REVIEW: Protein–Protein Interactions in DNA Base Excision Repair
N. A. Moor1 and O. I. Lavrik1,2*
1Institute of Chemical Biology and Fundamental Medicine, Siberian Branch of the Russian Academy of Sciences, 630090 Novosibirsk, Russia; E-mail: lavrik@niboch.nsc.ru2Novosibirsk State University, 630090 Novosibirsk, Russia
* To whom correspondence should be addressed.
Received November 15, 2017; Revision received December 5, 2017
The system of base excision repair (BER) ensures correction of the most abundant DNA damages in mammalian cells and plays an important role in maintaining genome stability. Enzymes and protein factors participate in the multistage BER in a coordinated fashion, which ensures repair efficiency. The suggested coordination mechanisms are based on formation of protein complexes stabilized via either direct or indirect DNA-mediated interactions. The results of investigation of direct interactions of the proteins participating in BER with each other and with other proteins are outlined in this review. The known protein partners and sites responsible for their interaction are presented for the main participants as well as quantitative characteristics of their affinity. Information on the mechanisms of regulation of protein–protein interactions mediated by DNA intermediates and posttranslational modification is presented. It can be suggested based on all available data that the multiprotein complexes are formed on chromatin independent of the DNA damage with the help of key regulators of the BER process – scaffold protein XRCC1 and poly(ADP-ribose) polymerase 1. The composition of multiprotein complexes changes dynamically depending on the DNA damage and the stage of BER process.
KEY WORDS: base excision repair, protein–protein interactions, DNA repairDOI: 10.1134/S0006297918040120
Abbreviations: AP site, apurinic/apyrimidinic site; APE1, AP endonuclease 1; APTX, aprataxin; BER, base excision repair; DNALigI/DNALigIIIα, DNA ligase I/IIIα; dRp, deoxyribose phosphate; FAM, 5(6)-carboxyfluorescein; FEN1, flap endonuclease 1; FRET, Förster resonance energy transfer; HR, homologous recombination; MMR, mismatch repair; NER, nucleotide excision repair; NHEJ, nonhomologous end joining; PAR, poly(ADP-ribose); PARP1/PARP2, poly(ADP-ribose) polymerase 1/2; PNKP, polynucleotide kinase/phosphatase; Polβ/Polδ/Polε, DNA polymerase β/δ/ε; PTM, posttranslational modification; TDP1, tyrosyl-DNA phosphodiesterase 1; TMR, 5(6)-carboxytetramethylrhodamine; XRD, X-ray diffraction analysis.
DNA repair defects are related to serious human hereditary diseases as
well as development of cancer and aging [1, 2]. Investigation of the repair mechanisms is an
important problem, because this area of molecular biology is directly
related both to understanding of fundamental mechanisms of maintaining
genetic stability of organisms and searching for optimal human
anticancer therapies [3]. Defects in the DNA repair
system are associated with cancerogenesis; and at the same time, when
oncotherapeutic approaches based on the targeted damage of DNA in
cancer cells are used, the DNA repair systems must be inhibited. Hence,
investigation of the repair mechanisms is necessary for the development
of effective treatment of cancer. The DNA damaging effects and the
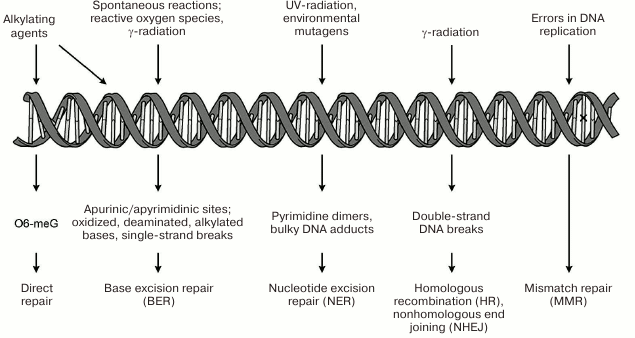
systems restoring its structure are presented schematically in
Fig. 1. The repair systems for damaged bases
(BER), bulk DNA damages (NER), double-strand DNA breaks (HR; NHEJ), and
of mismatched bases (MMR) are examples of such systems. One of the most
important DNA repair systems in human cells is base excision repair
(BER), which ensures correction of the most abundant damages –
modified nitrogenous bases and apurinic/apyrimidinic (AP) sites [3, 4]. Repair of single-strand DNA
breaks occurs with participation of the enzymes and factors of the BER
system and is considered as a separate pathway of this process [5].
Fig. 1. Genetic damages under the action of endogenous and exogenous factors and mechanisms of their correction. Mismatched bases in DNA (replication errors) marked with ×.
DNA BASE EXCISION REPAIR: MAIN STAGES AND PARTICIPANTS OF THE
PROCESS
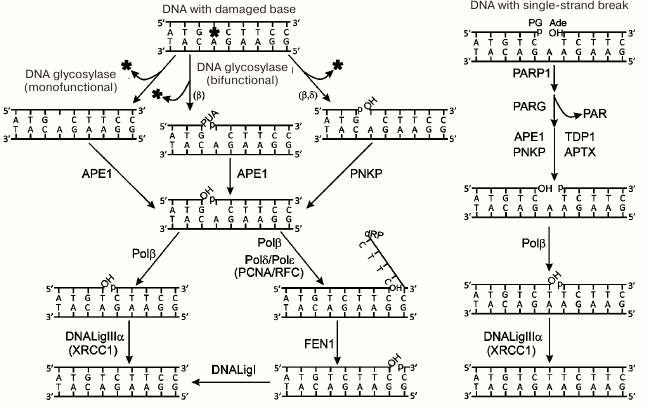
The processes of repair of damaged bases and breaks in one DNA strand are presented schematically in Fig. 2. The damaged bases are removed by DNA glycosylases specific to the certain type of damage. Intact or cleaved (according to mechanism of β- or β/δ-elimination) AP site is formed by the action of mono- or bifunctional DNA glycosylases, respectively [6]. The intact AP site is hydrolyzed by AP endonuclease 1 (APE1); next, DNA polymerase β (Polβ) excises the deoxyribose phosphate residue (dRp) at the 5′-end of the break by its dRp-lyase activity. Terminal blocking groups in the products of action of bifunctional DNA glycosylases are removed by the phosphatase activity of polynucleotide kinase/phosphatase (PNKP) or 3′-phosphatase and 3′-phosphodiesterase activities of APE1. The one-nucleotide gap is filled by DNA polymerase activity of Polβ. The last stage – restoration of chain integrity – is catalyzed by DNA ligase IIIα (DNALigIIIα) involving ATP. This main base repair pathway is known as a short-patch repair. In the case of modification of the 5′-dRp residue, which is impossible to remove by Polβ lyase activity, another pathway is realized – long-patch repair. DNA strand displacement synthesis is initiated by Polβ and continues catalyzed by replicative DNA polymerases δ and ε (Polδ, Polε). The flap structure produced in this synthesis is removed by the flap endonuclease 1 (FEN1) with its activity stimulated by the PCNA replication factor, and finally the break is ligated by DNA ligase I (DNALigI). Another mechanism of the gap translation has been suggested; in this case the extended flap is not formed, FEN1 sequentially removes nucleotides at the 5′-end of the break, and the formed gap is filled by activities of Polβ or Polλ [7, 8].
