Regulated Gene Expression as a Tool for Analysis of Heterochromatin Position Effect in Drosophila
A. S. Shatskikh1, O. M. Olenkina1, A. A. Solodovnikov1, and S. A. Lavrov1,a,b*
1Institute of Molecular Genetics, Russian Academy of Sciences, 123182 Moscow, Russia* To whom correspondence should be addressed.
Received December 6, 2017; Revision received December 25, 2017
Position effect variegation (PEV) is a perturbation of genes expression resulting from the changes in their chromatin organization due to the abnormal juxtaposition with heterochromatin. The exact molecular mechanisms of PEV remain enigmatic in spite of the long history of PEV studies. Here, we developed a genetic model consisting of PEV-inducing chromosome rearrangement and a reporter gene under control of the UAS regulatory element. Expression of the reporter gene could be regulated by adjustment of the GAL4 transactivator activity. Two UAS-based systems of expression control were tested – with thermosensitive GAL4 repressor GAL80ts and GAL4-based artificial transactivator GeneSwitch. Both systems were able to regulate the expression of the UAS-controlled reporter gene over a wide range, but GAL80ts repressed the reporter gene more efficiently. Measurements of the heterochromatin-mediated repression of the reporter gene in the GAL4+GAL80ts system point to the existence of a threshold level of expression, above which no PEV is observed.
KEY WORDS: heterochromatin, expression, Drosophila, Gal4, Gal80, MiMIC, GeneSwitchDOI: 10.1134/S0006297918050073
Abbreviations: PEV, position effect variegation; UAS-eGFP-Ret, insertion of the reporter gene UAS-eGFP in the Ret gene.
Position effects are disruptions of gene expression resulting from the
gene transfer to another genomic region without changes in its DNA
sequence. The existence of the effects of this type indicates that the
chromosomal environment influences the level of gene expression.
Position effect variegation (PEV) is a particular case of position
effect manifested as perturbations in the expression of euchromatin
genes after their relocation close to the heterochromatin as a result
of the chromosomal inversions, translocations, or transpositions.
Heterochromatin here is a type of chromatin organization in eukaryotes;
it consists of different types of DNA repeats and is characterized by a
specific set of non-histone proteins, histone modifications, and a high
level of compaction during interphase [1].
Chromosome regions close to centromeres (pericentromeric
heterochromatin) and telomeres are mainly composed of heterochromatin,
as well as chromosomes Y and 4 in Drosophila. Heterochromatin
contains a rather small number of genes, compared to euchromatin.
PEV in Drosophila melanogaster was discovered more than 60 years ago [2] and has been actively studied since then [1, 3, 4]. PEV-associated perturbations in the expression of euchromatin genes are caused by changes in the chromatin structure of the gene environment [5], the so-called heterochromatinization (the term introduced by Prokofieva-Belgovskaya [6]). Searches for the genes that affect PEV revealed a number of non-histone chromatin proteins and demonstrated the role of histone modifications in the processes of gene repression and activation [7]. It was found that the mechanisms of chromatin structure formation are quite similar among eukaryotes, non-histone proteins are conserved, and histone modifications have the same effects in yeast to mammals [8]. The most interesting aspect of the PEV is variegated repression – a situation when a gene is active in certain cells and repressed in others. This mosaic expression indicates the ability of the repressed/active state of chromatin to be inherited during mitosis (epigenetic inheritance).
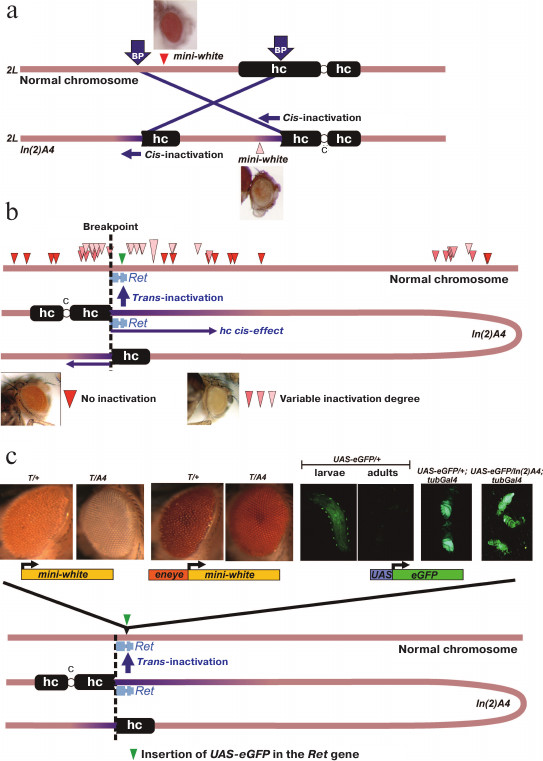
Earlier, we have created and studied the PEV-causing chromosomal rearrangement In(2)A4 [9-11]. This rearrangement is an inversion of a fragment of the chromosome 2 left arm with the breakpoints in the Mcm10 gene (region 39B) and in the left arm pericentromeric heterochromatin region containing the (AATAACATAG)n satellite (Fig. 1a). The In(2)A4 rearrangement affects genes located near the breakpoint (cis-effect), as well as the reporter transgenes located on the normal chromosome in regions homologous to those that were relocated closer to the heterochromatin in the inversion (trans-inactivation) (Fig. 1, a and b).
