REVIEW: Link Between Double-Strand DNA Break Hotspots and Transcription Regulation: Forum Domains – 50-250 kb Chromosome Regions Containing Coordinately Expressed Genes
N. A. Tchurikov*, Y. V. Kravatsky, and O. V. Kretova
Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, 119991 Moscow, Russia; E-mail: tchurikov@eimb.ru* To whom correspondence should be addressed.
Received November 9, 2017; Revision received December 11, 2017
The data on forum domains formed by DNA double-strand break (DSB) hotspots are reviewed including forum domain identification by pulsed-field gel electrophoresis, whole genome mapping of these domains using deep sequencing strategies, analysis of gene expression in forum domains, and binding of nuclear proteins to their boundaries. Earlier unpublished data by the authors are presented. The “piano playing” hypothesis is suggested based on coordinated active transcription in some of the forum domains and coordinated silencing in the majority of them. The data on the DSB hotspots in human ribosomal DNA gene clusters and their possible association with chromosomal translocations are presented. These clusters are the most actively transcribed DNA regions in cells, as well as the most fragile sites in human chromosomes. The need to revise the available data on DNase I-hypersensitive sites in various genomes, including endogenous DNA breaks of different nature, is discussed.
KEY WORDS: endogenous DNA double-strand breaks (DSB) hotspots, forum domains, epigenetics, coordinated expression, PARP1, HNRNPA2B1, chromosomal silencing, 4C, inter-chromosomal contacts, rDNA genes, translocations, DNase I-hypersensitive sites, H3K4me3, H3K27ac, CTCFDOI: 10.1134/S0006297918040144
Abbreviations: 4C (method), circular chromatin conformation capture; DSB, DNA double strand break; FT, forum termini; IGS, intergenic spacer; LAD, lamina-associated domain; LOCKs, large organized chromatin K9-modifications; Pc domains, Polycomb domains; RAFT, rapid amplification of forum domains termini; rDNA, ribosomal DNA; TAD, topologically associating domain.
The regulation of gene expression has been studied for several decades
including mechanisms of local regulation associated with promoters,
enhancers, insulators, and silencers. These elements are sites for
binding of complex protein associates and introduction of various
epigenetic marks. Such type of mitotically inherited regulation is very
well understood. There is also distant regulation, that involves large
chromosomal domains from tens kb to 1 Mb and above in length, including
Polycomb domains (Pc domains), forum domains, topologically associating
domains (TADs), lamina-associated domains (LADs), large organized
chromatin K9-modifications (LOCKs), and some others [1-5]. This still poorly understood
type of regulation is related to the formation of high-order
chromosomal structures. However, we have every reason to believe that
it is important for cell differentiation and development. Pc domains
are associated with the repressed chromatin H3K27me3 mark introduced by
the Polycomb repressive complex 2 (PRC2) containing histone
methyltransferase. TADs reflect the existence of extended compact
chromosomal regions up to several Mbs in length. LADs and LOCKs, as
follows from their names, are large segments of chromosomes (up to 30
Mb) linked to the nuclear lamina or large chromatin regions (up to
several Mb) containing the H3K9me2-marked repressed chromatin.
This review presents the results of studies of forum domains associated with double-strand DNA break (DSB) hotspots, DSBs in ribosomal DNA (rDNA) genes, and mechanisms ensuring coordinated gene expression in forum domains.
ISOLATION AND MAPPING OF FORUM DOMAINS
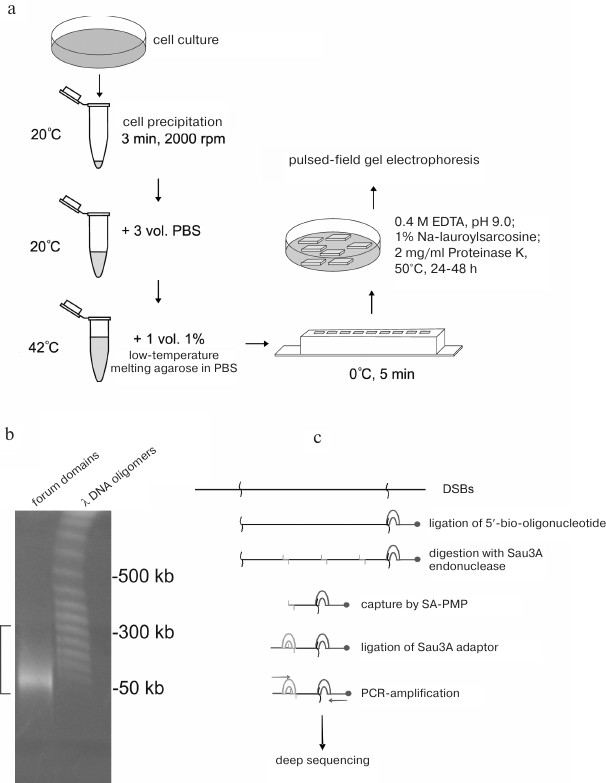
Forum domains were discovered during fractionation of eukaryotic DNA by pulsed-field gel electrophoresis [2, 6]. To avoid hydrodynamic DNA shearing, DNA samples were isolated in a solid phase. For this purpose, the cells were immobilized in 0.5% low-temperature-melting agarose, and DNA was isolated in agarose plugs by incubation in a solution containing 0.5 M EDTA, 1% SDS, and proteinase K (1 mg/ml) at 50°C for 48 h (Fig. 1a), which allowed detection of chromosomal DNA fragments of 50-300 kb (Fig. 1b). It was suggested that these DNA fragments were the results of non-random chromosome fragmentation and corresponded to some regular high-order chromosome structures delimited by DSBs existing in vivo [2, 6]. When fractionation was conducted in a regular 1% mini gel, the domains were observed as compact bands migrating in the region of poor separation (above 20 kb). Considering that these domains contained transcribed regions (i.e., contained genes), these domains were named “forum domains” by analogy with the ancient Rome square as a structural element of architecture.
