A Novel Fluorescent GFP Chromophore Analog-Based Dye for Quantitative PCR
A. A. Stakheev1,a*, D. Yu. Ryazantsev1, Yu. K. Zvezdina1, M. S. Baranov1, and S. K. Zavriev1
1Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, 117997 Moscow, Russia* To whom correspondence should be addressed.
Received February 20, 2018; Revision received April 20, 2018
This is the first report describing the possibility of using a green fluorescent protein chromophore synthetic analog, P-HOBDI-BF2, as a fluorescent dye for a linear hydrolysis probe used in qPCR. The study was carried out on a system for detection of the plant pathogenic fungus Fusarium avenaceum using a plasmid containing translation elongation factor 1α fragment as a template. To estimate fluorogenic properties of P-HOBDI-BF2, 6-FAM- and BDP-FL-labeled probes were used. It was demonstrated that a synthetic dye based on the P-HOBDI-BF2 chromophore can be used for labeling hydrolysis probes for qPCR, but fluorescence increase levels for P-HOBDI-BF2-labeled probes were slightly lower than those for 6-FAM-labeled ones. At the same time, the sensitivity of P-HOBDI-BF2-based assays remained high, and this fact together with acceptable fluorescence levels suggests that this dye can be considered as an efficient alternative for reporters traditionally used for fluorescence detection in the FAM channel.
KEY WORDS: quantitative PCR, fluorophore, probe, green fluorescent protein, fluorescence increaseDOI: 10.1134/S000629791807009X
Abbreviations: BDP-FL, boron-dipyrromethene-based dye for fluorescence detection on FAM channel; BHQ1 and 2, black hole quenchers 1 and 2; 6-FAM, 6-carboxyfluorescein; GFP, green fluorescent protein; JOE, 4′,5′-dichloro-2′,7′-dimethoxy-5(6)-carboxyfluorescein; P-HOBDI-BF2 ((5Z)-5-(difluoroboryl-4-hydroxyphenyl)-methylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one); qPCR, quantitative polymerase chain reaction; RBF, relative background fluorescence; RFA, relative fluorescence addition; RFI, relative fluorescence increase; RP-HPLC, reversed-phase high-performance liquid chromatography; TEF1α, translation elongation factor 1 alpha gene.
Quantitative or real-time PCR (qPCR) [1, 2] is one the most powerful tools of modern molecular
biology research. qPCR has been widely applied to solve fundamental and
practical problems such as gene expression estimation, medical
diagnostics, and identification of pathogens and GMOs in food and
agricultural products [3-5].
qPCR can also be used for increasing the sensitivity of such techniques
as enzyme immunoassay: the combination of these approaches is known as
immuno-PCR [6-10]. High
relevance and efficiency of qPCR are due to its high sensitivity and
specificity, the possibility of quantitative estimation of the genetic
material content, as well as significant decrease in risk of
false-negative results as signal detection occurs in a closed tube [11].
All known systems for the detection of amplification products during qPCR are based on measurement of reaction mix fluorescence. In the ideal case, fluorescence intensity should be proportional to the amount of PCR product, which could be detected either by nonspecific methods used to identify any double-stranded DNA or by specific systems able to register the accumulation of a particular amplification product. Among the specific approaches, the use of linear hydrolysis probes (TaqMan® [12]) is one of the most widely used methods. Usually, a probe of this type is labeled with two dyes: fluorophore and fluorescence quencher, which can be placed in terminal or internal positions. In the absence of template DNA, quencher and fluorophore are in close proximity, and the fluorescence is quenched. If a template is present in a reaction mix, probe that hybridizes to the amplicon is destroyed by 5′-exonuclease activity of DNA polymerase. Fluorescent signal intensity increases proportionally to the amount of amplified product.
