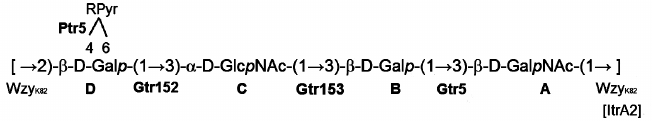Structure of the K82 Capsular Polysaccharide from Acinetobacter baumannii LUH5534 Containing a d-Galactose 4,6-Pyruvic Acid Acetal
A. A. Kasimova1,2,a*, J. J. Kenyon3,4,b, N. P. Arbatsky1, A. S. Shashkov1, A. V. Popova5,6,c, Y. A. Knirel1,d, and R. M. Hall3,7,e
1Zelinsky Institute of Organic Chemistry, Russian Academy of Sciences, 119991 Moscow, Russia2Higher Chemical College of the Russian Academy of Sciences, D. I. Mendeleev University of Chemical Technology of Russia, 125047 Moscow, Russia
3School of Molecular Bioscience, The University of Sydney, Sydney, NSW 2006, Australia
4Institute of Health and Biomedical Innovation, School of Biomedical Sciences, Faculty of Health, Queensland University of Technology, Brisbane, QLD 4059, Australia
5Moscow Institute of Physics and Technology, 141700 Dolgoprudny, Moscow Region, Russia
6State Research Center for Applied Microbiology and Biotechnology, 142279 Obolensk, Moscow Region, Russia
7School of Life and Environmental Sciences, The University of Sydney, Sydney, NSW 2006, Australia
* To whom correspondence should be addressed.
Received January 24, 2018; Revision received March 1, 2018
Type K82 capsular polysaccharide (CPS) was isolated from Acinetobacter baumannii LUH5534. The structure of a linear tetrasaccharide repeating unit of the CPS was established by sugar analysis along with one- and two-dimensional 1H and 13C NMR spectroscopy. Proteins encoded by the KL82 capsule gene cluster in the genome of LUH5534 were assigned to roles in the synthesis of the K82 CPS. In particular, functions were assigned to two new glycosyltransferases (Gtr152 and Gtr153) and a novel pyruvyltransferase, Ptr5, responsible for the synthesis of d-galactose 4,6-(R)-pyruvic acid acetal.
KEY WORDS: Acinetobacter baumannii, capsular polysaccharide structure, pyruvic acid acetal, K locus, genetics of capsule biosynthesisDOI: 10.1134/S0006297918070064
Abbreviations: COSY, correlation spectroscopy; CPS, capsular polysaccharide; HMBC, heteronuclear multiple-bond correlation; HSQC, heteronuclear single-quantum coherence; ROESY, rotating-frame nuclear Overhauser effect spectroscopy; TOCSY, total correlation spectroscopy; Und-P, undecaprenyl phosphate.
Acinetobacter baumannii has become one of the most widespread
agents causing nosocomial infections. Currently, the majority of A.
baumannii isolates display resistance to almost all therapeutically
suitable antibiotics [1].
A thick polysaccharide capsule (CPS) surrounds the A. baumannii cell and protects the bacterium from the action of immune system components, as well as disinfectants, desiccation, and some antimicrobial compounds. The CPS is composed of many oligosaccharide repeats (K units) and is characterized by high structural diversity mainly due to variability of the gene content at the chromosomal K locus (KL) driving the CPS biosynthesis. To date, more than 120 various gene clusters at the K locus have been recognized ([2, 3]; J. J. Kenyon, unpublished data). The chemical structures for about 40 different A. baumannii CPSs have been established ([4-8] and references cited in [4]) and form the basis for classification of strains of these bacteria. Most of these structures are consistent with putative functions of CPS synthesis genes located at the K locus.
In this work, we report on the structure and gene cluster of the CPS that is specific for A. baumannii LUH5534. The CPS gene cluster originally designated PSgc3 has been sequenced and genes have been annotated by comparison with sequences in available databases [3]. Here, we re-annotated this gene cluster using the established nomenclature system for A. baumannii CPS [2] and rename it KL82. Accordingly, the CPS of A. baumannii LUH5534 was assigned to the K82 type.
