Localization of Galectins within Glycocalyx
E. M. Rapoport1, V. K. Matveeva1, O. A. Vokhmyanina1, I. M. Belyanchikov1, H.-J. Gabius2,a, and N. V. Bovin1,b*
1Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, 117997 Moscow, Russia2Faculty of Veterinary Medicine, Institute of Physiological Chemistry, Ludwig-Maximilians-University, Veterinärstr. 13, D-80539 Munich, Germany
* To whom correspondence should be addressed.
Received December 27, 2017; Revision received February 9, 2018
Galectins are involved in various biological processes, e.g. cell–cell and cell–matrix adhesion and the transmission of cellular signals. Despite the diversity of functions, little is known about the nature of their physiological cognate ligands on the cell surface and the localization of galectins in the glycocalyx, although this information is important for understanding the functional activity of galectins. In this work, localization of endogenous and exogenously loaded galectins in the glycocalyx was studied. The following main conclusions are drawn: 1) galectins are not evenly distributed within the glycocalyx, they are accumulated in patches. Patching is not the result of a cross-linking of cellular glycans by galectins. Instead, patch-wise localization is the consequence of irregular distribution of glycans forming the glycocalyx; 2) galectins are accumulated in the inner zone of the glycocalyx rather than at its outer face or directly in vicinity of the cell membrane; 3) patches are not associated with cell rafts.
KEY WORDS: galectins, glycans, glycocalyx, mammalian cellDOI: 10.1134/S000629791806010X
Abbreviations: biot, biotin; BSA, bovine serum albumin; CRD, carbohydrate recognition domain; CTB, cholera toxin B-subunit; DSA, agglutinin from Datura stramonium; FITC, fluorescein isothiocyanate; Gal, galectin; PBS, phosphate-buffered saline, pH 7.2; PBA, phosphate-buffered saline, pH 7.2, containing 0.2% BSA; RPE, R-phycoerythrin; Str, streptavidin.
The term “glycocalyx” was introduced for the sugary coating
of cells (a “sweet husk”) [1]. In the
case of mammalian cells, it especially applies to a layer or surface
coat on the apical membrane of most cells [1-4], resulting from the organized meshwork of
membrane-bound and matrix component molecules, especially
glycoproteins, proteoglycans, and glycolipids. Changes in the
glycocalyx composition affect cell proliferation, differentiation,
mobility, and their capacity for adhesion. For example, decrease in
expression of O-glycans in acute monocytic leukemia THP-1 cells led to
increase in their adhesion to fibronectin [5];
aberrations in biosynthesis of N-glycans suppressed migration and
adhesion of MV3 human melanoma cells to extracellular matrix [6]; inhibition of biosynthesis of O-glycans in mucins
led to an increase in the adhesion of Staphylococcus aureus to
the epithelium and promoted the penetration causing bacterial infection
[7].
Electron transmission, electron paramagnetic resonance spectroscopy, and fluorescence microscopy, in particular confocal fluorescence laser scanning microscopy and two-photon laser scanning microscopy [3, 8-12], are widely used for the study of the structure of glycocalyx. Visualization of glycans by these methods is often carried out by using a panel of plant lectins, proteoglycan binding proteins, and antibodies to glycoproteins, sometimes in combination with application of glycosidases.
The thickness of the glycocalyx of animal cells usually varies from 30 to 500 nm, is minimal in erythrocytes (5-15 nm), whereas it can reach 4.5 μm in endothelial cells [1]. A glycocalyx is clearly revealed in separated cells at apical membranes, whereas it is difficult to distinguish the glycocalyx for the cells inside the tissue. The size of membrane proteins is about 10-12 nm, which is significantly less than the glycocalyx thickness. Thus, proteins are masked by glycans, likely preventing their interaction via protein–protein recognition with other cells. It is assumed that cell–cell adhesion is accompanied by mechanical deformation and decrease in the glycocalyx thickness at the place of contact of cells, this allowing the interaction between the proteins and their ligands [13].
The objects of our study are galectins, which mediate the processes of cell–cell and cell–extracellular matrix adhesion [14] and are involved in the transmission of intercellular signals. Despite the large variety of galectin functions, the structure of their counter-receptors and their particular glycan chains is known only in a small number of cases, such as ganglioside GM1 or the α5β1-integrin. Even less is known about the localization of galectins in the glycocalyx, though these data are helpful for the understanding of their functioning. In one case, it was shown that galectin-4 was detected in the basal layer of the membrane in T84 colon cancer cells and galectin-3 on the apical layer of the membrane [15]; galectins-8 and -9 binding sites were identified in the inner layer of the pellucida zone [16], and galectins-1 and -2 were seen to be accumulated in the upper layer of epithelium of trachea [17].
In general, galectins lack any structural motifs for anchoring in membranes. The only way of becoming a part of glycocalyx is binding with glycans. Almost all galectins have two carbohydrate recognition domains (CRDs). To function as a mediator of cell–cell adhesion, a galectin should anchor by one CRD to a glyco-molecule, whereas the second one must remain free for the binding to neighboring glycans. Of relevance, it was shown [18] that both CRDs are involved in the binding with glycocalyx, this seeming natural in a medium saturated with glycans. It is expected a priori that for an active interaction a galectin should be localized at the periphery of glycocalyx, where it is accessible for binding, but stronger (two-valent) binding is expected to proceed in the depth of glycocalyx.
The aim of this work was to reveal (i) where galectins are present in glycocalyx, i.e. whether in its upper or inner layer or both, and (ii) whether galectins are associated with glycocalyx regions that contact particular microdomains like membrane rafts.
MATERIALS AND METHODS
Reagents. Biotinylated lectin DSA-biot (agglutinin from Datura stramonium) was obtained from Vector Laboratories (USA); BSA, FITC-labeled streptavidin (Str-FITC), anti-rabbit IgG conjugated with RPE and FITC, FITC-labeled anti-mouse IgG, and Mowiol®4-88 (Mw = 31,000) were from Sigma (USA). Anti-rabbit IgG conjugated with Alexa Fluor®594 and cholera toxin B labeled with Alexa Fluor®488 were from Molecular Probes Invitrogen (USA). Culture medium RPMI-1640, fetal calf serum, glutamine, and antibiotic–antimycotic were obtained from Invitrogen (USA). The hybridoma line producing the Mac-2 monoclonal antibody (No. TIB-166) was obtained from ATCC (USA).