Fig. 1. Analysis of forum domains. a) Schematic representation of the procedure of DNA isolation in agarose plugs [16]; b) fractionation of DNA isolated in agarose plug. Square bracket indicates forum domains in the agarose gel. Bacteriophage λ DNA fragments were as DNA size markers [11]; c) schematic representation of RAFT (procedure for preparation of genome-wide libraries containing DSB regions) [7].
Genome-wide mapping of forum domain was performed by deep sequencing of forum domain short terminal regions. For this purpose, the 50-300 kb DNA domains were cut from the gel, eluted in a dialysis bag, and ligated with an excess of biotinylated oligonucleotide (Fig. 1c). Next, DNA was hydrolyzed with the high-frequency Sau3A restriction endonuclease, and the terminal regions were captured by streptavidin-modified paramagnetic particles (SA-PMPs). Following ligation of the Sau3A adapter, rapid amplification of forum domains termini (RAFT) was performed [7]. The amplified genome-wide set of forum domain terminal regions (forum termini, FT) was used for deep sequencing. Taking into account a high frequency of DSBs across the genome and the fact that some of them could be introduced into DNA during manipulations before DNA ligation to the biotinylated oligonucleotide, the obtained sequences were analyzed to reveal only DSB hotspots, i.e., sites with the highest occurrence of DNA breaks producing 50-300 kb DNA fragments. Using the RAFT method, DSB hotspots in human genome were mapped [8]. Regions located between the hotspots correspond to the forum domains.
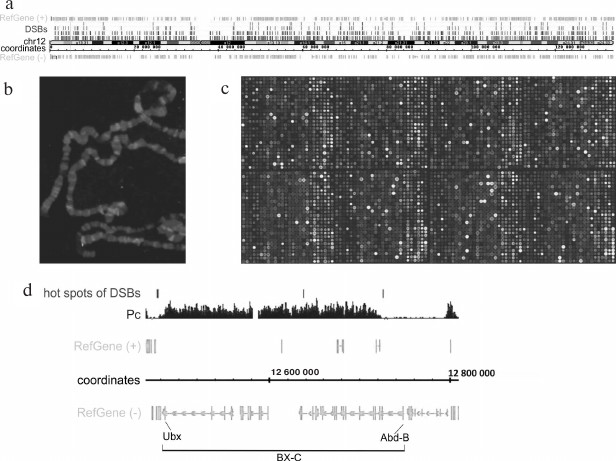
Figure 2 shows human Chromosome 12 and DSB hotspots with different thresholds of DSB frequency. It can be seen that the breaks are scattered over the entire chromosome length. Figure 2b demonstrates that fluorescently labeled RAFT preparations (i.e., FT) hybridize to certain regions of polytene chromosomes. Detailed analysis of the hybridization sites revealed that most of them are located in the regions of intercalary heterochromatin (p < 2.2·10–5) [7] characterized by late replication, ectopic pairing (inter-chromosomal contacts), and frequent chromosomal breaks [9, 10]. Hybridization of RAFT preparations to a microarray containing entire Drosophila genome showed that large forum domains encompass the Pc domains that include extended regions of repressed chromatin (Fig. 2, c and d) [7]. The Bithorax complex that contains homeotic genes and is repressed in embryos was found to be bordered by the DSB hotspots, i.e., this ~270-kb region containing inactive chromatin H3K27me3 marks most often breaks not within itself but at the adjacent DNA sequences.
Fig. 2. Mapping of forum domains. a) Distribution of DSB hotspots along Chr12 (DSB threshold frequencies are shown); b) in situ hybridization of Drosophila genome-wide library containing DSB regions (RAFT preparation); c) mapping of forum domains on a fragment of the Drosophila whole-genome microarray [7]. The brightest spots correspond to the sites of frequent DNA breaks; d) 3R region of Drosophila chromosome containing the Bithorax complex (BX-C). It can be seen that the 270-kb complex repressed by the Polycomb group proteins (Pc domain) is located inside the large forum domains [7].
PROPERTIES OF REGIONS CONTAINING DSB HOTSPOTS
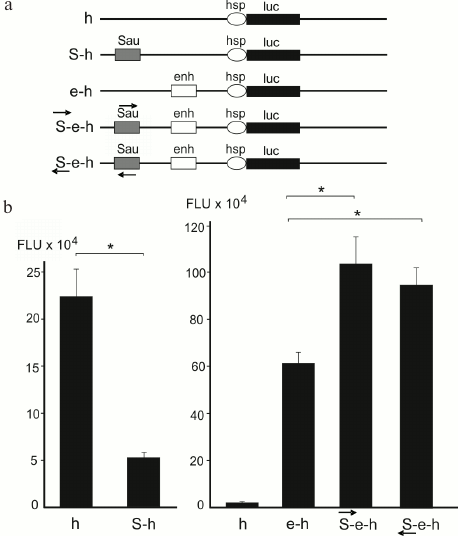
Silencers and enhancers. Mapping of DSB hotspots and forum domains bordered by them provides an opportunity to investigate genetic and epigenetic properties of these genome regions. The functions of DSB-containing short DNA fragments were elucidated using reporter gene constructs. The results of studies of one of the DSB hotspots in Drosophila chromosome X are presented in Fig. 3. It was found that this DNA fragment acted as a silencer and suppressed expression of the reporter luc gene [11]. When the same element was inserted into the construct containing an enhancer, the results were quite unexpected: in the presence of the active enhancer, this element acted as an enhancer further activating the reporter gene expression in both orientations. Such element displaying bivalent properties was termed oxymoron [12]. An oxymoron located in the forum domain terminal region could be either transcription repressor or activator. Hence, FT could play a role of regulatory elements.
Fig. 3. Investigation of the FT properties in Drosophila genome with the help of reporter gene constructs. a) Genetic constructs containing firefly luciferase reporter gene (luc), HSP70 promoter (h), copia element enhancer (enh or e), and a ~300 kb forum domain terminal region from the X-chromosome (Sau, or S) [11, 12]; b) gene activity in cells transfected with the indicated genetic constructs; * p < 0.05; FLU, firefly luciferase luminescence. Arrows indicate polarity of the forum domain terminal region (S).