Fig. 2. Schematic representation of repair pathways for damaged bases and single-strand DNA breaks. Protein designations are described in the text. Designations of blocking groups in DNA are as follows: PUA, 3′-phospho-α,β-unsaturated aldehyde; p, 3′-/5′-phosphate; OH, 3′-/5′-OH group; dRP, 5′-deoxyribose phosphate; PG, 3′-phosphoglycolate; Ade, 5′-aldehyde group.
Repair of single-strand breaks in DNA involves the following steps: 1) detection of the break; 2) removal of blocking groups; 3) filling the gap, and 4) ligation of the break (Fig. 2). Poly(ADP-ribose) polymerase 1 (PARP1) detects breaks in DNA; APE1, PNKP, aprataxin (APTX), and tyrosyl-DNA phosphodiesterase 1 (TDP1) participate in unblocking of 3′- and 5′-ends in breaks; gap filling and ligation are catalyzed by the same set of enzymes that participate in the respective steps of the short-patch repair of the damaged DNA bases. PARP1 is activated via interaction with the damaged DNA; it catalyzes the synthesis of poly(ADP-ribose) (PAR) and modification by covalent attachment of PAR polymer to the PARP1 itself and other proteins involved in the repair. The X-ray repair cross-complementing protein 1 (XRCC1) is considered as a main target of poly(ADP-ribosyl)ation in the BER process. It has been suggested that PARP1 plays the main role in attracting XRCC1 protein to the damages of chromosomal DNA [5, 9]. XRCC1 does not exhibit any enzymatic activity and is considered as a scaffold for organization of the BER complex and repair of single-strand breaks in DNA. PARP2 is another enzyme from the PARP family that catalyzes synthesis of poly(ADP-ribose) following binding with the single-strand break in DNA [10]. It was shown that parp1 gene knockout increased the sensitivity of cells to DNA-damaging agents [11], while parp1 and parp2 double knockouts caused early embryonic lethality [12]. These data indicate the important role of both enzymes in the repair processes. The role of PARP2 and possibility of synergetic action of PARP1 and PARP2 in BER processes has been the subject of intensive investigation recently. The synthesis of poly(ADP-ribose) is a regulated process: degradation of this polymer in cells is mediated by the enzyme poly(ADP-ribose) glycohydrolase (PARG) [13]. Another important role of PARP1 related to its participation in DNA repair is remodeling of chromatin structure via poly(ADP-ribosyl)ation of histones and binding of the remodeling proteins with the synthesized PAR polymer [14].
Coordinated action of the enzymes catalyzing separate stages of the multistep BER process is required for efficient repair of damaged DNA. One of the models suggested previously – the “passing the baton” – involves transfer of the damaged DNA during repair from one enzyme to another, which is likely accompanied by formation of dynamic protein complexes at the site of DNA damage [15, 16]. This model is based on numerous data on the mutual effects of BER enzymes on their activity [15, 17]. This model adequately describes the stimulating effect of APE1 on catalytic activity of DNA glycosylase OGG1, which was investigated in detail using kinetic methods [18]. Another mechanism of coordination suggests formation of multiprotein complexes (so-called repairosomes) involving enzymes of the repair process and proteins with scaffolding function [17]. XRCC1 is an example of such protein that does not have enzymatic activity. Most likely, both mechanisms are used for coordination of the repair process. Many experimental facts suggesting interactions of enzymes and protein factors involved in the BER process count in favor of the existence of “repairosomes” organized from proteins independent of the DNA damage.
DIRECT PROTEIN–PROTEIN INTERACTIONS OF THE MAIN BER
PARTICIPANTS
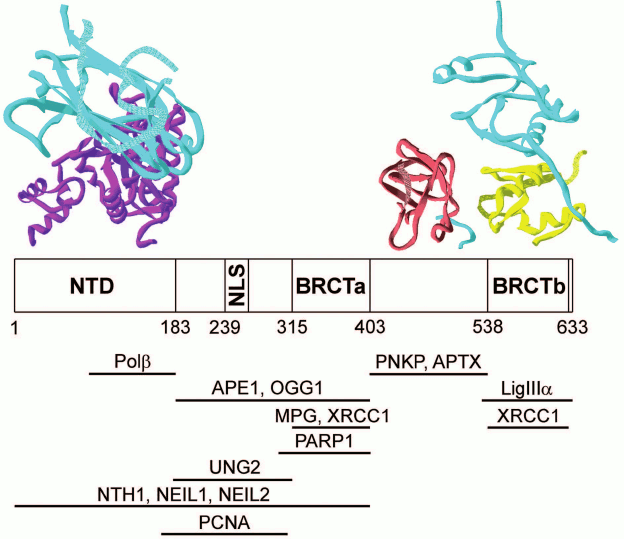
Direct physical interactions have been demonstrated for many proteins participating in BER, and their binding sites have been localized in structural domains (table). Interactions of the XRCC1 protein with partners have been characterized in the greatest detail. This protein consists of three structural domains (NTD, BRCTa, and BRCTb) linked with disordered fragments (linkers XL1 and XL2), one of which (XL1) contains a nuclear localization signal [22]. The availability of two BRCT domains with their main function of association with other proteins [53] and the DNA-binding domain (NTD) creates prerequisites for the main function of the XRCC1 protein as a structural organizer of “repairosomes”. Interestingly, the binding sites of four BER enzymes catalyzing different stages of the process – Polβ, APE1, PNKP, and LigIIIα – are localized on different structural modules (Fig. 3). At the same time, the DNA glycosylase binding sites overlap with the site for APE1 and/or Polβ and PARP1. It is likely that in the process of correction of the damaged bases, DNA glycosylases form dynamic contacts with XRCC1 and other enzymes in the “repairosome” composition. For example, the DNA glycosylase NEIL1 interacts directly with the enzymes of short- (PNKP, Polβ, and LigIIIα) and long-patch (PNKP, Polδ, FEN1, and LigI) repair, while the DNA glycosylase MYH forms a complex with the APE1 endonuclease (table). The multiprotein complexes with XRCC1 observed in many studies using recombinant proteins or cell extracts contain Polβ, PNKP, and LigIIIα as stable partners, and their presence enhances interaction of XRCC1 with DNA glycosylases [24, 26, 27, 45]. Another multiprotein complex including XRCC1 and TDP1 also contains PNKP and LigIIIα [46].
Interactions of proteins participating in BER

a Protein structural domains containing the binding site of
the partner protein are shown in brackets. Structural composition of
multidomain proteins: XRCC1: NTD 1-183, XL1 239-266, BRCTa
315-403, XL2 404-537, BRCTb 538-633 [50];
PARP1: ZnF1 1-96, ZnF2 97-206, NLS 207-240, ZnF3 241-366, BRCT
381-484, WGR 518-661, CD 662-1014 [51];
PARP2: NTD 1-63, WGR 64-198, CD 199-559 [31]; LigIIIα: ZnF 1-100, linker 101-170,
DBD 171-390, CD 391-836, BRCT 837-922 [52].
Designations: NTD/CTD domain, N-/C-terminal domain; CD, catalytic
domain; DBD, DNA-binding domain; XL1/XL2, linker 1/2 in XRCC1 protein;
NLS, nuclear localization signal; ZnF, zinc finger. All data were
produced for human recombinant proteins and mouse PARP2.
b Investigation methods: affinity coprecipitation [19, 23-31,
35, 36, 39-43, 48,
49], two-hybrid analysis [20,
26, 27, 30, 32, 41,
44-47], gel filtration [20, 21, 35,
37], ultracentrifugation [20,
43], immunoprecipitation [23-25, 27,
29, 31-35, 38, 41,
42, 45-47], fluorescence titration [33], fluorescence polarization [34], surface plasmon resonance (SPR) [35], small-angle X-ray scattering (SAXS) [37], XRD [22, 37, 40], NMR [48].