MATERIALS AND METHODS
Cultivation of bacteria. Acinetobacter baumannii strain LUH5534 was cultivated in 2×TY media for 24 h. Cells were harvested by centrifugation (10,000g, 20 min), washed with phosphate-buffered saline (pH 7.4), suspended in a 7 : 3 acetone–water mixture (v/v), precipitated, and dried.
Isolation of capsular polysaccharide. CPS was isolated by phenol–water extraction [9] of bacterial cells (1.12 g). The extract was dialyzed without layer separation, freed from insoluble contaminations by centrifugation, and proteins and nucleic acids were precipitated by aqueous 50% CCl3CO2H at 4°C. After dialysis against distilled water and centrifugation, the supernatant was fractionated by gel-permeation chromatography on a column (60 × 3.5 cm) of Sephadex G-50 in 0.1% CH3CO2H, and a high-molecular-mass fraction was freeze-dried to give a CPS preparation (65 mg).
Deacetalation. A CPS sample (16 mg) was treated with 4% CH3CO2H (1.5 ml, 100°C, 7 h), the products were fractionated by gel-permeation chromatography on a column (110 × 2 cm) of Sephadex G-25 in water, and a high-molecular-mass fraction (6 mg) was subjected to anion-exchange chromatography on a column (8 × 1 cm) of DEAE-Toyopearl TSK 650M in water to give a neutral modified polysaccharide (MPS) (1.5 mg).
Monosaccharide analysis. A CPS sample (0.5 mg) was hydrolyzed with 2 M CF3CO2H (120°C, 2 h). Monosaccharides were analyzed by GLC as the alditol acetates on a Maestro (Agilent 7820) chromatograph (Interlab, Russia) equipped with an HP-5 column (0.32 mm × 30 m) using a temperature program of 160°C (1 min) to 290°C at 7°C/min.
NMR spectroscopy. Samples were deuterium-exchanged by freeze-drying from 99.9% D2O and then examined as solutions in 99.95% D2O. Sodium 3-trimethylsilylpropanoate-2,2,3,3-d4 (δH 0, δC —1.6) was used as internal reference for calibration. NMR spectra were recorded on a Bruker Avance II 600 MHz spectrometer (Germany) at 60°C using standard Bruker software and Bruker TopSpin 2.1 program to acquire and process the NMR data. A 60-ms MLEV-17 spin-lock time and a 150-ms mixing time were used in TOCSY and ROESY experiments, respectively. A 60-ms delay was used for evolution of long-range couplings to optimize 1H,13C HMBC experiments for the coupling constant of JH,C 8 Hz.
Bioinformatics. The BLASTp database (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome) and the Pfam database (http://pfam.xfam.org/) were used to assign encoded proteins to CPS biosynthesis roles.
RESULTS AND DISCUSSION
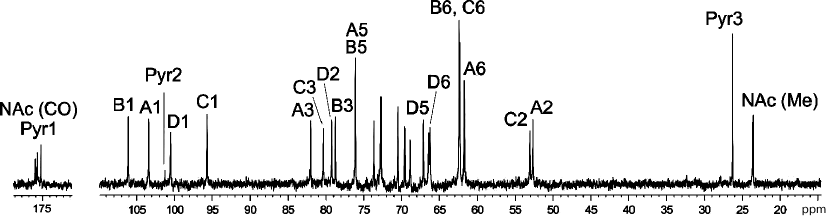
A crude CPS preparation was isolated from A. baumannii LUH5534 by phenol–water extraction. Sugar analysis of the CPS by GLC of the acetylated alditols revealed the presence of d-Gal, d-GlcNAc, and d-GalNAc in the ratio ~0.6 : 1 : 0.5, respectively. The CPS was studied by NMR spectroscopy, including one-dimensional 1H NMR and 13C NMR (Fig. 1) spectra and two-dimensional 1H,1H COSY, TOCSY, ROESY, 1H,13C HSQC, and HMBC experiments. Four sugar spin systems were identified, including those for β-GalpNAc (unit A), α-GlcpNAc (unit C), and two β-Galp residues (units B and D), all monosaccharides being in the pyranose form. The assigned 1H and 13C NMR chemical shifts of the CPSs are tabulated in Table 1. Therefore, the CPS has a tetrasaccharide K unit. Relatively large J1,2 coupling constants of 7-8 Hz indicated that the Galp and GalpNAc residues were β-linked, whereas an α-linked GlcpNAc was characterized by a smaller coupling constant (J1,2 < 4 Hz).