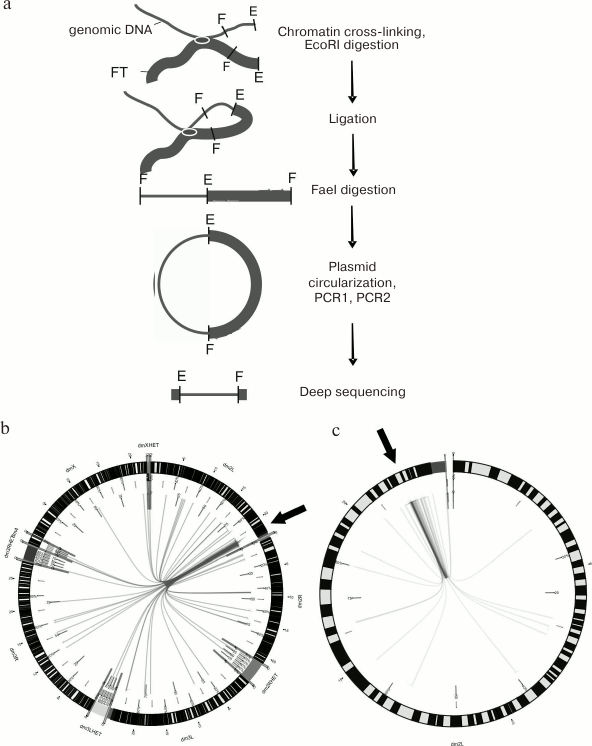
Chromosomal contacts. DSB hotspots in Drosophila polytene chromosomes are most frequently located in the intercalary heterochromatin known to be involved in inter-chromosomal contacts. Therefore, it was interesting to test whether short DNA fragments containing DSB hotspots demonstrate the same property. The 4C method (circular chromatin conformation capture) allows identification of contacts (either inter- or intra-chromosomal) between selected DNA fragment and any other chromosome region (see Fig. 4a for the scheme of 4C experiment). Thus, we found that in Drosophila chromosome 2L, the forum domain terminal region located at the boundary of the histone gene cluster forms multiple contacts both within the chromosome (most frequently, with the adjacent regions) and with different regions of other chromosomes, including heterochromatin regions (Fig. 4, b and c) [13]. Out of 80 most frequent contacts found, ~80% were with the intercalary heterochromatin. These data are in good agreement with the results of in situ hybridization that showed that the genome-wide FT library (RAFT preparation) hybridized most often with the intercalary heterochromatin regions [7]. Similar data were reported for several other FT from human and Drosophila genomes.
Fig. 4. FT form chromosomal contacts. a) Scheme of 4C experiment; E and F, recognition sites for EcoRI and FaeI restrictive endonucleases, respectively; the studied forum domain terminal region containing histone gene cluster (chromosome 2L) is marked with thick line; b) sites of contact of the studied forum domain terminal region (presented with the help of Circos program) [13]. Arrows indicate location of the investigated DNA fragment. Inter-chromosomal contacts are shown in the left panel; contacts within the chromosome 2L are shown in the right panel. It can be seen that multiple contacts exist around the studied forum domain terminal region sequence, which include both the contacts within the domain itself and with the neighboring domains of the chromosome 2L. The contacts with heterochromatin regions not yet introduced into the respective chromosomes are shown separately (for example, dm3LHET).
Hence, FT contain activators and repressors of gene expression and form a complex network of contacts in the genome, which provides them with a potential possibility to regulate cis- and trans-located genes.
DSB HOTSPOTS AND TRANSCRIPTION
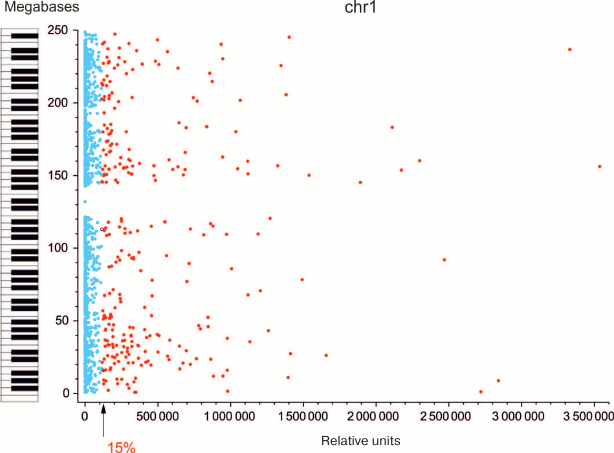
Most forum domains are either repressed or transcribed at a very low level. Since forum domains presumably correspond to certain structure-functional elements of chromosomes, we examined gene expression in these domains. Genome-wide mapping of DSB hotspots and forum domains located between them allowed us to study expression pattern of genes within individual domains. Using RNA-Seq and microarray data, median transcription activities for all forum domains were determined [8]. It was found that the portion of actively transcribed domains in different chromosomes was ~7-29% (actively transcribed domain in this case was defined as a domain with transcription level higher than the average transcription level for all domains in the chromosome). The data for human chromosome 1 are presented in Fig. 5. The piano keyboard in this figure illustrates our “playing piano” hypothesis. When one plays any piece on this instrument, only ≤11% of keys are active while all the other keys are “silent”. Similarly, in chromosomes only a minor portion of forum domains is actively transcribed, while up to 90% are silenced.
Fig. 5. Most of forum domains are either not transcribed or transcribed at a low level [11]; y-axis, coordinates of forum domains in Chr1; x-axis, relative transcription levels (according to RNA-Seq data); arrow, average expression level for the domains in Chr1. Blue dots correspond to non-transcribed or weakly transcribed domains (silencing); red dots indicate actively transcribed domains. Keyboard illustrates the “playing piano” hypothesis – regulation when most domains (keys) are silent (see text).