Fig. 1. Structure of the In(2)A4 inversion and manifestation of the reporter genes trans-inactivation. a) Structure of the In(2)A4 rearrangement. Rearrangement results from the breaks in euchromatin and pericentromeric heterochromatin (BP, breakpoints) and leads to the formation of two new euchromatin/heterochromatin borders. The reporter gene (mini-white) relocated to heterochromatin demonstrates the position effect (cis-inactivation). b) Trans-inactivation caused by the In(2)A4 inversion. The inversion causes trans-inactivation of the reporter genes in the homologous non-rearranged chromosome in heterozygous flies. Positions of the trans-inactivated mini-white reporter genes are shown with triangles of different color according to the degree of their repression: from the total absence (red) to strong repression (light pink) [9]. The inversion is represented as a loop so that regions of the normal chromosome and the respective regions of In(2)A4 would have the same orientation. c) Reporter constructions in the region of trans-inactivation caused by the In(2)A4 inversion. All constructions were site-specifically integrated into the Ret gene closest to the euchromatin/heterochromatin border. Constructions from left to right: mini-white reporter gene is strongly repressed in In(2)A4; mini-white gene under control of the eye-specific enhancer (eneye) is strongly expressed and its repression is not observed (T-reporter transgene mini-white or eneye-mini-white, respectively). The UAS-eGFP reporter is weakly expressed in larvae and is not expressed in the adults. Activation by the source of GAL4 under the tubulin promotor (tubGal4) results in the high level of eGFP expression, whereby the repression in In(2)A4 is not observed. Abbreviations: hc, heterochromatin; c, chromosome 2 centromere.
An example of cis-acting heterochromatin position effect is repression of the mini-white gene within the P-element located 55 kb from the eu-/heterochromatin boundary in In(2)A4 (Fig. 1a). The repression is manifested as the eye color variegation and demonstrates the specific features of heterochromatin position effect, such as the sensitivity to PEV modifier mutations and temperature [9, 11]. The trans-acting position effect (trans-inactivation) caused by the In(2)A4 inversion is manifested as an impaired expression of the mini-white reporter genes located in the homologous normal chromosome. Trans-inactivation is observed in the 500-kb region of the normal chromosome homologous to the region around the breakpoint in In(2)A4. Trans-inactivation demonstrates a complex pattern of spreading; the genomic regions with no repression are surrounded by trans-inactivated regions (Fig. 1b). Only a few examples of trans-acting position effect have been reported in the literature so far; the In(2)A4 inversion represents one of the two thoroughly studied cases (the first is the brownDominant mutation caused by the insertion of about 1.5 Mb of the AAGAG satellite into the coding region of the brown gene) [12-15]. Trans-inactivation results from the somatic conjugation (pairing) between the normal and rearranged chromosomes and translocation of a region of coupled chromosomes into the heterochromatin compartment of the nucleus via the sticking of heterochromatin blocks. According to our data, trans-inactivation of the reporter genes is a result of their de novo heterochromatinization, since the reporter gene on the normal chromosome can be inactivated, whereas the same region in the chromosome with the inversion remains transcriptionally active and does not contain heterochromatin marks [9].
Studies of molecular mechanisms of heterochromatin interaction with the gene transcription machinery require a genetic system in which a reporter gene of a known structure and with the adjustable expression level could be exposed to the position effect. Such system could be based on trans-inactivation-inducing In(2)A4 rearrangement; the reporter construction could be integrated into a normal chromosome which would be then combined with the rearranged chromosome in the same genome in order to study the position effect. In our work, transgene constructions bearing the UAS-eGFP and mini-white reporter genes were inserted into the region undergoing trans-inactivation using the phiC31 integrase-based site-specific integration system.
Here, we studied two transcription control systems based on the yeast GAL4 transactivator. The GAL4+GAL80ts-based system includes genomic sources of the GAL4 and the thermosensitive variant of the GAL80 protein (GAL4 inhibitor) [16, 17]. At low temperatures, GAL80ts inhibits GAL4 transactivation activity, whereas at high temperatures the GAL80ts degrades, allowing GAL4 to activate transcription. In the GeneSwitch system, transcription is activated by synthetic steroid hormone RU486. GeneSwitch is an artificial protein consisting of the GAL4 DNA-binding domain (recognizes UAS), the hormone-binding fragment of the human progesterone receptor, and the transactivation domain of the NF-κB protein [18, 19]. It was shown that both systems can activate the reporter gene expression hundred times. The GAL4+GAL80ts combination displays low background expression and is efficient in the situations when the low levels of the reporter gene expression are required.
Analysis of the influence of the In(2)A4 rearrangement on the level of the reporter genes transcription showed that high expression levels and the presence of additional regulatory elements prevented trans-inactivation. The use of the transcription regulation system allowed us to tweak the level of the reporter gene expression up to the threshold at which the heterochromatin repression is abolished. In the described system, the reporter gene expression should be 100 times higher than the background expression to suppress the PEV.
The aim of this work was to develop a system allowing to regulate the expression of the reporter gene affected by the heterochromatin position effect, and to use this system to study the relationship between the gene transcription and its repression in the heterochromatin environment.
MATERIALS AND METHODS
Fly stocks and transgenic constructions. Inversion In(2)A4 was produced from the fly strain carrying the mini-white reporter gene in the region 39B (5′-UTR of the Hr39 gene) of the chromosome 2. Inversion In(2)A4 reduces the viability of the flies and is maintained over the balancer chromosome Cy (In(2)A4/Cy).