Recombinant Gal-1, -3, -4, -8, -9 (including biotinylated proteins) and anti-galectin antibodies were produced and tested as described previously [19-25].
Cell cultures. The following three human cell lines were tested: Raji (B-lymphoma, ATCC® CCL-86™), Jurkat (T-leukemia, ATCC® TIB-152™) and HT-29 (colon adenocarcinoma, ATCC® HTB-38™). The cells were cultured in RPMI-1640 medium supplemented with 10% fetal calf serum, 2 mM glutamine, and 50 µg/ml of antimycotic at 37°C in an atmosphere of 5% CO2.
Loading of cell surfaces with galectins. Cells were washed three times with PBA to deplete the solution of glycoproteins from the serum using centrifugation at 4°C. Aliquots of the cell suspension (2·105 cells in 100 μl of PBA) were incubated with 50 μl of galectin-containing solution (0.04 or 0.4 mg/ml in PBA) for 30 min at 4°C under gentle agitation on a shaker in the wells of U-shape plates (Nunc, Denmark). To study kinetics of galectin distribution in the glycocalyx (0.04 mg/ml), the plate was incubated for periods of 1, 5, 10, or 20 min.
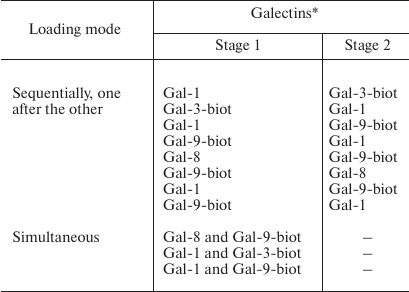
If two galectins were loaded on the cells, the following combinations were tested (table).
Galectins loading on Raji cells
* Concentration of galectins was 0.4 mg/ml.
Sample preparation for flow cytometry. Binding of galectins to cells in the presence of DSA. Cells loaded with Gal-1, -3, or -9 were incubated with DSA (0.01 mg/ml in PBA) at 4°C for 20 min. After that, the cells were washed by centrifugation and incubated with a solution of corresponding antibodies (0.02 mg/ml) in PBA for 20 min at 4°C. To detect fluorescence of cells, anti-rabbit (Gal-1 or Gal-9) or anti-mouse (Gal-3) IgG-FITC (dilution 1 : 50 in PBA) were added to the cell suspension followed by incubation for 20 min under the same conditions, washing three-times with PBA, transferring solution into tubes, and adding 2 ml of PBS. Flow cytometry was performed using a FACScan instrument (Becton-Dickinson, USA) equipped with Flowing Software [26] at 488 nm at room temperature. In each run, at least 5000 cells were analyzed.
Competition of galectins loaded on cells. Non-labeled galectins were detected with corresponding antibodies (0.02 mg/ml in PBA) by incubation for 20 min at 4°C. To determine fluorescence of cells, conjugates of anti-rabbit IgG-RPE (Gal-1 or Gal-8) were added (in dilution 1 : 50 in PBA). Biotinylated galectins (Gal-3, Gal-9) were detected with Str-FITC (dilution 1 : 50 in PBA). Cells were incubated for 20 min at 4°C with agitation on a shaker. The extent of binding of galectins to cells was analyzed as described above.
Sample preparation for confocal microscopy. Labeling of glycocalyx. Cells were fixed with 4% solution of formalin in PBS for 20 min at 37°C, followed by washing by centrifugation in PBS at 800 rpm for 3 min. To 50 μl of intact or fixed cells in suspension in PBA (1·106 cells/ml), 50 μl of DSA-biot-containing solution (0.01 mg/ml) was added. The cells were incubated for 20 min at 4°C, washed with PBA, and incubated with Str-FITC (dilution 1 : 50 in PBA) under the same conditions.
Labeling of galectins within glycocalyx of Raji cells. A solution of the corresponding antibodies (50 μl in PBA) was added into the wells of a plate containing Raji cells loaded with Gal-1 or Gal-9. To detect glycans, 50 μl of DSA-biot-containing solution (0.01 mg/ml in PBA) was added into wells. The plate was incubated for 20 min at 4°C with agitation. To detect galectins and bound DSA, conjugates of anti-rabbit IgG-Alexa Fluor®594 (dilution 1 : 200 in PBA) and Str-FITC (dilution 1 : 50 in PBA) were added to the wells of the plate, respectively.
Labeling of galectins in glycocalyx of HT-29 and Jurkat cells. The solution of antibodies (50 μl) to Gal-1 or Gal-9 and DSA-biot (50 μl) were added to the wells of a plate containing HT-29 or Jurkat (1·106 cells/ml) cells. The plate was incubated for 20 min at 4°C with agitation. DSA was detected as described above.
Labeling of Gal-9N. A 50-μl sample of solution of Gal-9N (0.04 mg/ml) was added into the wells of plates containing Raji and Jurkat cells. The plate was incubated for 20 min at 4°C under agitation. After that, the cells were washed with PBA as described above. To detect the galectin, the cells were incubated with rabbit antibodies against Gal-9 and then with anti-rabbit IgG-Alexa Fluor®594 (dilution 1 : 200 in PBA).
Labeling of galectins that were loaded on cells sequentially (one after the other) or simultaneously. Galectins were loaded on Raji cells as indicated in the table. To detect Gal-1 or Gal-8, the corresponding antibodies and anti-rabbit IgG-Alexa Fluor®594 were added to the wells. Gal-3-biot and Gal-9-biot were stained with Str-FITC.
Revealing of galectins in cellular rafts. To detect rafts, Raji cells were washed with PBS and incubated with cholera toxin B subunit conjugated with Alexa Fluor®488 (dilution of solution 1 : 5000 in PBS) on ice for 20 min. After that, the cells were washed with PBA and loaded with Gal-1, Gal-4, or Gal-9 as described above. The galectins were detected by their corresponding antibodies and anti-rabbit IgG-Alexa Fluor®594.
