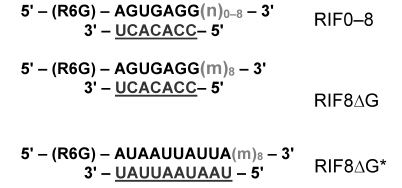Contribution of Eutrema salsugineum Cold Shock Domain Structure to the Interaction with RNA
V. V. Taranov1#, N. E. Zlobin1#, K. I. Evlakov1, A. O. Shamustakimova1,a*, and A. V. Babakov1,b
1All-Russia Research Institute of Agricultural Biotechnology, 127550 Moscow, Russia# These authors contributed equally to this work.
* To whom correspondence should be addressed.
Received June 5, 2018; Revision received July 24, 2018
Plant cold shock domain proteins (CSDPs) are DNA/RNA-binding proteins. CSDPs contain the conserved cold shock domain (CSD) in the N-terminal part and a varying number of the CCHC-type zinc finger (ZnF) motifs alternating with glycine-rich regions in the C-terminus. CSDPs exhibit RNA chaperone and RNA-melting activities due to their nonspecific interaction with RNA. At the same time, there are reasons to believe that CSDPs also interact with specific RNA targets. In the present study, we used three recombinant CSDPs from the saltwater cress plant (Eutrema salsugineum) – EsCSDP1, EsCSDP2, EsCSDP3 with 6, 2, and 7 ZnF motifs, respectively, and showed that their nonspecific interaction with RNA is determined by their C-terminal fragments. All three proteins exhibited high affinity to the single-stranded regions over four nucleotides long within RNA oligonucleotides. The presence of guanine in the single- or double-stranded regions was crucial for the interaction with CSDPs. Complementation test using E. coli BX04 cells lacking four cold shock protein genes (ΔcspA, ΔcspB, ΔcspE, ΔcspG) revealed that the specific binding of plant CSDPs with RNA is determined by CSD.
KEY WORDS: Eutrema salsugineum, Arabidopsis thaliana, cold shock domain proteins, cold shock domain, zinc fingers, RNA–protein interactionDOI: 10.1134/S000629791811007X
Abbreviations: a.a., amino acid; AtCSDP1-3, Arabidopsis thaliana cold shock domain proteins 1-3; CSD(P), cold shock domain (protein); EsCSDP1-3, Eutrema salsugineum cold shock domain proteins 1-3; R6G, fluorescent dye rhodamine; RIF, fluorescently-labelled RNA oligonucleotide; RNP, RNA-binding motif; ZnF, zinc finger motif.
Cold shock domain proteins (CSDPs) are widely spread among pro- and
eukaryotic organisms [1]. Bacterial CSDPs consist
only of the cold shock domains (CSDs) of about 69-74 a.a. and include
short conserved RNP1 (6 a.a.) and RNP2 (5 a.a.) motifs that determine
their ability to bind nucleic acids. Bacterial CSDPs are involved in
the regulation of transcription and translation and can function as RNA
chaperones [2-4].
Participation of CSDPs in the transcription and translation in animals and plants has been confirmed by numerous experimental data [5-7]. Lin28 protein that contains CSD in the N-terminal part and two CCHC-type zinc fingers (ZnFs) in the C-terminal part [8] is the closest to plant CSDPs in terms of structure. Its most studied molecular function in animal cells is inhibition of processing of the let-7 microRNA precursor [9] due to the ability of Lin28 to melt the hairpin structure of pre-let-7 [10].
CSDPs have been shown to participate in the molecular mechanisms providing plant resistance to abiotic stress [11] and regulation of plant development [12]. All plant CSDPs contain CSDs in the N-terminal parts of their molecules, while their C-terminal parts consist of several (from 1 to 7) CCHC-type ZnFs separated by glycine-rich regions. Genes coding CSDPs were found in the genomes of mono- and dicotyledons [11, 13].
The first plant CSDP was isolated from wheat (WCSP1) that had the CSD at the N-terminus and a glycine-rich region with three ZnF motifs at the C-terminus [14]. Later, three other CSDPs were found in wheat: WCSP2 with two ZnFs, WCSP3 with three ZnFs, and WCSP4 with four ZnFs [15]. There are two CSDPs in rice – OsCSP1 and OsCSP2 [16] that contain the CSDs and two or four ZnFs separated by glycine-rich regions. The genome of Arabidopsis thaliana has four genes coding for CSDPs [17]: two “long” genes – AtCSP1 and AtCSP3, with seven ZnFs each, and two “short” genes – AtCSP2 and AtCSP4, with two ZnFs each. The genome of the extremophyte Eutrema salsugineum (close relative of A. thaliana) also contains four genes coding for two “long” CSDPs (EsCSDP1 and EsCSDP3 with six and seven ZnFs, respectively) and two “short” CSDPs (EsCSDP2 and EsCSDP4 with two ZnFs each) [18]. Turnip (Brassica rapa) has five CSDPs: two “short” with two ZnFs and three “long” – one with six ZnFs and two with seven ZnFs each [19].