The following transgenic constructions in the chromosome 3 were used (Bloomington collection, http://flystocks.bio.indiana.edu): the P[w[+mC]=Act5C(–FRT)GAL4.Switch.PR]3 construction (hereafter pAct(GS)) containing the GeneSwitch gene under the Actin5C gene promoter (stock 9431); the P[tubP-GAL80ts]7 transgenic construction containing the Gal80ts gene under the α-tubulin gene promoter (stock 7018); and the P[tubP-GAL4]LL7 construction containing the Gal4 gene also under the α-tubulin gene promoter (stock 5138). The Actin5C and α-tubulin gene promoters provide the high level of gene expression in most of the tissues at all developmental stages.
To insert the reporter genes into specific genomic regions, we used the system of site-specific integration based on phiC31 integrase and Drosophila strains bearing transgenic MiMIC constructions [20-22]. The MiMIC construction is a landing site for specific integration of transgenes into the genome by phiC31 integrase. The vas-dPhiC31strain bearing the phiC31 gene under the control of the vasa gene promoter on the X chromosome [20] was used as an integrase source. The efficiency of integration in our system was approximately 70%.
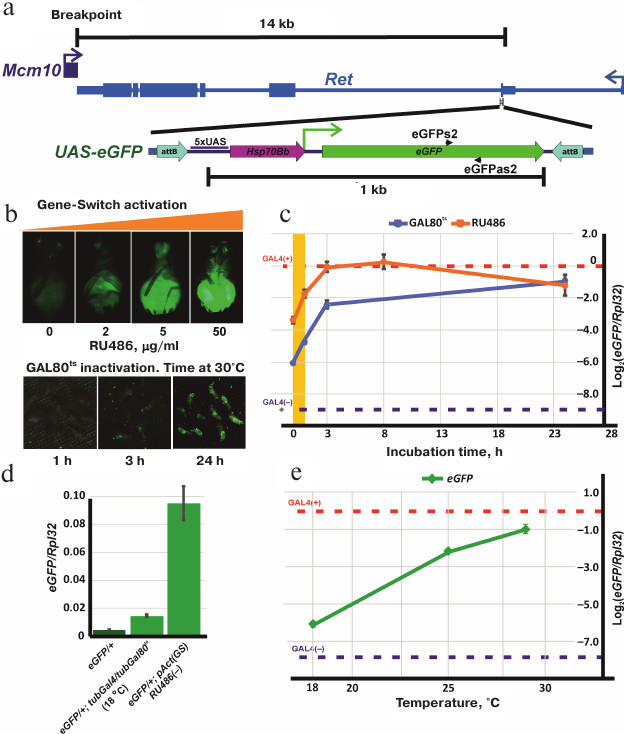
Plasmids attBs-Eneye-whited700-MCS-attBsrev-pSK=aeca containing the mini-white gene under control of the eye-specific eneye enhancer (eneye-mini-white) and attB recombination sites and pCaspew15-good-1RI with the mini-white gene without an enhancer were obtained from the Laboratory of Regulation of Genetic Processes, Institute of Gene Biology, Russian Academy of Sciences. To generate the transgene construction with mini-white reporter gene, the eneye-mini-white sequence in the attBs-Eneye-whited700-MCS-attBsrev-pSK=aeca plasmid was replaced by the mini-white gene sequence from pCaspew15 keeping the attB recombination sites intact. The construction containing the UAS-eGFP reporter gene flanked by the attB recombination sites (attBs-UASeGFP-attBsrev-pSK) was obtained by cloning the UAS-eGFP fragment into the vector with attB sites (attBs-Eneye-whited700-MCS-attBsrev-pSK=aeca). This construction was integrated into the genome by recombination with the MiMIC transgene located in the Ret gene (Drosophila stock 43099 from the Bloomington collection, genotype y[1]w[*]; Mi[y[+mDint2]=MIC]Ret[MI07200]/SM6a). The exact position, orientation, and structure of the UAS-eGFP transgene insertion (below referred as eGFP) are shown in Fig. 2a.
Fig. 2. Schematic representation of the transgenic eGFP construction and regulation of the UAS-eGFP expression using GAL4+GAL80ts and GeneSwitch. a) Position and orientation of the transgenic eGFP insertion relative to the Ret gene and the breakpoint of In(2)A4 inversion: attB, sites for specific recombination into the genome; 5xUAS, GAL4-binding site; Hsp70Bb, Hsp70 gene minimal promotor; eGFP, coding sequence of the eGFP reporter gene. Positions of primers eGFPs2 and eGFPas2 are shown. b) Activation of the UAS-eGFP reporter gene expression. Upper panel, phenotypes of eGFP/+; pAct(GS)/+ flies after incubation in the medium with increasing RU486 concentration; the lower panel, phenotypes of eGFP/+; tubGal4/tubGal80ts flies after incubation at 30°C for 1, 3, and 24 h. c) Profiles of eGFP expression in GAL4+GAL80ts and GeneSwitch+RU486 systems. Axis y, the logarithm of the ratio of eGFP expression to that of Rpl32 determined by quantitative PCR. Axis x, incubation time at 30°C for eGFP/+; tubGal4/tubGal80ts flies (blue graph) or hours after feeding with RU486-containing yeast mix for eGFP/+; pAct(GS)/+ flies (orange graph). Blue dashed horizontal line, the relative level of eGFP expression in eGFP/+ flies without transactivator (basic expression). Red dashed horizontal line, eGFP expression level in eGFP/+; tubGal4/+ flies (maximal activation of eGFP expression). Orange area, the period of feeding with the RU486-containing mix. d) eGFP expression levels without activation. From left to right: eGFP/+; tubGal80ts/+ flies (without a source of Gal4 in the genome); eGFP/+; tubGal80ts/tubGal4 flies at 18°C (maximum repression by Gal80ts); eGFP/+; pAct(GS)/+ flies (in the absence of RU486). e) Dependence of eGFP activation in eGFP/+; tubGal80ts/tubGal4 flies on temperature. The relative amount of eGFP mRNA was measured 24 h after incubation at 18, 25 and 29°C (for description, see Fig. 2c).