Most devices used for fluorescence detection from microarray scanners to fluorescent microscopes are equipped with a FAM-channel (green) to detect amplification products. Although most qPCR-based systems for different tasks are adapted for the detection in the FAM-channel, the search for novel compounds with high fluorogenic properties that might improve quality and efficiency of analysis is still important. Analysis of the list of commercially available compounds shows that most widely represented ones are dyes for nonspecific detection such as SYBR Green or derivatives of fluorescein [13], rhodamine [14], and boron-dipyrromethene (BODIPY [15]). 6-Carboxyfluorescein (6-FAM), a fluorescent dye with absorption wavelength of 494 nm and emission wavelength of 520 nm, is the most extensively used fluorophore in qPCR studies. BODIPY-FL, Alexa488, and their analogs that are less widely used but theoretically more photostable are also used.
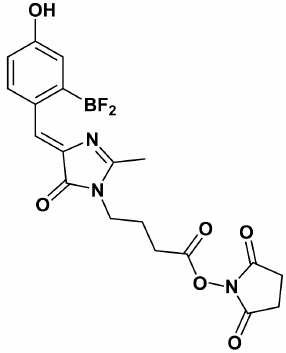
We believe that green fluorescent protein (GFP) chromophore analogs, such as P-HOBDI-BF2 and HOPyDI:Zn [16] synthesized in the Institute of Bioorganic Chemistry, Russian Academy of Sciences, are promising compounds to be used as fluorophores in qPCR. P-HOBDI-BF2 ((5Z)-5-(difluoroboryl-4-hydroxyphenyl)-methylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one) (Fig. 1) is the closest analog of the GFP chromophore from the known fluorescent compounds. Both protonated and deprotonated forms of P-HOBDI-BF2 are characterized by bright fluorescence. The dye has absorption wavelengths of 404 (neutral form) and 485 nm (anionic form) and dual emission at 486 and 528 nm. The goal of this study was to estimate the fluorogenic properties of linear hydrolysis probes labeled by P-HOBDI-BF2 and to compare their characteristics with those of probes labeled by the commonly used fluorophores 6-FAM and BDP-FL. Besides studying the properties of different dyes, we also compared fluorogenic properties of probes labeled in terminal and internal positions.
Fig. 1. Chemical structure of GFP chromophore synthetic analog P-HOBDI-BF2.
MATERIALS AND METHODS
Synthesis and modification of oligonucleotide probes. To compare characteristics of probes labeled by different dyes, the qPCR-based system for detection of the phytopathogenic fungus Fusarium avenaceum [17] was used. We applied this system earlier to test probes labeled by different types and numbers of fluorophores and quenchers [18-20]. The plasmid pTZ-Fat containing a specific fragment of the F. avenaceum strain 103100 TEF1α gene (GenBank accession number KJ508139.1) was used as a template. The structure of amplicon including the annealing sites of the primers and probe is shown in Fig. 2. Black hole quencher 1 (BHQ1) was used in all experiments as a fluorescence quencher.
Fig. 2. Nucleotide sequence of F. avenaceum TEF1α gene fragment. The primer annealing sites are underlined. The probe annealing site is highlighted with gray.
N-Hydroxysuccinimide ester of P-HOBDI-BF2 was used for modification of probes. One millimole of P-HOBDI-BF2 free acid [21] was diluted in 50 ml of tetrahydrofuran. Then, 1.5 mmol of triethylamine and 1.2 mmol of N,N,N′,N′-tetramethyl-O-(N-succinimidyl)-uronium tetrafluoroborate were added to the solution. The mixture was stirred at room temperature for 3 h, diluted with 100 ml of ethyl acetate, and rinsed twice with 25 ml of water and brine (saturated salt solution). The solution was dried over anhydrous sodium sulfate, evaporated, and separated using flash chromatography (ethyl acetate–tetrahydrofuran, 1 : 1), extracting the stained substance with Rf = 0.7. The residue was washed with 1 ml of chloroform and thrice with 5 ml of diethyl ester and then air-dried. NMR spectra were registered on a Bruker Avance III instrument (Bruker, Germany) (700 MHz) in DMSO-D6, using tetramethyl silane (TMS) as an internal standard.