Coordinated gene expression in forum domains. It could be seen in genome browsers that expression levels of genes located in the same domain are approximately equal. Hence, we conducted the experiment to reveal if it is also true for the whole genome. For this purpose, we used the well-known circular genomic permutation method [14]. All human chromosomes with marked forum domains were combined into a circle; then “positions” of the domains were permutated by 10,000 rounds of rotation by random values in both directions, and next expression levels (E) were estimated in the fragments of the same size but at random locations in the genome. The differences in the expression levels of genes located in the same domain were determined pairwise. It was found that the expression levels of genes in the same forum domain (D) differed (δD = <<|Ea – Eb|>>) much less than in the DNA fragments of the same size located at random genome sites (|z| > 4, p < 0.0001) [15]. The |z| values were calculated using the following equation:
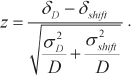
These data convincingly support the existence of strong relations between the transcription regulation and DSB hotspots and suggest that genes located in forum domains are expressed in a coordinated manner. Most forum domains are silenced, while only 7 to 29% of them (in different human chromosomes) are actively transcribed.
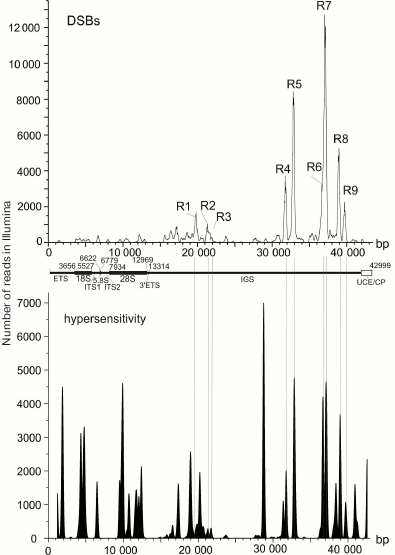
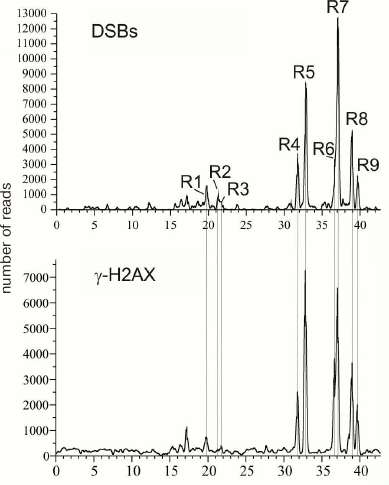
DSB hotspots in rDNA genes. rDNA genes are the most actively transcribed genes – about 80% of cell RNA is transcribed from these gene clusters. The data on strong correlation between the transcriptional activity and DSB hotspots in chromosomes suggest that rDNA genes should also contain DSB hotspots. So far, rDNA genes are not included in the sequenced part of the human genome and, hence, cannot be seen in gene browsers. Deep sequencing reads that correspond to the DNA breaks (RAFT preparations, Fig. 1c) were mapped in the 43-kb DNA sequence containing human rDNA genes. Nine DSB hotspots named Pleiades were identified that were located in the non-transcribed intergenic spacer (IGS) (Fig. 6) [15, 16]. Although the frequencies of breaks in Pleiades (R1-R9 in Fig. 6) differ, all Pleiades contain binding sites for the multifunctional CTCF protein and active chromatin H3K4me3 mark (not shown). The locations of the binding sites for these proteins are conserved and were found to be the same in human rDNA genes from different human cell lines [15].
Fig. 6. DSB hotspots in human rDNA genes. Profile of DSB distribution along the 43-kb rDNA gene is presented. R1-R9 are regions of nine DSB hotspots (Pleiades) located in the IGS [16, 22]. Lower panel shows distribution of the DNase I-hypersensitive sites.
DSB HOTSPOTS AND DNase I-HYPERSENSITIVE SITES
It is interesting that some sites hypersensitive to exogenous DNase I colocalize with Pleiades (Fig. 6). However, DNA breaks in Pleiades were mapped by the very fast DNA isolation procedure without using exogenous nucleases (Fig. 1). This means that DSB hotspots exist before the addition of the exogenous enzyme in the DNase-Seq procedure. Hence, mapping exogenous DNase I-hypersensitive sites, which involves longer procedures of isolation of nuclei (http://genome.ucsc.edu/ENCODE/protocols/general/Duke_DNase_protocol.pdf), results in mapping a mixture of preexisting endogenous DBS hotspots and true DNase I-hypersensitive sites [15]. Considering that the data on true DNase I-hypersensitive sites are important for studying open chromatin structures, the data currently available for various genomes must be revised and divided into two groups: related to true DNase I-hypersensitive sites formed upon addition of the exogenous enzyme and those related to endogenous DSB hotspots that have another origin. Based on the results presented in Fig. 6, only true DNase I-hypersensitive sites are located in the rDNA coding region, while the IGS regions mostly contain DSB hotspots.
rDNA genes are the most fragile regions in the human genome. The data on the density of mapped DSBs in the rDNA and in chromosomes are presented in the table [15]. Considering that human genome contains ~300 copies of rDNA genes [17], the number of mapped breaks per one gene is 4115 (1,234,439/300), which is almost 20-fold higher than the average level for the genome. Because not all clusters of human rDNA genes are active, this value is even higher in the operating clusters. R5 and R7 of the Pleiades (Fig. 6) that correspond to the peaks in the DSB profile in rDNA genes are the most fragile sites in the human genome. rDNA genes are the most actively transcribed genes. At the same time these genes contain the highest density of DSBs, which strongly suggests close correlation between transcription and DNA breaks.
DSB density in the 43-kb fragment of rDNA gene and in the sequenced part
of human genome
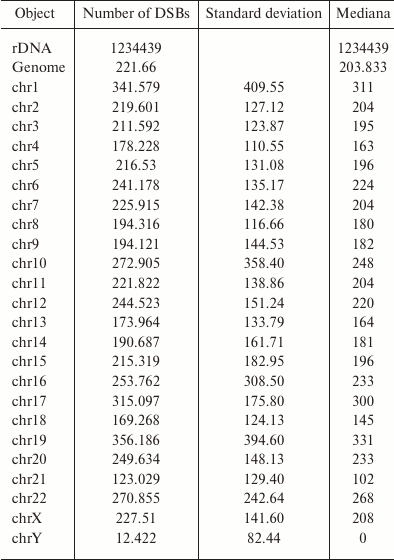
Note: DSB mapping in the rDNA gene and in the whole sequenced human
genome (hg19) was performed using the data on deep sequencing of DNA
library of fragments located at the DSB sites (RAFT preparations, see
Fig. 1c). The density of mapped DSB was scanned
using the window equal to the length of the rDNA gene (43 kb).