Confocal microscopy. The mixture (15 µl) containing 2.4 g of Mowiol®4-88, 6 g of glycerol, 6 ml of water, and 12 ml of 0.2 M Tris-HCl (pH 8.5) were dropped on slide followed by addition of cell suspension (10 μl). Images were obtained with an Eclipse TE-2000-E confocal microscope (Nikon, Japan). At least 10 randomly selected cells were analyzed in each experiment. Cell nuclei were stained with a solution of 4′,6-diamidino-2-phenylindole (DAPI), 1 μg/ml.
Analysis of images. Images were analyzed using programs ImageJ, Excel 2013 (Microsoft Office), and soft EZ-C1 (Nikon, Japan) supplied with the confocal microscope.
RESULTS AND DISCUSSION
The Raji cells were selected primarily because they do not express galectins on their surface, which has been demonstrated using antibodies to galectins [27]. First, we planned to determine whether galectins are evenly distributed throughout the entire glycocalyx or in restricted zones, and if in certain zones, then in which ones.
Staining of glycocalyx. To designate a glycocalyx area, we used agglutinin DSA [28, 29] that recognizes the Galβ-4GlcNAcβ disaccharide common for most cellular glycoproteins and glycolipids.
It was necessary to determine whether the addition of DSA to cells loaded with galectin would result in desorption of the latter, because the specificities of DSA and galectins overlap. To this end, Raji cells were loaded with proto- (Gal-1), chimera- (Gal-3), or tandem-repeat- (Gal-9) type galectins.
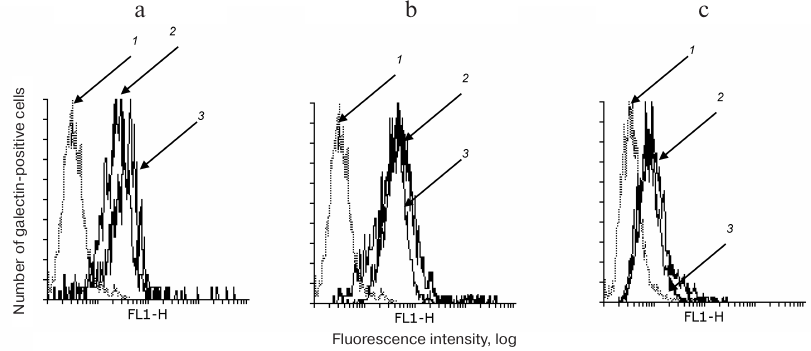
The binding of anti-galectin antibodies to cells in the presence or absence of DSA was comparable (Fig. 1, a-c), i.e. DSA did not displace the galectins.
Fig. 1. DSA does not displace galectins loaded on the cell. Gal-1 (a), Gal-3 (b), and Gal-9 (c) were loaded on Raji cells followed by incubation with DSA. After that, the cells were washed with PBA and incubated with antibodies to galectins as described in “Materials and Methods”. The logs of fluorescence intensity are plotted against cell number: 1) binding of cells with anti-rabbit or anti-mouse IgG-FITC; 2, 3) binding of galectin-loaded cells with antibodies in the presence or absence of DSA, correspondingly.
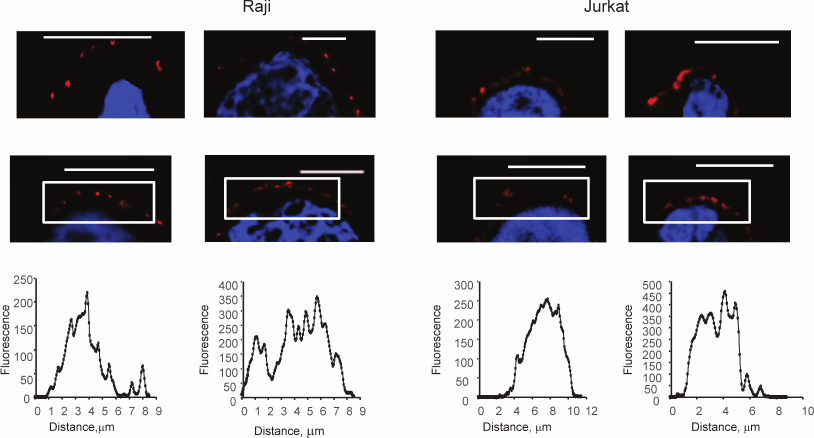
Then, the uniformity of the distribution of glycans was estimated; Raji, Jurkat, and HT-29 cells were studied. The last two were chosen because they express Gal-9 and Gal-1, respectively [30, 31].
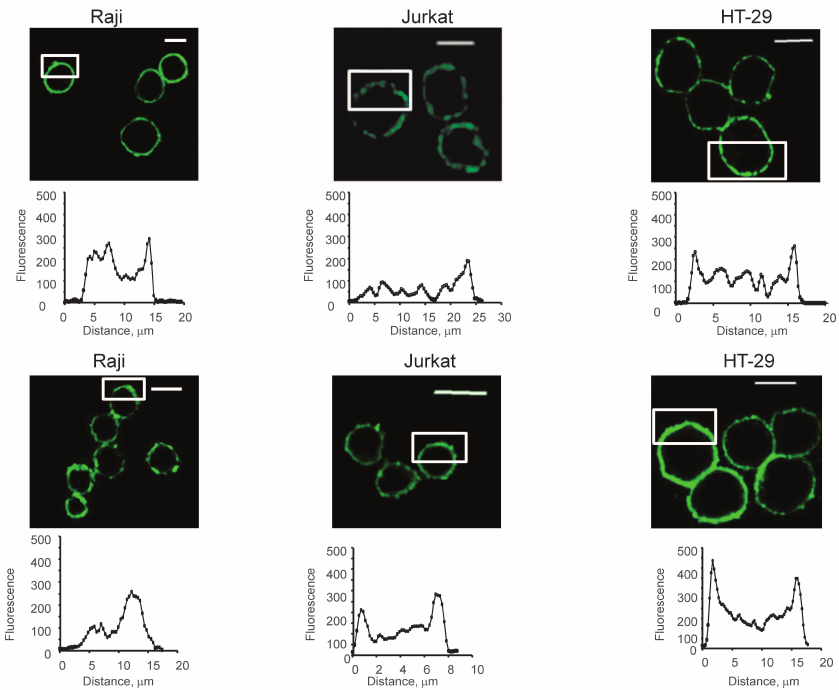
Both native (Fig. 2, upper panel) and fixed (by formalin) cells (Fig. 2, lower panel) were stained. As Fig. 2 shows, the colored zones have sizes from 2 to 5 μm. Uneven distribution of glycans was observed in both native and fixed cells. This led to the conclusion that the formation of the observed patches was not the result of cross-linking by two-subunit DSA.
