REVIEW: The 4D Nucleome: Genome Compartmentalization in an Evolutionary Context
T. Cremer1*, M. Cremer1, and C. Cremer2
1Biocenter, Department of Biology II, Ludwig Maximilian University (LMU), Munich, 82152 Martinsried, Germany; E-mail: thomas.cremer@lrz.uni-muenchen.de, marion.cremer@lrz.uni-muenchen.de2Institute of Molecular Biology (IMB) Mainz, 55128 Mainz, Germany; E-mail: c.cremer@imb-mainz.de
* To whom correspondence should be addressed.
Received November 29, 2017
4D nucleome research aims to understand the impact of nuclear organization in space and time on nuclear functions, such as gene expression patterns, chromatin replication, and the maintenance of genome integrity. In this review we describe evidence that the origin of 4D genome compartmentalization can be traced back to the prokaryotic world. In cell nuclei of animals and plants chromosomes occupy distinct territories, built up from ~1 Mb chromatin domains, which in turn are composed of smaller chromatin subdomains and also form larger chromatin domain clusters. Microscopic evidence for this higher order chromatin landscape was strengthened by chromosome conformation capture studies, in particular Hi-C. This approach demonstrated ~1 Mb sized, topologically associating domains in mammalian cell nuclei separated by boundaries. Mutations, which destroy boundaries, can result in developmental disorders and cancer. Nucleosomes appeared first as tetramers in the Archaea kingdom and later evolved to octamers built up each from two H2A, two H2B, two H3, and two H4 proteins. Notably, nucleosomes were lost during the evolution of the Dinoflagellata phylum. Dinoflagellate chromosomes remain condensed during the entire cell cycle, but their chromosome architecture differs radically from the architecture of other eukaryotes. In summary, the conservation of fundamental features of higher order chromatin arrangements throughout the evolution of metazoan animals suggests the existence of conserved, but still unknown mechanism(s) controlling this architecture. Notwithstanding this conservation, a comparison of metazoans and protists also demonstrates species-specific structural and functional features of nuclear organization.
KEY WORDS: 4D nucleome, nuclear architecture, evolution, chromatin domains, topologically associating domains (TADs), genome compartmentalizationDOI: 10.1134/S000629791804003X
Abbreviations: ANC-INC, active nuclear compartment/inactive nuclear compartment; CD, chromatin domain; CDC, chromatin domain cluster; CT, chromosome territory; DAPI, 4′,6-diamidino-2-phenylindole; IC, interchromatin compartment; PR, perichromatin region; SIM, structured illumination microscopy; TAD, topologically associating domain; TF, transcription factor.
We dedicate this review to Stanislav Fakan in recognition of his
pioneering contributions to nuclear architecture and its implications
for nuclear functions
The accumulation of evidence for a compartmentalized architecture of the cell nucleus with important functional implications has led to a new research field, called the 4D nucleome. In 2015 the National Institutes of Health (NIH) of the United States of America initiated a 4D nucleome program, which “aims to understand the principles behind the three-dimensional organization of the nucleus in space and time (the 4th dimension), the role nuclear organization plays in gene expression and cellular function, and how changes in the nuclear organization affect normal development as well as various diseases.” … “How does this architecture contribute to gene expression regulation? How does nuclear architecture change over time in the course of normal development? Do dysfunctional alterations in nuclear organization lead to disease, and/or could they be used to diagnose diseases?” (https://commonfund.nih.gov/4Dnucleome).
Major contributions to this new and rapidly expanding field have been made by researchers from many countries. The first 4D Nucleome Workshop was held in June 2013 by scientists from 14 countries in Mainz (Germany), followed by a second workshop held in December 2014 in Hiroshima (Japan). During these workshops a consensus was reached during these discussions “that, given the complexity and multi-faceted nature of the problem, large-scale collaborations amongst laboratories with distinct and complementary expertise around the world would be required to solve the nucleome problem” [1]. We still lack a generally accepted model able to integrate the complex organization and function of the cell nucleus into a common theoretical framework.
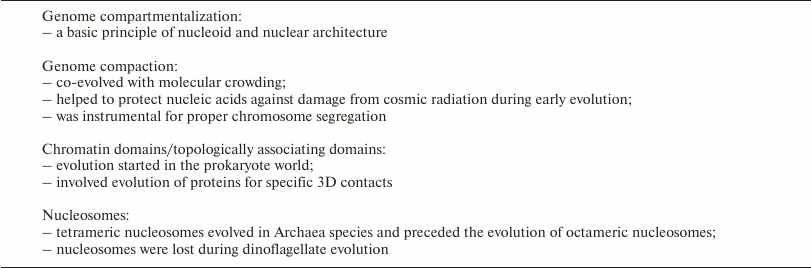
Studies of complex biological issues have always benefited greatly from an evolutionary context, but little work has been carried out so far to ascertain this context in the rising field of the 4D nucleome research. In this review we recapitulate structural features, which have been consistently noted in eukaryotes, and show that 3D genome compartmentalization already evolved in the prokaryotic world. Evidence for certain structural features of nuclear organization present in the most distant branches of the evolutionary tree would strongly suggest their profound and general biological importance, independent of the question, whether these features date back to the same evolutionary origin or whether they evolved independently in different branches of the evolutionary tree. Structures, which are found only in a distinct part of the tree may have a more limited, functional significance or no special, functional significance at all. The identification of this evolutionary context has implications for the planning of further research strategies and paves the way for future molecular studies of the underlying mechanisms. A brief overview of issues discussed below is provided in the table.
Features demonstrating the tightly linked evolution of genome structure
and function
CHROMOSOME TERRITORIES AND CHROMATIN DOMAINS FORM CO-ALIGNED
ACTIVE AND INACTIVE NUCLEAR COMPARTMENTS
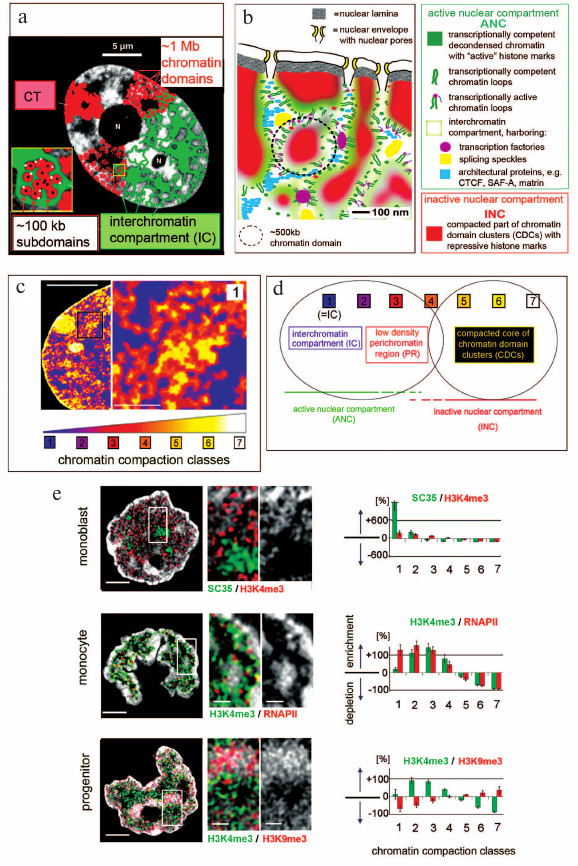
Chromosomes occupy distinct territories (CTs) in cell nuclei of animals and plants that are built up from ~1 Mb chromatin domains (CDs); ~1 Mb CDs in turn are composed of smaller chromatin subdomains (subCDs) and also form larger chromatin domain clusters (CDCs) (for review see [2-4]). Early microscopic evidence has led to the proposal of the chromosome territory – interchromatin compartment (CT-IC) model (Fig. 1a) [3, 4].