DSB HOTSPOTS FORM A NETWORK OF CHROMOSOMAL CONTACTS
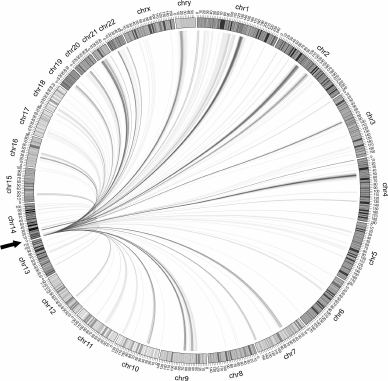
As mentioned above, FT (i.e., DSB hotspots) form chromosomal contacts in the Drosophila genome. The same is true for the DSB hotspots of human rDNA genes. The contacts for the R5 region are shown in Fig. 7. Since rDNA genes have not yet been included into the sequenced part of the genome, one copy of the gene was placed at the end of Chr14. The most frequent contacts with different chromosomes (>200 reads in rDNA-4C) are presented on the Circos circular interactions map. It can be seen that there are also contacts with the rDNA sequence itself at the end of Chr14. Detailed analysis allowed us to reveal contacts within and between the rDNA repeats [15].
Fig. 7. Contacts of the rDNA region containing DSB hotspots R4 and R5 of the Pleiades with different human chromosomes [15]. The most frequent contacts (number of reads in deep sequencing of rDNA-4C over 200) are visualized with the help of Circos program. One copy of the rDNA gene is placed at the end of Chr14 (y coordinate 0, marked with an arrow).
Figure 8 presents the genome region with rDNA gene contacts in more detail. This region of ~10 kb with scattered mapped rDNA contact sites contains both sense and antisense transcripts and an extended region of the H3K27ac histone mark that has been found in different cell lines and is characteristic of super-enhancers [18]. In addition, the region of rDNA contacts includes binding sites for the chromatin fragments that bind POL2 (ChIA-PET data). These results independently support the fact that this region participates in chromosomal contacts. The availability of binding sites for CTCF in the same region of rDNA contacts in embryonic stem cells (H1-hESC) and K562 cells also speaks in favor of this suggestion. The presence of chromatin marks H3K4me3 and H3K9ac, binding sites for the conserved transcription factors (TFBS Conserved track), and DNA methylation strongly supports functional significance of this Chr1 region in the formation of contacts with rDNA genes. Interestingly, different cell lines have different epigenetic marks in this region. More detailed information on the functional role of these contacts will be published separately.
Fig. 8. Epigenetic marks in rDNA contact sites in Chr1. The pericentromeric region (close to coordinate 121 Mb, hg19) with scattered rDNA contacts (IGB browser) is shown. Red frame indicates a 10-kb region displaying different epigenetic marks (UCSC browser).
DSB HOTSPOTS IN rDNA AND DISEASES
Detailed investigation of the contact sites in the R5 region revealed that DSB hotspot-containing sites in rDNA genes often form contacts with DSB hotspot-containing sites in other chromosomes. This creates the possibilities for the translocations involving rDNA genes [15] and explains why the so-called Robertsonian mutations occur only with participation of five acrocentric chromosomes – Chr13, 14, 15, 21, and 22. It is known that the rDNA gene clusters in, for example, Chr21 and Chr14, are located in the pericentromeric regions. Frequent breaks in Pleiades and contacts between them could result in erroneous DNA break repair and incorrect recombination of chromosomes. This can cause formation of chromosomes, in which the long arm of Chr14 is combined with the short arm of Chr21. This is the mechanism leading to the Down syndrome [der(14;21)(q10;q10)] and other Robertsonian translocations. It is also known that ~54% solid cancers are related to disorders in the rDNA genes that occur prior to the tumor cell clonal expansion stage [17]. DSB hotspots located in other genome regions can also potentially lead to chromosomal translocations and diseases. Hence, future studies in this area are essential for both understanding the mechanisms of gene expression regulation and developing new medical approaches.
PARP1 IS AN FT-BINDING PROTEIN
As described above, ChIP-Seq studies demonstrated that DSB hotspots have a number of characteristic epigenetic marks. In order to elucidate the mechanisms that provide coordinated gene expression in forum domains, we searched for the proteins that bind specifically at the domain boundaries. For this purpose, nuclear protein extracts were incubated with the library containing a genome-wide set of human FT produced by PCR with biotinylated primers (see Fig. 1). After incubation, nuclear proteins bound to the DNA fragments were isolated using streptavidin-coated paramagnetic beads. To prevent non-specific protein binding, excessive amounts of various competitors were used.
We found that under these conditions, two proteins (PARP1 and HNRNPA2B1) are bound to the DNA library, as well as to several individual FT [15] (Fig. 9). PARP1 has been extensively studied before. It has many functions in the cell including modification (PARylation) of transcription factors, thus affecting regulation of gene expression. PARP1 is involved in the global regulation of expression patterns and DNA methylation. It also binds to non-coding RNA [19, 20]. All these functions are in agreement with the notion of coordinated gene expression in the domains. Moreover, it is possible that that the inter-chromosomal contacts, which are common for the FT, might coordinate expression via interacting with enhancers or gene promoters. The functions of HNRNPA2B1 (heterogeneous nuclear ribonucleoprotein A2/B1) are still poorly understood. It is only known that HNRNPA2B1 is constitutively expressed and associated with pre-mRNAs, microRNAs, and their transporter, which also can be related to the coordination of gene expression [21]. Future studies would be able to clarify the role of these proteins in coordinated gene expression in forum domains.
