The spectrum of Congo Red in the absence of protein was subtracted from the obtained protein solution spectrum. The maximal spectral difference at 540 nm is indicative of the presence of amyloid fibrils [10].
Electrophoretic analysis of amyloid structures in agarose gel with sodium dodecyl sulfate (SDS). The presence of amyloids in the samples was confirmed by electrophoresis in agarose gel in the presence of 0.1% SDS according to the procedure [11]. The gel was stained with Zn/imidazole [12] and analyzed with the Totallab Quant software (http://totallab.com, Great Britain).
Ultracentrifugation. Using analytical ultracentrifugation, the distribution of molecular mass and sedimentation coefficients were determined for the BIF and N177S mutant samples in buffer with pH 7.2 immediately after dialysis (day 1 of incubation). Sedimentation measurements were performed at 20°C in a Beckman (USA) Optima XL-I analytical ultracentrifuge. The rotor speed was 40,000 rpm. Scanning conditions: 40 scans were registered with 4-min intervals and a resolution of 0.003 cm. Sedimentation was performed in two-sector cells using an An60 rotor. The sedimentation data were treated with SEDFIT [13] in the mode of continuous distribution of sedimentation coefficients c(S).
Small angle X-ray scattering (SAXS). SAXS measurements were performed in the small-angle BL-6A chamber at the Photon Factory (Tsukuba, Japan). Protein solution in the thermostated cell with mica windows was irradiated by X-rays with a wavelength of 1.503 Å at 23°C. The distance between the sample and the source was 2.35 m. The range of detectable scattering vectors Q was 0.008-0.2 Å–1 (Q = 4πsinθ/λ, where λ is the wavelength of X-rays, 2θ is the scattering angle). The data were registered with a PILATUS 100K two-dimensional CCD X-ray detector. The shape of particles was estimated from the slope (tanα) of logI dependence on logQ, where I is intensity of scattering, and Q is scattering vector [14].
AFM measurements. To prepare samples for AFM, 2 µl of the protein sample incubated at 37°C (0.1 mg/ml) was transferred to freshly cleaved mica and incubated for 5 min. The sample was then washed twice in a drop of distilled water for 30 s and dried in the air. All samples collected at different time points were prepared similarly.
To analyze the presence of fibrils after 30 days of incubation at 37°C, protein aliquots were taken, SDS was added to a final concentration of 0.09%, and the samples were incubated for two days at room temperature [15]. The samples were then loaded on freshly cleaved mica and analyzed by AFM.
AFM imaging was performed with an AFM Integra-Vita microscope (NT-MDT, Russia) in a noncontact (tapping) mode in air. The typical scan rate was 1 Hz. Measurements were carried out using NSG03 cantilevers with a resonance frequency of 47-150 kHz and ensured 10-nm tip curvature radius. The processing and presentations of AFM images were performed using Nova software (NT-MDT) and Gwyddion 2.44 software (http://gwyddion.net/, Czech Republic).
Congo Red birefringence assay. For the microscopic analysis, protein samples that were incubated at 37°C and stained with Congo Red for spectroscopic assay were transferred from the cuvette to a microtube and centrifuged at 12,000-14,000 rpm to pellet the aggregates. The pellet was washed with water, resuspended in a small amount of water, and placed on a cover glass. After drying for 24 h the sample was analyzed under polarized light at 40× magnification with an Olympus light microscope equipped with polarizers and a CCD camera.