It is believed that the function of CSDPs in plants is related to their RNA chaperone activity [11] including the ability to melt secondary structures of RNA and DNA confirmed in vivo and in vitro experiments [20-22]. As demonstrated in in vitro experiments, plant CSDPs have preference to single-stranded DNA fragments and their RNA chaperone and RNA- and DNA-melting activities are provided by the CSDP nonspecific interactions with RNA and DNA [23]. Identification of more than 6000 mRNAs co-immunoprecipitating with AtCSP1 from the lysate of A. thaliana leaves [24] also suggests considerable nonspecific RNA binding by plant CSDPs. Similar results were obtained for the Lin28 protein [25], which together with other data, point out the pleotropic mechanism of Lin28 action in animal cells that is realized via both specific interaction with pre-let-7 microRNA and nonspecific interactions with a large number of various RNA molecules.
So far, no specific RNA targets for plant CSDPs have been found; however, because of the structural similarity of CSDPs to the Lin28 protein their existence is quite probable. The molecules of plant CSDPs have several nucleic acid-binding sites, such as RNP1 and RNP2 motifs of the CSD in the N-terminal part and 2 to 7 ZnF motifs in the C-terminal part. The aim of this work was to reveal the contribution of the N- and C-terminal regions of E. salsugineum CSDPs to the nonspecific and specific RNA binding.
MATERIALS AND METHODS
Expression of recombinant proteins. Recombinant proteins EsCSDP1-3 and their fragments were purified using the HaloTag technology (Promega, USA) as described earlier [23]. The nucleotide sequence coding for the HaloTag protein and the linker with the TEV protease cleavage site were amplified from the pFC20K HaloTag® T7 SP6 Flexi® plasmid (Promega). The amplified HaloTag gene was cloned into the pET28a+ vector (Novagen, UK) using the NcoI and XhoI restriction sites. The nucleotide sequences for EsCSDP1-3, EsCSD1-3, EsZnF1, and CspA were amplified from the genomic DNA. The amplified DNA fragments were cloned in the pET28a+ vector using the NcoI and EcoRI restriction sites. To generate a construct coding for the mutant EsCSDP3m protein, we used synthetic DNA sequence bearing point mutations in the CSD (W16A, F25A, F36A) and ZnF motif (C95A, C127A, C157A, C191A, C222A, C250A, C277A) that affected the nucleic acid-binding activity of the CSDPs. The EsCSDP3m mutant gene was synthesized by the ATG Service Gene (Russia). All obtained genetic constructs were verified by sequencing and used to transform Escherichia coli BL21 cells.
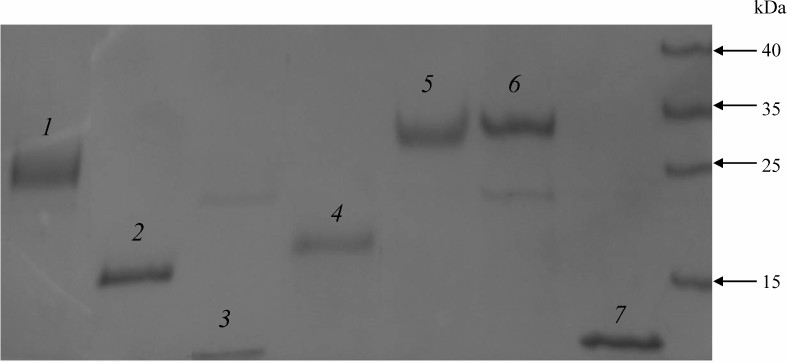
The recombinant proteins were expressed and purified using the HaloTag Protein Purification Kit (Promega) according to the manufacturer’s instructions. The proteins were purified from 50 ml of the overnight E. coli culture. The sonicated lysate was centrifuged for 30 min at 10,000g, and the supernatant was incubated with 1 ml of HaloLink resin (Promega) at room temperature for 1 h with continuous mixing. HaloLink resin with the bound recombinant protein was washed with the following buffers: 1) buffer 1 (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM DTT, 10 µM ZnSO4); 2) buffer 2 (50 mM HEPES, pH 7.5, 0.5 M NaCl, 1 mM DTT, 10 µM ZnSO4); 3) buffer 1 containing 2 mM ATP and 10 mM MgSO4; 4) buffer 1 (4 times). The recombinant proteins covalently bound to the HaloTag resin were recovered using TEV protease (Promega) according to the manufacturer’s instructions. The purified proteins were frozen in liquid nitrogen and stored at –70°C. Protein concentration was determined by the Bradford method. The obtained recombinant proteins were over 90% pure according to Laemmli’s SDS-PAGE (Fig. 1).