Fig. 9. Genome-wide library of human FT and individual fragment specifically bind two proteins PARP1 and HNRNPA2B1 [11].
EPIGENETIC MARKS IN DSB HOTSPOTS
It was mentioned above that DSB hotspots in human ribosomal genes corresponded to the main binding sites of the scaffold regulatory protein CTCF, as well as to the active chromatin mark H3K4me3 in various cell lines. According to the ChIP-Seq data, expression regulators KAP1, PARP1, TCF7L2, GATA3, T-bet are also associated with certain binding sites in Pleiades [22]. For example, GATA3 only binds to the R9 region. Hence, Pleiades are the binding sites of complex protein assemblies. Moreover, Pleiades are the sites of DNA methylation [15]. The epigenetic marks of DSB hotspots in other genome regions have been investigated to a lesser degree.
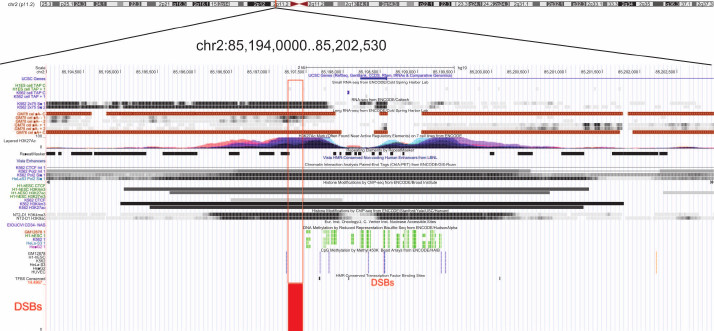
Figure 10 shows epigenetic marks characteristic for DSB hotspots in different genome regions in the ~8 kb fragment of Chr2. The DSB hotspot located close to the 5′-end of the KCMF1 gene corresponds to the region from both strands of which long non-coding RNA are transcribed. This region contains active chromatin marks H3K27ac, H3K9ac, and H3K4me3. Moreover, the multifunctional CTCF protein binds to this region only in embryonic stem cells. According to the ChIA-PET data, POL2 also binds to the region containing this FT. The latter fact strongly suggests involvement of this region in the formation of chromatin structures. Furthermore, this terminal fragment flanks a region of DNA methylation. Taken together, all these facts indicate that this DBS hotspot region is involved in the gene expression regulation. Currently, we perform genome-wide analysis of FT-containing regions that can have various epigenetic marks in human genome and play different roles in the mechanisms of coordinated gene expression.
Fig. 10. Epigenetic marks in the regions of DSB hotspots in Chr2. The euchromatin region with scattered DSBs (close to coordinate 85 Mb, hg19) is shown. Red frame indicates the region with various epigenetic marks (UCSC browser).
CAUSES OF FREQUENT DNA BREAKS
Integrity and fragmentation are natural properties of chromosomal DNA. DNA fragmentation occurs in various physiological cellular processes, such as chromatin remodeling in spermatids [23, 24], somatic V(D)J recombination in lymphocytes, and transposition of mobile genetic elements. Multiple transient DNA breaks have been reported in neurons [25]. Massive breaks have been observed in DNA in the phenomenon called chromothripsis [26]. Macronucleus formation in protozoans is accompanied by the programmed DNA fragmentation [27]. Therefore, both limited or massive fragmentation of chromosomal DNA can occur in different cell types during various physiological processes.
DNA breaks, including those in the hotspots, can emerge during replication. For example, the number of hotspots noticeably increases after cell treatment with hydroxyurea that induces replication stress [8]. The same has been observed during the heat shock, which causes global changes in the transcription patterns [8]. These data suggest that DSB hotspots can emerge during replication and transcription, although by yet poorly understood mechanisms. Recently, the role of RNA loops (R-loops) in the formation of DNA breaks has been actively discussed [28]. R-loops formed during transcription contain single-stranded DNA that could be a target for spontaneous deamination resulting in the dC/dU substitution followed by the DSB formation [29]. However, this does not explain the existence of Pleiades, because according to the RDIP data (DNA–RNA immunoprecipitation), the locations of RNA loops do not coincide with Pleiades [22, 30].
Most likely there are many (or several) major causes for the emergence of DSB hotspots. We believe that detailed genome-wide analysis of the DSB locations could help to answer the question what endogenous nucleases are responsible for these breaks and to elucidate biological processes associated with particular DSB hotspots. It cannot be ruled out that hotspots bordering a particular forum domain emerge as the results of different events in the nucleus, such as active transcription or conflict between replication and transcription.
DSB HOTSPOTS ARE GENERATED IN VIVO
One of the important questions related to the DSB origin is do DSBs appear as a result of certain in vivo processes or emerge during preparation of DNA samples, i.e., in vitro? Indeed, it is almost impossible to avoid hydrodynamic shearing of DNA during isolation, even when the procedure is very gentle, such as shown in Fig. 1. Such random DNA breaks would most likely distribute evenly along the chromosome. At the same time, the sites of most frequent (hundreds and thousands) breaks would most probably reflect the events occurring in vivo. To answer the question, we mapped the binding sites for the γ-H2AX histone in the human rDNA gene [22]. γ-H2AX is a reliable marker of DNA breaks in vivo [31]. Figure 11 demonstrates that the profile of Pleiades coincides with the profile of γ-H2AX-binding sites. This was characteristic for both resting cells and rapidly dividing cancer cells, which indicates that the observed breaks are most likely associated with transcription and not with replication [22]. Independent studies using by immunostaining with antibodies against γ-H2AX and FISH (hybridization with rDNA preparation) showed that in vivo DNA breaks are located in nucleoli [22]. These data confirm that rDNA genes are subjected to more breaks than other regions of human genome (table). Hence, DSB hotspots are generated in vivo. This conclusion is important for further investigation of DSB hotspots in different genomes.
Fig. 11. Pleiades correspond to in vivo DNA breaks in human rDNA genes as follows from the coincidence of the profile of Pleiades and the profile for the histone γ-H2AX marks [22, 31].