Fig. 3. Interactions of XRCC1 protein with other proteins participating in BER. Modular XRCC1 organization and interaction sites presented in the table are shown schematically. Structural models of complexes of XRCC1 individual domains/fragments (cyan) with the respective domains of Polβ, PNKP, and LigIIIα according to XRD data (accession codes to 3D protein structures in Protein Data Bank: 3K75, 2W3O, 3QVG) are shown at the top of the figure.
The PARP1 protein consists of multiple structural modules forming an N-terminal DNA-binding domain and a C-terminal catalytic domain in addition to the central BRCT domain [51, 54]. The coordinating function of the PARP1 protein can be realized via formation of either direct contact with some enzymes (PNKP, Polβ, LigIIIα, and TDP1) or indirect contacts mediated by its interaction with the XRCC1 protein. The main BER enzymes (Polβ and LigIIIα) and XRCC1 protein interact with the DNA binding and BRCT domains, while the TDP1 enzyme – with the catalytic domain of PARP1 (table). Formation of a stable ternary complex with PARP1 and XRCC1 was reported for TDP1 [46]. Overlapping of the binding sites for the majority of PARP1 partners creates conditions for dynamic contacts in the preformed multiprotein assemblies, which can be stabilized in the complex with auto-modified PARP1. Poly(ADP-ribose) acceptors were identified in all the structural domains of PARP1 [55], which expands significantly the platform for formation of the “repairosomes”. Many BER participants such as XRCC1, Polβ, PNKP, aprataxin, TDP1, LigIIIα, and LigI contain PAR-binding motifs [56, 57], and more efficient binding with PAR–PARP1 was demonstrated for XRCC1, LigIIIα, and TDP1 [30, 41, 42]. PARP2 does not contain DNA-binding and BRCT domains [58] and uses the non-conserved WGR domain for interaction with proteins (table). The function of PARP2 (similar to that of PARP1) in coordination of the DNA repair process can be mediated through its interaction with XRCC1 [59]. The enzyme of the final step of repair LigIIIα has direct binding partners among other BER enzymes (NEIL1, NEIL2, PNKP, and TDP1) using the BRCT domain for complex formation (table). Data reported recently indicate the ability of this enzyme to control the assembly of multiprotein complexes on single-strand DNA damages similarly to PARP1 [60].
Most studies on protein–protein interactions in BER have been conducted using the affinity coprecipitation, two-hybrid analysis, and immunoprecipitation techniques (table). The results of these investigations do not provide information on physicochemical, structural, and conformation characteristics of the complexes, leaving many questions on the mechanisms of their functioning unanswered, such as the relative contribution of the proteins to formation of macromolecular associates and their stoichiometry, role of dynamic interactions, conformation changes, and DNA intermediates in formation of the functional assemblies. Information on the structural organization of these complexes is very limited. The 3D structures of the NTD and BRCTb domains as well as of the XL2 fragment of the XRCC1 protein in complexes with the respective domains of three enzymes (Polβ, LigIIIα, and PNKP) were established using X-ray diffraction analysis (XRD) (Fig. 3). The structures of these complexes have been reviewed in detail [22]. It is interesting to note that the additional contact region of the XRCC1 protein with LigIIIα – a polypeptide consisting of hydrophobic amino acid residues adjacent to the N-terminus of the BRCTb domain – was revealed using XRD [37]. It is obvious that the sites localized in proteins by nonequilibrium methods participate in the most stable interactions. The available structural data are not sufficient for description of molecular mechanisms of coordination in the absence of data on the 3D structures of full-length XRCC1 and complexes of other BER participants.
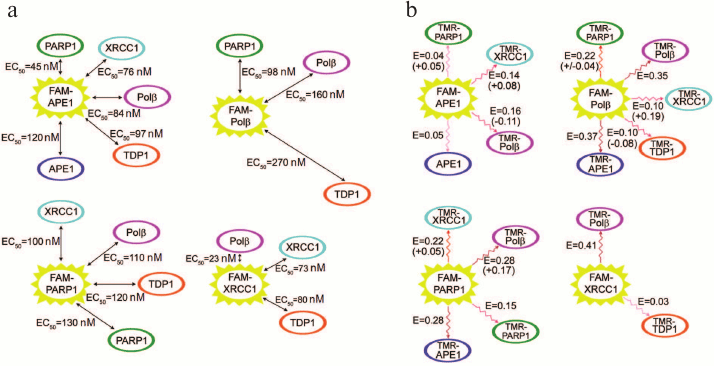
We have characterized various homo- and hetero-oligomeric complexes of BER proteins quantitatively using equilibrium methods of fluorescence titration and fluorescence (Förster) resonance energy transfer (FRET) [61]. N-hydroxysuccinimide esters of 5(6)-carboxyfluorescein (FAM) and 5(6)-carboxytetramethylrhodamine (TMR) were used for N-terminal fluorescent labeling of proteins. Direct (not mediated through DNA or other proteins) interactions of APE1 with Polβ, TDP1, and PARP1; Polβ with TDP1; as well as homo-oligomerization of APE1 were demonstrated for the first time. The apparent equilibrium dissociation constants of the complexes were in the range 23-270 nM (Fig. 4), which is comparable for the main participants with the value for the complex of XRCC1 with PNKP [62] determined using a similar method. The most stable complex was formed between Polβ and XRCC1, which was confirmed by the nonequilibrium method of multidetection size-exclusion chromatography (SEC-MALLS, size-exclusion chromatography coupled with multiangle laser light-scattering), based on measuring molecular mass of the material in the process of separation via detecting intensity of scattered light and refractive index [61]. It was found that the model DNAs that imitate DNA intermediates on different stages of BER modulated to a various degree the structure of protein complexes and their stability. The DNA-dependent effects on the protein affinity for each other were most pronounced for the complexes of APE1 with different protein partners. These results extend our notions on coordination and regulation mechanisms in BER. The dependence of the efficiency of APE1 interaction with Polβ on the type of DNA intermediate indicates that coordination of the functions of key enzymes is not only due to the differences in their affinity for DNA [63], but also due to the strength of their interaction with each other, which is controlled by DNA in different stages of repair. The higher affinity of APE1 for Polβ in the presence of DNA containing an AP site than in a complex with the cleavage product points to the fact that efficient repair is facilitated by the transfer of the intermediate directly in the process of the reaction catalyzed by APE1. The higher affinity of APE1 and Polβ for PARP1 than for each other in the presence of DNA with a single-strand break suggests that regulation of functions of the BER participants via DNA-dependent modulation of their affinity for each other represents a common mechanism for various proteins. At the same time, the stability of the complex of XRCC1 with Polβ does not depend on the presence of DNA intermediates, even though the most pronounced effect of different DNAs on the FRET signal, which reflects structural rearrangement of the complex, was recorded for this complex. These data indicate that this complex similarly to the complex of XRCC1 with LigIIIα serves not only for protection from proteasome degradation [64], but also can function as a stable component of the multiprotein assemblies over the duration of entire BER process. Synchronous colocalization of the XRCC1, Polβ, and LigIIIα proteins on the sites of DNA damage also speaks in favor of formation of the stable ternary complex of XRCC1 with Polβ and LigIIIα [29, 65].
