The Expression of Matryoshka Gene Encoding a Homologue of Kunitz Peptidase Inhibitor Is Regulated Both at the Level of Transcription and Translation
E. V. Sheshukova1, T. V. Komarova1,2, N. M. Ershova1,2, A. M. Bronstein3, and Y. L. Dorokhov1,2,a*
1Vavilov Institute of General Genetics, Russian Academy of Sciences, 119991 Moscow, Russia2Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119991 Moscow, Russia
3Sechenov First Moscow State Medical University, 119991 Moscow, Russia
* To whom correspondence should be addressed.
Received May 5, 2018; Revision received July 5, 2018
The gene for Kunitz peptidase inhibitor-like protein (KPILP) contains nested alternative open reading frame (aORF) that controls expression of the maternal mRNA. The content of NbKPILP mRNA in intact leaves of Nicotiana benthamiana plant is low but increases significantly upon extended dark exposure or when foreign nucleic acid is overexpressed in the cells. The NbKPILP gene promoter along with the expressed nested aORF are likely to play an important role in maintaining the levels of NbKPILP mRNA. To elucidate the role of NbKPILP promoter, we isolated a fragment of N. benthamiana chromosomal DNA upstream of the NbKPILP transcription start, sequenced it, and created constructs in which reporter E. coli uidA gene coding for β-D-glucuronidase (GUS) was placed under control of the NbKPILP promoter. By assessing the efficacy of uidA mRNA synthesis directed by the NbKPILP promoter and 35S promoter of the cauliflower mosaic virus in a transient expression system, we showed that the levels of GUS accumulation were comparable for both promoters. Prolonged incubation of the agroinjected plants in the darkness stimulated accumulation of the uidA mRNA directed by the NbKPILP promoter. Our experiments indicate that along with regulation at the transcriptional level, expression of NbKPILP mRNA can be affected by expression of the nested aORF controlled by the polypurine block (PPB) located upstream of its start codon, since introduction of mutations in the PPB resulted in significant accumulation of the NbKPILP mRNA. Nucleotide replacement in the aORF start codon led to the drastic increase in the amounts of NbKPILP mRNA and its protein product.
KEY WORDS: Agrobacterium tumefaciens, transient expression, Kunitz peptidase inhibitor, matryoshka gene, promoter, translation, abiotic factor, polypurine blockDOI: 10.1134/S0006297918100103
Abbreviations: aORF, nested alternative reading frame; GUS, β-D-glucuronidase; KPI, Kunitz peptidase inhibitor; (Nb)KPILP, (Nicotiana benthamiana) Kunitz peptidase inhibitor-like protein; PPB, polypurine block; proNbKPILP, NbKPILP promoter; RT-qPCR, quantitative reverse transcription polymerase chain reaction; 35S, cauliflower mosaic virus (CaMV) 35S RNA promoter; (s)ORF, (short) ORF; sPEP, peptide encoded by sORF; 3′-UTR, 3′-untranslated region.
In nature, plants are constantly affected by various biotic and abiotic
factors. When the action of stress factors ceases, the levels of mRNAs
coding for defense proteins drastically decrease. Stress-induced
responses including activation of transcription factors, chromatin
remodeling, and cytosine methylation in the promoter regions of some
genes have been extensively studied [1]. At the
same time, it has become evident that the variety of plant stress
responses cannot be limited to the regulation of gene transcription
alone. Until recently it had been believed that one RNA transcript
codes for one protein. However, latest proteogenomics studies
encountered several exceptions to this rule that have significantly
changed the traditional meaning of the term gene. The idea and
the topology of a gene with overlapping open reading frames (ORFs) are
well illustrated by a matryoshka toy (Russian nesting dolls) [2]. There are two levels in the
“matryoshka” genetic system. The first one is the
chromosomal level, i.e., when a nested gene is located in exons and/or
introns of a bigger chromosomal gene in either sense or antisense
orientation relatively to the main gene. The second level is a mature
mRNA molecule containing overlapping ORFs or ORFs with alternative
start codons (aORFs). Functional matryoshka genes can be identified,
first of all, by analysis of ORFs coding for short peptides (sPEPs).
Short ORFs (sORFs) are a common feature of all genomes. The coding
potential of such ORFs in most cases disregarded due to the lack of
information on the synthesis of corresponding sPEPs in the cell [3]. Plant mRNAs contain many sORFs located in the
5′-untranslated region (5′-UTR), as well as sORFs
overlapping with the main ORF [4-6]. About ~20% leader sequences of protein-encoding
genes in A. thaliana contain overlapping sORFs [7]. Similar to animal sORFs, sORFs located in
5′-UTRs of plant mRNAs are involved in the regulation of the main
ORF translation. An important role of such sORFs in the stress response
in plant cells has been confirmed by transcriptome-wide ribosome
profiling under normal and stress conditions [8-14]. On the contrary, expression of aORFs located in
the gene coding region was shown only for the Zm908 gene in
maize [15, 16].