Fig. 1. Proteins purified from the E. coli lysate and separated by SDS-PAGE: 1) EsCSDP1; 2) EsCSDP2; 3) EsCSD1; 4) C-terminal part of EsCSDP1; 5) EsCSDP3; 6) EsCSDPm; 7) EsCSD3.
Measurement of fluorescence anisotropy. Interaction of CSDPs with RNA was studied by measuring the fluorescence anisotropy upon binding of these proteins with fluorescently labelled RNA oligonucleotides [26].
RNA oligonucleotides labeled by the 5′-end with 6-carboxy-rhodamine (R6G) were synthesized by Sintol (Russia). Before the assay, fluorescently labelled and complementary oligonucleotides were mixed in buffer 1 at a 1 : 2 ratio, incubated for 5 min at 95°C, and transferred to ice. Fluorescence anisotropy was measured at 526/555 nm using an LS55 fluorimeter (Perkin Elmer, USA) equipped with a temperature-controlled cuvette holder connected to a water thermostat. Protein sample in the buffer 1 supplemented with 10% glycerol was added into a 100-μl cuvette with the optical path length of 10 mm, and then the oligonucleotides were added to a final concentration of 50 nM. The resulting solution was mixed and incubated for 2 min, and fluorescence anisotropy was measured (average of 10 consecutive measurements was recorded). Kd was calculated using the Sigmaplot 12 software. All measurements were done in triplicate; all obtained values are expressed as mean ± S.E.
Growth complementation of E. coli BX04 cells. Genetic constructs for studying growth complementation in bacterial cells were obtained using the pINIII vector [27]. Genes coding for EsCSD1-3 and AtCSD1-3 were amplified by PCR from the E. salsugineum and A. thaliana cDNAs, respectively, and cloned into the pINIII vector using the BamH1 and Nde1 restriction sites.
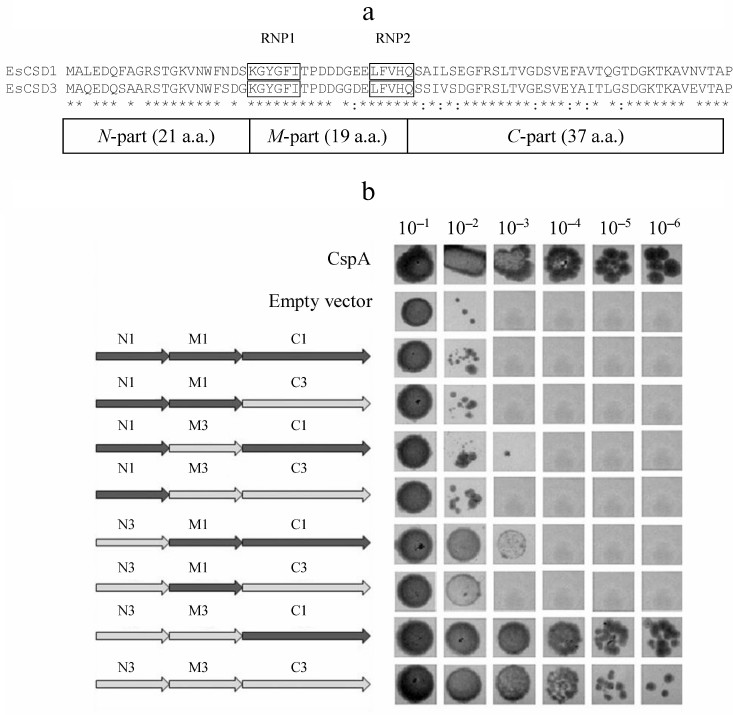
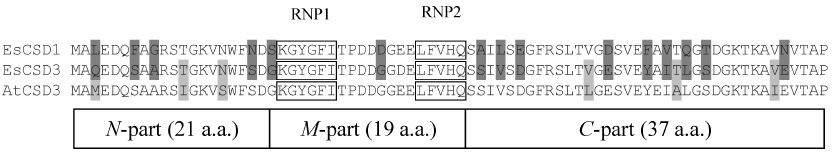
The constructs coding for chimeric CSDs consisting of a combination of fragments of CSDs from EsCSD1 and EsCSD3 were also generated. The CSDs were divided into three fragments: N-fragment (from the N-terminus to the RNP1 motif, 21 a.a.), M-fragment (from the beginning of RNP1 to the end of RNP2 motif, 19 a.a.) and C-fragment (from the RNP2 motif to the end of the domain, 37 a.a.) (Fig. 2a). These fragments were combined with each other in all possible ways, generating six different variants of chimeric domains (Fig. 2b).