Fig. 4. Complexes of BER proteins investigated by fluorescence titration (a) and FRET (b). a) Apparent equilibrium dissociation constants of complexes were determined as equilibrium concentrations of protein partners (EC50) that ensured 50% enhancement of the fluorescence of FAM-labeled protein in comparison with the maximum increase at saturating concentrations of partners; the sizes of arrows are proportional to the EC50 values. b) The values of efficiency of the fluorescence resonance energy transfer (E) for each pair of proteins in the complex of FAM-labeled protein (donor) with TMR-labeled partner (acceptor) are presented. The largest changes in the energy transfer efficiency (increase or decrease with + or – sign, respectively) recorded in the presence of DNA intermediates are presented in brackets.
Our investigations of the BER complexes revealed efficient interaction of PARP1 with Polβ and APE1 in the absence of DNA [61]. It can be suggested that these proteins interact also in the process of recognition and hydrolysis of an AP site as well as during the following stages catalyzed by Polβ (excision of 5′-dRp residue and filling the gap with the 2′-deoxynucleoside-5′-monophosphate residue). Interaction of PARP1, Polβ, and APE1 with the “central” DNA intermediate in BER was established by photoaffinity labeling of BER proteins in the cell extract [66], which indicated interaction of these proteins during repair synthesis catalyzed by Polβ. We showed later that PARP1 interacted with DNA containing an AP site [67]. All these data suggest that the interaction of PARP1 with APE1 is possible at the stage of recognition of the AP site in DNA and its following hydrolysis by APE1 (Fig. 2). Following hydrolysis of the AP site, PARP1 can catalyze the synthesis of poly(ADP-ribose). According to the initial hypothesis on the mechanism of its action, PARP1 dissociates from its complex with DNA after covalent attachment of the negatively charged PAR polymer. Recently, this hypothesis has been modified mainly with regards to the search for an active role of PAR in formation of repair complexes in different stages including the ones involving participation of such proteins as XRCC1. It was established that following poly(ADP-ribosyl)ation, PARP1 was capable of covalent binding to the photoreactive DNA intermediate [66], hence, the lifetime of the complex of poly(ADP-ribosyl)ated PARP1 with DNA could allow interaction of PARP1 with the damaged DNA. The lifetime of such complexes depends on both the size of covalently bound PAR and the initial affinity of PARP1 to the DNA damage. Complexes of poly(ADP-ribosyl)ated PARP1 with damaged DNA were detected by atomic force microscopy [68]. Hence, regulation of the formation of BER complexes on the damaged DNA can be realized via either poly(ADP-ribosyl)ation of proteins or their interactions with poly(ADP-ribose), synthesis of which is catalyzed by PARP1 and PARP2. Poly(ADP-ribose) is the most important cell regulator of many protein–protein and protein–nucleic acid interactions [69, 70].
PROTEIN COMPLEXES WITH NONCANONICAL FACTORS
Other proteins that were not previously considered as possible participants of BER might also be involved in this process. One such protein is the multifunctional protein YB-1. This positively charged protein has disordered structure. A proteolytic fragment of YB-1 protein, which is localized in the nucleus, is formed in response to DNA damage [71]. We established that YB-1 could be an acceptor for poly(ADP-ribosyl)ation [72]. It was shown previously that YB-1 interacted with poly(ADP-ribose) [56]. All these data combined may indicate the involvement of YB-1 into the process of repair of the damaged DNA. Examination of YB-1 as a noncanonical factor in BER by fluorescence titration showed that many proteins essential for this process (Polβ, NEIL1, PARP1, and PARP2) formed less strong complexes with YB-1 than with each other [73]. The apparent equilibrium dissociation constants are in the range 340-810 nM. The complex of APE1 with YB-1 demonstrating stability similar to the complexes of APE1 with Polβ, XRCC1, and PARP1 represents an exception. High affinity of two multifunctional proteins for each other could be an important factor for their cooperative action in transcription regulation [74]. Interactions of YB-1 protein with BER enzymes could ensure regulation of their activities: AP-endonuclease activity of APE1 and 5′-deoxyribose phosphate-lyase activity of Polβ are inhibited in the presence of YB-1, while the AP-lyase activity of NEIL1 is stimulated [73]. Moreover, it was found that YB-1 stimulated PARP1 activity via binding with poly(ADP-ribose) linked to PARP1, which increased the lifetime of this complex on DNA and the efficiency of poly(ADP-ribose) synthesis [73].
It was reported for some enzymes that they interact with protein factors involved in regulation of other cellular processes. For example, APE1 forms complex with the multifunctional protein nucleophosmin (NPM1), and this interaction plays an important role in regulation of activities of the multifunctional enzyme, its expression, and its intracellular localization [75]. In addition to multiple catalytic functions in DNA repair, APE1 is also a regulator of the transcription processes and RNA processing [76, 77]. DNA glycosylase OGG1 forms complexes with DNA-binding proteins hSSB1 and SATB1, thus enhancing its efficiency in recognition of DNA damage and its repair [78, 79]. It has been suggested that protein factors of unknown nature that are not participating in chromatin structure remodeling stimulate the activity of DNA glycosylase NTH1 in repair initiation [80]. Direct interactions of many DNA glycosylases (TDG, NEIL2, NTH1, OGG1, UNG2) and APE1 with the factors of nucleotide excision repair (XPC, XPG, CSB, RPA) and homologous recombination (Rad52) were observed, and it was shown that these interactions play a regulatory role in the overlapping repair pathways [81]. The HMGB1 protein – chromatin architecture factor – interacts directly with three BER enzymes (APE1, Polβ, and FEN1), modulates their catalytic activity in the process of DNA repair (including nucleosome one), and, hence, ensures regulation of the process via the short- or long-patch pathway [82-84]. In general, the functioning mechanism of the BER system at the level of chromatin is poorly understood [85], and its investigation could lead to discovery of new noncanonical factors.
REGULATION OF PROTEIN–PROTEIN INTERACTIONS
In addition to DNA-mediated regulation of protein–protein interactions in the BER system shown in our work [61], other mechanisms exist. Posttranslational modifications (PTM) of the proteins participating in BER regulate the level of their expression, intracellular localization, and degradation, as well as catalytic and DNA-binding activities of the enzymes either directly or indirectly via modulation of protein–protein interactions. The effect of poly(ADP-ribosyl)ation of PARP1 on its interaction with BER proteins has been described above. How such modification of other proteins affects their function in BER processes is still unknown. The most frequent PTMs were found for the multifunctional protein APE1: phosphorylation, acetylation, S-nitrosylation, S-glutathionylation, formation of disulfide bonds, and ubiquitination [81, 86]. Most modifications modulate functional activity of APE1 as a redox factor and transcription regulator. Only recently it was shown that acetylation of APE1 enhanced in vivo association of the enzyme with the XRCC1 protein and its complex with LigIIIα, ensuring efficient repair required for cell survival [87]. Acetylation of DNA glycosylase TDG1 reduces its interaction with APE1 and produces opposite effects on the excision activity of the enzyme towards various types of damages; repair of damages induced by the chemotherapeutic action of 5-fluorouracil is enhanced by modification [81]. It was suggested based on these data that the level of acetylation of TDG1 in tumor cells was the defining factor of chemotherapy efficiency. Phosphorylation of two residues (Thr6 and Tyr8) in the conformationally flexible N-terminus of DNA-glycosylase UNG2 increases its association with the RPA factor and suppresses interaction with the PCNA factor, which likely regulates formation of the tertiary complex with RPA and PCNA [88]. The most abundant evidence on regulation of protein–protein interactions mediated by PTM was produced for the XRCC1 protein. Phosphorylation of XRCC1 stimulates its binding to PNKP in vivo and affects the repair efficiency [32]. Phosphorylated and nonphosphorylated forms of XRCC1 react with different structural domains of PNKP, and the modified protein forms a stronger complex affecting mainly the kinase activity of the enzyme [33]. Phosphorylation sites in XRCC1 were identified that were responsible for regulation of the efficiency of complex formation with aprataxin, one of the functions of which is protection of the XRCC1 protein from intracellular degradation [34]. The oxidized form of XRCC1 stabilized by formation of a disulfide bond between residues Cys12 and Cys20 forms a more stable (in comparison with the reduced form) complex with Polβ; an increase in the number of intermolecular contacts because of structural reorganization of the complex was demonstrated by XRD analysis [22]. This form of XRCC1 exists in vivo and plays an essential role in repair of damages generated under conditions of oxidative stress [89].