Until recently, the influence of aORFs located within or upstream of the main ORF on gene expression, including mRNA stability under stress, had not been studied. Recently, we discovered a gene coding for Kunitz peptidase inhibitor-like protein (NbKPILP) in the Nicotiana benthamiana genome and in other representatives of Solanaceae family (potato, tomato, and tobacco) [17]. Unlike genes coding for Kunitz peptidase inhibitors (KPI) in animals, NbKPILP has no introns, thereby excluding the possibility of the regulation of its expression by alternative splicing [18]. The level of NbKPILP mRNA in leaves of intact N. benthamiana plants is low; however, it rapidly increases in the darkness or in the presence of foreign nucleic acid. Our studies of the mechanism of NbKPILP gene regulation showed that its mRNA contains a nested aORF encoding a 53-amino acid polypeptide (53-a.a. aORF) within the main ORF coding for NbKPILP. In order to demonstrate expression of the 53-a.a. aORF, we inserted into its reading frame a gene coding for the green fluorescent protein (GFP). Detection of the 53-a.a. polypeptide/GFP fusion protein in the cell membranes with anti-GFP antibodies proved the presence of the predicted transmembrane domain in the synthesized polypeptide [17].
It was found that the 53-a.a. polypeptide is synthesized from the same mRNA as NbKPILP (and not from a separate subgenomic mRNA directed by a cryptic promoter). It was assumed that synthesis of the membrane 53-a.a. polypeptide can reduce the level of maternal NbKPILP mRNA in intact leaves. Therefore, suppression of the 53-a.a. polypeptide synthesis should upregulate the level of maternal mRNA. To verify this hypothesis, the start codon (AUG) of the 53-a.a. aORF was changed to ACG leading, as expected, to a significant increase in the content of maternal mRNA [17]. We suggested a hypothetical scheme [17] that in intact leaves, ribosomes “ignore” the start codon of the NbKPILP mRNA, which is in an unfavorable Kozak’s context, and “prefer” the start codon of the 53-a.a. aORF. The ribosomes recognize this codon by the internal ribosomal entry mechanism, while the polypurine block (PPB) upstream of the 53-a.a. aORF plays a role of the internal ribosome entry site (IRES) [19]. The synthesis of the 53-a.a. polypeptide is suppressed under stress conditions, because translation initiation switches to the first start codon of the maternal mRNA, resulting in the upregulation of NbKPILP mRNA in leaves. Each step of this proposed mechanism still has to be proven experimentally. We wanted to answer the following questions. (i) What is the role of NbKPILP promoter in the gene regulation under stress conditions? (ii) How will the removal of the PPB affect the level of NbKPILP mRNA? (iii) Will the suppression of the 53-a.a. aORF lead to the accumulation of maternal NbKPILP mRNA and its protein product?
In this work, we studied the role of NbKPILP gene promoter (proNbKPILP) in the accumulation of mRNA under stress conditions. We found that prolonged dark exposure of plants stimulates accumulation of the mRNA for the reporter E. coli uidA gene, coding for β-D-glucuronidase (GUS), placed under control of proNbKPILP. Besides, the content of NbKPILP mRNA in the cell can be regulated by the PPB located upstream of the aORF, that can potentially drive the aORF translation. Finally, it was shown that replacement of the aORF start AUG codon with ACG significantly increases the cellular levels of the maternal NbKPILP mRNA and its protein product.
MATERIALS AND METHODS
Escherichia coli strain XL-1 and pCambia1300-based binary expression vector, pCambia-35S, containing promoter and terminator sequences of the cauliflower mosaic virus (CaMV) 35S RNA as well as pBIN19 binary vector were used in this work. Restriction endonucleases and other enzymes were from Evrogen (Russia), New England Biolabs (USA) and Fermentas (USA). DNA fragments were purified from agarose gel using commercial kits from Evrogen and ZymoResearch (USA); plasmid DNA was purified by a standard alkaline lysis method. Escherichia coli transformation, overnight cultivation, etc. were done according to standard techniques [20]. Transformation of plant cells was carried out using Agrobacterium tumefaciens (GV-3101 strain). Oligonucleotides were synthesized by the CCU Genome Company (Russia).