Fig. 2. a) Alignment of amino acid sequences of the CSDs from EsCSDP1 and EsCSDP3: RNP1 and RNP2 motifs are shown in frames; b) effect of chimeric CSDs obtained by combining EsCSD1 and EsCSD3 fragments on the growth of mutant E. coli BX04 cells at 17°C (dilutions of bacterial cell culture are shown above the images of bacterial clones).
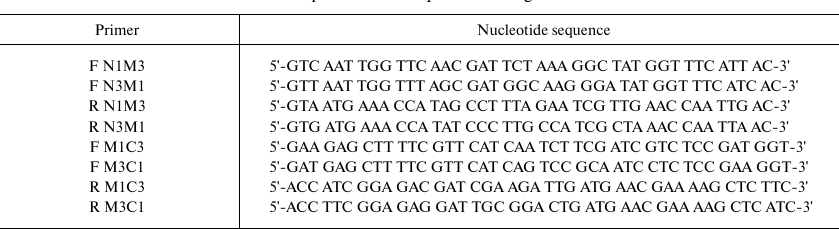
The nucleotide sequences of chimeric domains were amplified by two sequential PCRs. First, the N-, M- and C-fragments with overlapping sequences were amplified separately. In the second PCR, the nucleotide sequences encoding full-size chimeric domains were amplified using the flanking primers. The amplified sequences were cloned into the pINIII vector and verified by sequencing. Sequences of the primers used for the amplification are listed in the table.
Primers used for amplification of sequences coding for chimeric CSDs
Escherichia coli BX04 cells lacking four CSDP genes (ΔcspA, ΔcspB, ΔcspE, ΔcspG) were transformed with the obtained genetic constructs and grown in liquid LB medium supplemented with ampicillin (0.2 mg/ml) and kanamycin (0.05 mg/ml) to the optical density OD550 = 0.8. Then, 7 μl of the bacterial culture (10–1-10–6 dilutions) was spotted onto LB agar plates supplemented with ampicillin (0.2 mg/ml), kanamycin (0.05 mg/ml), and IPTG (0.2 mmol/ml). The plates were incubated at 17°C for 5 days in the darkness. The control samples were incubated at 37°C overnight. Escherichia coli BX04 cells transformed with the pINIII vector were used as a negative control. As a positive control, we used cells carrying the CspA gene coding for the main E. coli cold shock protein whose overexpression leads to the restoration of growth of E. coli BX04 cells at low temperatures.
RESULTS
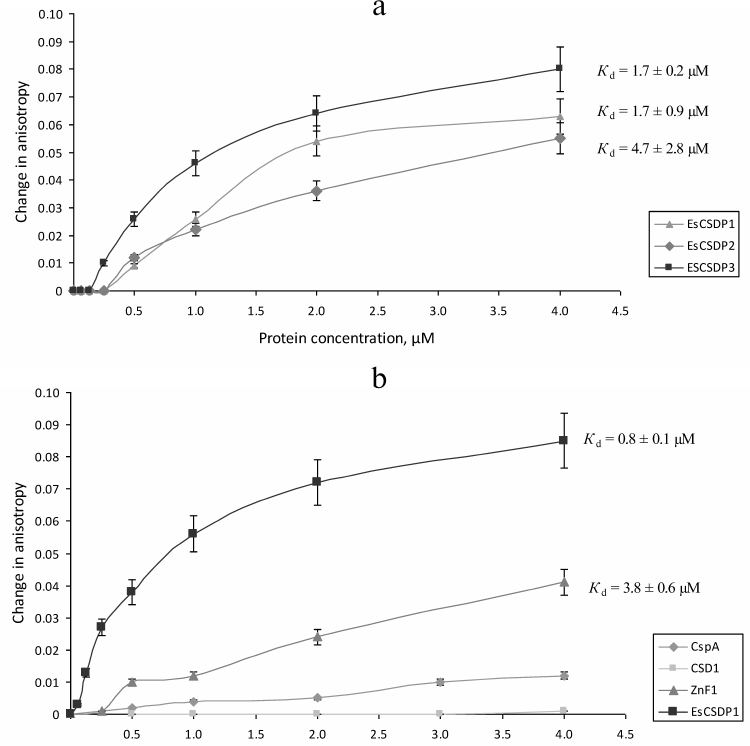
In order to compare the affinity of EsCSDPs to single- and double-stranded RNA fragments, we synthesized a set of fluorescently labelled RNA oligonucleotides (Fig. 3). Each oligonucleotide consisted of a 7-nucleotide fragment (shown in bold) fused to a degenerate nucleotide sequence of different length (shown in light grey) and the oligonucleotide complementary to the 7-nucleotide fragment (underlined). The random sequence of the 7-nucleotide fragment was generated with the NUPACK software (http://www.nupack.org). The affinity of the EsCSDP1-3 proteins and the EsCSDP3m mutant protein to the RIF0-8 nucleotides was analyzed by measuring an increase in the fluorescence anisotropy upon protein binding to the corresponding RNA structure. Figure 4 shows that EsCSDP3m exhibited no RNA-binding activity, whereas addition of any of the three EsCSDPs to RIF8 (Fig. 4a) or RIF0 (Fig. 4b) led to the increase in fluorescence anisotropy.