RESULTS
Immunoglobulin light chains can form monomers, dimers, and tetramers as a result of noncovalent association and SH/SS exchange, as well as complexes with individual CL- and VL-domains [16]. The process of protein self-association strongly depends on the protein concentration and microenvironment. An important question is what affects protein association and formation of aggregates of different morphology in multiple myeloma. The Bence-Jones protein BIF discovered by Solomon and co-authors had high amyloidogenicity at neutral pH [3], which, in the author’s opinion, was due to the substitution of Ser with Asn at position 177 of the protein constant domain. To analyze the contribution of this mutation to the process of amyloid formation, the recombinant protein N177S containing Ser at position 177 was obtained. The presence of Ser at position 177 corresponded to the known allotypes of κ chains of immunoglobulins in healthy humans. The aggregation properties of both proteins and the morphology of their aggregates under different conditions were compared by AFM. Three buffer systems were used in this study: (1) 50 mM phosphate buffer containing 0.1 M NaCl, pH 7.2; (2) 50 mM phosphate buffer containing 0.4 M NaCl and 0.4 M urea, pH 6.5; (3) 30 mM acetate buffer containing 0.245 M NaCl, pH 4.5. These buffer systems were chosen to reflect environments that immunoglobulin light chains are exposed to in the nephron [17]. Buffer 1 represents conditions expected during protein transport in the bloodstream and its filtration in the glomerulus. Buffer 2 contains urea and salt to emulate the microenvironment of the distal tubule. The salt concentration corresponds to the condition of partial dehydration, which results in the development of renal pathologies involving BJPs, and