Fig. 1. a) Chromosome territory – interchromatin compartment model (CT-IC) (adapted from [3, 4]). b) Active nuclear compartment – inactive nuclear compartment (ANC–INC) network model of nuclear organization. For details see text. c) Nuclear landscape shaped by different chromatin densities in a DAPI-stained female mouse C2C12 cell nucleus (adapted from [5]). Optical stacks from nuclei were recorded with 3D-structured illumination microscopy (3D-SIM). Voxels were attributed to seven classes with increasing DAPI intensities and served as proxies for increasing chromatin compaction levels (blue: lowest intensities close to background; white: highest chromatin compaction). Left: partial nucleus shows a network of chromatin domain clusters (CDCs) pervading the nuclear space with a cluster of constitutive heterochromatin (class 7, white) and a Barr body (class 6, yellow) adjacent to the nuclear envelope. The inset magnification presents an enlargement of a boxed area (1) and reveals a shell-like organization of CDCs with compact chromatin (class 6, yellow) in the interior and a peripheral zone of less dense chromatin with lower chromatin densities (class 2, purple; class 3, red). d) Interpretation of the nuclear color heat maps: classes 1-3 constitute the ANC: class 1 (IC) is lined by the perichromatin region (PR) (classes 2 and 3); class 4 presents an intermediate zone; the INC is represented by classes 5-7. For further details see text and references [6, 7]. e) Left: midplane sections from hematopoietic cell nuclei recorded with 3D-SIM from a monoblast (top), a monocyte (middle), and a progenitor cell (bottom) with DAPI-stained DNA (gray); scale bars, 2 μm. Additional two-color immunocytochemistry shows functionally relevant hallmarks. Monoblast nucleus: splicing speckles, which provide essential factors for co-transcriptional splicing, are immuno-stained with SC35 (green); H3K4me3 (red) is a marker of transcriptionally competent chromatin. Monocyte nucleus: H3K4me3 (green); RNA polymerase II with serine 5 phosphorylation (Ser5P) (red) is involved in the initiation of transcription. Progenitor cell nucleus: H3K4me3 (green); H3K9me3 (red), a marker of transcriptionally silent chromatin. Magnifications of boxed areas (scale bars, 0.5 μm) show SC35 stained splicing speckles located within the IC; H3K4me3 and RNA Pol II is seen in the periphery of chromatin domain clusters, whereas H3K9me3 is enriched in the CDC interior. Right: quantitative assignments of these hallmarks to the seven DAPI intensity classes. 3D evaluation of some 12 nuclei was performed for each panel and confirms SC35 stained splicing speckles almost exclusively in class 1, enrichments of H3K4m3 and RNA Pol II in classes 2 and 3, and H3K9me3 enrichments in classes 6 and 7 (adapted from [8]).
Essential features of this model have been consistently observed in all cell types studied to date notwithstanding the fact that major changes of higher order chromatin arrangements occur during development and cell differentiation [9-13]. Furthermore, a remarkable evolutionary conservation of such features could be demonstrated throughout the evolution of metazoan animals (see below). Chromatin domains were first considered as essential structural features by cytologists in the late 19th century (reviewed in [14]). One hundred years later it was proposed that transcription is controlled by chromatin packaging (reviewed in [15]). Domains with inactive genes arguably possess a compact structure, which hinders or even prevents the access of large macromolecules and protein complexes, while domains with active genes have an unfolded configuration and are easily accessible [16, 17]. A recent, more refined version of the CT-IC model integrates current knowledge of the structural organization of the cell nucleus into a framework of two structurally and functionally intertwined compartments, the active nuclear compartment (ANC) and the inactive nuclear compartment (INC) [6] (Fig. 1b). The ANC-INC model argues that the nucleus carries two spatially co-aligned, active and inactive nuclear compartments. The inactive compartment (INC) is composed of compact CDs located in the interior of CDCs and enriched with epigenetic marks for silent chromatin. In conventional terminology the INC may be considered as a part of the facultative heterochromatin. The active compartment (ANC) is formed by the IC together with less compacted, transcriptionally competent CDs located at the periphery of CDCs.
Seminal electron microscopic studies, pioneered by Wilhelm Bernhard and colleagues [18], combined with ultrastructural immuno-cytochemical analyses, demonstrated for the first time a clustering of the 3D genome into chromatin aggregations pervading the nuclear space and revealed a zone of transcriptionally active chromatin, called the perichromatin region (PR) (for reviews see [19-21]). Stan Fakan and colleagues first demonstrated that transcription and DNA replication are preferentially performed within the PR [22, 23]. In the terminology of the CT-IC model, the PR lines the IC and is enriched in coding and regulatory sequences of genes and epigenetic marks for transcriptionally active chromatin and serves as the major, though not exclusive nuclear subcompartment for transcription, DNA replication and repair. Interphase chromatin shows continuous, locally constrained movements [24, 25]. Initial evidence suggests movements of coding and regulatory sequences between the periphery and interior of CDCs depending on their transcriptionally active and silent state [26]. Rather than just a residual space between CDs, we consider the IC as a functional nuclear compartment of its own, which serves three functional requirements: 1) IC-channels allow facilitated movements of mRNPs along IC-channels toward the nuclear pores; 2) they provide preferential routes for functional proteins, such as transcription factors (TFs) entering the nuclear interior towards their DNA binding sites; 3) they allow a rapid intranuclear distribution of factors stored within nuclear bodies, as well as of regulatory, noncoding RNAs from sites were such RNAs are synthesized to sites where such factors and RNAs are needed. Compelling evidence for or against this hypothesis has not been provided to date.
For quantitative, microscopic analyses of the higher order chromatin landscape a procedure was developed with our colleague Volker Schmid based on the recording of 3D image stacks from nuclei stained with an appropriate DNA-specific fluorophore, e.g., DAPI, with structured illumination microscopy (SIM) and quantitative 3D image analysis [7]. SIM is a method of super-resolved fluorescence microscopy with a lateral resolution of ~100 nm and an axial resolution of ~300 nm. Bioinformatic tools allow the quantitative assessment of highly resolved 3D chromatin compaction levels in individual cell nuclei, which reflect functionally different regions [7]. Measured DNA densities serve as a proxy for differences in chromatin compaction. Figure 1c shows a heat map of a single SIM section through a mouse (C2C12) cell nucleus, where class 1 (blue) corresponds to the IC, whereas classes 2-7 classes reflect increasing chromatin compaction levels. These measurements suggest a layered organization of CDCs with CDs of higher compaction (classes 5-7) representing the INC located in the CDC interior, whereas CDs with less densely compacted chromatin (classes 2 and 3) line the IC as a part of the ANC (Fig. 1d). As examples for the application of this approach, Fig. 1e shows SIM sections through DAPI-stained nuclei from three human hematopoietic cell types, a monoblast (top), a monocyte (middle), and a progenitor cell (bottom) [8]. Prior to 3D SIM two color immuno-cytochemistry was used to stain SC35 (green), a marker for splicing speckles, and H3K4me3 (red), a marker for transcriptionally competent chromatin in the monoblast nucleus, RNA polymerase II (RNA Pol II) (red) and H3K4me3 (green) in the monocyte nucleus, and H3K4me3 (green) together with H3K9me3, a marker for transcriptionally silent chromatin (red) in the progenitor cell nucleus. 3D quantitative image analysis at each stage of differentiation demonstrates a strong enrichment of SC35-labeled splicing speckles within the IC. H3K4me3 shows a relative enrichment in the ANC (classes 2 and 3) and a relative depletion in the INC (classes 5-7). In contrast, H3K9me3 was relatively enriched in the INC, but depleted in the ANC. These features were observed in other normal cell types from several mammalian species (human, mouse, cattle) as well, including cells from bovine preimplantation embryos [27, 28], although nuclei of these cell types varied strongly with respect to size, shape, global chromatin arrangements, width of the largest IC-channels, and nuclear envelope invaginations. Remarkably, cancer cell nuclei also retain typical features of the ANC-INC organization despite profound differences between the morphology of cancer and normal cell nuclei with respect to shape, size, and chromatin texture, which have long been used for diagnostic purposes [29, 30]. In summary, these findings hint to the evolutionary conservation and functional importance of the ANC and INC at least in mammals.