Regulation of the reporter gene expression in the GeneSwitch system. The synthetic hormone RU486 (Sigma, USA) binds the GeneSwitch protein [18, 19] and transforms it into a strong transcription activator. RU486 penetrates the cell membrane, so experimental organisms can be treated simply by adding RU486 to the food or culturing medium or by incubation in RU486-containing solution. The concentrations of the hormone in the medium could vary from 2 to 200 µg/ml without effects on viability. Preliminary experiments showed that the maximal level of the eGFP expression was achieved at the RU486 concentration of 50 µg/ml and remained constant upon further increase of the hormone concentration (Fig. 2b). Therefore, we used 50 µg/ml RU486 in all our experiments.
Activation of the eGFP expression was studied in the eGFP/+; pAct(GS)/+ flies versus the control eGFP/+; +/+. The expression was activated at 25°C in starving adult flies (incubated overnight in tubes with agarose prepared on MilliQ water) by adding the RU486-containing yeast suspension as a food. The activation of eGFP expression was observed after 1 h of feeding (Fig. 2b). Addition of RU486 at the early developmental stages was lethal.
Regulation of the reporter gene expression in GAL4+GAL80ts system. To test the ability of the thermosensitive form of GAL80 protein (GAL80ts) to suppress activation of the UAS-eGFP reporter gene by GAL4 (encoded by tubGal4), flies with the following genotypes were used: eGFP/+; tubGal4/tubGal80ts (experiment), eGFP/+; tubGal4/+ and eGFP/+; +/tubGal80ts (positive and negative controls, respectively).
Drosophila flies were grown at 18, 25, and 30°C. No eGFP expression was visually observed at 18°C, while at 25°C, eGFP fluorescence was detected in almost all tissue types. Fluorescence in flies incubated at 30°C was very bright, however, a strong decrease in viability and fertility was observed. For further experiments, eGFP/+; tubGal4/tubGal80ts flies were grown at 18°C; adult flies were then incubated for different periods of time at 30°C for inactivation of GAL80ts and activation of GAL4 (Fig. 2b).
Regulation of the eGFP reporter gene transcription using the GAL4+GAL80ts and GeneSwitch systems. To analyze the profile of eGFP expression activation, the amounts of eGFP mRNA were measured in adult flies containing the eGFP reporter gene and the GeneSwitch or GAL4+GAL80ts sources. When using the GAL4+GAL80ts system, eGFP/+; tubGal4/tubGal80ts (experiment), eGFP/+; tubGal4/+ (positive control, contains only Gal4), and eGFP/+; tubGal80ts/+ (negative control, no Gal4) flies were grown at 18°C; 1- to 2-day-old adult flies were then incubated at 30°C for different times. mRNA was isolated from the flies before treatment and after incubation for 1, 3, and 24 h at high temperature (Fig. 2c). To determine the established level of eGFP expression at different temperatures, eGFP/+; tubGal4/tubGal80ts flies were grown at 18, 25, and 30°C during the entire period of development, and then mRNA was isolated from 1-2-day-old adults. Incubation at 30°C resulted in high lethality; however, some flies survived and were used for analysis (Fig. 2e).
When using the GeneSwitch system, mRNA was isolated from the eGFP/+; pAct(GS)/+ flies before and 1, 3, 8, 24 h after feeding (during 1 h) with the RU486-containing yeast paste. eGFP/+ flies (without GeneSwitch or Gal4) were used as a negative control (Fig. 2c).
Quantitative PCR. RNA from adult flies was isolated using Trizol reagent according to a standard protocol and treated with DNase (Thermo Fisher Scientific, USA) to eliminate the traces of DNA. Reverse transcription was performed using random hexanucleotides (Silex, Russia) and MINT reverse transcriptase (Eurogene, Russia). Relative quantities (ΔCq) of eGFP mRNA were estimated by real-time PCR with a DT-96 amplifier (DNA Technology, Russia); obtained mRNA values were normalized to the Rpl32 gene mRNA amount. The Rpl32 gene encodes one of the ribosomal proteins and is characterized by high and stable expression levels in the majority of tissues and at all developmental stages (gene profile in the FlyBase database (http://flybase.org); our data). The relative amounts of eGFP mRNA were measured using the RealTime_PCR software from the amplifier manufacturer.
The sequences of used primers are below (8.25 pmol of each primer per standard 30-µl PCR reaction):
for Rpl32 gene:
Rp49up: ATGACCATCCGCCCAGCATAC
Rp49rev2: GCTTAGCATATCGATCCGACTGG;
for eGFP gene:
eGFPs2: CCTGGGGCACAAGCTGGAGT
eGFPas2: GGGTAGCGGCTGAAGCACTGC.
Amplification protocol included denaturation for 5 min at 95°C; 45 cycles of amplification (15 s at 94°C, 10 s at 64°C, 10 s at 72°C); final elongation of PCR products for 5 min at 72°C before reading of the melting curves. Amplification was carried out using Hot Start Taq DNA polymerase and PCR buffer (SibEnzyme, Russia) in the presence of intercalating fluorescent dye SYTO13 (Thermo Fisher Scientific). The presented values are the result of at least three independent experiments.