the chosen urea concentration is considerably less than the concentration typically required to solubilize proteins. Buffer 3 with low pH simulates the conditions in the renal proximal tubule (the site of light chain catabolism, as well as urine acidification), because acidification is considered to be a contributing factor to the nephrotoxicity of BJPs. In addition, buffer 4 (25 mM bicarbonate buffer, pH 10.1) was used to estimate how pH shift into the alkaline range would affect the process of protein association. BIF and its N177S analog were obtained from inclusion bodies by the affinity chromatography and renaturation at pH 8.0. After renaturation, protein fractions were dialyzed against the appropriate buffer system. Some protein fractions were obtained in the presence of 5 mM DTT (reducing agent) to evaluate the influence of reducing conditions on the aggregation process; in the final step, these samples were dialyzed against buffer 1 (pH 7.2) containing 5 mM DTT.
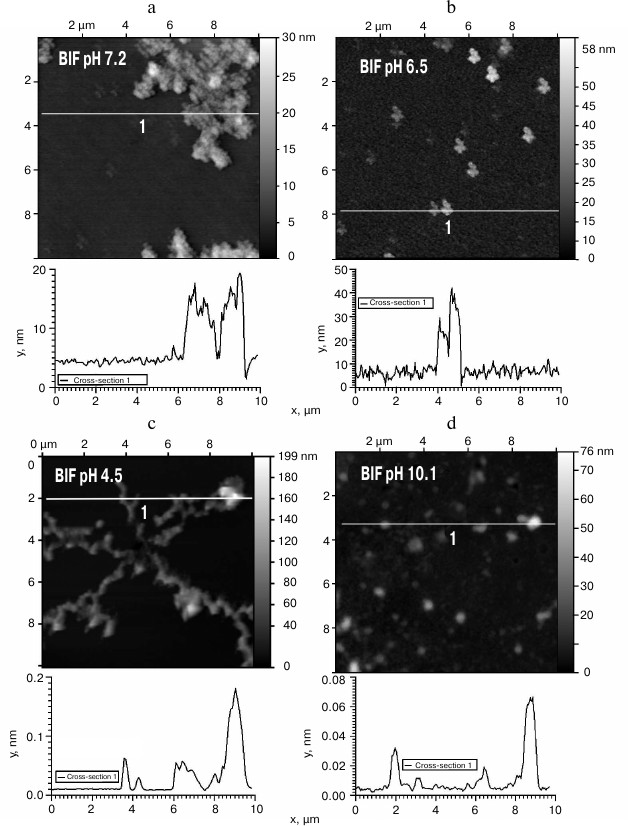
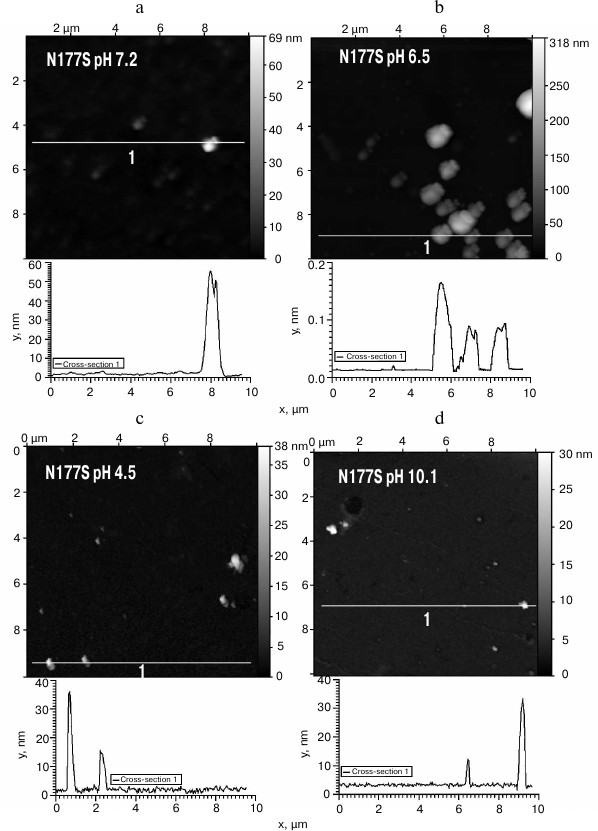
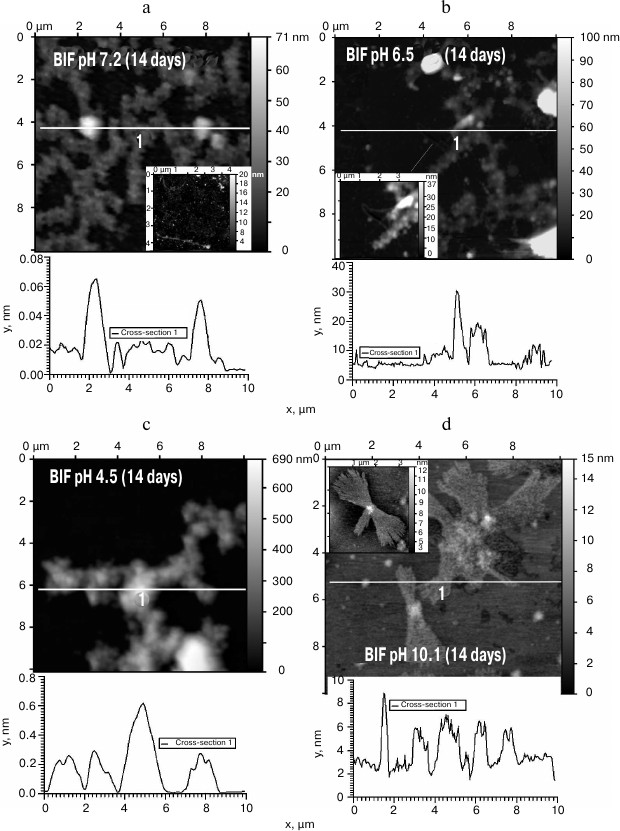
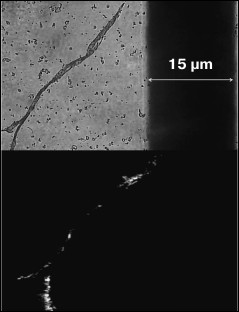
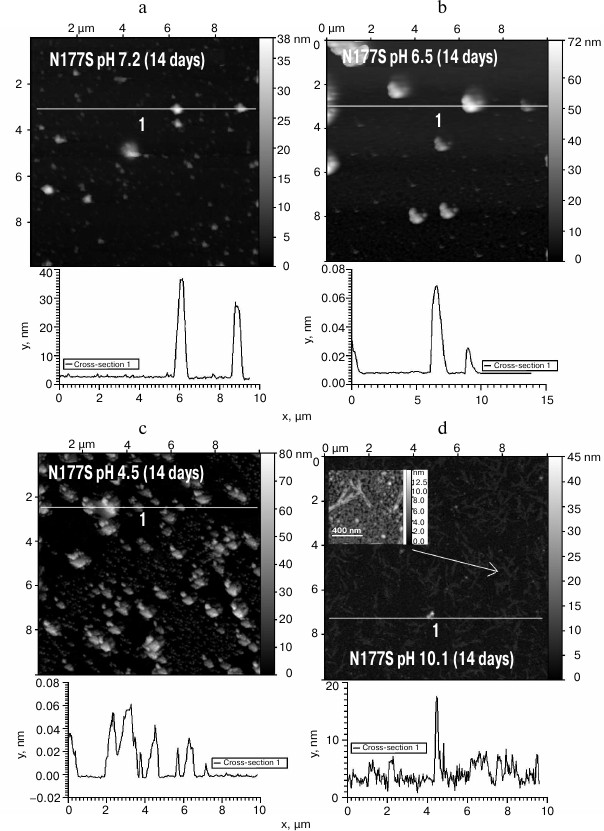
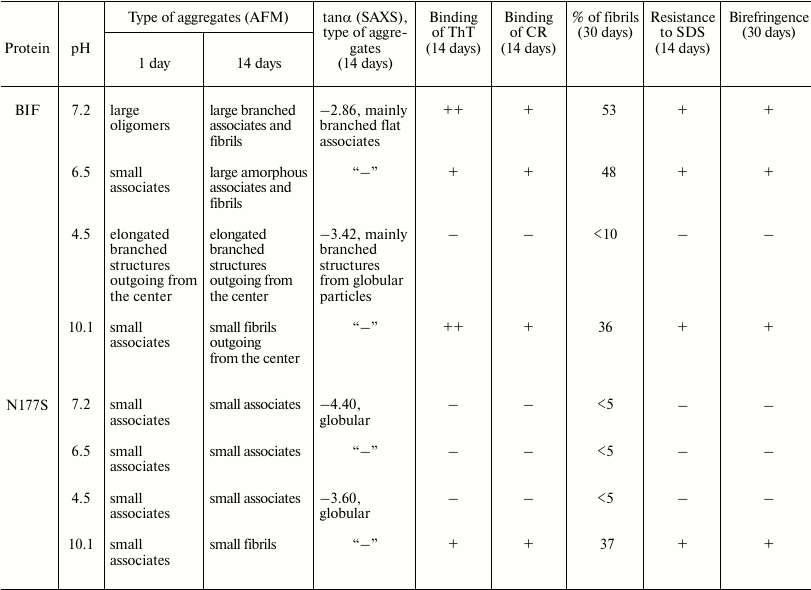
The obtained protein samples were analyzed by AFM immediately after isolation and refolding and after incubation at 37°C for 14 days. Already after dialysis (1 day of incubation), BIF formed various types of oligomers: large in buffer with pH 7.2, branched in buffer with pH 4.5, and small at pH 6.5 and 10.1 (Fig. 1). Under the same conditions, the N177S mutant formed small associates (Fig. 2). The light absorption for both proteins in the 400-nm region due to the Rayleigh light scattering indicated the presence in the protein samples of large aggregates