Oligonucleotides labeled by BDP FL-BHQ1, FAM-BHQ1, and NH2-BHQ1 were synthesized by Biotech Industry Ltd (Russia). The probes were modified by the N-hydroxysuccinimide ester of P-HOBDI-BF2 in two positions: terminal (3′) and one of the central nucleotides. In addition to fluorescence quencher, the oligonucleotide contained a free amino group. The modification reaction was conducted in 0.1 M solution of sodium hydrogen carbonate (pH 8.3) using 2 nmol of oligonucleotide and 40 nmol of activated fluorophore ester. The reaction mixture was incubated for 4 h at room temperature.
The modified oligonucleotides were washed free from unbound fluorophore by precipitation with lithium perchlorate. The pellet was washed thrice using 1 ml of acetone and diluted in 20 μl of deionized water (MilliQ). The probes were purified by reversed-phase high-performance liquid chromatography (RP-HPLC) on a Luna 5u C8(2) column (Phenomenex, USA) in an acetonitrile–water system with 50 mM of ammonium acetate in a linear gradient of acetonitrile (5-80%) for 40 min. The absorption spectrum was recorded using a NanoVue spectrophotometer (GE HealthCare, USA).
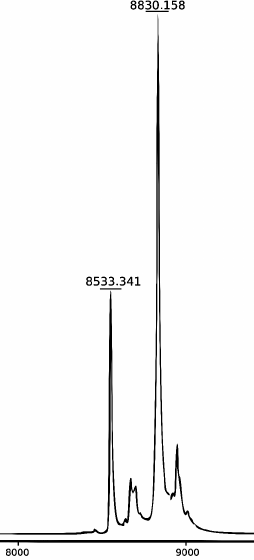
MALDI mass spectrometry. The mass spectra of the modified oligonucleotides were determined using a MicroFlex MALDI-TOF laser desorption/ionization time-of-flight mass spectrometer (Bruker) equipped with a nitrogen laser (337 nm) operating at repetition rate 60 Hz. Spectra were obtained in linear mode with detection of positive ions accelerated by voltage of 20 kV. 3-Hydroxypicolinic acid (10 mg/ml) mixed with ammonium citrate (1 mg/ml) was used as a matrix. The mass spectrum of the products of the FatT22-P-HOBDI-BF2 probe synthesis is shown in Fig. 3. In addition to the main peak, corresponding to the synthesized probe (calculated molecular weight 8829.15 Da), the spectrum contains a peak (8533.341 Da) corresponding to the oligonucleotide without a BHQ1 fragment (298 Da) formed by laser-induced dissociation.
Fig. 3. MALDI mass spectrum of the products of FatT22-P-HOBDI-BF2 probe synthesis.
qPCR. qPCR was carried out in a DT-96 thermal cycler (DNA-technology, Russia) using reaction mix (total volume 35 μl) containing 3.5 μl of 10× PCR buffer (750 mM Tris-HCl, pH 8.8, 200 mM ammonium sulfate, 25 mM magnesium chloride, 0.1% Tween 20), 2.5 U of Taq-polymerase, 5 μl of pTZ-Fat plasmid solution, 0.35 pmol of probe, 12.5 pmol of forward and reverse primers, and 12 mmol of dNTPs according to the following amplification program: 80°C – 60 s; 94°C – 90 s (1 cycle); 94°C – 30 s, 64°C – 15 s (5 cycles); 94°C – 10 s, 64°C – 15 s (45 cycles). Fluorescence was detected on the FAM channel (following the DT-96 manual, excitation wavelength 470 nm, fluorescent detection at 525 nm). For each of the analyzed probes, amplification was carried in four replicates. The threshold method was applied for interpretation of the results [11]. Ten-fold dilutions of the target plasmid (1000, 100, and 10 copies per reaction) were tested to estimate the system sensitivity.
Processing of results. To estimate the fluorogenic properties of the probes, the three following parameters were analyzed: (i) average fluorescence intensity in the initial cycles (background fluorescence) for each probe relative to a FAM-labeled corresponding probe (relative background fluorescence (RBF), see table), expressed as IoT/IoT(FAM), where IoT is background fluorescence of a probe labeled with P-HOBDI-BF2 or BDP-FL, and IoT(FAM) is background fluorescence of a corresponding probe labeled with FAM; (ii) relative fluorescence addition (RFA) expressed as the ratio between fluorescence intensity after reaching the plateau phase (IfT) to the background level; (iii) relative fluorescence increase (RFI) in comparison with a corresponding FAM-labeled probe expressed as (IfT – IoT)/(IT(FAM) – IoT(FAM)). Background fluorescence of each empty well was considered for calculation.