Microscopy. The images of eGFP fluorescence were obtained on Leica MZ6 binocular microscope (Leica Microsystems, Germany) equipped with a Leica DC300 camera. Fluorescence was excited at 450 nm (blue LED Luxeon Royal Blue 3W); the filter cut-off was <500 nm.
RESULTS
Trans-inactivation of the reporter constructions by the In(2)A4 inversion. As it was shown earlier [11], chromosomal rearrangement In(2)A4 causes trans-inactivation of the reporter genes located on the homologous non-rearranged chromosome (Fig. 1b). Taking into account the trans-inactivation distribution data, we chose the Ret gene genomic region as the place where the inserted reporter gene would be repressed with a high probability. The Ret gene is the closest to the breakpoint in In(2)A4; the fly stock 43099 (Bloomington) contains the MiMIC insertion within the first exon of the Ret at a distance of 14 kb from the breakpoint. The Ret encodes cadherin superfamily protein and is expressed at a low level and mainly in the nervous system. Insertion of a reporter gene in the Ret is not lethal, the resulting homozygous flies are viable.
The reporter genes eneye-mini-white (contains the mini-white gene under the control of the eye-specific enhancer) and mini-white (no additional regulatory elements) were inserted into the Ret gene by specific integration with the MiMIC element in the fly stock 43099. Analysis of the trans-effect of the In(2)A4 chromosomal rearrangement on the resulting constructions showed that the mini-white reporter gene is strongly inactivated, while the presence of the eye-specific enhancer prevents repression (Fig. 1c). Hence, the In(2)A4 causes the trans- acting position effect of the reporter genes inserted into the Ret region in the non-rearranged chromosome. At the same time, active expression of the mini-white gene triggered by the eye-specific enhancer prevents trans-inactivation.
To study the influence of heterochromatin on transcription activation, we’ve constructed flies where the eGFP reporter gene with Hsp70 promoter and UAS regulatory element was inserted into the Ret gene (Fig. 2a). In the absence of the GAL4 transactivator, the level of eGFP expression in these flies was low in both larvae and adults. In larvae, a specific pattern of expression associated with peripheral ganglia and, probably, corresponding to the expression pattern of the Ret gene was observed. In adult flies, no eGFP expression was visually detected (Fig. 1c). Insertion of the GAL4 source under the control of the α-tubulin promotor (tubGal4) into the genome of flies bearing the UAS-eGFP construction results in a high level of eGFP expression; at the same time, no trans-inactivation of the reporter gene was observed in eGFP/A4; tubGal4 flies comparing to eGFP/+; tubGal4 flies (Fig. 1c). Therefore, expression of the reporter gene at a high level prevents trans-inactivation (as in the case of the mini-white gene under the control of the eye-specific enhancer).
Regulation of the eGFP reporter gene expression in the GAL4+GAL80ts and GeneSwitch systems. The level of the UAS-eGFP expression can be regulated using the systems based on the modulation of the GAL4 transactivator activity. These systems allow to identify the threshold level of eGFP expression above which no trans-inactivation of the reporter gene is observed. To choose the optimal tool for the regulation of the reporter gene expression, we analyzed the GAL4+GAL80ts and GeneSwitch systems.
The results of our experiments are presented in the Fig. 2, c-e. Panels (d) and (e) (Fig. 2) show the levels of eGFP expression in control flies with blue (eGFP; Gal4(–), negative control) and red (eGFP; Gal4(+), positive control) dashed lines. The presence of Gal4 gene under the control of the tubulin promotor (tubGal4) in the genome leads to the 600-fold increase in the eGFP expression. The level of eGFP expression almost reaches the level of expression of the ribosomal protein Rpl32 gene used for normalization.
Inactivation of the GAL80ts protein occurs gradually from 18 to 30°C; when the temperature rises from 20 to 25°C, the reporter gene expression level increases 15 times (Fig. 2e). At the same time, the level of eGFP expression at 30°C in eGFP/+; tubGal4/tubGal80ts flies is two times lower than in eGFP/+; tubGal4/+ flies, i.e., GAL80ts partially retained its repressor activity. Higher temperatures have a negative effect on the viability of flies and were not tested. During the first hour of incubation of the flies at 30°C, the amount of eGFP mRNA increases 2.5 times and then continues to increase; after 3 h of incubation, the rate of mRNA accumulation drops down (Fig. 2c).
Activation of the GeneSwitch by the hormone RU486 in eGFP/+; pAct(GS)/+ flies increases the level of eGFP mRNA 3-fold during the first hour after feeding; then the mRNA accumulation continues. After 8 h, the mRNA level reaches its maximum which is close to the level of eGFP mRNA amount in the positive control flies (eGFP/+; tubGal4/+). The eGFP mRNA amount remains high 24 h after termination of hormone treatment (Fig. 2c). At the same time, the presence of GeneSwitch in the genome, even in the absence of RU486, results in a 47-fold increase in the reporter gene expression comparing to the background expression of eGFP in the flies without GeneSwitch. The level of eGFP expression in the presence of both tubGal4 and tubGal80ts in the genome at 18°C was only 3 times higher than the background expression (Fig. 2d). Therefore, eGFP expression can be regulated by both the GAL4+GAL80ts and GeneSwitch systems; however, GAL80ts is much more efficient in the suppression of the reporter gene than the GeneSwitch in the absence of RU486.