comparable in their size to the light wavelength. Using analytical ultracentrifugation of samples (1 day of incubation) in buffer with pH 7.2, we found that both proteins formed large number of aggregates with different molecular masses dominated by protein tetramers (data not shown). This is consistent with the data obtained earlier for another Bence-Jones protein, GRY, formed by immunoglobulin λ light chains [18]. Incubation of BIF protein at 37°C for 14 days resulted in the appearance of fibrils in samples at pH 7.2, 6.5, and 10.1 (Fig. 3, a, b, and d). The bulk of the aggregates at pH 7.2 was large branched associates, which was confirmed by the SAXS assay (table). Analysis of protein samples after incubation at 37°C for 30 days by electrophoresis in agarose gel in the presence of 0.1% SDS showed that the percent of fibrils in samples with pH 7.2 was up to 53% (table). The presence of fibrils was corroborated by the results of Congo Red binding assay and changes in the ThT fluorescence intensity in the 482-nm region (table). The fibrillar nature of the aggregates was confirmed by the apple-green birefringence in the polarized light of the Congo Red-stained samples (Fig. 4) and the resistance of fibrils to the incubation with 0.09% SDS for two days at room temperature, as demonstrated by the AFM (table). Incubation of BIF for 14 days at 37°C at acidic pH resulted in the formation of elongated branched structures (Fig. 3c), but the absence of birefringence in the polarized light after staining with Congo Red indicated that these aggregates were non-amyloid. The data obtained by SAXS show that the aggregates were predominantly globular branched structures (table). In addition to the large structures, BIF formed amorphous aggregates of various sizes in all buffers tested.