Sequences and fluorogenic properties of analyzed probes

Note: RBF values are calculated in relation to FAM.
RESULTS AND DISCUSSION
One of the key characteristics of linear hydrolysis probes is the intensity of their fluorescence in the initial phase (before the exponential phase of product accumulation), which can be named “background fluorescence”, and fluorescence intensity when the exponential phase is finished, i.e. after reaching a plateau. As shown earlier, decreasing of background fluorescence and increasing of its intensity at the final step can be achieved by including additional BHQ1 quenchers and/or fluorophores (FAM or its derivative 6-carboxy-4′,5′-dichloro-2′,7′-dimethoxyfluorescein (JOE)) [19, 22]. An alternative that does not require such complicated synthesis to introduce the additional dyes is to search for novel fluorophores with better fluorogenic properties. It was shown in a recent study [23] that quenchers BHQ1 and BHQ2 could be modified by a trace amount of ammonium persulfate during the electrophoretic purification of probes due to the presence of fluorescein in their structures. This effect was not observed when using rhodamine derivatives. In the current work, BDP-FL and P-HOBDI-BF2, two dyes promising in terms of optimization of fluorogenic properties of linear hydrolysis probes, were analyzed along with FAM. A derivative of GFP chromophore (P-HOBDI-BF2) was used as a fluorophore in qPCR for the first time. For each fluorophore, characteristics for either terminal (3′) and internal labeled probes were compared.
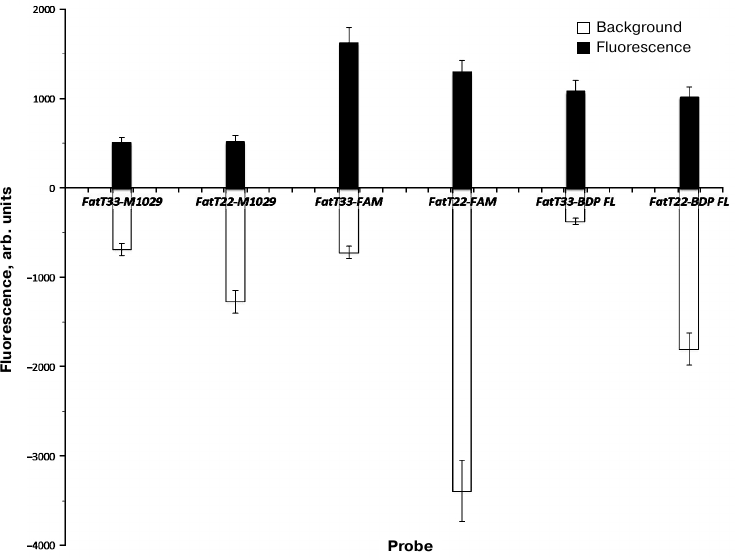
Relative fluorescence addition (RFA) and relative fluorescence increase (RFI) levels for internally labeled probes were, as expected, higher than for those labeled at 3′-end (table and Fig. 4). RFA values for the internally labeled probes were more than twice as high as those for terminally labeled ones (2.2 for FAM and 2.06 for BDP-FL). In contrast, the ratio for P-HOBDI-BF2 labeled probes was only 1.32. RFI of P-HOBDI-BF2-labeled probes was nearly three times less than the value for the FAM-labeled probe and two times less than for BDP-labeled one. However, it must be noted that this study was carried out on only one model of PCR thermal cycler (DT-96) characterized by excitation/fluorescence wavelengths 470/525 nm. The BDP-FL fluorophore has two close peaks of absorption and emission (503/509 nm). Higher sensitivity might be achieved using instruments equipped with monochromators that could be set to the absorption and emission maximums, eliminating the difficulty of estimation of fluorophore properties when detecting at constant wavelengths. The 6-FAM fluorophore, having similar spectral characteristics with P-HOBDI-BF2 (absorption/emission wavelengths 494/520 and 485/528 nm, correspondingly) demonstrated significantly higher RFI, although the background fluorescence level was also higher. However, P-HOBDI-BF2 is characterized by lower quantum yield (0.73) than 6-FAM and BDP-FL (0.95 and 0.97, correspondingly). It should be noted that the sensitivity of the system remained at the same level regardless of the probe used (10 copies of plasmid DNA per reaction) [17].