The mechanisms of excision repair of damaged bases and single-strand DNA breaks (BER) have been the subject of intensive studies in recent decades; impressive progress has been made in establishing the participants of the repair process, main pathways, and auxiliary mechanisms that become active when the main BER pathways are inefficient. In addition to the enzymes responsible for catalytic stages of the repair, the regulatory proteins were identified that actively participate in organization of the dynamic system for the repair of damaged bases and single-strand DNA breaks such as XRCC1, PARP1, PARP2, and others.
Multiprotein complexes of various compositions are formed without the involvement of DNA, but they are modulated by the damaged DNA in different stages of its repair. Interactions of individual BER enzymes with DNA substrates and products were investigated in detail by X-ray diffraction. At the same time, this method is of little use for investigation of dynamic supramolecular structures operating in DNA repair. The next step is required in investigation of the structure–function relationships of these protein machines that would provide clarification of the function of BER as a complex of interacting proteins associated with chromatin. It might be helpful to apply novel methods of structural analysis, such as electron microscopy, and to use more complex models imitating DNA repair in chromatin structure. Elucidation of molecular mechanisms underlying BER is an important aspect for understanding the origins of diseases because disruption of BER leads to pathological states.
Acknowledgments
This work was financially supported by the Russian Science Foundation (grant No. 14-24-00038).
REFERENCES
1.Mavragani, I. V., Nikitaki, Z., Souli, M. P., Aziz,
A., Nowsheen, S., Aziz, K., Rogakou, E., and Georgakilas, A. G. (2017)
Complex DNA damage: a route to radiation-induced genomic instability
and carcinogenesis, Cancers (Basel), 9, 91.
2.Talhaoui, I., Matkarimov, B. T., Tchenio, T.,
Zharkov, D. O., and Saparbaev, M. K. (2017) Aberrant base excision
repair pathway of oxidatively damaged DNA: implications for
degenerative diseases, Free Radic. Biol. Med., 107,
266-277.
3.Poletto, M., Legrand, A. J., and Dianov, G. L.
(2017) DNA base excision repair: the Achilles’ heel of tumor
cells and their microenvironment? Curr. Pharm. Des., doi:
10.2174/1381612823666170710123602.
4.Whitaker, A. M., Schaich, M. A., Smith, M. R.,
Flynn, T. S., and Freudenthal, B. D. (2017) Base excision repair of
oxidative DNA damage: from mechanism to disease, Front. Biosci.
(Landmark Ed.), 22, 1493-1522.
5.Abbotts, R., and Wilson III, D. M. (2017)
Coordination of DNA single strand break repair, Free Radic. Biol.
Med., 107, 228-244.
6.Berti, P. J., and McCann, J. A. B. (2006) Toward a
detailed understanding of base excision repair enzymes: transition
state and mechanistic analyses of N-glycoside hydrolysis and
N-glycoside transfer, Chem. Rev., 106, 506-555.
7.Liu, Y., Beard, W. A., Shock, D. D., Prasad, R.,
Hou, E. W., and Wilson, S. H. (2005) DNA polymerase beta and flap
endonuclease 1 enzymatic specificities sustain DNA synthesis for long
patch base excision repair, J. Biol. Chem., 280,
3665-3674.
8.Lebedeva, N. A., Rechkunova, N. I., Dezhurov, S.
V., Khodyreva, S. N., Favre, A., Blanco, L., and Lavrik, O. I. (2005)
Comparison of functional properties of mammalian DNA polymerase lambda
and DNA polymerase beta in reactions of DNA synthesis related to DNA
repair, Biochim. Biophys. Acta, 1751, 150-158.
9.Caldecott, K. W. (2014) DNA single-strand break
repair, Exp. Cell. Res., 329, 2-8.
10.Amé, J. C., Rolli, V., Schreiber, V.,
Niedergang, C., Apiou, F., Decker, P., Muller, S., Höger, T.,
Ménissier-de Murcia, J., and de Murcia, G. (1999) PARP-2, a
novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase, J.
Biol. Chem., 274, 17860-17868.
11.De Murcia, J. M., Niedergang, C., Trucco, C.,
Ricoul, M., Dutrillaux, B., Mark, M., Oliver, F. J., Masson, M.,
Dierich, A., LeMeur, M., Walztinger, C., Chambon, P., and de Murcia, G.
(1997) Requirement of poly(ADP-ribose) polymerase in recovery from DNA
damage in mice and in cells, Proc. Natl. Acad. Sci. USA,
94, 7303-7307.
12.Ménissier-de Murcia, J., Ricoul,
M., Tartier, L., Niedergang, C., Huber, A., Dantzer, F.,
Schreiber, V., Amé, J. C., Dierich, A., LeMeur, M., Sabatier,
L., Chambon, P., and de Murcia, G. (2003) Functional interaction
between PARP1 and PARP2 in chromosome stability and embryonic
development in mouse, EMBO J., 22, 2255-2263.
13.Pascal, J. M., and Ellenberger, T. (2015) The
rise and fall of poly(ADP-ribose): an enzymatic perspective, DNA
Repair (Amst.), 32, 10-16.
14.Ray Chaudhuri, A., and Nussenzweig, A. (2017) The
multifaceted roles of PARP1 in DNA repair and chromatin remodeling,
Nat. Rev. Mol. Cell Biol., 18, 610-621.
15.Prasad, R., Beard, W. A., Batra, V. K., Liu, Y.,
Shock, D. D., and Wilson, S. H. (2011) A review of recent experiments
on step-to-step “hand-off” of the DNA intermediates in
mammalian base excision repair pathways, Mol. Biol.
({{anchor|GoBack}} Moscow), 45, 586-600.
16.Kim, Y.-J., and Wilson III, D. M. (2012) Overview
of base excision repair biochemistry, Curr. Mol. Pharmacol.,
5, 3-13.
17.Dutta, A., Yang, C., Sengupta, S., Mitra, S., and
Hegde, M. L. (2015) New paradigms in the repair of oxidative damage in
human genome: mechanisms ensuring repair of mutagenic base lesions
during replication and involvement of accessory proteins, Cell. Mol.
Life Sci., 72, 1679-1698.
18.Esadze, A., Rodriguez, G., Cravens, S. L., and
Stivers, J. T. (2017) AP-endonuclease 1 accelerates turnover of human
8-oxoguanine DNA glycosylase by preventing retrograde binding to the
abasic-site product, Biochemistry, 56, 1974-1986.
19.Kubota, Y., Nash, R. A., Klungland, A.,
Schär, P., Barnes, D. E., and Lindahl, T. (1996) Reconstitution of
DNA base excision-repair with purified human proteins: interaction
between DNA polymerase β and the XRCC1 protein, EMBO J.,
15, 6662-6670.