Fig. 1. The morphology of BIF protein aggregates at pH 7.2 (a), 6.5 (b), 4.5 (c), and 10.1 (d) (AFM images obtained in tapping mode).
Fig. 2. The morphology of N177S protein aggregates at pH 7.2 (a), 6.5 (b), 4.5 (c), and 10.1 (d) (AFM images obtained in tapping mode).
Fig. 3. The morphology of BIF protein aggregates after the incubation for 14 days at 37°C at pH 7.2 (a), 6.5 (b), 4.5 (c), and 10.1 (d) (AFM images obtained in tapping mode).
Fig. 4. The Congo Red birefringence assay in the polarized light for BIF protein in a buffer with pH 6.5 after incubation at 37°C for 14 days.
The comparative characteristics of BIF and N177S protein aggregates
Notes: ThT, thioflavin T; CR, Congo Red; “—”, no
data.
The N177S mutant after incubation for 14 days at 37°C under the studied conditions formed granular aggregates at pH 7.2, 6.5, and 4.5 (Fig. 5, a-c). The structures formed at pH 7.2 and 4.5 were globular, as shown by SAXS (table). At alkaline pH, small fibrils were found along with amorphous aggregates present in the solution (Fig. 5d). The results were confirmed by the Congo Red and ThT assays, Congo Red birefringence assay, electrophoresis in agarose gel in the presence of 0.1% SDS, and analysis of aggregate resistance to 0.09% SDS by AFM (table).
Fig. 5. The morphology of N177S protein aggregates after incubation for 14 days at 37°C at pH 7.2 (a), 6.5 (b), 4.5 (c), and 10.1 (d) (AFM images obtained in tapping mode).
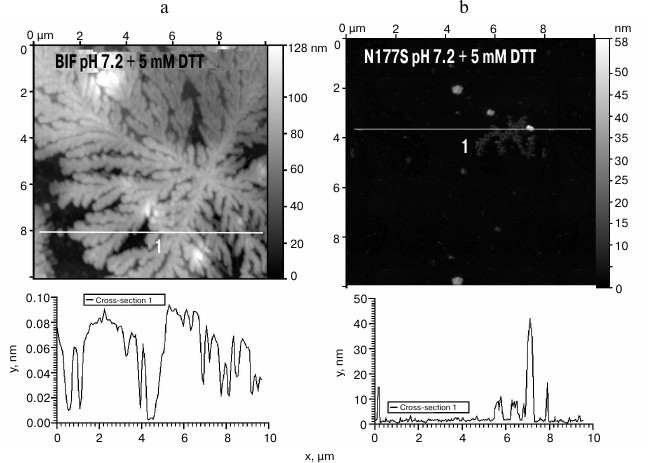
Since BIF sequence contains four cysteine residues, we studied the influence of the disulfide bond-reducing agent DTT on the aggregation process. In the presence of 5 mM DTT, both the BIF protein and the N177S mutant started to form branched elongated structures in the process of refolding, but the aggregation ability of BIF was significantly higher, and it formed large aggregates immediately during the isolation process (Fig. 6, a and b).
Fig. 6. The morphology of BIF (a) and N177S (b) protein aggregates formed in the presence of 5 mM DTT (AFM images obtained in tapping mode).