TOPOLOGICALLY ASSOCIATING DOMAINS AND BOUNDARIES
Microscopic evidence for this higher order chromatin landscape was strengthened by chromosome conformation capture experiments (reviewed in [31, 32]). This ingenious molecular approach is based on the identification of DNA–DNA “contact” frequencies in cis and trans and was first described by Job Dekker and colleagues [33]. Subsequent improvements of this approach (in particular Hi-C [34]; for review see [35]) allowed for the first time genome-wide studies of contact frequencies and demonstrated topologically associating domains (TADs) with a DNA content of ~1 Mb in mammalian cell nuclei, separated by boundaries [36, 37]. In line with ∼1 Mb CDs, subCDs, and CDCs, ∼1 Mb TADs contain smaller subTADs and also form larger chromatin complexes, called metaTADs [38]. Notwithstanding the apparent relationship between microscopically observed CDs and TADs, it must be emphasized that the organization of TADs with a best “resolution” of about 1 kb [39] is based on contact frequencies measured in millions of cells. Recently, it has become possible to carry out Hi-C of single cell nuclei [40-43], but this approach suffers from a very limited resolution [44]. Hi-C has provided the major advantage to assign chromatin modifications and architectural proteins identified along the linear DNA sequence directly to the 3D TADs [45, 46]. For example, DNA sequence motifs required for the binding of the architectural protein CTCF are enriched at TAD boundaries [47]. Another architectural protein, cohesin, acts as a molecular motor in chromatin loop formation [48], loss of cohesin prevents loop formation [49]. Structural variants (deletions, duplications, and inversions) that eliminate boundaries can lead to the formation of neoTADs with pathological interactions between enhancers and genes, resulting in malformations [50, 51] and cancer [52, 53]. In the near future, we expect the publication of many more examples that demonstrate the importance of pathological 3D chromatin structures affecting the regulatory genome landscape in a large variety of health issues. Clusters of evolutionarily conserved noncoding elements (CNEs) coincide with boundaries of TADs in humans and Drosophila [54]. The evolution of higher order chromatin landscapes with TADs and boundaries dates back to the origin of bilateralia, revealing a regulatory architecture conserved over hundreds of millions of years [55].
Details of the higher order chromatin landscape, such as the average size of TADs, vary between species that are more remote from each other in the evolutionary tree [54, 56]. In plants some species, e.g., rice, show a distinct TAD-pattern [57], whereas TADs are not an obvious feature of Arabidopsis nuclei [57, 58]. Far from being exhaustive, these few hints emphasize remarkable interspecies differences of chromatin landscapes (see also below).
EVOLUTIONARILY CONSERVED 3D ARRANGEMENTS OF GENE-DENSE AND
GENE-POOR CHROMATIN
The nonrandom radial location of gene-poor chromatin at the nuclear periphery and gene-rich chromatin within the nuclear interior was first observed in human cell types [59-62]. Subsequent studies demonstrated that this organization is an evolutionarily conserved feature of the 3D nuclear landscape. The gene-dense human chromosome 19 (HSA19), for example, is typically located in the interior of human cell nuclei, whereas the gene-poor HSA18 has a peripheral location [62, 63]. Despite major evolutionary changes of primate karyotypes, the same preferential internal or peripheral location was observed for orthologous segments in other primate species [64]. Moreover, syntenic regions of HSA19 have been assigned to gene-dense chicken microchromosomes, located in the nuclear interior, whereas syntenic regions of HSA18 have been assigned to gene-poor chicken macrochromosomes 2 and Z, located at the nuclear periphery [65]. These observations support an evolutionary conservation of nonrandom radial chromatin arrangements for at least 300 million years. A study of the nuclear architecture of the polyp Hydra, which belongs to the earliest metazoan phylum separated from mammals by at least 600 million years, revealed CTs with striking similarities to the replication labeling patterns in mammalian nuclei [66]. These patterns reflect a persistent nuclear arrangement of early, mid-, and late replicating chromatin foci. The conservation of fundamental features of higher order chromatin arrangements throughout the evolution of metazoan animals suggests the existence of conserved, but still unknown mechanism(s) controlling this architecture.
Evidence for chromosome territories and chromatin domains in animal cell nuclei has triggered a wealth of studies in plant nuclei [67-76]. A detailed comparison of the findings on nuclear organization in animal and plant species is beyond the scope of this review. Suffice it to say that the literature describes structural features common to all animals and plants studied to date, such as CTs and chromatin domains, despite a great deal of structural diversity between species. This is what one would expect for 4D nucleomes of species adapted to an enormous variety of ecological niches. A recent review on the effects of light on nuclear architecture in plants provides a case in point of special functional needs of plant species [77]. Light regulates many aspects of plant development, and photoreversible changes in nuclear organization correlate with transcriptional regulation patterns.
The mammalian retina provides an example for the evolutionary impact of light on the nuclear architecture of photoreceptor cells [13, 78]. Rod nuclei of diurnal retinas possess the typical architecture of mammalian cell types with euchromatin residing in the nuclear interior and significant amounts of heterochromatin situated at the nuclear periphery. Rod nuclei of retinas from mammalian species with a nocturnal life style, however, show an inverted pattern, where nearly all constitutive and facultative heterochromatin localizes in the interior, whereas euchromatin in association with nascent transcripts and the splicing machinery, lines the nuclear border [13, 79, 80]. Computer simulations have indicated that rod nuclei with the inverted pattern act as collecting lenses, which help to channel light efficiently toward the light-sensing rod outer segments [13, 81, 82]. This hypothesis was supported by evidence that the mass of centrally localized heterochromatin has a higher refractive index compared with the less compacted nuclear periphery. Profound changes of rod cell nuclear architecture in mice occur during the postnatal terminal differentiation of rods, but it is not clear whether light is necessary as a direct stimulus or whether these changes would also occur in the absence of light. The latter hypothesis is supported by the fact that these changes are already under way before a newborn mouse opens its eyes. Rod cell nuclear architecture provides a prominent example for a global nuclear reorganization that occurs during postmitotic terminal differentiation to facilitate specialized functions. Some other pertinent examples concern changes of nuclear architecture, including heterochromatin clustering, during terminal differentiation of Purkinje cells [12], mouse myoblasts [10], and Drosophila wing cells [83]. Many others might be detected in future studies.
A layer of constitutive peripheral heterochromatin beneath the nuclear lamina has been noted in a wide variety of animal and plant nuclei. In animal cell nuclei this layer is formed by transcriptionally repressed chromatin regions, termed lamina-associated domains (LADs) [84, 85]. Although less well studied so far in plant cells, a recent report demonstrates a similar chromatin organization at the nuclear periphery of Arabidopsis thaliana nuclei [86]. The identified chromatin domains are enriched with silenced protein-coding genes, transposable element genes, and heterochromatic marks. Unlike LADs in animals, however, these domains are neither gene-poor nor A/T-rich. Why a nuclear subcompartment with largely repressed genes evolved at the nuclear periphery is not clear. At face value such an organization does not seem obvious as an advantageous evolutionary adaptation. The positioning of most or all transcriptionally active genes in the nuclear periphery would provide much shorter routes for transcription factors to their target genes and for messenger RNAs to nuclear pores. Rod cell nuclei described above provide a case in point that such an organization is functionally possible, but this organization may come at a cost.
In 1975 T. C. Hsu hypothesized that constitutive peripheral heterochromatin beneath the nuclear lamina provides a shield that may protect central euchromatin from chemical mutagens and X-ray radiation [87]. Using immunofluorescence with antibodies specific for single- and double-strand breaks (SSB and DSBs), the Bickmore group [88] performed a microscopic analysis of the nuclear distribution of such breaks following hydrogen peroxide treatment or UV-C irradiation of cell cultures. Contrary to the expectation of the bodyguard hypothesis, the authors detected even an excess of damage in the nuclear interior. In contrast, another recent study presented evidence in human melanoma and lung squamous cell carcinoma cohorts “that the nuclear periphery, compared with the core, had a larger mutation burden and also displayed mutation signatures consistent with greater exposure to external mutagens” [89]. Still other data also support a protective role of peripheral heterochromatin (for review see [90]).
Whether a bodyguard of peripheral heterochromatin shields nuclear DNA against reactive oxygen species (ROS) seems even less certain. ROS are permanently produced in mitochondria but are short-lived and typically move only over very short distances (up to a few hundred nanometer) until they react with other proteins, lipids, and ribonucleic acids, including mtDNA [91, 92]. At this point the validity of Hsu’s bodyguard hypothesis remains an open question. The ANC-INC model (Fig. 1b) may partly explain the preference of the induction of DSBs in active chromatin [93] if we assume that DNA-breaking substances, which enter through nuclear pores, may preferentially follow IC channels. In this case these substances would first reach active chromatin exposed in the PR.