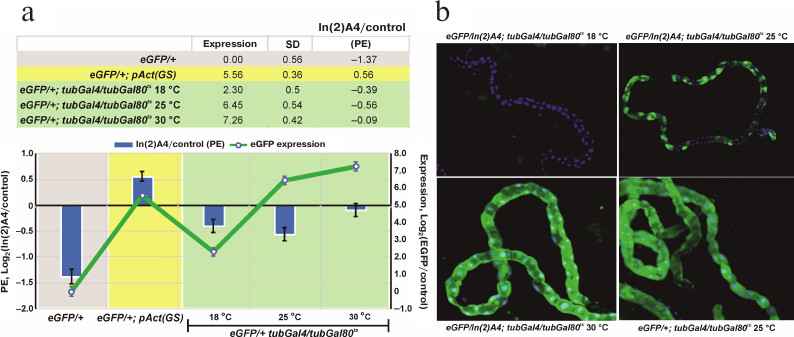
The level of reporter gene expression affects the degree of heterochromatin-mediated repression. The transgenic eGFP construction locates in the area affected by In(2)A4-caused trans-inactivation. The influence of heterochromatin on a euchromatin gene could be modulated by the level of expression of this gene at different developmental stages. Using the GAL4+GAL80ts system, we found the temperature range (and consequently, the level of eGFP expression) at which the significant trans-inactivation of the reporter gene is observed (Fig. 3). Comparison of the eGFP expression in eGFP/+; tubGal4/+ (control) and eGFP/In(2)A4; tubGal4/+ flies (trans-inactivation condition) shows that the high expression levels block trans-inactivation of UAS-eGFP like in the case of the eneye-mini-white reporter gene (Fig. 1c). In the flies without GAL4 (eGFP/+ and eGFP/In(2)A4), the eGFP fluorescence is not visually detectable; however, its expression level could be determined by quantitative PCR. We found that the background level (without GAL4 transactivator) of eGFP expression in eGFP/In(2)A4 flies was ~3 times lower than in eGFP/+ flies. Therefore, in absence of specific activation of the expression, eGFP is trans-inactivated by the In(2)A4 rearrangement (Fig. 3a). Insertion of the sources of GAL4 transactivator and GAL80ts repressor into the genome allows to regulate the level of eGFP expression by changing the incubation temperature. Comparison of the eGFP expression levels in eGFP/+; tubGal4/tubGal80ts and eGFP/In(2)A4; tubGal4/tubGal80ts flies at 18, 25, and 30°C shows that at 18 and 25°C, when the levels of eGFP expression are 3 and 87 times higher than the background, the ~1.5- to 2.0-fold repression of reporter by the In(2)A4 is observed. No inactivation is detected at 30°C (where the level of eGFP expression is 153 times higher than the background level) (Fig. 3a). Hence, the ~100-time increase in the gene expression prevents heterochromatin repression. In some types of tissues (malpighian tubes), heterochromatin repression of the eGFP reporter gene leads to the classical mosaic PEV phenotype (Fig. 3b). The high background level of the reporter gene transcription in the flies with GeneSwitch source (eGFP/In(2)A4; pAct(GS)/+) prevents trans-inactivation (Fig. 3a).
Fig. 3. The influence of the eGFP reporter gene expression level on the degree of its trans-inactivation by the In(2)A4 inversion. a) eGFP expression level in the GeneSwitch and GAL4+GAL80ts systems (graph, right scale) and the repression degree under the influence of In(2)A4 (bars, left scale). Expression is presented as a logarithm of the ratio of the eGFP expression level in a given genotype to that in eGFP/+ flies (with the reporter gene, but without the transactivator GAL4). The degree of trans-inactivation (In(2)A4/control) is presented as a logarithm of the ratio of the eGFP reporter expression in eGFP/In(2)A4 flies to that in eGFP/+ flies (control); negative values indicate trans-inactivation. eGFP/+; pAct(GS) – flies with the reporter gene and GeneSwitch in the absence of activation with RU486; eGFP/+; tubGal4/tubGal80ts – flies with eGFP reporter gene in the presence of GAL4 and GAL80ts; flies were grown at 18, 25, and 30°C. b) Expression of the eGFP reporter gene in Malpighian tubules of eGFP/In(2)A4; tubGal4/tubGal80ts and eGFP/+; tubGal4/tubGal80ts (control) flies at different temperatures; no eGFP expression is observed at 18°C; mosaic inactivation of the eGFP expression is observed at 25°C; at 30°C the eGFP expression level is high, and no significant position effect (PE) is observed. Green fluorescence, eGFP; blue fluorescence, DAPI-stained nuclei.
DISCUSSION
Chromosomal rearrangement In(2)A4 affects both the expression of euchromatin genes translocated near the heterochromatin (cis-position effect) and reporter genes in the corresponding regions of homologous non-rearranged chromosome (trans-inactivation), providing an opportunity to study and to compare the mechanisms of cis- and trans-inactivation. Earlier, we have found a number of unexpected features of the cis-position effect in the case of In(2)A4 rearrangement. The effect of heterochromatin on the juxtaposed euchromatin (cis-effect) is not limited to the repression of genes located there. Only a small part of genes translocated to heterochromatin changed their level of expression, and both repression and activation of genes were observed. Moreover, in some cases, the character of heterochromatin influence on a gene depends on the developmental stage; the cis-inactivation of a gene at the late larval stage could be switched to the activation of its transcription in adults [9-11].
The different sensitivity of euchromatin genes to the heterochromatin position effect was reported for the position effect caused by In(1)wm4h inversion. In this case, detectable repression was observed only for the white gene, and the authors [23] suggested that this gene is specifically sensitive to heterochromatin. This hypothesis, however, contradicts a large number of PEV cases described in the literature for different genes [3].