20.Marintchev, A., Robertson, A., Dimitriadis, E.
K., Prasad, R., Wilson, S. H., and Mullen, G. P. (2000) Domain specific
interaction in the XRCC1-DNA polymerase β complex, Nucleic
Acids Res., 28, 2049-2059.
21.Marintchev, A., Gryk, M. R., and Mullen, G. P.
(2003) Site-directed mutagenesis analysis of the structural interaction
of the single-strand-break repair protein, X-ray cross-complementing
group 1, with DNA polymerase β, Nucleic Acids Res.,
31, 580-588.
22.London, R. E. (2015) The structural basis of
XRCC1-mediated DNA repair, DNA Repair (Amst.), 30,
90-103.
23.Fan, J., Otterlei, M., Wong, H. K., Tomkinson, A.
E., and Wilson III, D. M. (2004) XRCC1 co-localizes and physically
interacts with PCNA, Nucleic Acids Res., 32,
2193-2201.
24.Akbari, M., Solvang-Garten, K., Hanssen-Bauer,
A., Lieske, N. V., Pettersen, H. S., Pettersen, G. K., Wilson III, D.
M., Krokan, H. E., and Otterlei, M. (2010) Direct interaction between
XRCC1 and UNG2 facilitates rapid repair of uracil in DNA by XRCC1
complexes, DNA Repair (Amst.), 9, 785-795.
25.Campalans, A., Marsin, S., Nakabeppu, Y.,
O’Connor, T. R., Boiteux, S., and Radicella, J. P. (2005) XRCC1
interactions with multiple DNA glycosylases: a model for its
recruitment to base excision repair, DNA Repair (Amst.),
4, 826-835.
26.Wiederhold, L., Leppard, J. B., Kedar, P.,
Karimi-Busheri, F., Rasouli-Nia, A., Weinfeld, M., Tomkinson, A. E.,
Izumi, T., Prasad, R., Wilson, S. H., Mitra, S., and Hazra, T. K.
(2004) AP endonuclease-independent DNA base excision repair in human
cells, Mol. Cell, 15, 209-220.
27.Das, A., Wiederhold, L., Leppard, J. B., Kedar,
P., Prasad, R., Wang, H., Boldogh, I., Karimi-Busheri, F., Weinfeld,
M., Tomkinson, A. E., Wilson, S. H., Mitra, S., and Hazra, T. K. (2006)
NEIL2-initiated, APE-independent repair of oxidized bases in DNA:
evidence for a repair complex in human cells, DNA Repair
(Amst.), 5, 1439-1448.
28.Marsin, S., Vidal, A. E., Sossou, M.,
Ménissier-de Murcia, J., Le Page, F., Boiteux, S., de Murcia,
G., and Radicella, J. P. (2003) Role of XRCC1 in the coordination and
stimulation of oxidative DNA damage repair initiated by the DNA
glycosylase hOGG1, J. Biol. Chem., 278, 44068-44074.
29.Hanssen-Bauer, A., Solvang-Garten, K., Gilljam,
K. M., Torseth, K., Wilson III, D. M., Akbari, M., and Otterlei, M.
(2012) The region of XRCC1 which harbors the three most common
nonsynonymous polymorphic variants, is essential for the scaffolding
function of XRCC1, DNA Repair (Amst.), 11, 357-366.
30.Masson, M., Niedergang, C., Schreiber, V.,
Muller, S., Ménissier-de Murcia, J., and de Murcia, G. (1998)
XRCC1 is specifically associated with poly(ADP-ribose) polymerase and
negatively regulates its activity following DNA damage, Mol. Cell.
Biol., 18, 3563-3571.
31.Schreiber, V., Amé, J. C., Dolle, P.,
Schultz, I., Rinaldi, B., Fraulob, V., Ménissier-de Murcia, J.,
and de Murcia, G. (2002) Poly(ADP-ribose) polymerase-2 (PARP-2) is
required for efficient base excision DNA repair in association with
PARP-1 and XRCC1, J. Biol. Chem., 277, 23028-23036.
32.Loizou, J. I., El-Khamisy, S. F., Zlatanou, A.,
Moore, D. J., Chan, D. W., Qin, J., Sarno, S., Meggio, F., Pinna, L.
A., and Caldecott, K. W. (2004) The protein kinase CK2 facilitates
repair of chromosomal DNA single-strand breaks, Cell,
117, 17-28.
33.Lu, M., Mani, R. S., Karimi-Busheri, F., Fanta,
M., Wang, H., Litchfeld, D. W., and Weinfeld, M. (2010) Independent
mechanisms of stimulation of polynucleotide kinase/phosphatase by
phosphorylated and non-phosphorylated XRCC1, Nucleic Acids Res.,
38, 510-521.
34.Luo, H., Chan, D. W., Yang, T., Rodriguez, M.,
Chen, B. P., Leng, M., Mu, J. J., Chen, D., Songyang, Z., Wang, Y., and
Qin, J. (2004) A new XRCC1-containing complex and its role in cellular
survival of methyl methanesulfonate treatment, Mol. Cell. Biol.,
24, 8356-8365.
35.Beernink, P. T., Hwang, M., Ramirez, M., Murphy,
M. B., Doyle, S. A., and Thelen, M. P. (2005) Specificity of protein
interactions mediated by BRCT domains of the XRCC1 DNA repair protein,
J. Biol. Chem., 280, 30206-30213.
36.Nash, R. A., Caldecott, K. W., Barnes, D. E., and
Lindahl, T. (1997) XRCC1 protein interacts with one of two distinct
forms of DNA ligase III, Biochemistry, 36, 5207-5211.
37.Cuneo, M. J., Gabel, S. A., Krahn, J. M., Ricker,
M. A., and London, R. E. (2011) The structural basis for partitioning
of the XRCC1/DNA ligase III-α BRCT-mediated dimer complexes,
Nucleic Acids Res., 39, 7816-7827.
38.Plo, I., Liao, Z. Y., Barceló, J. M.,
Kohlhagen, G., Caldecott, K. W., Weinfeld, M., and Pommier, Y. (2003)
Association of XRCC1 and tyrosyl DNA phosphodiesterase (Tdp1) for the
repair of topoisomerase I-mediated DNA lesions, DNA Repair
(Amst.), 2, 1087-1100.
39.Dantzer, F., de la Rubia, G., Ménissier-de
Murcia, J., Hostomsky, Z., de Murcia, G., and Schreiber, V. (2000) Base
excision repair is impaired in mammalian cells lacking poly(ADP-ribose)
polymerase-1, Biochemistry, 39, 7559-7569.
40.Ali, A. A. E., Timinszky, G., Arribas-Bosacoma,
R., Kozlowski, M., Hassa, P. O., Hassler, M., Ladurner, A. G., Pearl,
L. H., and Oliver, A. W. (2012) The zinc-finger domains of PARP1
cooperate to recognize DNA strand breaks, Nat. Struct. Mol.
Biol., 19, 685-692.
41.Leppard, J. B., Dong, Z., Mackey, Z. B., and
Tomkinson, A. E. (2003) Physical and functional interaction between DNA
ligase IIIα and poly(ADP-ribose) polymerase 1 in DNA
single-strand break repair, Mol. Cell. Biol., 23,
5919-5927.
42.Das, B. B., Huang, S. Y., Murai, J., Rehman, I.,
Amé, J. C., Sengupta, S., Das, S. K., Majumdar, P., Zhang, H.,
Biard, D., Majumder, H. K., Schreiber, V., and Pommier, Y. (2014)
PARP1-TDP1 coupling for the repair of topoisomerase I-induced DNA
damage, Nucleic Acids Res., 42, 4435-4449.