COMPARTMENTALIZATION: A PRINCIPLE OF GENOME ORGANIZATION WITH
DIFFERENT EVOLUTIONARY SOLUTIONS
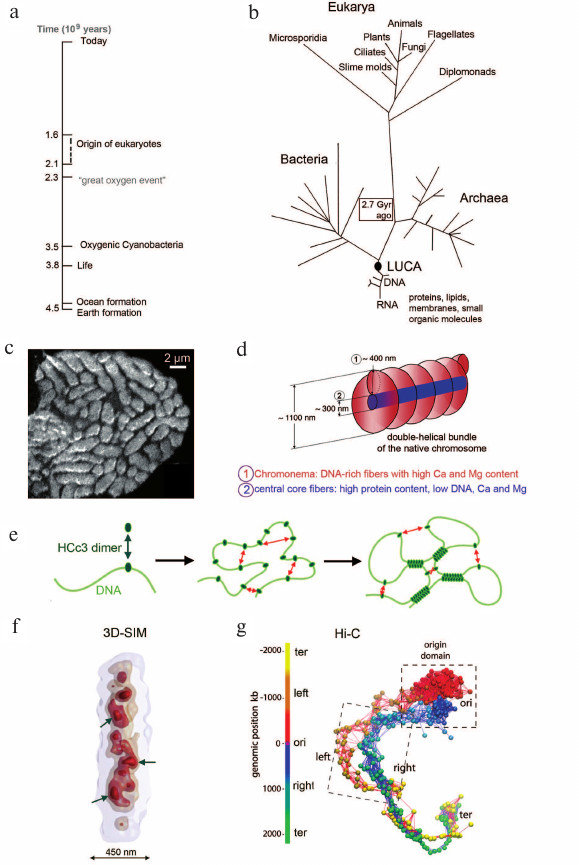
Life’s still unfolding story of chance and necessity, luck and disaster, has been a topic of intense debate in evolution biology. The evolutionary origin of life started some 3.8 billion (3.8·109) years ago, likely in hydrothermal vents or land-based volcanic hot springs [94-96] (Fig. 2, a and b). The origin of eukaryotes has been dated some 2 billion years ago with evolutionary roots reaching back to the origin of archaea. In addition, endosymbiosis with certain bacteria played an important role in the evolution of eukaryotes [96, 97]. Mitochondria and chloroplasts have remained as telling evidence of such endosymbiotic events. Nucleosomes appeared first as tetramers in the Archaea kingdom [98, 99] and later evolved to the octamers typical for Eucarya built up from two H2A, two H2B, two H3, and two H4 proteins [100].
Nuclear architecture of single cell eukaryotes. The evolution of the wide range of metazoans started with the emergence of single cell eukaryotes or protists. Present-day Protista constitute an extremely diverse group of eukaryotic microorganisms with respect to both genome and nuclear size. Each of them passed through a complex evolutionary history of its own. It is currently not known to which extent all protists share features of the spatial organization of mammalian cell nuclei. Alternatively, it seems possible that evolution “found” different solutions for functional nuclear architectures (see the case of dinoflagellates below). Within single cell eukaryotes, nuclear architecture has been extensively studied in yeast. Among the more than 1000 described yeast species the fission yeast Schizosaccharomyces pombe and the budding yeast Saccharomyces cerevisiae can be easily grown in the laboratory and have been studied extensively. A recent Hi-C study of the 3D organization of the fission yeast genome demonstrated condensin-mediated TADs. TADs form larger units of 300 kb-1 Mb during mitosis, which gradually diminishes during interphase [101].
Intra- and inter-chromosomal interaction patterns based on genome-wide chromosome conformation capture studies were also determined for budding yeast and led to the proposal of a multi-chromosome constrained self-avoiding chromatin model [102]. Chromosome painting of fission yeast revealed chromosome territories [103]. Overlap between CTs is cell cycle dependent and becomes largest during S-phase. CT structure is significantly compromised by condensin mutations. Pioneering studies of the 4D structure and function of the S. cerevisiae genome have been performed by Susan Gasser and collaborators [104, 105]. While these studies indicate evolutionarily conserved mechanisms in the compartmentalization and function of yeast and mammalian genomes, they also point to profound differences. The about 1000-fold smaller volume of a yeast nucleus results in profound differences of the necessary extent of chromatin motion of two loci with a functional need to meet in either a mammalian or budding yeast nuclei [106]. Homologous DSB repair provides a case in point. It requires the pairing of a damaged DNA sequence with its intact, homologous counterpart. In case of a DSB formed in a single copy sequence, one needs to take into account that homologous CTs are often widely separated in a mammalian cell nucleus (diameter ca. 10 μm). To overcome this separation in order to allow pairing of intact and damaged sequences located in homologous CTs would require an elaborate mechanism. Under which conditions and to which extent CTs can undergo large scale movements in somatic mammalian cell types is still a controversial issue. Constraints of CT movements may explain why homologous DSB repair does not take place during G1 in contrast to the yeast nuclei (diameter 1-2 μm), where damaged and intact homologous sequences can readily sample the entire nuclear space in search for homologous pairing during the entire interphase. In mammalian cell nuclei a situation fit for a homologous search occurs only after replication of a given chromatin domain. The fact that the resulting sister CDs can be separated by a few hundred nanometers [107, 108] implies major unsolved problems, how sister CDs can move together, decondense, allow pairing of intact and damaged homologous sequences, and finally recombine. A necessity for such movements was demonstrated in yeast nuclei [109, 110].
Fig. 2. a) Timeline from the beginning of life to the present time (adapted from [96]). b) Life presumably started some 4 billion years ago in hydrothermal vents or land-based volcanic hot springs. LUCA, last universal common ancestor. Image adapted from [111, 112]. c) Image of a cell nucleus from the dinoflagellate Prorocentrum micans recorded by secondary ion mass spectrometry (SIMS) of DNA-bound Ca2+ [113]. Dinoflagellate chromosomes lack nucleosomes and persist as compact entities during interphase (image adapted from [113]). d) Liquid crystal model of dinoflagellate chromosome architecture: chromosomes are organized as a stack of flat liquid crystals in which a single DNA filament is packed in zig-zag with a cholesteric organization. Image adapted from [113]. e) Mechanism of HCc3-induced DNA condensation. Left: DNA strand represented by a green line. HCc3, a DNA-binding protein, forms a dimer and is in equilibrium as a free solution form and DNA-bound form. Middle: at high HCc3 concentration, a single DNA strand may acquire multiple molecules of HCc3 dimers. Right: interactions between multiple “seeding” points result in a condensed, network-like pattern. Image adapted from [114]. f) Higher order organization of the 4.2 Mb sized nucleoid of Bacillus subtilis. Arrows point to high density regions (HDRs), which are surrounded by less dense chromatin at the nucleoid periphery. Image adapted from [115]. g) 3D reconstruction of the B. subtilis chromosome calculated from Hi-C data. The chromosome is represented as a chain of beads (1 bead = 4 kb). The color-coding of the beads reflects their linear genomic position along the chromosome. Image adapted from [115].
Dinoflagellates – chromatin without nucleosomes. Notably, nucleosomes were lost during the evolution of the Dinoflagellata phylum, its >2000 species represent one of the largest groups of marine eukaryotes. Dinoflagellates carry large genomes (2-200 pg) distributed in many chromosomes, which remain permanently condensed chromosomes during the entire cell cycle (Fig. 2c). Due to the absence of nucleosomes, chromosome architecture is radically different from the architecture of other eukaryotes [113, 116] (Fig. 2d). HCc proteins, a family of DNA-binding proteins with homologies to bacterial and eukaryotic histone H1, are involved in chromatin compaction [117] (Fig. 2e).
Bacterial nucleoids. Studies of bacterial nucleoids demonstrate the presence of genome compartmentalization in bacteria and archaea species [118-122]. For example, Fig. 2f shows the 3D reconstruction of a DAPI-stained, 4.2 Mb sized nucleoid of Bacillus subtilis recorded with 3D SIM. This reconstruction reveals a nucleoid landscape with about 20 chromosomal interaction domains (CDRs) ranging in size from 50 to 300 kb, which form high density regions (HDRs) surrounded by less dense chromatin at the nucleoid periphery [123]. Notably, the nucleoid landscape of B. subtilis showed the same 3D configuration when the nucleoid was visualized with GFP-tagged nucleoid-binding protein HBsu. Figure 2g provides the 3D chromosome structure of B. subtilis calculated from Hi-C data [115]. Further studies are required in order to explore whether the 3D configuration of bacterial nucleoids and of CDs and CDCs show similarities to an extent that argues for a common evolutionary origin of certain structural features. Notably, the circular DNAs present in mitochondria and chloroplasts, respectively, have maintained the nucleoid organization of their prokaryotic ancestors [124], emphasizing again the importance of a compact genome organization throughout evolutionary times.