The data presented above do not correspond to traditional expectations on the influence of heterochromatin on euchromatin, according to which the position effect is a result of nonspecific repression of genes caused by the chromatin compaction. Heterochromatin interacts in a complex manner with the transcriptional machinery, and the result of such interactions varies for different genes. Heterochromatin environment can prevent chromatin remodeling (changes in the histone code and nucleosome positioning under the influence of promoter-associated chromatin-remodeling protein complexes) during transcription induction. This model suggests that the effect of heterochromatin environment is manifested at the moment of expression activation, whereas the established level of expression does not change.
Expression of some genes is induced during metamorphosis, and comparison of gene expression profiles at the pupal stage in flies with PEV and in control wild-type flies could reveal the influence of heterochromatin on the expression activation. We verified this suggestion in experiments with the genes potentially exposed to the position effects of inversions In(2)A4 and In(1)wm4h. It was shown that the genes, whose expression does not change at the pupal stage, are not subjected to the heterochromatin position effect in the In(2)A4 and In(1)wm4h inversions, while the genes whose expression was activated at the pupal stage experienced the position effect, and their activation was perturbed (preliminary data).
To study the link between trans-inactivation and expression, a genetic system containing the reporter gene with adjustable expression located in the trans-inactivated region of a normal chromosome was developed. Constructions carrying the mini-white or mini-white under the control of the eye-specific enhancer (eneye) were inserted into the same position in the region affected by trans-inactivation. It was shown that the mini-white reporter gene underwent strong trans-inactivation by In(2)A4, while the presence of an enhancer in the transgene construction significantly increases expression of the mini-white gene and suppresses trans-inactivation (Fig. 1c). Two possible explanations of the observed effect of the enhancer can be proposed: either the high level of gene expression prevents position effect or the enhancer itself blocks heterochromatinization of a neighbor gene. To verify these hypotheses, the transgene construction containing the eGFP gene under the control of the regulatory UAS element was inserted into the same position as mini-white-containing transgenes, and the fly stocks with combination of UAS-eGFP, In(2)A4 inversion, and a system of the UAS-eGFP expression control were generated.
We controlled the reporter gene expression using the GAL4 activator and its thermosensitive repressor GAL80ts or the artificial protein GeneSwitch, which is able to bind UAS and activate the expression in the presence of RU486. Both systems can burst the expression of the reporter gene hundreds of times, however, the GeneSwitch protein significantly activates transcription even in the absence of RU486 (Fig. 2). Analysis of the changes in eGFP expression in the presence of In(2)A4 shows that the trans-inactivation is suppressed when the reporter gene expression is adjusted to a high level by GAL4+GAL80ts system. At lower expression levels, the position effect is observed; the eGFP expression level in eGFP/In(2)A4 flies is 1.5-2.0 times lower than in eGFP/+ flies (Fig. 3). Therefore, strong activation of the reporter gene expression is enough to prevent heterochromatin repression. The position effect is observable in a wide range of expression levels (from 3 to 87 times over the background; Fig. 3) but disappears when the expression is ~150 times higher than the background. This indicates the existence of some threshold level of transcription above which the gene is resistant to heterochromatinization.
Interestingly, when using the GeneSwitch-based system (eGFP/In(2)A4, pAct(GS)/+ flies), no position effect was observed despite rather low levels of the reporter gene expression (Fig. 3). This observation can be possibly explained by the fact that GeneSwitch is under the Actin5C gene promoter, whereas the Gal4 of the GAL4-GAL80ts system is under the α-tubulin promotor. Although both promotors control housekeeping genes and provide high levels of transcription, α-tubulin promotor shows lower activity (compared to the Actin5C promotor) at the late embryonic stages and in the middle of metamorphosis at the pupal stage (FlyBase data). A higher level of expression of the GeneSwitch and, as a result, of the reporter eGFP gene at these stages can lead to the complete suppression of trans-inactivation.
Authors of [24] studied trans-inactivation of the UAS-eGFP reporter gene in the brownDominant system. In this case, the expression of the reporter gene was driven by the sources of GAL4 under the control of tissue-specific enhancers [24]. Unlike the results of our study, the degree of the UAS-eGFP trans-inactivation in [24] was not affected by the level of its expression. We believe that tissue-specificity and complex expression profiles of the GAL4 sources during development (see [24]) complicated the identification of correlations between the level of the reporter gene expression and the degree of its inactivation under the influence of heterochromatin.
The experimental system including the UAS-eGFP reporter gene and the system to control its expression using a combination of GAL4+GAL80ts can be used in the studies of interconnections between the chromatin structure and gene expression, e.g., for identification of critical steps in the formation of transcription-inactive chromatin structure in ontogenesis and understanding of how the changes in the gene expression influence this process. It would be interesting to study the competition between heterochromatin components and transcription factors for binding to the promoter and track the possible changes in the intranuclear reporter gene localization in the case of trans-inactivation.
In this work, the system of the regulated reporter gene expression was used for the first time to study heterochromatin repression. In the future, this system might be used to investigate the dynamics of interactions between the gene transcriptional machinery and its heterochromatin environment.
Acknowledgments
Authors are grateful to V. A. Gvozdev for valuable comments and help in the preparation of this manuscript and to P. G. Georgiev for kindly providing the plasmids and fly stocks.
This work was supported by the Russian Foundation for Basic Research (projects 17-04-01984 and 17-00-00282).
REFERENCES
1.Elgin, S. C., and Reuter, G. (2013) Position-effect
variegation, heterochromatin formation, and gene silencing in
Drosophila, Cold Spring Harb. Perspect. Biol., 5,
a017780.