DISCUSSION
Insoluble fibrils and amorphous deposits of immunoglobulin light chains play the key role in the development of renal nephropathy in MM. According to the current views, the aim for the therapy of any type of amyloidosis is the development of procedures to decrease the amount of protein precursors that promote disease development. The environment strongly influences the process of amyloid formation and can favor generation of either amorphous aggregates or amyloid fibrils. Amyloidosis accompanied by accumulation in kidneys of fibrils formed by full-length immunoglobulin light chains, products of light chain proteolytic degradation, or separate VL- or CL-domains is mostly unfavorable in myeloma, whereas deposition of amorphous aggregates of light chains is less dangerous and has better clinical prognosis [2].
It was shown previously for immunoglobulin light chain variable domains (SMAs) [19] that depending on the microenvironment, these proteins can aggregate through partially folded intermediates into ordered structures (protofibrils and fibrils) or disordered amorphous structures. Amorphous aggregates are formed much more quickly than fibrils. There are two mechanisms for the fibril formation: growth from amorphous aggregates and assembly from oligomers. It should be noted that some non-fibrillar oligomeric forms are also toxic. Therefore, the questions remain: what triggers the aggregation of light chains and what is the fundamental factor determining the morphology of resulting aggregates?
So far, most of information on the mechanism of amyloid aggregation has been obtained for the light chain variable domains and BJPs, whose variable domain also participates in the amyloid aggregate formation [18-20]. However, Solomon and co-authors were able to isolate the BJP BIF that formed amyloid deposits by aggregation of the light chain CL-domains. It was proposed that amyloidogenic properties of this protein were related to the substitution of Ser177 with Asn in the amino acid sequence of the CL-domain, and that this mutation was an inherited one [3]. Until now, the role of this substitution in BIF amyloidosis and the influence of environment on this process have remained unclear.
To elucidate these questions, we obtained recombinant BJP BIF and its mutant N177S analog that corresponded to the healthy genotype without hereditary predisposition for MM with renal nephropathy. AFM was used to compare the morphology of protein aggregates formed under different conditions that reflected microenvironments in separate parts of the renal nephron. The known procedures for recombinant antibody isolation allow to purify them from inclusion bodies at a concentration of 0.1-0.8 mg/ml only [8], which considerably limits the number of methods that could be applied to studying protein aggregates. To obtain more information on the nature of obtained aggregates, we used Congo Red and ThT assays, Congo Red birefringence assay, SAXS, analytical ultracentrifugation, and electrophoresis in agarose gel in the presence of 0.1% SDS.
We found that BIF has a high aggregation activity, and that the type of the aggregates formed strongly depends on pH. When the protein was incubated at 37°C for 14 days, amorphous aggregates of different sizes were formed at all tested pH values. At pH close to neutral (pH 7.2 and 6.5), fibrils also formed, as shown by the increase in the ThT fluorescence intensity and birefringence of the Congo Red-stained samples. Our results are in a good agreement with the data of Solomon et al. [3] and Kim et al. [21] for BIF isolated from patient’s urine in a buffer with pH 7.0. When this protein was incubated under these conditions at 37°C, it started to aggregate already within 24 h of incubation; after 14 days of incubation >75% of BIF formed insoluble aggregates that exhibited green birefringence in the polarized light after Congo Red binding. Similar results were obtained for the Bence-Jones protein GRY [18] that is composed of λ light chains. This protein started to form fibrils in the buffer systems with pH 7.0 and 6.5 already on day 4 of incubation. At acidic pH, BIF formed elongated non-amyloid globular structures after 14 days of incubation. This observation correlates with the data for GRY [18], which is capable of the granular aggregate formation at pH 3.5. Despite the fact that BIF did not form fibrils at acidic pH, we cannot exclude that other aggregates formed by this protein are also toxic. At alkaline pH values, small fibrillar structures were detected in BIF solutions after 14 days of incubation.