Chromatin compaction – maintenance of genome integrity. We do not know why the evolution of genomes with increasing size was associated early on with a compartmentalized, compact architecture in both prokaryotes and eukaryotes. But it seems reasonable to argue that genome structure and function have been tightly linked from the first (proto)cells to the rich variety of today’s species. Based on experimental evidence that chromatin compaction protects genomic DNA from radiation damage, Hideaki Takata and colleagues suggested that genomic DNA compaction plays an important role in maintaining genomic integrity [125]. Taking into account a lack of sophisticated repair mechanisms during the early evolution of genomes, further studies of this hypothesis seem of great interest.
REFERENCES
1.Tashiro, S., and Lanctot, C. (2015) The
International Nucleome Consortium, Nucleus, 6, 89-92.
2.Cremer, T., and Cremer, M. (2010) Chromosome
territories, Cold Spring Harb. Perspect. Biol., 2,
a003889.
3.Cremer, T., Kreth, G., Koester, H., Fink, R. H.,
Heintzmann, R., Cremer, M., Solovei, I., Zink, D., and Cremer, C.
(2000) Chromosome territories, interchromatin domain compartment, and
nuclear matrix: an integrated view of the functional nuclear
architecture, Crit. Rev. Eukaryot. Gene Expr., 10,
179-212.
4.Cremer, T., and Cremer, C. (2001) Chromosome
territories, nuclear architecture and gene regulation in mammalian
cells, Nat. Rev. Genet., 2, 292-301.
5.Smeets, D., Markaki, Y., Schmid, V. J., Kraus, F.,
Tattermusch, A., Cerase, A., Sterr, M., Fiedler, S., Demmerle, J.,
Popken, J., Leonhardt, H., Brockdorff, N., Cremer, T., Schermelleh, L.,
and Cremer, M. (2014) Three-dimensional super-resolution microscopy of
the inactive X chromosome territory reveals a collapse of its active
nuclear compartment harboring distinct Xist RNA foci, Epigenetics
Chromatin, 7, 8.
6.Cremer, T., Cremer, M., Hubner, B., Strickfaden,
H., Smeets, D., Popken, J., Sterr, M., Markaki, Y., Rippe, K., and
Cremer, C. (2015) The 4D nucleome: evidence for a dynamic nuclear
landscape based on co-aligned active and inactive nuclear compartments,
FEBS Lett., 589, 2931-2943.
7.Schmid, V. J., Cremer, M., and Cremer, T. (2017)
Quantitative analyses of the 3D nuclear landscape recorded with
super-resolved fluorescence microscopy, Methods, 123,
33-46.
8.Hubner, B., Lomiento, M., Mammoli, F., Illner, D.,
Markaki, Y., Ferrari, S., Cremer, M., and Cremer, T. (2015) Remodeling
of nuclear landscapes during human myelopoietic cell differentiation
maintains co-aligned active and inactive nuclear compartments,
Epigenetics Chromatin, 8, 47.
9.Borsos, M., and Torres-Padilla, M. E. (2016)
Building up the nucleus: nuclear organization in the establishment of
totipotency and pluripotency during mammalian development, Genes
Dev., 30, 611-621.
10.Brero, A., Easwaran, H. P., Nowak, D., Grunewald,
I., Cremer, T., Leonhardt, H., and Cardoso, M. C. (2005) Methyl
CpG-binding proteins induce large-scale chromatin reorganization during
terminal differentiation, J. Cell Biol., 169,
733-743.
11.Mayer, R., Brero, A., von Hase, J., Schroeder,
T., Cremer, T., and Dietzel, S. (2005) Common themes and cell type
specific variations of higher order chromatin arrangements in the
mouse, BMC Cell Biol., 6, 44.
12.Solovei, I., Grandi, N., Knoth, R., Volk, B., and
Cremer, T. (2004) Positional changes of pericentromeric heterochromatin
and nucleoli in postmitotic Purkinje cells during murine cerebellum
development, Cytogenet. Genome Res., 105, 302-310.
13.Solovei, I., Kreysing, M., Lanctot, C., Kosem,
S., Peichl, L., Cremer, T., Guck, J., and Joffe, B. (2009) Nuclear
architecture of rod photoreceptor cells adapts to vision in mammalian
evolution, Cell, 137, 356-368.
14.Cremer, T., and Cremer, C. (2006) Rise, fall and
resurrection of chromosome territories: a historical perspective. Part
I. The rise of chromosome territories, Eur. J. Histochem.,
50, 161-176.
15.Razin, S. V., and Vassetzky, Y. S. (2017) 3D
genomics imposes evolution of the domain model of eukaryotic genome
organization, Chromosoma, 126, 59-69.
16.Bodnar, J. W. (1988) A domain model for
eukaryotic DNA organization: a molecular basis for cell differentiation
and chromosome evolution, J. Theor. Biol., 132,
479-507.
17.Goldman, M. A. (1988) The chromatin domain as a
unit of gene regulation, Bioessays, 9, 50-55.
18.Monneron, A., and Bernhard, W. (1969) Fine
structural organization of the interphase nucleus in some mammalian
cells, J. Ultrastruct. Res., 27, 266-288.
19.Fakan, S. (2004) The functional architecture of
the nucleus as analysed by ultrastructural cytochemistry, Histochem.
Cell Biol., 122, 83-93.
20.Fakan, S., and van Driel, R. (2007) The
perichromatin region: a functional compartment in the nucleus that
determines large-scale chromatin folding, Semin. Cell Dev.
Biol., 18, 676-681.
21.Rouquette, J., Cremer, C., Cremer, T., and Fakan,
S. (2010) Functional nuclear architecture studied by microscopy:
present and future, Int. Rev. Cell Mol. Biol., 282,
1-90.
22.Cmarko, D., Verschure, P. J., Martin, T. E.,
Dahmus, M. E., Krause, S., Fu, X. D., van Driel, R., and Fakan, S.
(1999) Ultrastructural analysis of transcription and splicing in the
cell nucleus after bromo-UTP microinjection, Mol. Biol. Cell,
10, 211-223.
23.Jaunin, F., Visser, A. E., Cmarko, D., Aten, J.
A., and Fakan, S. (2000) Fine structural in situ analysis of nascent
DNA movement following DNA replication, Exp. Cell Res.,
260, 313-323.
24.Bornfleth, H., Edelmann, P., Zink, D., Cremer,
T., and Cremer, C. (1999) Quantitative motion analysis of
subchromosomal foci in living cells using four-dimensional microscopy,
Biophys. J., 77, 2871-2886.
25.Strickfaden, H., Zunhammer, A., van
Koningsbruggen, S., Kohler, D., and Cremer, T. (2010) 4D chromatin
dynamics in cycling cells: Theodor Boveri’s hypotheses revisited,
Nucleus, 1, 284-297.
26.Cremer, M., Schmid, V. J., Kraus, F., Markaki,
Y., Hellmann, I., Maiser, A., Leonhardt, H., John, S.,
Stamatoyannopoulos, J., and Cremer, T. (2017) Initial high-resolution
microscopic mapping of active and inactive regulatory sequences proves
non-random 3D arrangements in chromatin domain clusters, Epigenetics
Chromatin, 10, 39.
27.Popken, J., Brero, A., Koehler, D., Schmid, V.
J., Strauss, A., Wuensch, A., Guengoer, T., Graf, A., Krebs, S., Blum,
H., Zakhartchenko, V., Wolf, E., and Cremer, T. (2014) Reprogramming of
fibroblast nuclei in cloned bovine embryos involves major structural
remodeling with both striking similarities and differences to nuclear
phenotypes of in vitro fertilized embryos, Nucleus,
5, 555-589.