Chromosome walking for isolating and cloning proNbKPILP. Nicotiana benthamiana genomic DNA was isolated using Diatom™ DNA Prep set kit (Galart Diagnosticum, Russia) as recommended by the manufacturer. Cloning of the genome fragment adjacent to the 5′-end of the KPILP gene region and its sequencing were done according to the protocol from GenomeWalker™ Universal Kit (Clontech, USA) with modifications [21-23]. First, genomic DNA (500 ng per reaction) was treated in separate reactions with seven rare-cutting restriction endonucleases generating blunt ends: SmaI, PvuII, EcoRV, DraI, SnaBI, ScaI, and HpaI. Adapters (GenWal/Adapt and GenWal/supp primers mix; Evrogen) were added to each of the restriction products using T4 DNA ligase and the resulting DNA fragments were amplified with the Adaptor/T7 (GTAATACGACTCACTATAGGGC) and KPILP-out (TAGTAGGTGTAACCTGGACGGA) primers. In the case of DraI and SnaBI digestion products, well distinguished PCR products over 500 bp long were obtained. To enrich these PCR products with the gene-specific sequences, the second PCR was carried out using KPILP-in primer (GAACTGGTTCAGCTTTTACCTGG) and step-out primer mix (Evrogen). The resulting products were cloned into pAL2-T vector (Evrogen). Two variants of the NbKPILP promoter region – DraI_proNbKPILP and SnaBI_proNbKPILP – were obtained.
Genetic constructs. To generate the proNbKPILP-GUS construct, PCR was carried out with the forward primers AAGCTTAAATGAGAATTTACTTAAG or AAGCTTGTACAAAATATTTAAGCACAAAG for both promoter variants and reverse primer CCATGGTGTTAAGGATATGGTTAATG (sequences shown in bold in the forward and reverse primers are recognition sites for HindIII and NcoI restriction endonucleases, respectively). Each PCR product was treated with HindIII and NcoI endonucleases and cloned into the 35S-GUS construct (earlier obtained in our laboratory) by replacing the 35S-promoter by the HindIII and NcoI sites. The resulting plasmids are referred to as DraI_proNbKPILP-GUS and SnaBI_proNbKPILP-GUS.
To generate the 35S-NbKPILP-STOP_3xFLAG construct, PCR was performed with the CCATGGAGATCATATCAAGGA (forward) and TCTAGATTAAACCTTCTTGAACAC (reverse) primers (NcoI and XbaI sites are shown in bold, respectively). The resulting product containing the NbKPILP gene with a stop codon was treated with NcoI and XbaI endonucleases and cloned into the 35S-GUS plasmid treated with NcoI and SalI endonucleases together with a sequence coding for 3xFLAG generated by annealing of oligonucleotides (the areas of complementation are highlighted by underlining)
TCTAGAGGAGACTATAAAGACGACGATGATAAAGACTACAAGGATGACGATAAG and GTCGACTATTTATCGTCGTCGTCCTTATAATCCTTATCGTCATCCTTGTAGTC followed by completion using Taq-polymerase and treatment with XbaI and SalI endonucleases (XbaI and SalI sites are shown in bold, respectively).
To obtain the 35S-NbKPILP-PPBmut-STOP_3xFLAG construct with mutations in the PPB, we first generated two NbKPILP gene fragments using CCATGGAGATCATATCAAGGA (forward)/GAGAAGAGAAAGGAGAAAAACGGTGGATTTATTGG (reverse) and TTCTCCTTTCTCTTCTCTTTGTGAGCACTGGCGG (forward)/TCTAGATTAAACCTTCTTGAACAC (reverse) primer pairs. The full-length NbKPILP-PPBmut sequence was generated by the overlap extension PCR method using CCATGGAGATCATATCAAGGA (forward)/TCTAGATTAAACCTTCTTGAACAC (reverse) primers (NcoI and XbaI sites are shown in bold). The resulting product was treated with NcoI and XbaI endonucleases and cloned into the 35S-NbKPILP-STOP_3xFLAG construct by the same sites.
To obtain the 35S-NbKPILP_ACG construct, in which the start codon of the 53-a.a. aORF was replaced with ACG, a fragment containing the CaMV 35S promoter and the 5′-terminal region of the NbKPILP sequence and a second fragment containing the 3′-terminal part of NbKPILP and the 35S terminator were excised from the 35S-NbKPILP(ACG) construct generated earlier [17] using restriction endonucleases HindIII/SalI and SalI/EcoRI, respectively. These two fragments were cloned into the pBIN19 vector treated with HindIII and EcoRI. The 35S-NbKPILP construct in pBIN was obtained by the same method.
Agroinjection. Agrobacterium tumefaciens (GV-3101 strain) was grown at 28°C in LB media supplemented with corresponding antibiotics (50 µg/ml rifampicin, 25 µg/ml gentamycin and 50 µg/ml kanamycin or carbenicillin, depending on the plasmid used). The buffer for agroinjection containing 10 mM MES (pH 5.5) and 10 mM MgCl2 was added to an aliquot of the overnight A. tumefaciens culture to a final OD600 of 0.1. The resulting bacterial suspension was injected in the leaves of N. benthamiana plants using a syringe without a needle.
GUS enzymatic activity. GUS enzymatic activity in plant extracts was estimated with the substrate 4-methylumbelliferyl-β-D-glucuronide (mug). The fluorescence of MUG cleavage product was analyzed with a Turner Quantech fluorimeter (Barnstead International, USA) using an NB455 (narrow-band) excitation filter and an NB520 emission filter. All measurements were done according to the standard protocol described in [24].