Fig. 2. Visualization of glycocalyx of native (upper panel) and fixed (lower panel) cells, confocal microscopy, the bar corresponds to 10 μm. The cells were fixed with a solution of 4% formalin in PBS as described in “Materials and Methods”. To stain glycans, the cells were incubated with DSA-biot and Str-FITC (green color). The graphs show the distribution of fluorescence in the marked white rectangles areas; the y-axis shows fluorescence intensity; the x-axis – length of the marked area, μm.
Localization of galectins-1 and -9 in cellular glycocalyx. Unlabeled and biotinylated galectins were used in the work. In the first case, galectins were stained by antibodies, in the second by fluorescent streptavidin. Identical results were obtained for Gal-1 and Gal-9 loaded on Raji cells (not shown). Endogenous lectins can be revealed only by corresponding antibodies, so we used unlabeled proteins for loading on Raji cells, which were also visualized using corresponding antibodies.
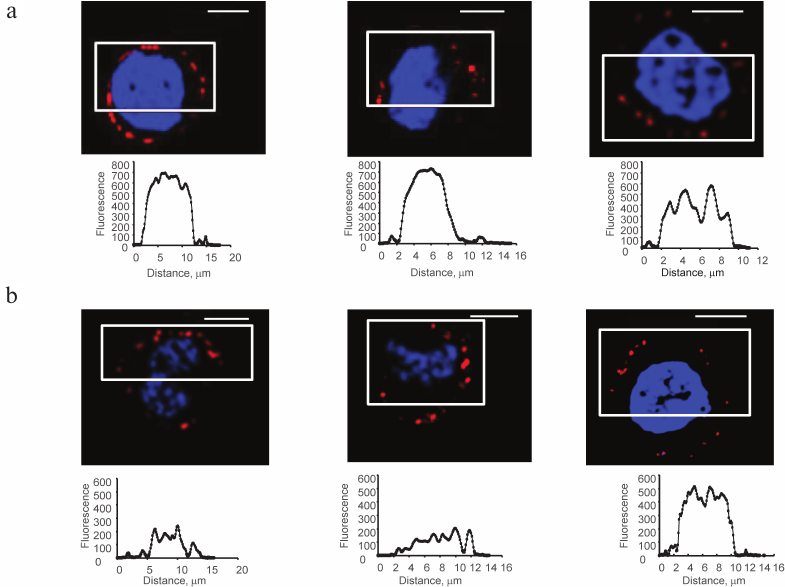
Galectins-1 and -9 exogenously loaded on Raji cells are localized in patches, their size varying from 2 to 8 μm (Fig. 3), the size of some patches being comparable with the thickness of the glycocalyx (Fig. 4, upper panel).
Fig. 3. Localization of Gal-1 (a) and Gal-9 (b) loaded on Raji cells, confocal microscopy, the bar corresponds to 6 μm. Raji cells loaded with Gal-1 or Gal-9 (0.04 mg/ml) were incubated with the corresponding antibodies and anti-IgG-Alexa Fluor®594 (red); nuclei are stained with DAPI (blue). In each case, three typical images are shown. The graphs (below) show the fluorescence distribution in the marked areas (white rectangles); the y-axis is fluorescence intensity; the x-axis is the size of the stained area, μm.
Fig. 4. Localization of Gal-1 and Gal-9 within glycocalyx of Raji (upper panel), HT-29 and Jurkat cells (lower panel), confocal microscopy (magnification ×600), the bar corresponds to 6 µm. Raji cells loaded by Gal-1 or Gal-9 in concentration of 0.4 or 0.04 mg/ml, as well as HT-29 and Jurkat cells expressing the same galectins endogenously were incubated with corresponding antibodies and IgG-Alexa Fluor®594 (red). Glycocalyx was detected by DSA (green) as described in “Materials and Methods”, nuclei are stained with DAPI (blue). The areas of galectin localization are indicated with arrows.
On HT-29 and Jurkat cells, endogenous Gal-1 and Gal-9 were seen to be present also in patches (Fig. 4, lower panel); there were no clearly visible individual features for the discussed galectins. Mostly, the patches are localized in the middle zone of the glycocalyx; tendency to localization near membranes was not observed (though it was expected for a galectin bound to glycolipid). Comparing to endogenous galectins, the total area of the stained zones and the size of single patches were bigger in the case of exogenously loaded proteins; this can be explained by the high (0.4 mg/ml) loading concentration of the galectins.
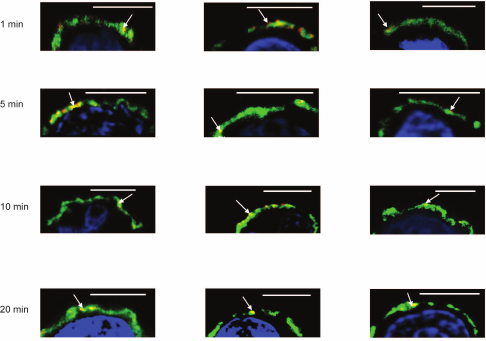
To reveal how fast galectins are accumulated into the glycocalyx layer, the kinetics of deposition was studied. Incubation of galectins with cells in the first 5 min led to association of the protein and its insertion into a depth of glycocalyx; in 20 min galectin was detected in the inner zone (Fig. 5).
Fig. 5. Kinetics of loading of Gal-1 onto Raji cells, confocal microscopy, bar corresponds to 6 μm. Gal-1 (0.04 mg/ml) was incubated with the cells for 1, 5, 10, or 20 min. After washing, the cells were incubated with corresponding antibodies and IgG-Alexa Fluor®594 (red). Glycocalyx was stained with DSA (green) as described in “Materials and Methods”; nuclei are stained with DAPI (blue). In each case, three typical images are shown. The areas of galectin localization are indicated with arrows.