Table 1. 1H and 13C
NMR chemical shifts (δ, ppm) in K82 CPS from A. baumannii
LUH55534
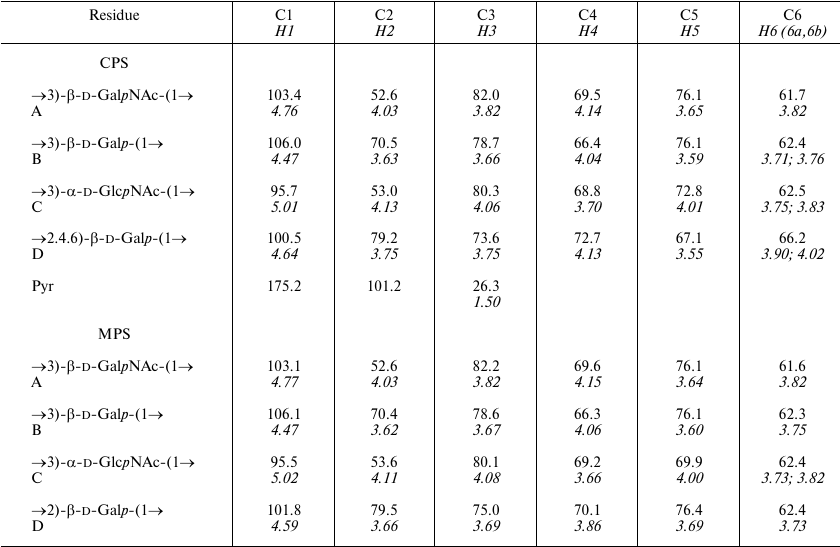
Notes: 1H NMR chemical shifts are italicized. Chemical shifts
for N-acetyl groups are: δC 23.4-23.6 (Me) and
175.2-176.0 (CO), δH 2.01-2.13. MPS, modified
polysaccharide.
Fig. 1. 13C NMR spectrum of the K82 CPS from A. baumannii LUH5534. Numbers refer to carbons in Pyr and sugar residues denoted by letters as shown in Table 1 and Fig. 2.
The NMR spectra also showed signals for a pyruvic acid acetal (Pyr). They were identified by C/H correlations in the 1H,13C HSQC spectrum (Table 1) and C1/H3 and C2/H3 correlations at δ 175.0/1.51 and 101.2/1.51, respectively, in the 1H,13C HMBC spectrum.
Low-field positions at δ 82.0, 78.7, 80.3 and 79.2 of the signals for C3 of units A, B, and C, and C2 of unit D, respectively, showed that the CPS is linear, the first three monosaccharide residues in the repeating unit being 3-substituted and residue D 2-substituted. The signals for C4 and C6 of β-Galp of D unit were shifted downfield to δ 72.7 and 66.2, respectively, and the C5 signal was shifted upfield to δ 67.1, as compared with their positions in non-substituted β-Galp [10], showing that Pyr is attached at positions 4 and 6 of this monosaccharide. The chemical shift of δ 26.3 for C3 of Pyr indicated that the acetal has the (R)-configuration [11].
The sequence of the monosaccharides in the repeating unit was determined by the 1H,1H ROESY and 1H,13C HMBC experiments, which showed correlations of the anomeric protons and carbons with atoms of the neighboring sugar residues (Table 2). These data also confirmed the substitution pattern in the K unit.