Western blotting. Total protein samples were prepared on day 3 after agroinjection. Briefly, leaf fragments (~10 mg) from the infiltration zone were ground with Celite in 50 µl of phosphate-buffered saline (PBS). Proteins were separated in 12% polyacrylamide gel and transferred to a Hybond-P PVDF membrane (GE Healthcare, USA). Western blotting was carried out as described earlier [17]. The membrane was blocked with 5% fat-free milk in tris-buffered saline (TBS) supplemented with 0.01% Tween-20 and incubated with primary anti-KPILP antibodies (polyclonal mouse serum; 1 : 15,000 dilution in TBS) and then with anti-mouse antibodies conjugated with horseradish peroxidase (1 : 15,000 dilution; Rockland, USA). Protein bands were visualized with an ECL kit (GE Healthcare).
mRNA expression assay (RT-qPCR). Total RNA from plant tissues was extracted with TRI Reagent (MRC, USA) according to the manufacturer’s instructions. cDNA was synthesized in a reaction mixture containing 2 µg of total RNA, 2 pmol of oligo-dT18 primer, 0.1 µg of random hexamer mix, and Superscript II reverse transcriptase (Invitrogen, USA). Quantitative PCR was performed with an iCycler iQ real-time qPCR detection system (Bio-Rad, USA) and Eva Green master mix (Syntol, Russia). Each sample was run in triplicate. The results of RT-qPCR were evaluated using the Pfaffl algorithm [25]. The levels of mRNA expression for the genes of interest were normalized to 18S rRNA. The following primers pairs were used (forward/reverse): ACGGCTACCACATCCAAG/ACTCATTCCAATTACCAGACTC for 18S; CCTGCGTCAATGTAATGTTC/CGTATTCGGTGATGATAATCG for GUS; and GATTGTTGTGTGGGTATATTGG/CATCGTCGTCTTTATAGTCTCC for KPILP-FLAG.
RESULTS AND DISCUSSION
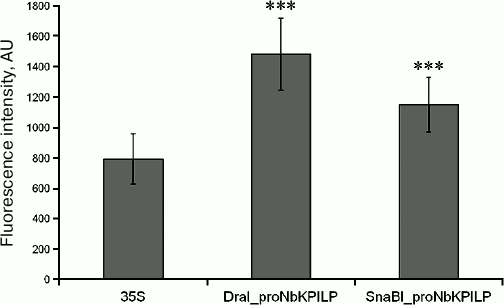
NbKPILP gene promoter region (proNbKPILP) directs the synthesis of mRNA for the reporter E. coli uidA gene encoding β-D-glucuronidase (GUS). It is possible that the NbKPILP gene promoter (proNbKPILP) plays an important role in the maintenance of NbKPILP mRNA levels in the cell. To evaluate the contribution of proNbKPILP to the NbKPILP expression regulation, the 1652-bp region of N. benthamiana chromosomal DNA was isolated and sequenced (ENA #LT882590). Partial nucleotide sequence of proNbKPILP including the TATA-box (TATATAAAACAC) and some predicted cis-elements involved in plant response to light, damage, and pathogen infection is shown on Fig. 1a. The core region of proNbKPILP is highly homologous to the KPILP gene promoters of other Nicotiana species (Fig. 1b). The nucleotide sequence of the core region of the CaMV 35S RNA promoter (35S) used as a control is shown for comparison (Fig. 1c). The 35S promoter [26-28] is one of the constitutive promoters of viral origin commonly used in research and generation of transgenic plants. The 35S promoter contains cis-elements, such as homologs of the circadian rhythm element of Lycopersicon esculentum (gattccattg) and light-responsive G-box element (cacgtc) of Zea mays (Fig. 1c). To analyze the functional role of proNbKPILP, two fragments of the promoter region were isolated from the chromosomal DNA: 1652-bp fragment (DraI_proNbKPILP) and 488-bp fragment (SnaBI_proNbKPILP) obtained by chromosomal DNA digestion with the restriction endonucleases DraI and SnaBI, respectively. These fragments where used to generate constructs, in which the reporter E. coli uidA gene coding for β-D-glucuronidase (GUS) was placed under control of proNbKPILP or the 35S promoter (control). Analysis of transient expression of these constructs in N. benthamiana leaves revealed high levels of GUS production in plant cells (Fig. 2). We concluded that proNbKPILP is able to efficiently direct the synthesis of mRNA for the reporter uidA gene upon transient expression. The activity of the longer construct DraI_proNbKPILP was higher, so it was chosen for further experiments.