Localization of galectins loaded together. Typically, several galectins are expressed on the cell. They can compete with each other for binding to glycans [32, 33]. We studied the localization of two galectins loaded onto cells simultaneously or one by one (according to the scheme given in the table).
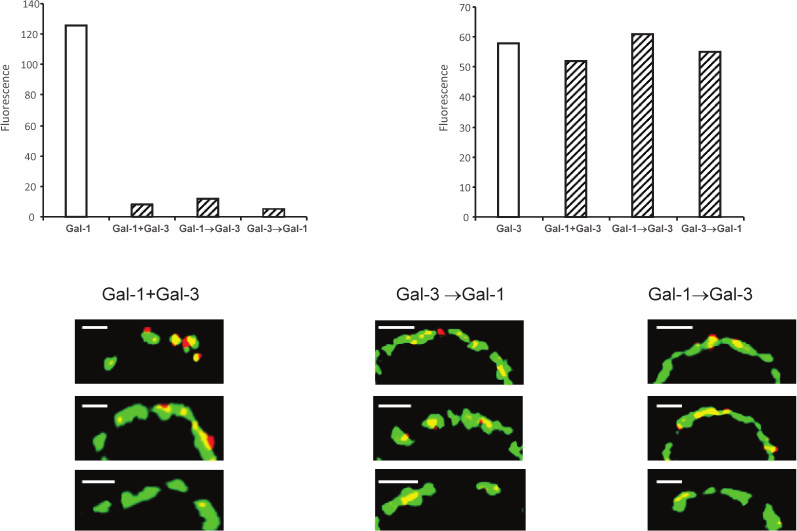
Unlike the divalent proto- or tandem-type galectins (which have two CRDs) [32], Gal-3 has formally only one CRD, but its N-domain is able to form multimers, presumably pentamers in the presence of a suited carbohydrate “grid” [34]. Due to potentially higher affinity of a multimeric galectin, it is assumed that Gal-3 will displace bivalent galectin. Indeed, the fluorescence intensity of Gal-1 in the absence of Gal-3 proved to be higher (Fig. 6, upper panel). Gal-1 did not interfere with Gal-3 binding. As a rule, Gal-1 and Gal-3 are colocalized (Fig. 6, bottom panel); however, in some cases Gal-1 is localized separately. This is understandable; their specificity to the glycans is only partly overlapping.
Fig. 6. Localization of galectins-1 and -3 on Raji cells. Gal-3-biot and non-labeled Gal-1 were loaded on the cells simultaneously (indicated as “+” on the x-axis) or sequentially, one after the other (indicated as “→” on the x-axis) as described in the table; Gal-1 was detected with corresponding antibodies and IgG-Alexa®594 (red), Gal-3-biot – with Str-FITC (green). The bar corresponds to 2 µm. The binding of galectins to cells analyzed by flow cytometry is shown on the upper panel; fluorescence was calculated as [(Fi/F0)·100] – 100, where Fi and F0 – fluorescence intensity of galectin-loaded or galectin-free cells after incubation with antibodies to Gal-1 or Str-FITC, correspondingly. Three typical images are presented in each case.
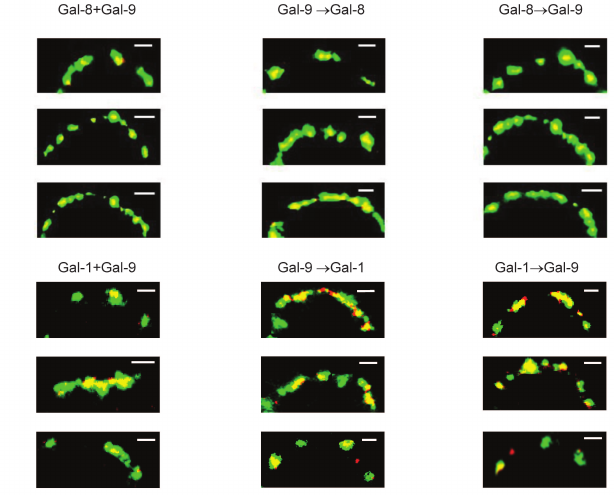
Regardless of the loading regimen, the level of binding of divalent Gal-1 and Gal-8 (or -8 and -9) in the case of simultaneous loading remained the same as in the case of loading only one protein (results not shown). Colocalization of Gal-8 and Gal-9 was observed (Fig. 7). Gal-1, loaded on the cells before or after Gal-9, was located in the glycocalyx together with Gal-9 or separate from it (Fig. 7). Their localization was observed in the case of simultaneous loading.
Fig. 7. Localization of galectins-8 and -9 (upper panel), or -1 and -9 (lower panel) on Raji cells. Galectins were loaded simultaneously or sequentially (one by one) as described in the table; Gal-1 and Gal-8 were stained with corresponding antibodies and IgG-Alexa®594 (red), Gal-9-biot – with Str-FITC (green). The bar corresponds to 2 μm. Three typical images are shown.
Presentation of galectins is organized in patches, as shown in Figs. 3-7. Presumably, patching is the result of cross-linking carbohydrate ligands by galectins, initially situated far from each other [35]. To verify this, the localization of galectins containing only a single CRD was studied.
Localization of Gal-9N. Unlike the full-length proteins, single CRDs are monovalent: so, they cannot cause cross-linking. We have studied the distribution of the N-domain of Gal-9 (Gal-9N) in the glycocalyx of Raji and Jurkat cells. As shown on the histograms of fluorescent distribution (Fig. 8) Gal-9N similar to full-size bivalent protein localized in patches, and the patches varied by their size.
Fig. 8. Localization of Gal-9N into glycocalyx of Raji and Jurkat cells, confocal microscopy (magnification ×600). The bar corresponds to 6 µm. Gal-9N (0.04 mg/ml) was loaded onto the cells followed by incubation with corresponding antibodies and IgG-Alexa®594 (red); nuclei are stained in blue. The graphs show the distribution of fluorescence in the marked white rectangles areas; the y-axis shows fluorescence intensity; the x-axis shows the size of the marked area, µm. Four typical images are presented in each case.