Fig. 4. Relative background fluorescence (gray) during the initial cycles and fluorescence addition after reaching the plateau (black). Average fluorescence intensity values for four replicates, expressed in relative units, before and after the exponential phase are given for each probe.
Thus, the possibility of using of the GFP chromophore synthetic analog P-HOBDI-BF2 as a fluorescent dye of a linear hydrolysis probe for qPCR has been demonstrated. P-HOBDI-BF2-labeled probes are characterized by relatively low background fluorescence, but their fluorescence increase was 2-3 times lower than that of analogous probes labeled with 6-FAM and BDP-FL.
Acknowledgments
We thank Artem V. Maerle from the Institute of Immunology, Federal Medical and Biological Agency of Russia, for help in obtaining mass spectra of the analyzed oligonucleotides.
This work was supported by the Russian Science Foundation (grant No. 16-14-00136).
REFERENCES
1.Kubista, M., Andrade, J. M., Bengtsson, M.,
Forootan, A., Joniak, J., Lind, K., Sindelka, R., Sjoback, R.,
Sjogreen, B., Strombom, L., Stahlberg, A., and Zoric, N. (2006) The
real-time polymerase chain reaction, Mol. Aspects Med.,
27, 95-125.
2.Bustin, S. A., Benes, V., Garson, J. A., Hellemans,
J., Huggert, J., Kubista, M., Mueller, R., Nolan, T., Pfaffl, M. W.,
Shipley, G. L., Vandersompele, J., and Wittwer, C. T. (2009) The MIQE
guidelines: minimum information for publication of quantitative
real-time PCR experiments, Clin. Chem., 55, 611-622.
3.Gasparic, M. B., Tengs, T., La Paz, J. L.,
Holst-Jensen, A., Pla, M., Esteve, T., Zei, J., and Gruden, K. (2010)
Comparison of nine different real-time PCR chemistries for qualitative
and quantitative applications in GMO detection, Anal. Bioanal.
Chem., 396, 2023-2029.
4.Kralik, P., and Ricchi, M. (2017) A basic guide to
real time PCR in microbial diagnostics: definitions, parameters, and
everything, Front. Microbiol., 8, 108.
5.Maurin, M. (2012) Real-time PCR as a diagnostic
tool for bacterial diseases, Expert Rev. Mol. Diagn., 12,
731-754.
6.Maerle, A. V., Sergeev, I. V., and Alekseev, L. P.
(2014) Immuno-PCR method: prospect of application, Immunology
(Moscow), 1, 44-48.
7.Ryazantsev, D. Y., Voronina, D. V., and Zavriev, S.
K. (2016) Immuno-PCR: achievements and perspectives, Biochemistry
(Moscow), 81, 1754-1770.
8.He, X., and Patfield, S. A. (2015) Immuno-PCR assay
for sensitive detection of proteins in real time, Methods Mol.
Biol., 1318, 139-148.
9.Niemeyer, C. M., Adler, M., and Wacker, R. (2007)
Detecting antigens by quantitative immuno-PCR, Nat. Protoc.,
2, 1918-1930.
10.Sano, T., Smith, T. L., and Cantor, C. R. (1992)
Immuno-PCR: very sensitive detection by means of specific
antibody–DNA conjugates, Science, 258, 120-122.
11.Rebrikov, D. V., Samatov, G. A., Trofimov, D. Y.,
Semenov, P. A., Savilova, A. M., Kofiadi, I. A., and Abramov, D. D.
(2009) Real-Time PCR [in Russian], BINOM, Laboratoriya Znanii,
Moscow.