Fig. 1. Comparative analysis of the NbKPILP gene promoter (proNbKPILP) and CaMV 35S promoter. a) Partial nucleotide sequence of the chromosomal DNA region upstream of the NbKPILP gene. The TATA-box (TATATAAAACAC) predicted by the YAPP service (http://www.bioinformatics.org/yapp/cgi-bin/yapp.cgi) is shown in bold, ATG start codon is underlined. Some predicted cis-elements are shown (underlined lowercase letters in italic): 1) homolog of the light-responsive GT1 element of Solanum tuberosum; 2) homolog of the circadian rhythm element of Lycopersicon esculentum; 3) homolog of the Box-W1 element of Petroselinum crispum involved in plant response to damage and pathogen infection. Arrow shows transcription start identified earlier by the method of rapid amplification of cDNA ends (5′RACE) [17]. b) Alignment of 3′-terminal nucleotide sequences of the NbKPILP gene promoter (ENA LT882590) and the corresponding DNA regions from two other Nicotiana species: N. glutinosa (NgKPILP) (ENA DI091817) and N. tabacum (NtKPILP) (ENA FH845044). Nucleotides are numbered starting from the predicted NbKPILP transcription start site [17]. c) Nucleotide sequence of the CaMV 35S promoter from the pRT100 vector [28] used for analysis and generation of the 35S-GUS vector. The homologs of the circadian rhythm element of L. esculentum (GATTCCATTG) and light-responsive G-box element (CACGTC) of Zea mays are shown in capital bold letters.
Fig. 2. Comparative analysis of the ability of NbKPILP promoter regions to drive GUS expression in N. benthamiana leaves. Fluorescence intensity after incubation with MUG was measured in extracts of N. benthamiana leaves agroinjected with the plasmids coding GUS under control of the 35S promoter or DraI_proNbKPILP (1652 bp) and SnaBI_proNbKPILP (488 bp) variants of proNbKPILP; *** p < 0.001 (Student’s t- test) for comparison of GUS activity for the 35S promoter and proNbKPILP.
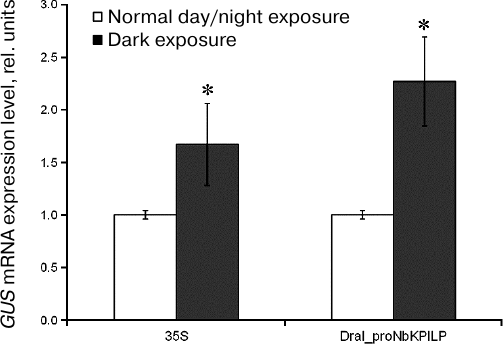
Effect of extended dark exposure of plants agroinjected with plasmids encoding GUS under control of proNbKPILP. The role of proNbKPILP in the stimulation of NbKPILP mRNA accumulation by extended dark exposure of plants has been proposed earlier [17]. This hypothesis was supported by bioinformatic analysis that revealed the presence of potential light-responsive elements in the proNbKPILP sequence. To study directly the influence of darkness on the activity of proNbKPILP, the expression level of the reporter uidA gene placed under control of proNbKPILP was studied after incubation of agroinjected plants in the darkness for three days (the period of dark exposure was chosen based on the previously obtained data [17]).
The relative amount of GUS mRNA was estimated by RT-qPCR (Fig. 3). We found that dark exposure stimulated expression of the reporter gene placed under the control of proNbKPILP. Similar effect of darkness was observed in the plants agroinjected with the 35S-GUS vector.
Fig. 3. Effect of extended dark exposure of plants agroinjected with 35S-GUS or DraI_proNbKPILP-GUS plasmids on the accumulation of GUS mRNA in the leaves as measured by RT-qPCR. The level of GUS mRNA in the leaves of plants kept under optimal day/night conditions was used as a control (taken as 1); * p < 0.05 (Student’s t-test) when compared to the control.
These results showed that stress stimulates the activity of proNbKPILP. In principle, light- and stress-dependent proNbKPILP regulation alone can explain extremely low levels of NbKPILP mRNA in intact leaves. However, our previous studies [17] and this work demonstrated that regulation of the NbKPILP mRNA content in leaves is not limited to the transcriptional level.
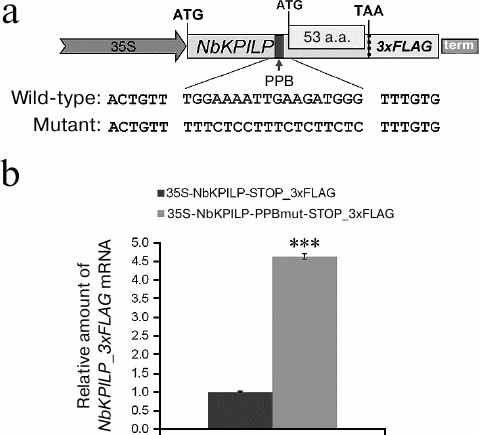
The role of the NbKPILP PPB in the accumulation of NbKPILP mRNA. In order to prove our conclusion that the NbKPILP mRNA level in leaves is mainly determined by translation of the nested aORF [17], we studied the role of the NbKPILP PPB in accumulation of NbKPILP mRNA in the cell. To estimate the ability of PPB to direct aORF translation resulting in the synthesis of the 53-a.a. polypeptide, we replaced purine nucleotides with pyrimidine nucleotides in the PPB sequence (Fig. 4a). Since the level of plant’s endogenous NbKPILP mRNA increases in response to stress including agroinjection, to evaluate the expression of NbKPILP mRNA synthesized from the exogenous plasmids, we introduced a sequence coding for the 3xFLAG tag into the 3′-untranslated regions (3′-UTR) of the wild-type and mutated NbKPILP genes. Both NbKPILP variants were placed under control of the 35S promoter. Introduction of mutations into the PPB led to a significant increase in the level of the corresponding mutant NbKPILP_3xFLAG mRNA (Fig. 4b), which can be due to the decrease in the efficiency of 53-a.a. polypeptide synthesis and stabilization of this mRNA.