Fig. 3. Structure of fluorescently labelled RNA oligonucleotides; n, any nucleotide (A, U, G, C); m, any nucleotide except G; numbers outside the brackets, length of the single-stranded region; 7-nucleotide sequences are shown in bold; degenerated sequences are shown in light grey; complementary fragments are underlined.
Fig. 4. Affinity of EsCSDP1-3 and EsCSDP3m to RNA oligonucleotides with different single-stranded region length: a) 8 nucleotides (RIF8); b) 0 nucleotides (RIF0); c) 0, 2, 4, 6, and 8 nucleotides (RIF0, 2, 4, 6, and 8, respectively).
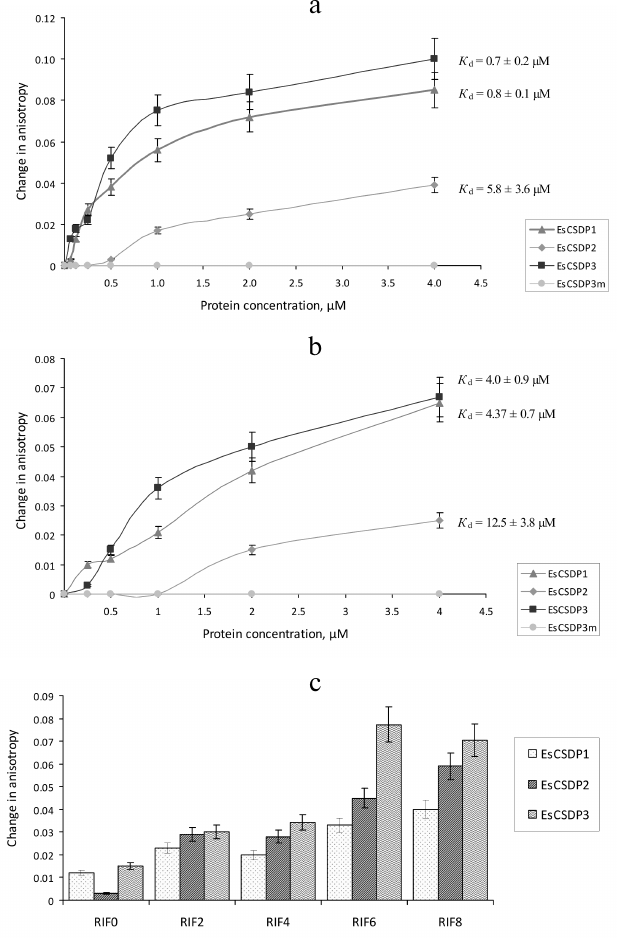
The Kd values calculated from our experimental data are presented in Figs. 4 and 5. Since the increase in the anisotropy of RIF0 did not reach the plateau within the range of used protein concentrations (especially with EsCSDP2), to calculate Kd we used the maximal anisotropy increase value determined for the binding of EsCSDP1-3 to RIF8. Figure 4 shows that for all the three proteins, the Kd values of RIF0 binding (Fig. 4b) was much higher than those observed for RIF8 binding (Fig. 4a). This fact indicates that the affinity of EsCSDP1-3 to the single-stranded RNA is significantly higher than their affinity to the double-stranded RNA. Moreover, the obtained Kd values show that the affinity of EsCSDP2 to RIF8 is ~5 times lower than the affinities of EsCSDP1 and EsCSDP3 for the same nucleotide. Shortening of the single-stranded region from 8 to 6 nucleotides does not alter the anisotropy increase; however, further decrease in the nucleotide length has a much more pronounced effect (Fig. 4c).
Fig. 5. Affinity of EsCSDP1-3 and isolated domains to RNA oligonucleotides (a) with no guanine nucleotides in the single-stranded region (RIF8ΔG) and (b) with guanine nucleotides in both double- and single-stranded regions (RIF8).
Removal of guanine from the single-stranded region of RIF8 (RIF8ΔG) leads to a ~2.5-fold increase in the Kd values for EsCSDP1 and EsCSDP3 but only slightly affects the Kd value for EsCSDP2 (Fig. 5a). When guanine was removed from both single- and double-stranded regions of RIF8ΔG*, no increase in the anisotropy was detected (data not shown).