28.Popken, J., Graf, A., Krebs, S., Blum, H.,
Schmid, V. J., Strauss, A., Guengoer, T., Zakhartchenko, V., Wolf, E.,
and Cremer, T. (2015) Remodeling of the nuclear envelope and lamina
during bovine preimplantation development and its functional
implications, PLoS One, 10, e0124619.
29.Chow, K. H., Factor, R. E., and Ullman, K. S.
(2012) The nuclear envelope environment and its cancer connections,
Nat. Rev. Cancer, 12, 196-209.
30.Zink, D., Fischer, A. H., and Nickerson, J. A.
(2004) Nuclear structure in cancer cells, Nat. Rev. Cancer,
4, 677-687.
31.Razin, S. V., and Ulianov, S. V. (2017) Gene
functioning and storage within a folded genome, Cell. Mol. Biol.
Lett., 22, 18.
32.Ulianov, S. V., Gavrilov, A. A., and Razin, S. V.
(2015) Nuclear compartments, genome folding, and
enhancer–promoter communication, Int. Rev. Cell Mol.
Biol., 315, 183-244.
33.Dekker, J., Rippe, K., Dekker, M., and Kleckner,
N. (2002) Capturing chromosome conformation, Science,
295, 1306-1311.
34.Lieberman-Aiden, E., van Berkum, N. L., Williams,
L., Imakaev, M., Ragoczy, T., Telling, A., Amit, I., Lajoie, B. R.,
Sabo, P. J., Dorschner, M. O., Sandstrom, R., Bernstein, B., Bender, M.
A., Groudine, M., Gnirke, A., Stamatoyannopoulos, J., Mirny, L. A.,
Lander, E. S., and Dekker, J. (2009) Comprehensive mapping of
long-range interactions reveals folding principles of the human genome,
Science, 326, 289-293.
35.Le Dily, F., Serra, F., and Marti-Renom, M. A.
(2017) 3D modeling of chromatin structure: is there a way to integrate
and reconcile single cell and population experimental data? WIREs
Comput. Mol. Sci., 7, e1308.
36.Dixon, J. R., Gorkin, D. U., and Ren, B. (2016)
Chromatin domains: the unit of chromosome organization, Mol.
Cell, 62, 668-680.
37.Dixon, J. R., Selvaraj, S., Yue, F., Kim, A., Li,
Y., Shen, Y., Hu, M., Liu, J. S., and Ren, B. (2012) Topological
domains in mammalian genomes identified by analysis of chromatin
interactions, Nature, 485, 376-380.
38.Fraser, J., Ferrai, C., Chiariello, A. M.,
Schueler, M., Rito, T., Laudanno, G., Barbieri, M., Moore, B. L.,
Kraemer, D. C., Aitken, S., Xie, S. Q., Morris, K. J., Itoh, M.,
Kawaji, H., Jaeger, I., Hayashizaki, Y., Carninci, P., Forrest, A. R.,
FANTOM Consortium, Semple, C. A., Dostie, J., Pombo, A., and Nicodemi,
M. (2015) Hierarchical folding and reorganization of chromosomes are
linked to transcriptional changes in cellular differentiation, Mol.
Syst. Biol., 11, 852.
39.Rao, S. S., Huntley, M. H., Durand, N. C.,
Stamenova, E. K., Bochkov, I. D., Robinson, J. T., Sanborn, A. L.,
Machol, I., Omer, A. D., Lander, E. S., and Aiden, E. L. (2014) A 3D
map of the human genome at kilobase resolution reveals principles of
chromatin looping, Cell, 159, 1665-1680.
40.Furlan-Magaril, M., Varnai, C., Nagano, T., and
Fraser, P. (2015) 3D genome architecture from populations to single
cells, Curr. Opin. Genet. Dev., 31, 36-41.
41.Nagano, T., Lubling, Y., Stevens, T. J.,
Schoenfelder, S., Yaffe, E., Dean, W., Laue, E. D., Tanay, A., and
Fraser, P. (2013) Single-cell Hi-C reveals cell-to-cell variability in
chromosome structure, Nature, 502, 59-64.
42.Nagano, T., Lubling, Y., Yaffe, E., Wingett, S.
W., Dean, W., Tanay, A., and Fraser, P. (2015) Single-cell Hi-C for
genome-wide detection of chromatin interactions that occur
simultaneously in a single cell, Nat. Protoc., 10,
1986-2003.
43.Stevens, T. J., Lando, D., Basu, S., Atkinson, L.
P., Cao, Y., Lee, S. F., Leeb, M., Wohlfahrt, K. J., Boucher, W.,
O'Shaughnessy-Kirwan, A., Cramard, J., Faure, A. J., Ralser, M.,
Blanco, E., Morey, L., Sanso, M., Palayret, M. G. S., Lehner, B., Di
Croce, L., Wutz, A., Hendrich, B., Klenerman, D., and Laue, E. D.
(2017) 3D structures of individual mammalian genomes studied by
single-cell Hi-C, Nature, 544, 59-64.
44.Ulianov, S. V., Tachibana-Konwalski, K., and
Razin, S. V. (2017) Single-cell Hi-C bridges microscopy and genome-wide
sequencing approaches to study 3D chromatin organization,
Bioessays, 39, No. 10; doi: 10.1002/bies.201700104; Epub
2017 Aug 9.
45.Fullwood, M. J., and Ruan, Y. (2009) ChIP-based
methods for the identification of long-range chromatin interactions,
J. Cell. Biochem., 107, 30-39.
46.Ji, X., Dadon, D. B., Powell, B. E., Fan, Z. P.,
Borges-Rivera, D., Shachar, S., Weintraub, A. S., Hnisz, D., Pegoraro,
G., Lee, T. I., Misteli, T., Jaenisch, R., and Young, R. A. (2016) 3D
chromosome regulatory landscape of human pluripotent cells, Cell
Stem Cell, 18, 262-275.
47.Ong, C. T., and Corces, V. G. (2014) CTCF: an
architectural protein bridging genome topology and function, Nat.
Rev. Genet., 15, 234-246.
48.Sanborn, A. L., Rao, S. S., Huang, S. C., Durand,
N. C., Huntley, M. H., Jewett, A. I., Bochkov, I. D., Chinnappan, D.,
Cutkosky, A., Li, J., Geeting, K. P., Gnirke, A., Melnikov, A.,
McKenna, D., Stamenova, E. K., Lander, E. S., and Aiden, E. L. (2015)
Chromatin extrusion explains key features of loop and domain formation
in wild-type and engineered genomes, Proc. Natl. Acad. Sci. USA,
112, E6456-6465.
49.Rao, S. S. P., Huang, S. C., Glenn St Hilaire,
B., Engreitz, J. M., Perez, E. M., Kieffer-Kwon, K. R., Sanborn, A. L.,
Johnstone, S. E., Bascom, G. D., Bochkov, I. D., Huang, X., Shamim, M.
S., Shin, J., Turner, D., Ye, Z., Omer, A. D., Robinson, J. T.,
Schlick, T., Bernstein, B. E., Casellas, R., Lander, E. S., and Aiden,
E. L. (2017) Cohesin loss eliminates all loop domains, Cell,
171, 305-320.
50.Franke, M., Ibrahim, D. M., Andrey, G.,
Schwarzer, W., Heinrich, V., Schopflin, R., Kraft, K., Kempfer, R.,
Jerkovic, I., Chan, W. L., Spielmann, M., Timmermann, B., Wittler, L.,
Kurth, I., Cambiaso, P., Zuffardi, O., Houge, G., Lambie, L., Brancati,
F., Pombo, A., Vingron, M., Spitz, F., and Mundlos, S. (2016) Formation
of new chromatin domains determines pathogenicity of genomic
duplications, Nature, 538, 265-269.
51.Lupianez, D. G., Spielmann, M., and Mundlos, S.
(2016) Breaking TADs: how alterations of chromatin domains result in
disease, Trends Genet., 32, 225-237.
52.Achinger-Kawecka, J., and Clark, S. J. (2017)
Disruption of the 3D cancer genome blueprint, Epigenomics,
9, 47-55.
53.Flavahan, W. A., Drier, Y., Liau, B. B.,
Gillespie, S. M., Venteicher, A. S., Stemmer-Rachamimov, A. O., Suva,
M. L., and Bernstein, B. E. (2016) Insulator dysfunction and oncogene
activation in IDH mutant gliomas, Nature, 529,
110-114.