Table 2. Correlations for H1 and C1 in the
two-dimensional 1H,1H ROESY and
1H,13C HMBC spectra of the K82 CPS from A.
baumannii LUH55534
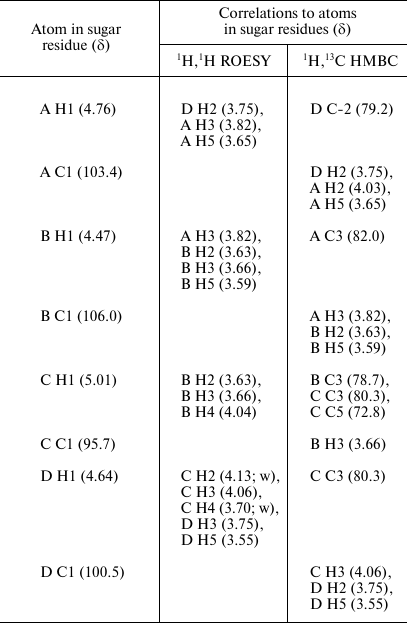
Note: w, weak.
The K82 CPS structure was corroborated by cleavage of Pyr by mild acid hydrolysis followed by structure elucidation of the resulting modified polysaccharide (MPS) by NMR spectroscopy as described above for the CPS (the assigned 1H and 13C NMR chemical shifts are tabulated in Table 1).
Based on these data, it was concluded that the K82 CPS from A. baumannii LUH5534 has the structure shown in Fig. 2. A peculiar feature of this CPS is the presence of (R)-configured d-galactose 4,6-pyruvic acid acetal. Earlier, a similar acetal but on a d-GalpNAc residue was reported in the K4 CPS from A. baumannii D78 [12].
Fig. 2. Structure of the K82 CPS from A. baumannii LUH5534. RPyr indicates (R)-1-carboxyethylidene (pyruvic acid acetal). Transferases are shown near the linkage they are predicted to catalyze the formation of.
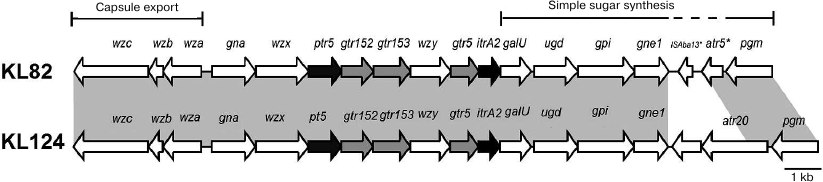
The KL82 gene cluster in A. baumannii LUH5534 (GenBank accession number KC526908) between conserved genes gna and galU contains genes involved specifically with the synthesis of the K82 CPS (Fig. 3).
The itrA2 gene in the KL82 locus encodes an initiating transferase, ItrA2 (WeeH in GenPept, accession number AHB32565.1), which is 99% identical to ItrA2 of A. baumannii KL2 (AGK44809.1 in GenPept) shown to use d-GalNAc as the initiating sugar in the K-unit assembly [13, 14]. ItrA2 in KL82 is 99% identical to ItrA2 of A. baumannii ATCC 17978 (KL3). Hence, d-GalNAc is the first sugar of the K82 unit, and the Wzy polymerase (AHB32563.1 in GenPept) would catalyze formation of the β-d-GalpNAc-(1→3)-d-Galp linkage between the K units (Fig. 2).
Fig. 3. Comparison of the A. baumannii KL82 and KL124 capsule biosynthesis gene clusters. Shading between gene clusters indicates shared regions of >95% nucleotide sequence identity.
Glycosyltransferase Gtr5 encoded by KL82 (WafH in GenPept, accession number AHB32564.1) is 95% identical to Gtr5a from A. baumannii KL2 (GenPept accession number AHM95430.1), which is responsible for formation of the β-d-Galp-(1→3)-d-GalpNAc linkage in the K2 CPS [15]. This linkage is the first in the K82 unit, and Gtr5 was assigned accordingly.