CSD-containing proteins have two RNA-binding sites. One is the CSD itself, and the second is the C-terminal fragment with ZnFs. Using EsCSDP1 as an example, we attempted to evaluate to what extent each of these two domains influences the affinity of EsCSDP to RNA. For this purpose, we purified the recombinant EsCSD1 and the C-terminal domain EsZnF1 with six ZnFs. Figure 5b shows the dependence between the protein concentration and anisotropy increase upon RIF8 interaction with EsCSD1 and EsZnF1. Similar correlations for EsCSDP1 and bacterial protein CspA are shown for comparison. Our data demonstrated that EsZnF1 binds to RIF8, although according to the calculated Kd, its affinity to RIF8 should be several times lower compared to that of the full-length EsCSDP1. No increase in the RIF8 anisotropy was observed upon its binding to EsCSD1, whereas the addition of CspA (whose molecular mas is similar to that of EsCSD1) to RIF8 resulted in the anisotropy increase. It should be mentioned that no increase in the RIF8 anisotropy was observed after addition of EsCSD2 or EsCSD3.
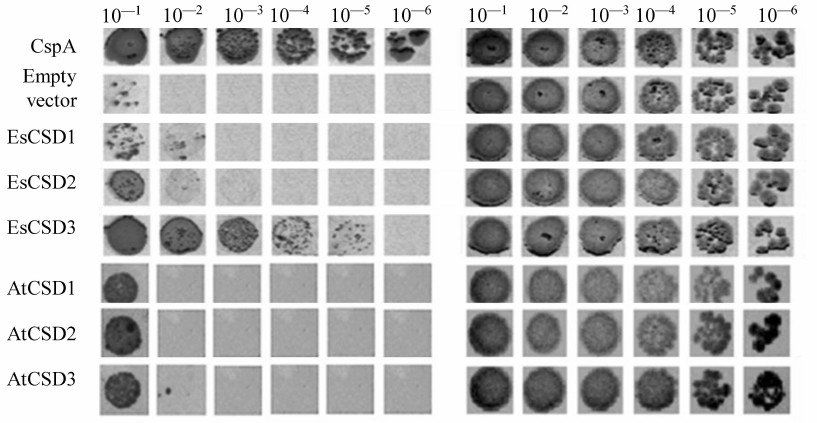
To assess the interactions between CSDs and RNAs, we used the complementation growth test estimating the ability of the studies proteins to restore the growth of E. coli BX04 cells at low temperatures. Escherichia coli BX04 cells normally cannot grow at low temperatures because of the deletion of four endogenous cold shock protein genes (ΔcspA, ΔcspB, ΔcspE, ΔcspG). This test is commonly used for assessing the RNA chaperone activity of proteins [28], and the positive results in this test indicate the ability of a studied protein to interact with RNA. We compared three CSDs of E. salsugineum (EsCSD1-3) and homologous CSDs from A. thaliana (AtCSD1-3). In our experiments (Fig. 6), all tested CSDs were active in the growth complementation test on E. coli BX04 cells compared to the empty vector. However, the activity of most CSDs was much lower than the activity of CspA (positive control). The only CSD whose activity in the complementation test was comparable to that of CspA was EsCSD3.
Fig. 6. Effect of EsCSDP1-3 and AtCSDP1-3 on the growth of mutant E. coli BX04 cells: left panel, 17°C; right panel, 37°C; numbers above the images of bacterial colonies, dilutions of bacterial culture.
It should be mentioned that despite considerable differences in the functional activity, CSDs of different proteins possess significant structural similarity. In order to study which amino acid is responsible for the differences in the activity of the studied CSDs in the complementation test, we generated and tested chimeric domains composed of different combinations of EsCSD1 and EsCSD3 fragments (see “Materials and Methods” and Fig. 2). Analysis of the activity of the obtained chimeric domains in E. coli BX04 growth complementation test showed that the most important region determining the difference in the activities of EsCSD3 and AtCSD3 is the N-terminal fragment (Fig. 2). Thus, all cells expressing chimeric domains with the EsCSD3 N-terminal fragment, regardless of the origin of the M- and C-fragments, grew at low temperatures much faster than the cells with the N-terminal fragment from the EsCSD1. Another important region is the central fragment of the EsCSD3 that differs from that of EsCSD1 by a single amino acid (glycine in EsCSD3 vs. aspartate in EsCSD1) (Fig. 7). Exchange of the C-terminal fragments has the minimal impact on the protein activity; although, unlike the N- and M-fragments, the C-terminal fragment of EsCSD1 (but not EsCSD3) provided higher functional activity.