54.Harmston, N., Ing-Simmons, E., Tan, G., Perry,
M., Merkenschlager, M., and Lenhard, B. (2017) Topologically
associating domains are ancient features that coincide with metazoan
clusters of extreme noncoding conservation, Nat. Commun.,
8, 441.
55.Acemel, R. D., Maeso, I., and Gomez-Skarmeta, J.
L. (2017) Topologically associated domains: a successful scaffold for
the evolution of gene regulation in animals, Wiley Interdiscip. Rev.
Dev. Biol., 6, No. 3; doi: 10.1002/wdev.265; Epub 2017 Mar
2.
56.Rowley, M. J., Nichols, M. H., Lyu, X.,
Ando-Kuri, M., Rivera, I. S. M., Hermetz, K., Wang, P., Ruan, Y., and
Corces, V. G. (2017) Evolutionarily conserved principles predict 3D
chromatin organization, Mol. Cell., 67, 837-852.
57.Liu, C., Cheng, Y. J., Wang, J. W., and Weigel,
D. (2017) Prominent topologically associated domains differentiate
global chromatin packing in rice from Arabidopsis, Nat.
Plants, 3, 742-748.
58.Feng, S., Cokus, S. J., Schubert, V., Zhai, J.,
Pellegrini, M., and Jacobsen, S. E. (2014) Genome-wide Hi-C analyses in
wild-type and mutants reveal high-resolution chromatin interactions in
Arabidopsis, Mol. Cell., 55, 694-707.
59.Bickmore, W. A. (2013) The spatial organization
of the human genome, Annu. Rev. Genom. Hum. Genet., 14,
67-84.
60.Bolzer, A., Kreth, G., Solovei, I., Koehler, D.,
Saracoglu, K., Fauth, C., Muller, S., Eils, R., Cremer, C., Speicher,
M. R., and Cremer, T. (2005) Three-dimensional maps of all chromosomes
in human male fibroblast nuclei and prometaphase rosettes, PLoS
Biol., 3, e157.
61.Boyle, S., Gilchrist, S., Bridger, J. M., Mahy,
N. L., Ellis, J. A., and Bickmore, W. A. (2001) The spatial
organization of human chromosomes within the nuclei of normal and
emerin-mutant cells, Hum. Mol. Genet., 10, 211-219.
62.Cremer, M., von Hase, J., Volm, T., Brero, A.,
Kreth, G., Walter, J., Fischer, C., Solovei, I., Cremer, C., and
Cremer, T. (2001) Non-random radial higher-order chromatin arrangements
in nuclei of diploid human cells, Chromosome Res., 9,
541-567.
63.Croft, J. A., Bridger, J. M., Boyle, S., Perry,
P., Teague, P., and Bickmore, W. A. (1999) Differences in the
localization and morphology of chromosomes in the human nucleus, J.
Cell Biol., 145, 1119-1131.
64.Tanabe, H., Muller, S., Neusser, M., von Hase,
J., Calcagno, E., Cremer, M., Solovei, I., Cremer, C., and Cremer, T.
(2002) Evolutionary conservation of chromosome territory arrangements
in cell nuclei from higher primates, Proc. Natl. Acad. Sci. USA,
99, 4424-4429.
65.Habermann, F. A., Cremer, M., Walter, J., Kreth,
G., von Hase, J., Bauer, K., Wienberg, J., Cremer, C., Cremer, T., and
Solovei, I. (2001) Arrangements of macro- and microchromosomes in
chicken cells, Chromosome Res., 9, 569-584.
66.Alexandrova, O., Solovei, I., Cremer, T., and
David, C. N. (2003) Replication labeling patterns and chromosome
territories typical of mammalian nuclei are conserved in the early
metazoan Hydra, Chromosoma, 112, 190-200.
67.Baroux, C., Pecinka, A., Fuchs, J., Kreth, G.,
Schubert, I., and Grossniklaus, U. (2017) Non-random chromosome
arrangement in triploid endosperm nuclei, Chromosoma,
126, 115-124.
68.Fransz, P., and de Jong, H. (2011) From
nucleosome to chromosome: a dynamic organization of genetic
information, Plant J., 66, 4-17.
69.Grob, S., and Grossniklaus, U. (2017) Chromosome
conformation capture-based studies reveal novel features of plant
nuclear architecture, Curr. Opin. Plant Biol., 36,
149-157.
70.Heslop-Harrison, J. S., and Schwarzacher, T.
(2011) Organisation of the plant genome in chromosomes, Plant
J., 66, 18-33.
71.Liu, C., and Weigel, D. (2015) Chromatin in 3D:
progress and prospects for plants, Genome Biol., 16,
170.
72.Pawlowski, W. P. (2010) Chromosome organization
and dynamics in plants, Curr. Opin. Plant Biol., 13,
640-645.
73.Schubert, I., and Shaw, P. (2011) Organization
and dynamics of plant interphase chromosomes, Trends Plant.
Sci., 16, 273-281.
74.Schubert, V., Meister, A., Tsujimoto, H., Endo,
T. R., and Houben, A. (2011) Similar rye A and B chromosome
organization in meristematic and differentiated interphase nuclei,
Chromosome Res., 19, 645-655.
75.Vergara, Z., and Gutierrez, C. (2017) Emerging
roles of chromatin in the maintenance of genome organization and
function in plants, Genome Biol., 18, 96.
76.Wang, C., Liu, C., Roqueiro, D., Grimm, D.,
Schwab, R., Becker, C., Lanz, C., and Weigel, D. (2015) Genome-wide
analysis of local chromatin packing in Arabidopsis thaliana,
Genome Res., 25, 246-256.
77.Perrella, G., and Kaiserli, E. (2016) Light
behind the curtain: photoregulation of nuclear architecture and
chromatin dynamics in plants, New Phytol., 212,
908-919.
78.Joffe, B., Peichl, P., Hendrickson, A.,
Leonhardt, H., and Solovei, I. (2014) Diurnality and nocturnality in
primates: an analysis from the rod photoreceptor nuclei perspective,
Evol. Biol., 41, 1-11.
79.Kizilyaprak, C., Spehner, D., Devys, D., and
Schultz, P. (2010) In vivo chromatin organization of mouse rod
photoreceptors correlates with histone modifications, PLoS One,
5, e11039.
80.Solovei, I., Wang, A. S., Thanisch, K., Schmidt,
C. S., Krebs, S., Zwerger, M., Cohen, T. V., Devys, D., Foisner, R.,
Peichl, L., Herrmann, H., Blum, H., Engelkamp, D., Stewart, C. L.,
Leonhardt, H., and Joffe, B. (2013) LBR and lamin A/C sequentially
tether peripheral heterochromatin and inversely regulate
differentiation, Cell, 152, 584-598.
81.Blaszczak, Z., Kreysing, M., and Guck, J. (2014)
Direct observation of light focusing by single photoreceptor cell
nuclei, Opt. Express, 22, 11043-11060.
82.Kreysing, M., Boyde, L., Guck, J., and Chalut, K.
J. (2010) Physical insight into light scattering by photoreceptor cell
nuclei, Opt. Lett., 35, 2639-2641.
83.Ma, Y., and Buttitta, L. (2017) Chromatin
organization changes during the establishment and maintenance of the
postmitotic state, Epigenetics Chromatin, 10, 53.
84.Bickmore, W. A., and van Steensel, B. (2013)
Genome architecture: domain organization of interphase chromosomes,
Cell, 152, 1270-1284.
85.Van Steensel, B., and Belmont, A. S. (2017)
Lamina-associated domains: links with chromosome architecture,
heterochromatin, and gene repression, Cell, 169,
780-791.
86.Bi, X., Cheng, Y. J., Hu, B., Ma, X., Wu, R.,
Wang, J. W., and Liu, C. (2017) Nonrandom domain organization of the
Arabidopsis genome at the nuclear periphery, Genome Res.,
27, 1162-1173.
87.Hsu, T. C. (1975) A possible function of
constitutive heterochromatin: the bodyguard hypothesis,
Genetics, 79 (Suppl.), 137-150.
88.Gazave, E., Gautier, P., Gilchrist, S., and
Bickmore, W. A. (2005) Does radial nuclear organisation influence DNA
damage? Chromosome Res., 13, 377-388.