Two further glycosyltransferase genes, gtr152 and gtr153, were also identified in KL82 (Fig. 3) and are evidently responsible for adding the next two sugar residues to complete the assembly of the tetrasaccharide K unit (Fig. 2). Gtr153 (WafK in GenPept, accession number AHB32562.1) is 54% identical to Gtr58 from A. baumannii KL27 (ALL34866.1 in GenPept) that was previously predicted to form an α-D-GlcpNAc-(1→3)-D-Galp linkage [16]. Accordingly, Gtr153 would catalyze this linkage, which also is present in the K82 CPS, and Gtr152 (WafJ in GenPept, accession number AHB32561.1) was assigned to the last β-D-Galp-(1→3)-α-D-GlcpNAc linkage (Fig. 2).
The product of the gene located immediately downstream of the wzx gene was identified as a pyruvyl transferase belonging to protein family (Pfam) PF04230 and was named Ptr5 (WafI in GenPept, accession number AHB32560.1). Ptr5 is not significantly related to Ptr1 from A. baumannii KL4 (GenPept accession number JN409449.3) putatively involved with formation of a D-GalNAc4,6Pyr [16], though it would be responsible for the similar attachment of pyruvic acid to the Gal residue (D) in the K82 CPS.
The generally conserved galU-ugd-gpi-gne1-pgm arrangement in A. baumannii is interrupted in KL82 by a partial copy of ISAba13 (138-1016 of 1039 bp) and a gene encoding a putative acyltransferase (GenPept accession number AHB32571.1 previously annotated as CgmA) that belongs to Pfam PF01757. The first 142 amino acids of the acyltransferase (201 a.a. totally) is 82% identical to Atr5 (142 a.a.) from A. baumannii KL4 (GenPept accession number ACJ39541.2), and neither appears to modify the K4 [12] or K82 CPS. An additional gene cluster KL124 differs from KL82 only in this region (Fig. 3).
Finally, the occurrence in KL82 of genes for flippase Wzx and polymerase Wzy indicated that, as for all other known CPSs of A. baumannii, the K82 CPS is synthesized by the Wzx/Wzy-dependent pathway. Therefore, the content of the KL82 gene cluster and predicted gene functions are consistent with the K82 CPS structure established in this work.
Acknowledgments
This work was supported by the Russian Foundation for Basic Research (project No. 17-04-01254) and the National Health and Medical Research Council (project No. 1026189).
REFERENCES
1.World Health Organization (2017) Global priority
list of antibiotic-resistant bacteria to guide research, discovery, and
development of new antibiotics
(http://www.who.int/medicines/publications/global-priority-list-antibioticresistant-bacteria/en/).
2.Kenyon, J. J., and Hall, R. M. (2013) Variation in
the complex carbohydrate biosynthesis loci of Acinetobacter
baumannii genomes, PLoS One, 8, e62160.
3.Hu, D., Liu, B., Dijkshoorn, L., Wang, L., and
Reeves, P. R. (2013) Diversity in the major polysaccharide antigen of
Acinetobacter baumannii assessed by DNA sequencing, and
development of a molecular serotyping scheme, PLoS One,
8, e70329.
4.Kenyon, J. J., Kasimova, A. A., Shneider, M. M.,
Shashkov, A. S., Arbatsky, N. P., Popova, A. V., Miroshnikov, K. A.,
Hall, R. M., and Knirel, Y. A. (2017) The KL24 gene cluster and a
genomic island encoding a Wzy polymerase contribute genes needed for
synthesis of the K24 capsular polysaccharide by the multiply antibiotic
resistant Acinetobacter baumannii isolate RCH51,
Microbiology, 163, 355-363.
5.Kasimova, A. A., Shneider, M. M., Arbatsky, N. P.,
Popova, A. V., Shashkov, A. S., Miroshnikov, K. A., Balaji, V., Biswas,
I., and Knirel, Y. A. (2017) Structure and gene cluster of the capsular
polysaccharide of Acinetobacter baumannii B11911 containing
5-N-acetyl-7-N-[(R)-3-hydroxybutanoyl]pseudaminic
acid, Biochemistry (Moscow), 82, 483-489.