43.Dimitriadis, E. K., Prasad, R., Vaske, M. K.,
Chen, L., Tomkinson, A. E., Lewis, M. S., and Wilson, S. H. (1998)
Thermodynamics of human DNA ligase I trimerization and association with
DNA polymerase β, J. Biol. Chem., 273,
20540-20550.
44.Bennett, R. A., Wilson III, D. M., Wong, D., and
Demple, B. (1997) Interaction of human apurinic endonuclease and DNA
polymerase β in the base excision repair pathway, Proc. Natl.
Acad. Sci. USA, 94, 7166-7169.
45.Whitehouse, C. J., Taylor, R. M., Thistlethwaite,
A., Zhang, H., Karimi-Busheri, F., Lasko, D. D., Weinfeld, M., and
Caldecott, K. W. (2001) XRCC1 stimulates human polynucleotide kinase
activity at damaged DNA termini and accelerates DNA single-strand break
repair, Cell, 104, 107-117.
46.El-Khamisy, S. F., Saifi, G. M., Weinfeld, M.,
Johansson, F., Helleday, T., Lupski, J. R., and Caldecott, K. W. (2005)
Defective DNA single-strand break repair in spinocerebellar ataxia with
axonalneuropathy-1, Nature, 434, 108-113.
47.Chiang, S. C., Carroll, J., and El-Khamisy, S. F.
(2010) TDP1 serine 81 promotes interaction with DNA ligase IIIα
and facilitates cell survival following DNA damage, Cell Cycle,
9, 588-595.
48.Luncsford, P. J., Manvilla, B. A., Patterson, D.
N., Malik, S. S., Jin, J., Hwang, B. J., Gunther, R., Kalvakolanu, S.,
Lipinski, L. J., Yuan, W., Lu, W., Drohat, A. C., Lu, A. L., and Toth,
E. A. (2013) Coordination of MYH DNA glycosylase and APE1 endonuclease
activities via physical interactions, DNA Repair (Amst.),
12, 1043-1052.
49.Hegde, P. M., Dutta, A., Sengupta, S., Mitra, J.,
Adhikari, S., Tomkinson, A. E., Li, G. M., Boldogh, I., Hazra, T. K.,
Mitra, S., and Hegde, M. L. (2015) The C-terminal domain (CTD) of human
DNA glycosylase NEIL1 is required for forming BERosome repair complex
with DNA replication proteins at the replicating genome: dominant
negative function of the CTD, J. Biol. Chem., 290,
20919-20933.
50.El-Khamisy, S. F., Masutani, M., Suzuki, H., and
Caldecott, K. W. (2003) A requirement for PARP-1 for the assembly or
stability of XRCC1 nuclear foci at sites of oxidative DNA damage,
Nucleic Acids Res., 31, 5526-5533.
51.Langelier, M. F., Planck, J. L., Roy, S., and
Pascal, J. M. (2012) Structural basis for DNA damage-dependent
poly(ADP-ribosyl)ation by human PARP-1, Science, 336,
728-732.
52.Cotner-Gohara, E., Kim, I. K., Tomkinson, A. E.,
and Ellenberger, T. (2008) Two DNA-binding and nick recognition modules
in human DNA ligase III, J. Biol. Chem., 283,
10764-10772.
53.Gerloff, D. L., Woods, N. T., Farago, A. A., and
Monteiro, A. N. (2012) BRCT domains: a little more than kin, and less
than kind, FEBS Lett., 586, 2711-2716.
54.Eustermann, S., Wu, W. F., Langelier, M. F.,
Yang, J. C., Easton, L. E., Riccio, A. A., Pascal, J. M., and Neuhaus,
D. (2015) Structural basis of detection and signaling of DNA
single-strand breaks by human PARP-1, Mol. Cell, 60,
742-754.
55.Gagné, J. P., Ethier, C., Defoy, D.,
Bourassa, S., Langelier, M. F., Riccio, A. A., Pascal, J. M., Moon, K.
M., Foster, L. J., Ning, Z., Figeys, D., Droit, A., and Poirier, G. G.
(2015) Quantitative site-specific ADP-ribosylation profiling of
DNA-dependent PARPs, DNA Repair (Amst.), 30, 68-79.
56.Gagné, J. P., Isabelle, M., Lo, K. S.,
Bourassa, S., Hendzel, M. J., Dawson, V. L., Dawson, T. M., and
Poirier, G. G. (2008) Proteome-wide identification of
poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated
protein complexes, Nucleic Acids Res., 36, 6959-6976.
57.Teloni, F., and Altmeyer, M. (2016) Readers of
poly(ADP-ribose): designed to be fit for purpose, Nucleic Acids
Res., 44, 993-1006.
58.Bock, F. J., and Chang, P. (2016) New directions
in poly(ADP-ribose) polymerase biology, FEBS J., 28,
4017-4031.
59.Hanzlikova, H., Gittens, W., Krejcikova, K.,
Zeng, Z., and Caldecott, K. W. (2017) Overlapping roles for PARP1 and
PARP2 in the recruitment of endogenous XRCC1 and PNKP into oxidized
chromatin, Nucleic Acids Res., 45, 2546-2557.
60.Abdou, I., Poirier, G. G., Hendzel, M. J., and
Weinfeld, M. (2015) DNA ligase III acts as a DNA strand break sensor in
the cellular orchestration of DNA strand break repair, Nucleic Acids
Res., 43, 875-892.
61.Moor, N. A., Vasil’eva, I. A., Anarbaev, R.
O., Antson, A. A., and Lavrik, O. I. (2015) Quantitative
characterization of protein–protein complexes involved in base
excision DNA repair, Nucleic Acids Res., 43,
6009-6022.
62.Mani, R. S., Fanta, M., Karimi-Busheri, F.,
Silver, E., Virgen, C. A., Caldecott, K. W., Cass, C. E., and Weinfeld,
M. (2007) XRCC1 stimulates polynucleotide kinase by enhancing its
damage discrimination and displacement from DNA repair intermediates,
J. Biol. Chem., 282, 28004-28013.
63.Liu, Y., Prasad, R., Beard, W. A., Kedar, P. S.,
Hou, E. W., Shock, D. D., and Wilson, S. H. (2007) Coordination of
steps in single-nucleotide base excision repair mediated by
apurinic/apyrimidinic endonuclease 1 and DNA polymerase β, J.
Biol. Chem., 282, 13532-13541.
64.Fang, Q., Inanc, B., Schamus, S., Wang, X. H.,
Wei, L., Brown, A. R., Svilar, D., Sugrue, K. F., Goellner, E. M.,
Zeng, X., Yates, N. A., Lan, L., Vens, C., and Sobol, R. W. (2014)
HSP90 regulates DNA repair via the interaction between XRCC1 and DNA
polymerase β, Nat. Commun., 5, 5513.
65.Lan, L., Nakajima, S., Oohata, Y., Takao, M.,
Okano, S., Masutani, M., Wilson, S. H., and Yasui, A. (2004) In
situ analysis of repair processes for oxidative DNA damage in
mammalian cells, Proc. Natl. Acad. Sci. USA, 101,
13738-13743.
66.Lavrik, O. I., Prasad, R., Sobol, R. W., Horton,
J. K., Ackerman, E. J., and Wilson, S. H. (2001) Photoaffinity labeling
of mouse fibroblast enzymes by a base excision repair intermediate.