Fig. 4. Introduction of mutations into PPB leads to upregulation of NbKPILP_3xFLAG mRNA expression under control of the 35S promoter. a) Schematic representation of the 35S-NbKPILP_3xFLAG construct; mutations introduced to the PPB to generate the 35S-NbKPILP-PPBmut_3xFLAG construct are highlighted; 53 a.a., nested aORF. b) Expression levels of mutant and wild-type NbKPILP_3xFLAG mRNAs in N. benthamiana leaves as measured by RT-qPCR; *** p < 0.001 (Student’s t-test).
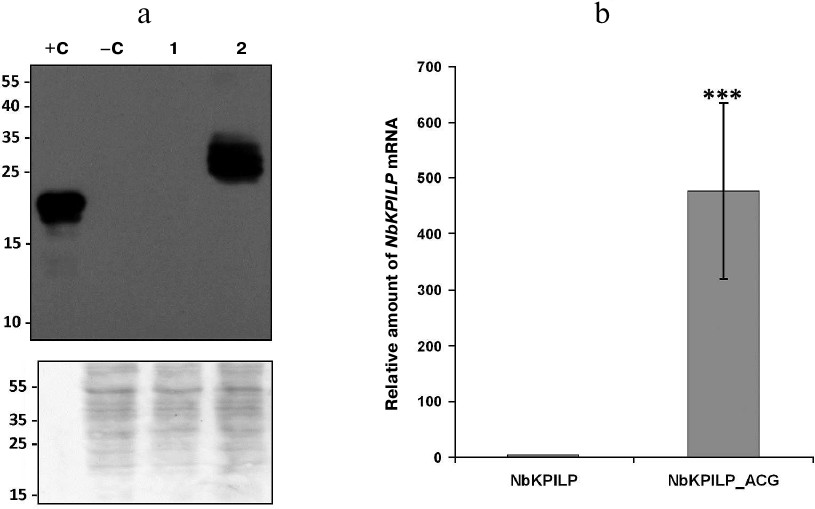
Replacement of the AUG start codon in the aORF with ACG significantly upregulates NbKPILP expression. We also tested the influence of the AUG start codon replacement with ACG on accumulation of NbKPILP. The choice of this codon was dictated, on the one hand, by the need to avoid amino acid changes in the main protein, and, on the other hand, by the low activity of ACG as a start codon in mammals [29] and yeast (0.4-0.5% of the AUG activity) [30]. Figure 5a shows that this mutation led to a dramatic increase in the amount of NbKPILP in the leaves, as detected with specific antibodies. We believe that the obvious reason for NbKPILP accumulation was a significant increase in the levels of NbKPILP mRNA (Fig. 5b), which has been also shown earlier [17].
Fig. 5. Replacement of the AUG start codon in the aORF coding for the 53-a.a. polypeptide nested within the NbKPILP gene with ACG leads to the significant increase in the levels of both NbKPILP mRNA and its protein product. a) Western blot analysis of total protein extracts from leaves injected with 35S-NbKPILP or 35S-NbKPILP_ACG; +C, 60 ng of recombinant NbKPILP; –C, extract of intact leaves. Antibodies against recombinant NbKPILP were used. Lower panel, loading control (membrane stained with Amido Black); molecular weight markers are shown on the left. b) Expression of NbKPILP mRNA on day 3 after injection of leaves with 35S-NbKPILP or 35S-NbKPILP_ACG measured by RT-qPCR; *** p < 0.001 (Student’s t-test) compared to the control.
We conclude that replacement of the aORF start codon AUG significantly increases the levels of NbKPILP mRNA and its protein product.