6.Kenyon, J. J., Shashkov, A. S., Senchenkova, S. N.,
Shneider, M. M., Liu, B., Popova, A. V., Arbatsky, N. P., Miroshnikov,
K. A., Wang, L., Knirel, Y. A., and Hall, R. M. (2017) Acinetobacter
baumannii K11 and K83 capsular polysaccharides have the same
6-deoxy-l-talose-containing pentasaccharide K units but different
linkages between the K units, Int. J. Biol. Macromol.,
103, 648-655.
7.Shashkov, A. S., Liu, B., Kenyon, J. J., Popova, A.
V., Shneider, M. M., Senchenkova, S. N., Arbatsky, N. P., Miroshnikov,
K. A., Wang, L., and Knirel, Y. A. (2017) Structures of the K35 and K15
capsular polysaccharides of Acinetobacter baumannii LUH5535 and
LUH5554 containing amino and diamino uronic acids, Carbohydr.
Res., 448, 28-34.
8.Kenyon, J. J., Kasimova, A. A., Notaro, A.,
Arbatsky, N. P., Speciale, I., Shashkov, A. S., De Castro, C., Hall, R.
M., and Knirel, Y. A. (2017) Acinetobacter baumannii K13 and K73
capsular polysaccharides differ only in K-unit side branches of novel
non-2-ulosonic acids: di-N-acetylated forms of either
acinetaminic acid or 8-epiacinetaminic acid, Carbohydr. Res.,
452, 149-155.
9.Westphal, O., and Jann, K. (1965) Bacterial
lipopolysaccharides. Extraction with phenol–water and further
applications of the procedure, Methods Carbohydr. Chem.,
5, 83-91.
10.Lipkind, G. M., Shashkov, A. S., Knirel, Y. A.,
Vinogradov, E. V., and Kochetkov, N. K. (1988) A computer-assisted
structural analysis of regular polysaccharides on the basis of
13C NMR data, Carbohydr. Res., 175,
59-75.
11.Garegg, P. J., Lindberg, B., and Kvarnstrom, I.
(1980) Preparation and NMR studies of pyruvic acid and related acetals
of pyranosides: configuration at the acetal carbon atoms, Carbohydr.
Res., 77, 71-78.
12.Kenyon, J. J., Speciale, I., Hall, R. M., and De
Castro, C. (2016) Structure of repeating unit of the capsular
polysaccharide from Acinetobacter baumannii D78 and assignment
of the K4 gene cluster, Carbohydr. Res., 408, 12-17.
13.Iwashkiw, J. A., Seper, A., Weber, B. S., Scott,
N. E., Vinogradov, E. V., Stratilo, C., Reiz, B., Cordwell, S. J.,
Whittal, R., Schild, S., and Feldman, M. F. (2012) Identification of a
general O-linked protein glycosylation system in Acinetobacter
baumannii and its role in virulence and biofilm formation, PLoS
Pathog., 8, e1002758.
14.Lees-Miller, R. G., Iwashkiw, J. A., Scott, N.
E., Seper, A., Vinogradov, E., Schild, S., and Feldman, M. F. (2013) A
common pathway for O-linked protein glycosylation and synthesis of
capsule in Acinetobacter baumannii, Mol. Microbiol.,
89, 816-830.
15.Kenyon, J. J., Marzaioli, A. M., Hall, R. M., and
De Castro, C. (2014) Structure of the K2 capsule associated with the
KL2 gene cluster of Acinetobacter baumannii,
Glycobiology, 24, 554-563.
16.Shashkov, A. S., Kenyon, J. J., Senchenkova, S.
N., Shneider, M. M., Popova, A. V., Arbatsky, N. P., Miroshnikov, K.
A., Volozhantsev, N. V., Hall, R. M., and Knirel, Y. A. (2016)
Acinetobacter baumannii K27 and K44 capsular polysaccharides
have the same K unit but different structures due to the presence of
distinct wzy genes in otherwise closely related K gene clusters,
Glycobiology, 26, 501-508.