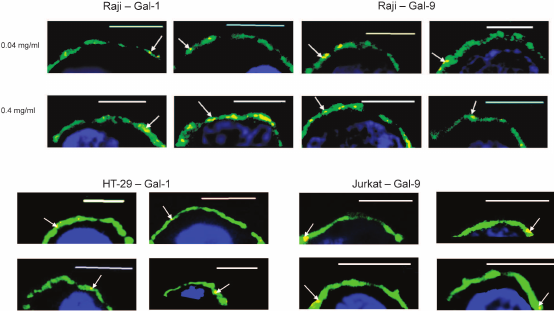
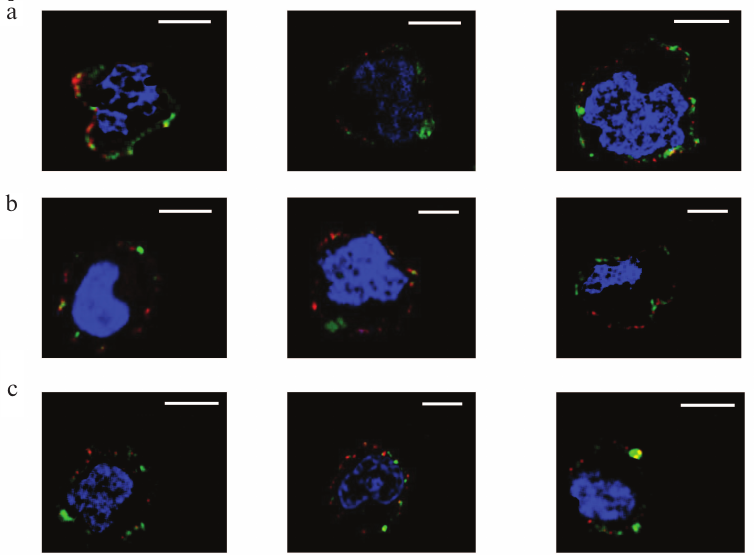
Localization of galectins as related to rafts. Rafts are well known as well-structured microdomains of the plasma membrane [36]. The rafts are constituted by glycosphingolipids and glycoproteins [36, 37], some of which like ganglioside GM1 being capable of binding galectins [38-42]. The repertoire of these glycoconjugates in rafts and outside of the rafts is significantly different. We studied the selectivity of binding of galectins-1, -4, and -9 to rafts in comparison with neighboring glycocalyx regions. Cholera toxin B-subunit (CTB), which binds ganglioside GM1, an obligatory part of rafts [37], has been used for flagging the rafts. The results of the simultaneous detection of rafts and galectins (Gal-1, Gal-4, or Gal-9) using the appropriate antibodies are shown in Fig. 9. Although sporadic colocalization has been observed, it is likely to be of a statistical, not causal nature, i.e. there is no explicit direct or indirect association of galectins and raft glycoconjugates.
Fig. 9. Revealing of Gal-1 (a), or Gal-4 (b), or Gal-9 (c) and rafts on Raji cells by confocal microscopy. The bar corresponds to 6 µm. Rafts were visualized with CTB-FITC (green), galectins were detected with the corresponding antibodies and IgG-Alexa®594 (red), and nuclei are stained in blue. Three typical images are presented in each case.
Galectins do not contain an N-terminal signal peptide sequence and are translocated to cytoplasm in a nonclassical way without passage through endoplasmic reticulum and Golgi apparatus [43]. These proteins can be exported to the surface of the cell in exosomes, formed by protrusion of the membrane region in the place of accumulation of galectin. At present, the mechanism of the transfer is unknown. As mentioned above, galectins lack a special domain for anchoring in the membrane, so their retention on the cell surface is due to the binding to glycocalyx. In this work, we studied the localization in the glycocalyx of galectins, both endogenous and loaded artificially.
To function as a mediator of cell–cell adhesion, i.e. to bind to glycans of adjacent cells, it is logical to assume that a galectin should be localized in the upper layer of the glycocalyx. On the contrary, considering the pathway of galectins from the cytosol through the glycocalyx (packed with galectin ligands), one can expect their presence in the lower layer of the glycocalyx. In fact, these assumptions have not been confirmed: both endogenous and artificially loaded galectins are detected in the depth of the glycocalyx. In the case of exogenous loading, reaching the depth proceeds rapidly, within a few minutes. According to a study carried out by electron microscopy, glycocalyx is a quasi-ordered 3D-structure having distances up to 100 nm between glycans organized in clusters [44, 45]. For a galectin, a rather small-sized protein (10-12 nm), it will likely not be difficult to penetrate this “net”. An answer as to why galectin is present deep in the glycocalyx can be obtained by the determination of the 3D glycome of the glycocalyx, i.e. when spatial mapping of ligands of the galectins is carried out.
In summary, galectins accumulate in patches, the reason being irregular distribution of glycans in glycocalyx. Bivalent galectins do not strictly compete with each other for binding to glycans in glycocalyx, typically they are localized together. Accumulation of galectins in rafts was not observed even in the case of Gal-4, for which the possibility of binding not only to glycolipids but also to cholesterol was demonstrated [39].
Acknowledgments
The authors are thankful to Alexander Tuzikov and Elena Kovalenko for valuable discussion of the material of this study.
This study was supported financially by the Russian Foundation for Basic Research (grant Nos. 13-04-00096, 16-04-01084, and 16-54-76023).
REFERENCES
1.Bennett, H. S. (1963) Morphological aspects of
extracellular polysaccharides, J. Histochem. Cytochem.,
11, 14-23.
2.Martins, M. F., and Barios, V. A. (2002) Glycocalyx
in lung epithelial cells, Int. Rev. Cytol., 216,
131-173.
3.Weinbaum, S., Tarbell, J. M., and Damiano, E. R.
(2007) The structure and function of endothelial glycocalyx layer,
Annu. Rev. Biomed. Eng., 9, 121-167.
4.Kovbasnjuk, O. N., and Spring, K. R. (2000) The
apical membrane glycocalyx of MDCK cells, J. Membr. Biol.,
176, 19-29.
5.Sabri, S., Soler, M., Foa, C., Pierres, A.,
Benoliel, A.-M., and Bongrand, P. (2000) Glycocalyx modulation is a
physiological means of regulating cell adhesion, J. Cell Sci.,
113, 1589-1600.