Therefore, our experiments showed that along with the regulation at the transcription level, the amount of NbKPILP mRNA in a cell can be adjusted at the stage of aORF translation regulated by the PPB within the NbKPILP mRNA. Although we do not have data that mutations in the PPB reduce the synthesis of the 53-a.a. polypeptide, we believe that translation of the aORF is responsible for the mRNA stability. Our conviction is based, in particular, on the fact that mutations eliminating the amphipathic helix in this protein increase the stability of maternal mRNA [17]. One can think of alternative explanations of the results of PPB replacement by a polypyrimidine block in the NbKPILP mRNA. Although increase in the mRNA stability, stimulation of gene transcription, and increase in the template copy number are not very probable, they cannot be completely ruled out. We believe that the level of NbKPILP mRNA in intact leaves is controlled, mainly, by the aORF expression and synthesis of the “toxic” 53-a.a. polypeptide [17]. The pronounced effect of replacement of the canonical AUG start codon with less efficient ACG codon (Fig. 5) also demonstrated an important role of the aORF in the regulation of NbKPILP gene expression in intact leaves.
In conclusion, our results show that different levels of NbKPILP expression under normal and stress conditions are achieved by coordination of processes at both transcriptional and translation levels. More detailed assessment of the relative contribution of transcriptional and translational mechanisms to the regulation of NbKPILP mRNA levels requires further studies. The described gene regulation mechanism is the only known example when aORF can control the levels of maternal mRNA. Taking into account the increasing interest in peptides coded by aORFs in mammals [31], we are convinced that other examples of genes with similar expression mechanism will be discovered in the nearest future.
Acknowledgements
Authors are grateful to anonymous reviewers for their valuable remarks.
Funding
This work was supported by the Russian Science Foundation (project a16-14-00002; studies of the NbKPILP gene transcriptional activity) and Russian Foundation for Basic Research (project 17-29-08012; bioinformatic analysis of proNbKPILP).
Author’s Contributions
DYuL and KTV have conceived and designed the experiments. Most of experiments were carried out by ShEV; ENM and BAM performed some experiments. DYuL, ShEV and KTV evaluated the data and drafted the outline of the manuscript. DYuL and KTV revised and finalized the manuscript. All the authors read and approved the manuscript.
Conflict of Interests
The authors declare that this research was conducted in the absence of any commercial or financial relationship that could be construed as a potential conflict of interests.
REFERENCES
1.Neto, J. R. C. F., da Silva, M. D., Pandolfi, V.,
Crovella, S., Benko-Iseppon, A. M., and Kido, E. A. (2016) Epigenetic
signals on plant adaptation: a biotic stress perspective, Curr.
Protein Pept. Sci., 8, 352-367.
2.Ribrioux, S., Brungger, A., Baumgarten, B., Seuwen,
K., and John, M. R. (2008) Bioinformatics prediction of overlapping
frameshifted translation products in mammalian transcripts, BMC
Genomics, 9, 122.
3.Andrews, S. J., and Rothnagel, J. A. (2014)
Emerging evidence for functional peptides encoded by short open reading
frames, Nat. Rev. Genet., 15, 193-204.
4.Hayden, C. A., and Jorgensen, R. A. (2007)
Identification of novel conserved peptide uORF homology groups in
Arabidopsis and rice reveals ancient eukaryotic origin of select
groups and preferential association with transcription factor-encoding
genes, BMC Biol., 5, 32.
5.Tran, M. K., Schultz, C. J., and Baumann, U. (2008)
Conserved upstream open reading frames in higher plants, BMC
Genomics, 9, 361.
6.Vaughn, J. N., Ellingson, S. R., Mignone, F., and
Arnim, A. (2012) Known and novel post-transcriptional regulatory
sequences are conserved across plant families, RNA, 18,
368-384.
7.Jorgensen, R. A., and Dorantes-Acosta, A. E. (2012)
Conserved peptide upstream open reading frames are associated with
regulatory genes in angiosperms, Front. Plant Sci., 3,
191.
8.Bailey-Serres, J., and Ma, W. (2017) Plant biology:
an immunity boost combats crop disease, Nature, 545,
420-421.
9.Juntawong, P., Girke, T., Bazin, J., and
Bailey-Serres, J. (2014) Translational dynamics revealed by genome-wide
profiling of ribosome footprints in Arabidopsis, Proc. Natl.
Acad. Sci. USA, 111, 203-212.
10.Schepetilnikov, M., and Ryabova, L. A. (2017)
Auxin signaling in regulation of plant translation reinitiation,
Front. Plant Sci., 8, 1014.
11.Sesma, A., Castresana, C., and Castellano, M. M.
(2017) Regulation of translation by TOR, eIF4E and eIF2α in
plants: current knowledge, challenges and future perspectives,
Front. Plant Sci., 8, 644.
12.Tanaka, M., Sotta, N., Yamazumi, Y., Yamashita,
Y., Miwa, K., Murota, K., Chiba, Y., Hirai, M. Y., Akiyama, T.,
Onouchi, H., Naito, S., and Fujiwara, T. (2016) The minimum open
reading frame, AUG-stop, induces boron-dependent ribosome stalling and
mRNA degradation, Plant Cell, 28, 2830-2849.