The substitution of Asn177 with Ser significantly altered the aggregation properties of the protein. Under the same conditions, the N177S mutant did not form elongated structures during the incubation period (14 days at 37°C) but associated with the formation of small granular aggregates. Only at alkaline pH, the N177S mutant formed fibrillar structures that could be detected by the Congo Red and ThT assays. These observations are in agreement with the data presented in [21]. In the case of the non-amyloidogenic GAL protein, whose variable domain amino acid sequence differs from that of BIF in 15 positions and who has Ser instead of Asn at the position 177 in the constant domain, fibrils were not found after incubation in a buffer with pH 7.0 at 37°C for 14 days.
Refolding of both recombinant proteins in the presence of the reducing agent (5 mM DTT) promoted formation of elongated and branched structures for both proteins; however, in the case of BIF, this process proceeded rapidly and resulted in the formation of very large structures. Because of the presence of four cysteines in the BIF amino acid sequence, the importance of disulfide bonds in proper folding of the BIF protein molecule is obvious, and disruption of these bonds causes irreversible outcomes.
In conclusion, our results show that a single mutation (Ser177Asn) in the sequence of immunoglobulin light chain CL-domain results in the changes in the aggregation properties and appearance of amyloidogenic potential of the protein. The morphology of formed aggregates strongly depends on protein microenvironment. It is known that the key factor promoting amyloid formation in neurodegenerative diseases is the presence of multiple repeats of Asn and Gln residues in the protein sequence [22]. No analogous studies have been carried out for the BJPs. It is possible that the appearance of an additional asparagine residue at position 177 triggers the process of amyloid formation by the same principle. However, the mechanism of the protein association initiation and factors determining formation of certain protein aggregate forms at different pH require further investigations.
Acknowledgments
The authors are grateful to I. V. Kolesnikov and M. G. Sharapov for creation of protein constructs, to Prof. H. Kihara for help with SAXS experiments, and to G. A. Enin for AFM consultations.
This work was supported by the Russian Foundation for Basic Research (projects Nos. 14-04-01682-a and 16-04-01027-a).
REFERENCES
1.Korbet, S. M., and Schwartz, M. M. (2006) Multiple
myeloma, J. Am. Soc. Nephrol., 17, 2533-2545.
2.Pozzi, C., and Locatelli, F. (2002) Kidney and
liver involvement in monoclonal light chain disorders, Semin.
Nephrol., 22, 319-330.
3.Solomon, A., Weiss, D. T., Murphy, C. L., Hrncic,
R., Wall, J. S., and Schell, M. (1998) Light chain-associated amyloid
deposits comprised of a novel kappa constant domain, Proc. Natl.
Acad. Sci. USA, 95, 9547-9551.
4.Mukherjee, S., Pondaven, S. P., and Jaroniec, C. P.
(2011) Conformational flexibility of a human immunoglobulin light chain
variable domain by relaxation dispersion nuclear magnetic resonance
spectroscopy: implications for protein misfolding and amyloid assembly,
Biochemistry, 50, 5845-5857.
5.Wilkins-Stevens, P., Raffen, R., Hanson, D. K.,
Deng, Y. L., Berrios-Hammond, M., Westholm, F. A., Murphy, C., Eulitz,
M., Wetzel, R., Solomon, A., Schiffer, M., and Stevens, F. J. (1995)
Recombinant immunoglobulin variable domains generated from synthetic
genes provide a system for in vitro characterization of
light-chain amyloid proteins, Protein Sci., 4,
421-432.
6.Maniatis, T., Fritisch, E. F., and Sambrok, J.
(1982) in Molecular Cloning: A Laboratory Manual, Cold Spring
Harbor Laboratory Press, N. Y.
7.Laemmli, U. K. (1970) Cleavage of structural
proteins during the assembly of the head of bacteriophage T4,
Nature, 227, 680-685.
8.Dubnovitsky, A. P., Kravchuk, Z. I., Chumanevich,
A. A., Cozzi, A., Arosio, P., and Martsev, S. P. (2000) Expression,
refolding, and ferritin-binding activity of the isolated
VL-domain of monoclonal antibody F11, Biochemistry
(Moscow), 65, 1011-1018.