6.Krahling, H., Mally, S., Eble, J. A., Noel, J.,
Schwab, A., and Stock, Ch. (2009) The glycocalyx maintains a cell
surface pH nanoenvironment crucial for integrin-mediated migration of
human melanoma cells, Eur. J. Physiol., 458,
1069-1083.
7.Guzman-Aranguez, A., and Argueso, P. (2010)
Structure and biological roles of mucin-type O-glycans at the ocular
surface, Ocul. Surf., 8, 8-17.
8.Chappell, D., Jacob, M., Paul, O., Rehm, M.,
Welsch, U., Stoeckelhuber, M., Conzen, P., and Becker, B. F. (2009) The
glycocalyx of the human umbilical vein endothelial cell, Circ.
Res., 104, 1313-1317.
9.Reitsma, S., Oude Egbrink, M. G. A., Vink, H., van
den Berg, B. M., Passos, V. L., Engels, W., Slaaf, D. W., and van
Zandvoort, M. A. M. J. (2011) Endothelial glycocalyx structure in the
intact carotid artery: a two-photon laser scanning microscopy study,
J. Vasc. Res., 48, 297-306.
10.Reitsma, S., Dick, W., Slaaf, D. W., Vink, H.,
van Zandvoort, M. A. M. J., and Oude Egbrink, M. G. A. (2007) The
endothelial glycocalyx: composition, functions, and visualization,
Eur. J. Physiol., 454, 345-359.
11.Strancar, J., Schara, M., and Pefiar, S. (2003)
New EPR method for cellular surface characterization, J. Membr.
Biol., 193, 15-22.
12.Megens, R. T. A., Reitsma, S., Schiffers, P. H.
M., Hilgers, R. H. P., DeMey, J. G. R., Slaaf, D. W., Oude Egbrink, M.
G. A., and van Zandvoort, M. A. M. J. (2007) Two-photon microscopy of
vital murine elastic and muscular arteries, J. Vasc. Res.,
44, 87-98.
13.Paszek, M. J., Boettiger, D., Weaver, V. M.,
Daniel, A., and Hammer, D. A. (2009) Integrin clustering is driven by
mechanical resistance from the glycocalyx and the substrate,
PLoS, http://dx.doi.org/10.1371/journal.pcbi.1000604.
14.Rapoport, E. M., and Bovin, N. V. (2015)
Specificity of human galectins on cell surfaces, Biochemistry
(Moscow), 80, 1010-1022.
15.Huflejt, M. E., Jordan, E. T., Gitt, M. A.,
Barondes, S. H., and Leffler, H. (1997) Strikingly different
localization of galectin-3 and galectin-4 in human colon adenocarcinoma
T84 cells. Galectin-4 is localized at sites of cell adhesion, J.
Biol. Chem., 272, 14294-14303.
16.Habermann, F. A., Andre, S., Kaltner, H., Kubler,
D., Sinowatz, F., and Gabius, H.-J. (2011) Galectins as tools for
glycan mapping in histology: comparison of their binding to the bovine
zona pellucida by confocal laser scanning microscopy, Histochem.
Cell Biol., 135, 539-552.
17.Grendel, T., Sokolsky, J., Vascakova, A., Hudak,
V., Chovanec, M., Sabol, F., Andre, S., Kaltner, H., Gabius, H.-J.,
Frankovicova, M., Lences, P., Betka, J., Smetana, K., and Gal, P.
(2012) Early stages of trachea healing process:
(immuno/lectin)histochemical monitoring of selected markers and
adhesion/growth-regulatory endogenous lectins, Folia Biol.
(Praha), 58, 135-143.
18.Vokhmyanina, O. A., Rapoport, E. M., Andre, S.,
Severov, V. V., Ryzhov, I. M., Pazynina, G. V., Korchagina, E. Ju.,
Gabius, H.-J., and Bovin, N. V. (2012) Comparative study of the glycan
specificities of cell-bound human tandem repeat-type galectins-4, -8
and -9, Glycobiology, 22, 1207-1217.
19.Gabius, H.-J. (1990) Influence of type of linkage
and spacer on the interaction of β-galactoside-binding proteins
with immobilized affinity ligands, Anal. Biochem., 189,
91-94.
20.Kopitz, J., Andre, S., von Reitzenstein, C.,
Versluis, K., Kaltner, H., and Pieters, R. J. (2003) Homodimeric
galectin-7 (p53-induced gene 1) is a negative growth regulator for
human neuroblastoma cells, Oncogene, 22, 6277-6288.
21.Lensch, M., Lohr, M., Russwurm, R., Vidal, M.,
Kaltner, H., Andre, S., and Gabius, H.-J. (2006) Unique sequence and
expression profiles of rat galectins-5 and -9 as a result of
species-specific gene divergence, Int. J. Biochem. Cell Biol.,
38, 1741-1758.
22.Andre, S., Sansone, F., Kaltner, H., Casnati, A.,
Kopitz, J., Gabius, H. J., and Ungaro, R. (2008) Calix[n]arene-based
glycoclusters: bioactivity of thiourea-linked galactose/lactose
moieties as inhibitors of binding of medically relevant lectins to a
glycoprotein and cell-surface glycoconjugates and selectivity among
human adhesion/growth-regulatory galectins, ChemBioChem,
9, 1649-1661.
23.Solis, D., Mate, M. J., Lohr, M., Ribeiro, J. P.,
Lopez-Merino, L., Andre, S., Buzamet, E., Canada, J., Kaltner, H.,
Lensch, M., Ruiz, F. M., Haroske, G., Wollina, U., Kloor, M., Kopitz,
J., Saiz, J. L., Menendez, M., Jimenez-Barbero, J., Romero, A., and
Gabius, H.-J. (2010) N-domain of human
adhesion/growth-regulatory galectin-9: preference for distinct
conformers and non-sialylated N-glycans and detection of ligand-induced
structural changes in crystal and solution, Int. J. Biochem. Cell
Biol., 42, 1019-1029.