Studies of DSB hotspots and forum domains result in several important conclusions. It is necessary to revise DNase I-hypersensitive sites in different genomes and to identify true sites that are introduced by exogenously added enzyme. In vivo breaks in the chromosomal DNA are required for yet unknown cellular mechanisms of gene expression regulation. Replication and active transcription are associated with the formation of DSBs that occur more frequently in particular genome regions. The most frequently transcribed genes – rDNA genes – are the most fragile sites in the human genome. Genome regions containing DSB hotspots can contain enhancers, silencers, and oxymorons. These regions participate frequently in chromosomal contacts and display a set of epigenetic marks related to the gene activation or repression. Genes in the forum domains are coordinately expressed. Up to 90% of forum domains are silenced. Proteins can bind specifically at the forum domain boundaries. A high frequency of DSBs in the genome requires highly efficient repair systems. Hence, stress and unfavorable external factors (bad ecological state of the environment, unhealthy food that might contain ingredients whose safety has not been confirmed by consumption for several generation, unhealthy life style, and others) can potentially decrease the efficiency of DNA repair and lead to the development of diseases. Further studies could elucidate the role of rDNA genes in cancerogenic translocations and Robertsonian mutations. One of the important directions of future studies should be revealing the role of rDNA gene contacts in global regulation of gene expression and carcinogenesis.
Acknowledgments
The authors are grateful to S. V. Razin for invitation to write this review and to V. R. Chechetkin for his help in mathematical data processing.
This work was supported by the Molecular and Cell Biology Program of the Presidium of the Russian Academy of Sciences and by the Russian Foundation for Basic Research (projects 15-04-00299, 17-04-02152, 18-04-00198, and 18-04-00680).
The results presented on Figs. 5 and 11 were obtained with financial support by the Program of Fundamental Research for State Academics for 2013-2020 years (No. 0103-2014-0005). The data presented on Fig. 8 and 10 were obtained with financial support by the Russian Science Foundation (project 18-14-00122).
REFERENCES
1.Guelen, L., Pagie, L., Brasset, E., Meuleman, W.,
Faza, M. B., Talhout, W., Eussen, B. H., De Klein, A., Wessels, L., De
Laat, W., and Van Steensel, B. (2008) Domain organization of human
chromosomes revealed by mapping of nuclear lamina interactions,
Nature, 453, 948-951.
2.Tchurikov, N. A., and Ponomarenko, N. A. (1992)
Detection of DNA domains in Drosophila, human and plant
chromosomes possessing mainly 50- to 150-kilobase stretches of DNA,
Proc. Natl. Acad. Sci. USA, 89, 6751-6755.
3.Dixon, J. R., Gorkin, D. U., and Ren, B. (2016)
Chromatin domains: the unit of chromosome organization, Mol.
Cell, 62, 668-680.
4.Van Steensel, B., and Belmont, A. S. (2017)
Lamina-associated domains: links with chromosome architecture,
heterochromatin, and gene repression, Cell, 169,
780-791.
5.Wen, B., Wu, H., Shinkai, Y., Irizarry, R. A., and
Feinberg, A. P. (2009) Large histone H3 lysine 9 dimethylated chromatin
blocks distinguish differentiated from embryonic stem cells, Nat.
Genet., 41, 246-250.
6.Tchurikov, N. A., Ponomarenko, N. A., and Airich,
L. G. (1988) Isolation and characterization of specific fraction of
human chromosomal DNA – forum DNA, Dokl. Akad. Nauk USSR,
303, 491-493.
7.Tchurikov, N. A., Kretova, O. V., Sosin, D. V.,
Zykov, I. A., Zhimulev, I. F., and Kravatsky, Y. V. (2011) Genome-wide
profiling of forum domains in Drosophila melanogaster,
Nucleic Acids Res., 39, 3667-3685.
8.Tchurikov, N. A., Kretova, O, V., Fedoseeva, D. M.,
Sosin, D. V., Grachev, S. A., Serebraykova, M. V., Romanenko, S. A.,
Vorobieva, N. V., and Kravatsky, Y. V. (2013) DNA double-strand breaks
coupled with PARP1 and HNRNPA2B1 binding sites flank coordinately
expressed domains in human chromosomes, PLoS Genet., 9,
e1003429.
9.Zhimulev, I. F., Belyaeva, E. S., Makunin, I. V.,
Pirrotta, V., Volkova, E. I., Alekseyenko, A. A., Andreyeva, E. N.,
Makarevich, G. F., Boldyreva, L. V., Nanayev, R. A., and Demakova, O.
V. (2003) Influence of the SuUR gene on intercalary
heterochromatin in Drosophila melanogaster polytene chromosomes,
Chromosoma, 111, 377-398.
10.Tchurikov, N. A., Kretova, O. V., Chernov, B. K.,
Golova, Y. B., Zhimulev, I. F., and Zykov, I. A. (2004) SuUR protein
binds to the boundary regions separating forum domains in Drosophila
melanogaster, J. Biol. Chem., 279, 11705-11710.
11.Sosin, D. V., Kretova, O. V., and Tchurikov, N.
A. (2006) A study of the locus control region (LCR) of the cut
locus of Drosophila melanogaster using luciferase expressing
genetic constructs, Dokl. Biochem. Biophys., 409,
248-252.
12.Tchurikov, N. A., Sosin, D. V., Kretova, O. V.,
and Moiseeva, E. D. (2007) Functional analysis of LCR sequences from
the cut locus of Drosophila melanogaster, Dokl.
Biochem. Biophys., 415, 217-221.
13.Sosin, D. V., Moiseeva, E. D., and Tchurikov, N.
A. (2010) Study of distant interactions of LCR from the Drosophila
melanogaster cut locus, Dokl. Biochem. Biophys., 432,
102-105.