13.Xu, G., Greene, G. H., Yoo, H., Liu, L., Marques,
J., Motley, J., and Dong, X. (2017) Global translational reprogramming
is a fundamental layer of immune regulation in plants, Nature,
545, 487-490.
14.Xu, G., Yuan, M., Ai, C., Liu, L., Zhuang, E.,
Karapetyan, S., Wang, S., and Dong, X. (2017) uORF-mediated translation
allows engineered plant disease resistance without fitness costs,
Nature, 545, 491-494.
15.Dong, X., Wang, D., Liu, P., Li, C., Zhao, Q.,
Zhu, D., and Yu, J. (2013) Zm908p11, encoded by a short open reading
frame (sORF) gene, functions in pollen tube growth as a profilin ligand
in maize, J. Exp. Bot., 64, 2359-2372.
16.Hanada, K., Higuchi-Takeuchi, M., Okamoto, M.,
Yoshizumi, T., Shimizu, M., Nakaminami, K., Nishi, R., Ohashi, C.,
Iida, K., Tanaka, M., Horii, Y., Kawashima, M., Matsui, K., Toyoda, T.,
Shinozaki, K., Seki, M., and Matsui, M. (2013) Small open reading
frames associated with morphogenesis are hidden in plant genomes,
Proc. Natl. Acad. Sci. USA, 110, 2395-2400.
17.Sheshukova, E. V., Komarova, T. V., Ershova, N.
M., Shindyapina, A. V., and Dorokhov, Y. L. (2017) An alternative
nested reading frame may participate in the stress-dependent expression
of a plant gene, Front. Plant Sci., 8, 2137.
18.Corley, M., Solem, A., Phillips, G., Lackey, L.,
Ziehr, B., Vincent, H. A., Mustoe, A. M., Ramos, S. B. V., Weeks, K.
M., Moorman, N. J., and Laederach, A. (2017) An RNA structure-mediated,
posttranscriptional model of human α-1-antitrypsin expression,
Proc. Natl. Acad. Sci. USA, 114, 10244-10253.
19.Dorokhov, Y. L., Skulachev, M. V., Ivanov, P. A.,
Zvereva, S. D., Tjulkina, L. G., Merits, A., Gleba, Y. Y., Hohn, T.,
and Atabekov, J. G. (2002) Polypurine (A)-rich sequences promote
cross-kingdom conservation of internal ribosome entry, Proc. Natl.
Acad. Sci. USA, 99, 5301-5306.
20.Maniatis, T., Fritsch, E., and Sambroock, D.
(1984) in Methods of Genetic Engineering. Molecular Cloning
[Russian translation], Mir, Moscow.
21.Lukyanov, S. A., Gurskaia, N. G.,
Luk’ianov, K. A., Tarabykin, V. S., and Sverdlov, E. D. (1994)
Highly-effective subtractive hybridization of cDNA, Bioorg.
Khim., 20, 701-704.
22.Lukyanov, K. A., Gurskaya, N. G., Bogdanova, E.
A., and Lukyanov, S. A. (1998) Selective suppression of polymerase
chain reaction, Bioorg. Khim., 25, 163-170.
23.Siebert, P. D., Chenchik, A., Kellogg, D. E.,
Lukyanov, K. A., and Lukyanov, S. A. (1995) An improved PCR method for
walking in uncloned genomic DNA, Nucleic Acids Res., 23,
1087-1088.
24.Jefferson, R. (1987) Assaying chimeric genes in
plants: the GUS gene fusion system, Plant Mol. Biol. Rep.,
5, 387-405.
25.Pfaffl, M. W. (2001) A new mathematical model for
relative quantification in real-time RT-PCR, Nucleic Acids Res.,
29, e45.
26.Benfey, P. N., Ren, L., and Chua, N. H. (1990)
Combinatorial and synergistic properties of CaMV 35S enhancer
subdomains, EMBO J., 9, 1685-1696.
27.Odell, J. T., Knowlton, S., Lin, W., and Mauvais,
C. J. (1988) Properties of an isolated transcription stimulating
sequence derived from the cauliflower mosaic virus 35S promoter,
Plant Mol. Biol., 10, 263-272.
28.Topfer, R., Prols, M., Schell, J., and
Steinbiß, H. H. (1988) Transient gene expression in tobacco
protoplasts: II. Comparison of the reporter gene systems for CAT, NPT
II, and GUS, Plant Cell Rep., 7, 225-228.
29.Ivanov, I. P., Loughran, G., and Atkins, J. F.
(2008) uORFs with unusual translational start codons autoregulate
expression of eukaryotic ornithine decarboxylase homologs, Proc.
Natl. Acad. Sci. USA, 105, 10079-10084.
30.Asano, K. (2014) Why is start codon selection so
precise in eukaryotes? Translation, 2, e28387.
31.Chugunova, A., Navalayeu, T., Dontsova, O., and
Sergiev, P. (2018) Mining for small translated ORFs, J. Proteome
Res., 17, 1-11.