Fig. 7. Comparison of amino acid sequences of the CSDs from EsCSDP1, EsCSDP3, and AtCSDP3: RNP1 and RNP2 motifs are shown in frames; vertical lines show the borders of the N-terminal, central and C-terminal fragments; dark grey, amino acids different in EsCSD1 and EsCSD3; light grey, amino acids different in EsCSD3 and AtCSD3.
DISCUSSION
RNA oligonucleotides used in this work consisted of a double-stranded fragment and a random nucleotide sequence fused to a degenerate nucleotide sequence of varying length. These oligonucleotides are characterized by the lack of specific secondary structure in the single-stranded region and the absence of specificity to the studied proteins in the double-stranded region. Therefore, our results on the EsCSDP binding to RNAs describe exclusively nonspecific RNA–protein interactions that, according to the studies of homologous Lin28 protein from animal cells [25], can play an essential role in the functioning of plant CSDPs.
Comparison of Kd values presented in Figs. 4a and 4b shows that shortening of the single-stranded region of the RNA oligonucleotide RIF8 and transition to the double-stranded RNA oligonucleotide RIF0 decreased the affinities of EsCSDP1, EsCSDP2, and EsCSDP3 to RNA (5-, 2-, and 6-fold, respectively), which suggested higher affinity of EsCSDP1-3 to the single-stranded RNA. At the same time, EsCSDP1-3 proteins did not bind DNA oligonucleotides lacking the single-stranded regions [23]. The absence of RNA-binding properties in EsCSD1 (Fig. 5b) indicates the dominant role of the EsCSDP1 C-terminal fragment with six ZnFs in the nonspecific RNA binding. The CSDs from EsCSDP2 and EsCSDP3 did not bind RIF8; therefore, the conclusion on the essential role of the C-termini of EsCSDP2 and EsCSDP3 in nonspecific RNA binding can probably be also applied to these proteins. ZnFs play the major role in binding of the C-termini of EsCSDPs to nucleic acids; hence, the correlation between the decrease in Kd and increase in the number of ZnFs in the protein (Fig. 4, a and b) is not surprising: the lowest Kd was observed for EsCSDP3 with seven ZnFs; the highest – for EsCSDP2 with two ZnFs. In other words, the more ZnFs are present in the protein C-terminal part, the higher is the protein affinity of its nonspecific interaction with RNA.
Interestingly, guanine plays a special role in the binding of plant CSDPs to nucleic acids. In absence of guanine in the single-stranded RNA oligonucleotide region, the affinity of EsCSDP1 and EsCSDP3 to RNA decreased more than two times (Figs. 4a and 5a) (EsCSDP2 is an exception). However, when guanine was absent from both the single- and double-stranded regions of RNA oligonucleotide, no binding of EsCSDP1-3 was detected. Our in vitro experiments confirm the earlier obtained in vivo data showing that guanine-enriched RNAs co-immunoprecipitate with AtCSP1 from the lysate of A. thaliana leaves [24].
It should be mentioned that the length of the single-stranded RNA region required for the efficient binding of EsCSDP1-3 has to be over 4 nucleotides (Fig. 4c). Another interesting feature was discovered when we compared the binding of RNA oligonucleotides by EsCSDP1-3 and their fragments. The affinity of the full-length EsCSDP1 toward RIF8 was ~4.5 times higher than that of ZnF1, while CSD1 alone demonstrated no RNA-binding activity in our experimental conditions. The data presented in Fig. 5b correlate with the data from Figs. 4b and 5a. In the first case, combination of the inactive CSD1 with ZnF1 in the EsCSDP1 molecule results in the increase in the protein affinity to RNA; in the second case, addition of the single-stranded guanine-lacking region to the double-stranded fragment that originally did not bind to CSDPs also increased the affinity of the protein–RNA interaction (compare Figs. 4b and 5a). So far, we cannot explain the observed effects. It also remains unclear why the affinity of EsCSDP2 (Figs. 4a and 5a) does not depend on the presence of guanine in the single-stranded region (unlike the affinity of EsCDP1 and EsCSDP3). This phenomenon can probably be explained by a higher relative affinity of EsCSDP2 to the double-stranded RNA regions compared to the affinity of “long” EsCDP1 and EsCSDP3.
According to the X-ray studies of the complex between animal Lin28 and pre-let-7 RNA, Lin28 binds the GGAG motif though its two ZnFs [29]. Later, the nucleotide sequence of this motif was found to be GNNG [30]. According to our data, the presence of G in the binding motif is required for the binding of EsCSDP1-3. The CCHC-type ZnF is a 14-a.a. motif with the consensus sequence CNNCNNNNHNNNNC (amino acids coordinating zinc ion are highlighted in bold). Amino acid sequences of ZnFs are not identical; the most variable residues are residues 2, 3, 5, and 6. ZnFs within the same protein are different, as well as ZnFs in different CSDPs. If the specificity of the CCHC-type ZnF interaction with RNA does not depend on the ZnF primary structure, a question arises of what can hide behind the variety of the ZnF primary structures? So far, there is no well-founded answer to this question. A large number of AtCSDP3 interaction partners have been found in A. thaliana that presumably modulate the functional activity of this protein [31]. Therefore, it is possible that each plant CSDP has a specific set of partner proteins that is defined by the number and the primary structures of CSDP ZnFs.