89.Smith, K. S., Liu, L. L., Ganesan, S., Michor,
F., and De, S. (2017) Nuclear topology modulates the mutational
landscapes of cancer genomes, Nat. Struct. Mol. Biol.,
24, 1000-1006.
90.Qiu, G. H. (2015) Protection of the genome and
central protein-coding sequences by non-coding DNA against DNA damage
from radiation, Mutat. Res. Rev. Mutat. Res., 764,
108-117.
91.Belousov, V. V., Enikolopov, G. N., and Mishina,
N. M. (2013) Compartmentalization of ROS-mediated signal transduction,
Bioorg. Khim., 39, 383-399.
92.Yakes, F. M., and Van Houten, B. (1997)
Mitochondrial DNA damage is more extensive and persists longer than
nuclear DNA damage in human cells following oxidative stress, Proc.
Natl. Acad. Sci. USA, 94, 514-519.
93.Obe, G., Pfeiffer, P., Savage, J. R., Johannes,
C., Goedecke, W., Jeppesen, P., Natarajan, A. T., Martinez-Lopez, W.,
Folle, G. A., and Drets, M. E. (2002) Chromosomal aberrations:
formation, identification and distribution, Mutat. Res.,
504, 17-36.
94.Martin, W., Baross, J., Kelley, D., and Russell,
M. J. (2008) Hydrothermal vents and the origin of life, Nat. Rev.
Microbiol., 6, 805-814.
95.Mulkidjanian, A. Y., Bychkov, A. Y., Dibrova, D.
V., Galperin, M. Y., and Koonin, E. V. (2012) Origin of first cells at
terrestrial, anoxic geothermal fields, Proc. Natl. Acad. Sci.
USA, 109, E821-830.
96.Speijer, D. (2015) Birth of the eukaryotes by a
set of reactive innovations: new insights force us to relinquish
gradual models, Bioessays, 37, 1268-1276.
97.Postberg, J., Lipps, H. J., and Cremer, T. (2010)
Evolutionary origin of the cell nucleus and its functional
architecture, Essays Biochem., 48, 1-24.
98.Pereira, S. L., Grayling, R. A., Lurz, R., and
Reeve, J. N. (1997) Archaeal nucleosomes, Proc. Natl. Acad. Sci.
USA, 94, 12633-12637.
99.Mattiroli, F., Bhattacharyya, S., Dyer, P. N.,
White, A. E., Sandman, K., Burkhart, B. W., Byrne, K. R., Lee, T., Ahn,
N. G., Santangelo, T. J., Reeve, J. N., and Luger, K. (2017) Structure
of histone-based chromatin in Archaea, Science, 357,
609-612.
100.Luger, K., Mader, A. W., Richmond, R. K.,
Sargent, D. F., and Richmond, T. J. (1997) Crystal structure of the
nucleosome core particle at 2.8 Å resolution, Nature,
389, 251-260.
101.Tanizawa, H., Kim, K. D., Iwasaki, O., and
Noma, K. I. (2017) Architectural alterations of the fission yeast
genome during the cell cycle, Nat. Struct. Mol. Biol.,
24, 965-976.
102.Gursoy, G., Xu, Y., and Liang, J. (2017)
Spatial organization of the budding yeast genome in the cell nucleus
and identification of specific chromatin interactions from
multi-chromosome constrained chromatin model, PLoS Comput.
Biol., 13, e1005658.
103.Iwasaki, O., Corcoran, C. J., and Noma, K.
(2016) Involvement of condensin-directed gene associations in the
organization and regulation of chromosome territories during the cell
cycle, Nucleic Acids Res., 44, 3618-3628.
104.Gasser, S. M. (2002) Visualizing chromatin
dynamics in interphase nuclei, Science, 296,
1412-1416.
105.Taddei, A., and Gasser, S. M. (2012) Structure
and function in the budding yeast nucleus, Genetics, 192,
107-129.
106.Chubb, J. R., and Bickmore, W. A. (2003)
Considering nuclear compartmentalization in the light of nuclear
dynamics, Cell, 112, 403-406.
107.Selig, S., Okumura, K., Ward, D. C., and Cedar,
H. (1992) Delineation of DNA replication time zones by fluorescence in
situ hybridization, EMBO J., 11, 1217-1225.
108.Yalon, M., Gal, S., Segev, Y., Selig, S., and
Skorecki, K. L. (2004) Sister chromatid separation at human telomeric
regions, J. Cell Sci., 117, 1961-1970.
109.Amitai, A., Seeber, A., Gasser, S. M., and
Holcman, D. (2017) Visualization of chromatin decompaction and break
site extrusion as predicted by statistical polymer modeling of
single-locus trajectories, Cell Rep., 18, 1200-1214.
110.Seeber, A., and Gasser, S. M. (2017) Chromatin
organization and dynamics in double-strand break repair, Curr. Opin.
Genet. Dev., 43, 9-16.
111.Shen, Y., Buick, R., and Canfield, D. E. (2001)
Isotopic evidence for microbial sulphate reduction in the early
archaean era, Nature, 410, 77-81.
112.Knoll, A. H. (1999) A new molecular window on
early life, Science, 285, 1025-1026.
113.Levi-Setti, R., Gavrilov, K. L., and Rizzo, P.
J. (2008) Divalent cation distribution in dinoflagellate chromosomes
imaged by high-resolution ion probe mass spectrometry, Eur. J. Cell
Biol., 87, 963-976.
114.Chan, Y. H., and Wong, J. T. (2007)
Concentration-dependent organization of DNA by the dinoflagellate
histone-like protein HCc3, Nucleic Acids Res., 35,
2573-2583.
115.Marbouty, M., Le Gall, A., Cattoni, D. I.,
Cournac, A., Koh, A., Fiche, J. B., Mozziconacci, J., Murray, H.,
Koszul, R., and Nollmann, M. (2015) Condensin- and replication-mediated
bacterial chromosome folding and origin condensation revealed by Hi-C
and super-resolution imaging, Mol. Cell, 59, 588-602.
116.Gornik, S. G., Ford, K. L., Mulhern, T. D.,
Bacic, A., McFadden, G. I., and Waller, R. F. (2012) Loss of
nucleosomal DNA condensation coincides with appearance of a novel
nuclear protein in dinoflagellates, Curr. Biol., 22,
2303-2312.
117.Wong, J. T., New, D. C., Wong, J. C., and Hung,
V. K. (2003) Histonelike proteins of the dinoflagellate
Crypthecodinium cohnii have homologies to bacterial DNA-binding
proteins, Eukaryot. Cell, 2, 646-650.
118.Eltsov, M., and Zuber, B. (2006) Transmission
electron microscopy of the bacterial nucleoid, J. Struct. Biol.,
156, 246-254.
119.Feijoo-Siota, L., Rama, J. L. R.,
Sanchez-Perez, A., and Villa, T. G. (2017) Considerations on bacterial
nucleoids, Appl. Microbiol. Biotechnol., 101,
5591-5602.
120.Fisher, J. K., Bourniquel, A., Witz, G.,
Weiner, B., Prentiss, M., and Kleckner, N. (2013) Four-dimensional
imaging of E. coli nucleoid organization and dynamics in living
cells, Cell, 153, 882-895.
121.Macvanin, M., and Adhya, S. (2012)
Architectural organization in E. coli nucleoid, Biochim.
Biophys. Acta, 1819, 830-835.
122.Peeters, E., Driessen, R. P., Werner, F., and
Dame, R. T. (2015) The interplay between nucleoid organization and
transcription in archaeal genomes, Nat. Rev. Microbiol.,
13, 333-341.
123.Smits, W. K., Goranov, A. I., and Grossman, A.
D. (2010) Ordered association of helicase loader proteins with the
Bacillus subtilis origin of replication in vivo, Mol.
Microbiol., 75, 452-461.
124.Archibald, J. M. (2011) Origin of eukaryotic
cells: 40 years on, Symbiosis, 54, 69-86.
125.Takata, H., Hanafusa, T., Mori, T., Shimura,
M., Iida, Y., Ishikawa, K., Yoshikawa, K., Yoshikawa, Y., and Maeshima,
K. (2013) Chromatin compaction protects genomic DNA from radiation
damage, PLoS One, 8, e75622.