12.Holland, P. M., Abramson, R. D., Watson, R., and
Gelfand, D. H. (1991) Detection of specific polymerase chain reaction
product by utilizing the 5′-3′ exonuclease activity of
Thermus aquaticus DNA polymerase, Proc. Natl. Acad. Sci.
USA, 88, 7276-7280.
13.Crockett, A. O., and Wittwer, C. T. (2001)
Fluorescein-labeled oligonucleotides for real-time PCR: using the
inherent quenching of deoxyguanosine nucleotides, Anal.
Biochem., 290, 89-97.
14.Wang, G., Becker, E., and Mesa, C. (2007)
Optimization of 6-carboxy-X-rhodamine concentration for real-time
polymerase chain reaction using molecular beacon chemistry, Can. J.
Microbiol., 53, 391-397.
15.Kurata, S., Kanagawa, T., Yamada, K., Torimura,
M., Yokomaku, T., Kamagata, Y., and Kurane, R. (2001) Fluorescent
quenching-based quantitative detection of specific DNA/RNA using a
BODIPY® FL-labeled probe or primer, Nucleic Acids Res.,
29, e34.
16.Baranov, M. S., Lukyanov, K. A., Borissova, A.
O., Shamir, J., Kosenkov, D., Slipchenko, L. V., Tolbert, L. M.,
Yampolsky, I. V., and Solntsev, K. M. (2012) Conformationally locked
chromophores as models of excited-state proton transfer in fluorescent
proteins, J. Am. Chem. Soc., 134, 6025-6032.
17.Stakheev, A. A., Ryazantsev, D. Yu., Gagkaeva, T.
Yu., and Zavriev, S. K. (2011) PCR detection of Fusarium fungi
with similar profiles of the produced mycotoxins, Food Control,
22, 462-468.
18.Ryazantsev, D. Y., Kvach, M. V., Tsybulsky, D.
A., Prokhorenko, I. A., Stepanova, I. A., Martynenko, Y. V., Gontarev,
S. V., Shmanai, V. V., Zavriev, S. K., and Korshun, V. A. (2014) Design
of molecular beacons: 3′-couple quenchers improve fluorogenic
properties of a probe in real-time PCR assay, Analyst,
139, 2867-2872.
19.Ryazantsev, D. Y., Tsybulsky, D. A., Prokhorenko,
I. A., Kvach, M. V., Martynenko, Y. V., Philipchenko, P. M., Shmanai,
V. V., Korshun, V. A., and Zavriev, S. K. (2012) Two-dye and one- or
two-quencher DNA probes for real-time PCR assay: synthesis and
comparison with TaqMan™ probe, Anal. Bioanal. Chem.,
404, 59-68.
20.Tsybulsky, D. A., Kvach, M. V., Ryazantsev, D.
Y., Aparin, I. O., Stakheev, A. A., Prokhorenko, I. A., Martynenko, Y.
V., Gontarev, S. V., Formanovsky, A. A., Zatsepin, T. S., Shmanai, V.
V., Korshun, V. A., and Zavriev, S. K. (2016) Molecular beacons with
JOE dye: influence of linker and 3′-couple quencher, Mol.
Cell. Probes, 30, 285-290.
21.Frizler, M., Yampolsky, I. V., Baranov, M. S.,
Stirnberg, M., and Gutschow, M. (2013) Chemical introduction of the
green fluorescence: imaging of cysteine cathepsins by an irreversibly
locked GFP fluorophore, Org. Biomol. Chem., 11,
5913-5921.
22.Tsybulsky, D. A., Kvach, M. V., Stepanova, I. A.,
Korshun, V. A., and Shmanai, V. V. (2012)
4′,5′-Dichloro-2′,7′-dimethoxy-5(6)-carboxyfluorescein
(JOE): synthesis and spectral properties of oligonucleotide conjugates,
J. Org. Chem., 77, 977-984.
23.Ryabinin, V. A., Kostina, E. V., and Sinyakov, A.
N. (2017) Unexpected transformation of black hole quenchers in
electrophoretic purification of the fluorescein-containing TaqMan
probes, Nucleosides Nucleotides Nucleic Acids, 36,
418-427.