24.Andre, S., Kojima, S., Yamazaki, N., Fink, C.,
Kaltner, H., Kayser, K., and Gabius, H.-J. (1999) Galectins-1 and -3
and their ligands in tumor biology. Non-uniform properties in
cell-surface presentation and modulation of adhesion to matrix
glycoproteins for various tumor cell lines, in biodistribution of free
and liposome-bound galectins and in their expression by breast and
colorectal carcinomas with/without metastatic propensity, J. Cancer
Res. Clin. Oncol., 125, 461-474.
25.Goehler, A., Andre, S., Kaltner, H., Sauer, M.,
Gabius, H.-J., and Doose, S. (2010) Hydrodynamic properties of human
adhesion/growth-regulatory galectins studied by fluorescence
correlation spectroscopy, Biophys. J., 98, 3044-3053.
26.http://flowingsoftware.btk.fi
27.Rapoport, E. M., Matveeva, V. K., Kaltner, H.,
Andre, S., Vokhmyanina, O. A., Pazynina, G. V., Severov, V. V., Ryzhov,
I. M., Korchagina, E. Y., Belyanchikov, I. M., Gabius, H.-J., and
Bovin, N. V. (2015) Comparative lectinology: delineating
glycan-specificity profiles of the chicken galectins using
neoglycoconjugates in a cell assay, Glycobiology, 25,
726-734.
28.Nishimoto, K., Tanaka, K., Murakami, T.,
Nakashita, H., Sakamoto, H., and Oguri, S. (2015) Datura
stramonium agglutinin: cloning, molecular characterization and
recombinant production in Arabidopsis thaliana,
Glycobiology, 25, 157-169.
29.Saitoh, O., Wang, W. C., Lotan, R., and Fukuda,
M. (1992) Differential glycosylation and cell surface expression of
lysosomal membrane glycoproteins in sublines of a human colon cancer
exhibiting distinct metastatic potentials, J. Biol. Chem.,
267, 5700-5711.
30.Chabot, S., Kashio, Y., Seki, M., Shirato, Y.,
Nakamura, K., Nishi, N., Nakanura, T., Matsumoto, R., and Hirashima, M.
(2002) Regulation of galectin-9 expression and release in Jurkat T cell
line cells, Glycobiology, 12, 111-118.
31.Satelli, A., Rao, P. S., Gupta, P. K., Lockman,
P. R., Srivenugopal, K. S., and Rao, U. S. (2008) Varied expression and
localization of multiple galectins in different cancer cell lines,
Oncol. Rep., 19, 587-594.
32.Kaltner, H., Toegel, S., Caballero, G. G.,
Manning, J. C., Ledeen, R. W., and Gabius, H.-J. (2017) Galectins:
their network and roles in immunity/tumor growth control, Histochem.
Cell Biol., 147, 239-256.
33.Thijssen, V. L., Heusschen, R., Caers, J., and
Griffioen, A. W. (2015) Galectin expression in cancer diagnosis and
prognosis: a systematic review, Biochim. Biophys. Acta,
1855, 235-247.
34.Ahmad, N., Gabius, H.-J., Andre, S., Kaltner, H.,
Sabesan, S., Roy, R., Liu, B., Macaluso, F., and Brewer, C. F. (2004)
Galectin-3 precipitates as a pentamer with synthetic multivalent
carbohydrates and forms heterogeneous cross-linked complexes, J.
Biol. Chem., 279, 10841-10847.
35.Brewer, C. F. (2002) Binding and cross-linking of
galectins, Biochim. Biophys. Acta, 1572, 255-282.
36.Levental, I., and Veatch, S. L. (2016) The
continuing mystery of lipid rafts, J. Mol. Biol., 428,
4749-4764.
37.Nichols, B. J. (2003) GM1-containing lipid rafts
are depleted within clathrin-coated pits, Curr. Biol., 13,
686-690.
38.Boscher, C., Zheng, Yu. Z., Lakshminarayan, R.,
Johannes, L., Dennis, J. W., Foster, L. J., and Nabi, I. R. (2012)
Galectin-3 protein regulates mobility of N-cadherin and GM1 ganglioside
at cell–cell junctions of mammary carcinoma cells, J. Biol.
Chem., 287, 32940-32952.
39.Stechly, L., Morelle, W., Dessein, A. F., Andre,
S., Grard, G., Trinel, D., Dejonghe, M. J., Leteurtre, E., Drobecq, H.,
Trugnan, G., Gabius, H.-J., and Huet, G. (2009) Galectin-4-regulated
delivery of glycoproteins to the brush border membrane of
enterocyte-like cells, Traffic, 10, 438-450.
40.Delacour, D., Gouyer, V., Zanetta, J. P.,
Drobecq, H., Leteurtre, E., Grard, G., Moreau-Hannedouche, O., Maes,
E., Pons, A., Andre, S., Le Bivic, A., Gabius, H.-J., Manninen, A.,
Simons, K., and Huet, G. (2005) Galectin-4 and sulfatides in apical
membrane trafficking in enterocyte-like cells, J. Cell Biol.,
169, 491-501.
41.Tanikawa, R., Tanikawa, T., Okada, Y., Nakano,
K., Hirashima, M., Yamauchi, A., Hosokawa, R., and Tanaka, Y. (2008)
Interaction of galectin-9 with lipid rafts induces osteoblast
proliferation through the c-Src/ERK signaling pathway, J. Bone Min.
Res., 23, 278-286.
42.Ideo, H., Seko, A., and Yamashita, K. (2007)
Recognition mechanism of galectin-4 for cholesterol 3-sulfate, J.
Biol. Chem., 282, 21081-21089.
43.Nikel, W. (2003) The mystery of nonclassical
protein secretion. A current view on cargo proteins and potential
export routes, Eur. J. Biochem., 270, 2109-2119.
44.Squire, J. M., Chew, M., Nneji, G., Neal, Ch.,
Barry, J., and Michel, Ch. (2001) Quasi-periodic substructure in the
microvessel endothelial glycocalyx: a possible explanation for
molecular filtering? J. Str. Biol., 136, 239-255.
45.Arkill, K. P., Neal, C. R., Mantell, J. M.,
Michel, C. C., Qvortrup, K., Rostgaard, J., Bates, D. O., Knupp, C.,
and Squire, J. M. (2012) 3D reconstruction of the glycocalyx structure
in mammalian capillaries using electron tomography,
Microcirculation, 19, 343-351.