14.Cabrera, C. P., Navarro, P., Huffman, J. E.,
Wright, A. F., Hayward, C., Campbell, H., Wilson, J. F., Rudan, I.,
Hastie, N. D., Vitart, V., and Haley, C. S. (2012) Uncovering networks
from genome-wide association studies via circular genomic permutation,
G3 (Bethesda), 2, 1067-1075.
15.Tchurikov, N. A., Fedoseeva, D. M., Sosin, D. V.,
Snezhkina, A. V., Melnikova, N. V., Kudryavtseva, A. V., Kravatsky, Y.
V., and Kretova, O. V. (2015) Hotspots of DNA double-strand breaks and
genomic contacts of human rDNA units are involved in epigenetic
regulation, J. Mol. Cell Biol., 7, 366-382.
16.Tchurikov, N. A., Kretova, O. V., Fedoseeva, D.
M., Chechetkin, V. R., Gorbacheva, M. A., Karnaukhov, A. A.,
Kravatskaya, G. I., and Kravatsky, Y. V. (2015) Mapping of genomic
double-strand breaks by ligation of biotinylated oligonucleotides to
forum domains: analysis of the data obtained for human rDNA units,
Genomics Data, 3, 15-18.
17.Stults, D. M., Killen, M. W., Williamson, E. P.,
Hourigan, J. S., Vargas, H. D., Arnold, S. M., Moscow, J. A., and
Pierce, A. J. (2009) Human rRNA gene clusters are recombinational
hotspots in cancer, Cancer Res., 69, 9096-9104.
18.Hnisz, D., Abraham, B. J., Lee, T. I., Lau, A.,
Saint-Andre, V., Sigova, A. A., Hoke, H. A., and Young, R. A. (2013)
Super-enhancers in the control of cell identity and disease,
Cell, 155, 934-947.
19.Frizzell, K. M., Gamble, M. J., Berrocal, J. G.,
Zhang, T., Krishnakumar, R., Cen, Y., Sauve, A. A., and Kraus, W. L.
(2009) Global analysis of transcriptional regulation by
poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in
MCF-7 human breast cancer cells, J. Biol. Chem.,
284, 33926-33938.
20.Guetg, C., Scheifele, F., Rosenthal, F.,
Hottiger, M. O., and Santoro, R. (2012) Inheritance of silent rDNA
chromatin is mediated by PARP1 via noncoding RNA, Mol. Cell,
45, 790-800.
21.Villarroya-Beltri, C., Gutierrez-Vazquez, C.,
Sanchez-Cabo, F., Perez-Hernandez, D., Vazquez, J., Martin-Cofreces,
N., Martinez-Herrera, D. J., Pascual-Montano, A., Mittelbrunn, M., and
Sanchez-Madrid, F. (2013) Sumoylated hnRNPA2B1 controls the sorting of
miRNAs into exosomes through binding to specific motif, Nat.
Commun., 4, 2980.
22.Tchurikov, N. A., Yudkin, D. V., Gorbacheva, M.
A., Kulemzina, A. I., Grischenko, I. V., Fedoseeva, D. M., Sosin, D.
V., Kravatsky, Y. V., and Kretova, O. V. (2016) Hotspots of DNA
double-strand breaks in human rDNA units are produced in vivo,
Sci. Rep., 6, 25866.
23.Marcon, L., and Boissonneault, G. (2004)
Transient DNA strand breaks during mouse and human spermiogenesis: new
insights in stage specificity and link to chromatin remodeling,
Biol. Reprod., 70, 910-918.
24.Leduc, F., Maquennehan, V., Nkoma, G. B., and
Boissonneault, G. (2008) DNA damage response during chromatin
remodeling in elongating spermatids of mice, Biol. Reprod.,
78, 324-332.
25.Blondet, B., Ait-Ikhlef, A., Murawsky, M., and
Rieger, F. (2001) Transient massive DNA fragmentation in nervous system
during the early course of a murine neurodegenerative disease,
Neurosci. Lett., 305, 202-206.
26.Stephens, P. J., Greenman, C. D., Fu, B., Yang,
F., Bignell, G. R., Mudie, L. J., Pleasance, E. D., Lau, K. W., Beare,
D., Stebbings, L. A., McLaren, S., Lin, M. L., McBride, D. J., Varela,
I., Nik-Zainal, S., Leroy, C., Lia, M., Menzies, A., Butler, A. P.,
Teque, J. W., Quail, M. A., Burton J., Swerdlow, H., Carter, N. P.,
Morsberger, L. A., Lacobuzio-Donahue, C., Follows, G. A., Green, A. R.,
Flagan, A. M., Stratton, M. R., Futreal, P. A., and Campbell, P. J.
(2011) Massive genomic rearrangement acquired in a single catastrophic
event during cancer development, Cell, 144, 27-40.
27.Mochizuki, K. (2010) DNA rearrangements directed
by non-coding RNAs in ciliates, Wiley Interdiscip. Rev. RNA,
1, 376-387.
28.Aguilera, A., and Garcia-Muse, T. (2012) R-loops:
from transcription byproducts to threats to genome stability, Mol.
Cell, 46, 115-124.
29.Skourti-Stathaki, K., and Proudfoot, N. J. (2014)
A double-edged sword: R-loops as threats to genome integrity and
powerful regulators of gene expression, Genes Dev., 28,
1384-1396.
30.Nadel, J., Athanasiadou, R., Lemetre, C.,
Wijetunga, N. A., O’Broin, P., Sato, H., Zhang, Z., Jeddeloh, J.,
Montagna, C., Golden, A., Seoighe, C., and Greally, J. M. (2015)
RNA:DNA hybrids in the human genome have distinctive nucleotide
characteristics, chromatin composition, and transcriptional
relationships, Epigenetics Chromatin, 8, 46.
31.Seo, J., Kim, S. C., Lee, H. S., Kim, J. K.,
Shon, H. J., Salleh, N. L., Desai, K. V., Lee, J. H., Kang, E. S., Kim,
J. S., and Choi, J. K. (2011) Genome-wide profiles of H2AX and
γ-H2AX differentiate endogenous and exogenous DNA damage hotspots
in human cells, Nucleic Acids Res., 40, 5965-5974.