9.LeVine, H., 3rd. (1993) Thioflavine T interaction
with synthetic Alzheimer’s disease beta-amyloid peptides:
detection of amyloid aggregation in solution, Protein Sci.,
2, 404-410.
10.Klunk, W. E., Jacob, R. F., and Mason, R. P.
(1999) Quantifying amyloid beta-peptide (Abeta) aggregation using the
Congo Red-Abeta (CR-abeta) spectrophotometric assay, Anal.
Biochem., 266, 66-76.
11.Kushnirov, V. V., Alexandrov, I. M., Mitkevich,
O. V., Shkundina, I. S., and Ter-Avanesyan, M. D. (2006) Purification
and analysis of prion and amyloid aggregates, Methods,
39, 50-55.
12.Vishnyakov, I. E., Borchsenius, S. N., Basovskii,
Y. I., Levitskii, S. A., Lazarev, V. N., Snigirevskaya, E. S., and
Komissarchik, Y. Y. (2009) Localization of the division protein FTSZ in
micoplasma cells, Cell Tissue Biol., 3, 254-263.
13.Schuck, P. (2000) Size-distribution analysis of
macromolecules by sedimentation velocity ultracentrifugation and Lamm
equation modeling, Biophys. J., 78, 1606-1619.
14.Timchenko, A., Timchenko, M., Shinjo, M., and
Kihara, H. (2015) SAXS study of N177S mutant of Bence-Jones protein
BIF, Photon Factory Activity Report, 2014, 32,
366.
15.Pertinhez, T. A., Bouchard, M., Smith, R. A.,
Dobson, C. M., and Smith, L. J. (2002) Stimulation and inhibition of
fibril formation by a peptide in the presence of different
concentrations of SDS, FEBS Lett., 529, 193-197.
16.Stevens, F. J., Westholm, F. A., Solomon, A., and
Schiffer, M. (1980) Self-association of human immunoglobulin kappa I
light chains: role of the third hypervariable region, Proc. Natl.
Acad. Sci. USA, 77, 1144-1148.
17.Myatt, E. A., Westholm, F. A., Weiss, D. T.,
Solomon, A., Schiffer, M., and Stevens, F. J. (1994) Pathogenic
potential of human monoclonal immunoglobulin light chains: relationship
of in vitro aggregation to in vivo organ deposition,
Proc. Natl. Acad. Sci. USA, 91, 3034-3038.
18.Bliznyukov, O. P., Kozmin, L. D., Vysotskaya, L.
L., Golenkov, A. K., Tishchenko, V. M., Samoylovich, M. P., and
Klimovich, V. B. (2005) Human immunoglobulin light chains λ form
amyloid fibrils and granular aggregates in solution, Biochemistry
(Moscow), 70, 458-466.
19.Zhu, M., Souillac, P. O., Ionescu-Zanetti, C.,
Carter, S. A., and Fink, A. L. (2002) Surface-catalyzed amyloid fibril
formation, J. Biol. Chem., 277, 50914-50922.
20.Davis, D. P., Gallo, G., Vogen, S. M., Dul, J.
L., Sciarretta, K. L., Kumar, A., Raffen, R., Stevens, F. J., and
Argon, Y. (2001) Both the environment and somatic mutations govern the
aggregation pathway of pathogenic immunoglobulin light chain, J.
Mol. Biol., 313, 1021-1034.
21.Kim, Y., Wall, J. S., Meyer, J., Murphy, C.,
Randolph, T. W., Manning, M. C., Solomon, A., and Carpenter, J. F.
(2000) Thermodynamic modulation of light chain amyloid fibril
formation, J. Biol. Chem., 275, 1570-1574.
22.Jiang, Y., Li, H., Zhu, L., Zhou, J. M., and
Perrett, S. (2004) Amyloid nucleation and hierarchical assembly of
Ure2p fibrils. Role of asparagine/glutamine repeat and nonrepeat
regions of the prion domains, J. Biol. Chem., 279,
3361-3369.