Evidence for the role of poly(ADP-ribose) polymerase-1 in DNA repair,
J. Biol. Chem., 276, 25541-25548.
67.Khodyreva, S. N., Prasad, R., Ilina, E. S.,
Sukhanova, M. V., Kutuzov, M. M., Liu, Y., Hou, E. W., Wilson, S. H.,
and Lavrik, O. I. (2010) Apurinic/apyrimidinic (AP) site recognition by
the 5′-dRP/AP lyase in poly(ADP-ribose) polymerase-1 (PARP-1),
Proc. Natl. Acad. Sci. USA, 107, 22090-22095.
68.Sukhanova, M. V., Abrakhi, S., Joshi, V., Pastre,
D., Kutuzov, M. M., Anarbaev, R. O., Curmi, P. A., Hamon, L., and
Lavrik, O. I. (2016) Single molecule detection of PARP1 and PARP2
interaction with DNA strand breaks and their poly(ADP-ribosyl)ation
using high-resolution AFM imaging, Nucleic Acids Res., 44, e60.
69.Alemasova, E. E., and Lavrik, O. I. (2017) At the
interface of three nucleic acids: the role of RNA-binding proteins and
poly(ADP-ribose) in DNA repair, Acta Naturae, 9, 4-16.
70.Altmeyer, M., Neelsen, K. J., Teloni, F.,
Pozdnyakova, I., Pellegrino, S., Grøfte, M., Rask, M. B.,
Streicher, W., Jungmichel, S., Nielsen, M. L., and Lukas, J. (2015)
Liquid demixing of intrinsically disordered proteins is seeded by
poly(ADP-ribose), Nat. Commun., 6, 8088.
71.Sorokin, A. V., Selyutina, A. A., Skabkin, M. A.,
Guryanov, S. G., Nazimov, I. V., Richard, C., Th’ng, J., Yau, J.,
Sorensen, P. H., Ovchinnikov, L. P., and Evdokimova, V. (2005)
Proteasome-mediated cleavage of the Y-box-binding protein 1 is linked
to DNA-damage stress response, EMBO J., 24,
3602-3612.
72.Alemasova, E. E., Pestryakov, P. E., Sukhanova,
M. V., Kretov, D. A., Moor, N. A., Curmi, P. A., Ovchinnikov, L. P.,
and Lavrik, O. I. (2015) Poly(ADP-ribosyl)ation as a new
posttranslational modification of YB-1, Biochimie, 119,
36-44.
73.Alemasova, E. E., Moor, N. A., Naumenko, K. N.,
Kutuzov, M. M., Sukhanova, M. V., Pestryakov, P. E., and Lavrik, O. I.
(2016) Y-box-binding protein 1 as a non-canonical factor of base
excision repair, Biochim. Biophys. Acta, 1864,
1631-1640.
74.Sengupta, S., Mantha, A. K., Mitra, S., and
Bhakat, K. K. (2011) Human AP endonuclease (APE1/ref-1) and its
acetylation regulate YB-1-p300 recruitment and RNA polymerase II
loading in the drug-induced activation of multidrug resistance gene
MDR1, Oncogene, 30, 4482-4493.
75.Poletto, M., Lirussi, L., Wilson III, D. M., and
Tell, G. (2014) Nucleophosmin modulates stability, activity, and
nucleolar accumulation of base excision repair proteins, Mol. Biol.
Cell, 25, 1641-1652.
76.Tell, G., Fantini, D., and Quadrifoglio, F.
(2010) Understanding different functions of mammalian AP endonuclease
(APE1) as a promising tool for cancer treatment, Cell. Mol. Life
Sci., 67, 3589-3608.
77.Dyrkheeva, N. S., Lebedeva, N. A., and Lavrik, O.
I. (2016) AP endonuclease 1 as a key enzyme in repair of
apurinic/apyrimidinic sites, Biochemistry (Moscow), 81,
951-967.
78.Paquet, N., Adams, M. N., Leong, V., Ashton, N.
W., Touma, C., Gamsjaeger, R., Cubeddu, L., Beard, S., Burgess, J. T.,
Bolderson, E., O’Byrne, K. J., and Richard, D. J. (2015) hSSB1
(NABP2/OBFC2B) is required for the repair of 8-oxo-guanine by the
hOGG1-mediated base excision repair pathway, Nucleic Acids Res.,
43, 8817-8829.
79.Kaur, S., Coulombe, Y., Ramdzan, Z. M., Leduy,
L., Masson, J. Y., and Nepveu, A. (2016) Special AT-rich
sequence-binding protein 1 (SATB1) functions as an accessory factor in
base excision repair, J. Biol. Chem., 291,
22769-22780.
80.Maher, R. L., Marsden, C. G., Averill, A. M.,
Wallace, S. S., Sweasy, J. B., and Pederson, D. S. (2017) Human cells
contain a factor that facilitates the DNA glycosylase-mediated excision
of oxidized bases from occluded sites in nucleosomes, DNA Repair
(Amst.), 57, 91-97.
81.Limpose, K. L., Corbett, A. H., and Doetsch, P.
W. (2017) BERing the burden of damage: pathway crosstalk and
posttranslational modification of base excision repair proteins
regulate DNA damage management, DNA Repair (Amst.), 56,
51-64.
82.Prasad, R., Liu, Y., Deterding, L. J.,
Poltoratsky, V. P., Kedar, P. S., Horton, J. K., Kanno, S., Asagoshi,
K., Hou, E. W., Khodyreva, S. N., Lavrik, O. I., Tomer, K. B., Yasui,
A., and Wilson, S. H. (2007) HMGB1 is a cofactor in mammalian base
excision repair, Mol. Cell, 27, 829-841.
83.Liu, Y., Prasad, R., and Wilson, S. H.
(2010) HMGB1: roles in base excision repair and related
function, Biochim. Biophys. Acta, 1799, 119-130.
84.Balliano, A., Hao, F., Njeri, C., Balakrishnan,
L., and Hayes, J. J. (2017) HMGB1 stimulates activity of polymerase
β on nucleosome substrates, Biochemistry, 56,
647-656.
85.Menoni, H., Di Mascio, P., Cadet, J., Dimitrov,
S., and Angelov, D. (2017) Chromatin associated mechanisms in base
excision repair – nucleosome remodeling and DNA transcription,
two key players, Free Radic. Biol. Med., 107,
159-169.
86.Almeida, K. H., and Sobol, R. W. (2007) A unified
view of base excision repair: lesion-dependent protein complexes
regulated by post-translational modification, DNA Repair
(Amst.), 6, 695-711.
87.Roychoudhury, S., Nath, S., Song, H., Hegde, M.
L., Bellot, L. J., Mantha, A. K., Sengupta, S., Ray, S., Natarajan, A.,
and Bhakat, K. K. (2017) Human apurinic/apyrimidinic endonuclease
(APE1) is acetylated at DNA damage sites in chromatin, and acetylation
modulates its DNA repair activity, Mol. Cell. Biol., 37,
e00401-16.
88.Weiser, B. P., Stivers, J. T., and Cole, P. A.
(2017) Investigation of N-terminal phospho-regulation of uracil DNA
glycosylase using protein semisynthesis, Biophys. J.,
113, 393-401.
89.Horton, J. K., Seddon, H. J., Zhao, M. L.,
Gassman, N. R., Janoshazi, A. K., Stefanick, D. F., and Wilson, S. H.
(2017) Role of the oxidized form of XRCC1 in protection against extreme
oxidative stress, Free Radic. Biol. Med., 107,
292-300.