2.Muller, H. J. (1930) Types of visible variations
induced by X-rays in Drosophila, J. Genet., 22,
299.
3.Spofford, J. B. (1976) Position-effect variegation
in Drosophila, in The Genetics and Biology of Drosophila,
Academic Press.
4.Weiler, K. S., and Wakimoto, B. T. (1995)
Heterochromatin and gene expression in Drosophila, Annu. Rev.
Genet., 29, 577-605.
5.Zhimulev, I. F., Belyaeva, E. S., Fomina, O. V.,
Protopopov, M. O., and Bolshakov, V. N. (1986) Cytogenetic and
molecular aspects of position effect variegation in Drosophila
melanogaster, Chromosoma, 94, 492-504.
6.Prokofyeva-Belgovskaya, A. A. (1945)
Heterochromatinization as a change of chromosome cycle, Zh. Obshch.
Biol., 4, 93-124 (Russian version); Prokofyeva-Belgovskaya,
A. A. (1947) Heterochromatinization as a change of chromosome cycle,
J. Genet., 48, 80-98 (English version).
7.Reuter, G., and Spierer, P. (1992) Position effect
variegation and chromatin proteins, Bioessays, 14,
605-612.
8.Singh, J., Freeling, M., and Lisch, D. (2008) A
position effect on the heritability of epigenetic silencing, PLoS
Genet., 4, e1000216.
9.Abramov, Y. A., Shatskikh, A. S., Maksimenko, O.
G., Bonaccorsi, S., Gvozdev, V. A., and Lavrov, S. A. (2016) The
differences between cis- and trans-gene inactivation
caused by heterochromatin in Drosophila, Genetics,
202, 93-106.
10.Lavrov, S. A., Shatskikh, A. S., Kibanov, M. V.,
and Gvozdev, V. A. (2013) Correlation on a cellular level of gene
transcriptional silencing and heterochromatin compartment dragging in
case of PEV-producing eu-heterochromatin rearrangement in Drosophila
melanogaster, Mol. Biol. (Moscow), 47, 286-291.
11.Abramov, Y. A., Kibanov, M. V., Gvozdev, V. A.,
and Lavrov, S. A. (2011) Genetic and molecular analysis of gene
trans-inactivation caused by homologous eu-heterochromatic chromosome
rearrangement in Drosophila melanogaster, Dokl. Biochem.
Biophys., 437, 72-76.
12.Csink, A. K., Bounoutas, A., Griffith, M. L.,
Sabl, J. F., and Sage, B. T. (2002) Differential gene silencing by
trans-heterochromatin in Drosophila melanogaster,
Genetics, 160, 257-269.
13.Csink, A. K., and Henikoff, S. (1996) Genetic
modification of heterochromatic association and nuclear organization in
Drosophila, Nature, 381, 529-531.
14.Talbert, P. B., Leciel, C. D., and Henikoff, S.
(1994) Modification of the Drosophila heterochromatic mutation
brownDominant by linkage alterations, Genetics,
136, 559-571.
15.Henikoff, S., and Dreesen, T. D. (1989)
Trans-inactivation of the Drosophila brown gene: evidence
for transcriptional repression and somatic pairing dependence, Proc.
Natl. Acad. Sci. USA, 86, 6704-6708.
16.Mcguire, S. E., Le, P. T., Osborn, A. J.,
Matsumoto, K., and Davis, R. L. (2003) Spatiotemporal rescue of memory
dysfunction in Drosophila, Science, 302,
1765-1768.
17.Fujimoto, E., Gaynes, B., Brimley, C. J., Chien,
C. B., and Bonkowsky, J. L. (2011) Gal80 intersectional regulation of
cell-type specific expression in vertebrates, Dev. Dyn.,
240, 2324-2334.
18.Osterwalder, T., Yoon, K. S., White, B. H., and
Keshishian, H. (2001) A conditional tissue-specific transgene
expression system using inducible GAL4, Proc. Natl. Acad. Sci.
USA, 98, 12596-12601.
19.Roman, G., Endo, K., Zong, L., and Davis, R. L.
(2001) P[Switch], a system for spatial and temporal control of gene
expression in Drosophila melanogaster, Proc. Natl.
Acad. Sci. USA, 98, 12602-12607.
20.Bischof, J., Maeda, R. K., Hediger, M., Karch,
F., and Basler, K. (2007) An optimized transgenesis system for
Drosophila using germ-line-specific phiC31 integrases, Proc.
Natl. Acad. Sci. USA, 104, 3312-3317.
21.Groth, A. C., Fish, M., Nusse, R., and Calos, M.
P. (2004) Construction of transgenic Drosophila by using the
site-specific integrase from phage phiC31, Genetics, 166,
1775-1782.
22.Venken, K. J., Schulze, K. L., Haelterman, N. A.,
Pan, H., He, Y., Evans-Holm, M., Carlson, J. W., Levis, R. W.,
Spradling, A. C., Hoskins, R. A., and Bellen, H. J. (2011) MiMIC: a
highly versatile transposon insertion resource for engineering
Drosophila melanogaster genes, Nat. Methods, 8,
737-743.
23.Vogel, M. J., Pagie, L., Talhout, W., Nieuwland,
M., Kerkhoven, R. M., and Van Steensel, B. (2009) High-resolution
mapping of heterochromatin redistribution in a Drosophila
position-effect variegation model, Epigenetics Chromatin,
2, 1.
24.Sage, B. T., Wu, M. D., and Csink, A. K. (2008)
Interplay of developmentally regulated gene expression and
heterochromatic silencing in trans in Drosophila,
Genetics, 178, 749-759.