In conclusion, our data suggest that the nonspecific interactions of CSDPs with RNA are provided by the ZnF motifs located in the C-terminal regions of EsCSDP1-3. This suggestion is confirmed by the loss of the nucleic acid-binding ability of EsCSDP3 upon the introduction of the C/A mutation into all seven ZnFs. However, despite the nonspecific character of RNA binding by CSDPs, there are no reasons to exclude that these proteins can also interact with specific RNA targets in plant cells. For example, it was shown recently that cold shock domain protein PpCSP1 from the moss Physcomitrella patens regulates reprogramming of differentiated leaf cells to chloronema apical stem cells [32]. Lin28, whose amino acid sequence and domain structure are close to those of PpCSP1 participates in the reprogramming of human fibroblasts into induced pluripotent stem cells [33]; moreover, the molecular mechanism of this process includes inhibition of let-7 microRNA precursor processing induced by its specific binding to Lin28 [29].
The possibility of specific interactions between of E. salsugineum proteins with nucleic acids is supported by the studies of different animal CSDPs, the majority of which are multifunctional proteins characterized by both highly and low-specific functional interactions with nucleic acids [34]. At the same time, there is almost no information on specific RNA targets of plant CSDPs. It is well known that the animal Lin28 protein specifically interacts with the pre-let-7 RNA and blocks its processing [29, 30]. The domain structure of this protein is very close to that of plant CSDPs: it has the CSD in the N-terminal part of the molecule and two CCHC-type ZnF motifs in the C-terminal part.
The results of growth complementation test in E. coli BX04 cells demonstrated that specific interactions of plant CSDPs with nucleic acids are provided by CSDs. This hypothesis is corroborated by, first, the unique ability of the EsCSD3 to restore the growth of E. coli BX04 cells at low temperatures. Out of six CSDs of A. thaliana and E. salsugineum CSDPs, only EsCSD3 showed activity in the growth complementation test in E. coli BX04 cells that was comparable to the activity of bacterial CspA. Second, using the combinations of EsCSD1 and EsCSD3 fragments, we showed that the activity of EsCSD3 in the complementation test was tightly related to the primary structure of its N-terminal and central fragments. EsCSD1 and EsCSD3 contain only five non-homologous amino acid substitutions in their N-terminal fragments, while EsCSD3 and AtCSD3 contain only three substitutions (Q3M, T12I, and N16S) (Fig. 7). The central fragments of EsCSD1 and EsCSD3 differ only in two positions: D32G and E34D. If we neglect the conserved E34D replacement, then only a single substitution of aspartate D32 by glycine leads to the increase in the activity of the chimeric N3M3C3 domain in the growth complementation test as compared to the N3M1C3 domain activity (Fig. 2). It is necessary to emphasize that all substitutions mentioned above are located outside of the RNP1 and RNP2 motifs essential for the RNA binding.
All CSDPs, as well as CspA, bear two conserved RNP1 and RNP2 motifs that directly interact with RNA. However, according to our results, the specificity of interaction with RNA is very sensitive to the amino acid environment of these motifs. This fact allows us to expect that structurally close CSDPs from the same species, as well as homologous CSDPs from different species, will interact with different RNA targets.
Funding
This work was supported by the State program no. 0574-2014-0017 and Russian Foundation for Basic Research (project 14-04-32260). We used the scientific equipment of the Biotechnology Center for Collective Use (agreement RFMEFI62114×0003) at the Russian Research Institute of Agricultural Biotechnology, Moscow.
Acknowledgements
The authors are grateful to M. Inouye for kindly providing the mutant BX04 strain and the pINIII vector.
Conflict of Interests
Authors declare no conflict of interests.
Ethical Approval
The present article does not contain any description of experiments on humans or animals.
REFERENCES
1.Horn, G., Hofweber, R., Kremer, W., and Kalbitzer,
H. R. (2007) Structure and function of bacterial cold shock proteins,
Cell. Mol. Life Sci., 64, 1457-1470.
2.Bae, W., Xia, B., Inouye, M., and Severinov, K.
(2000) Escherichia coli CspA-family RNA chaperones are
transcription antiterminators, Proc. Natl. Acad. Sci. USA,
97, 7784-7789.
3.Jiang, W., Hou, Y., and Inouye, M. (1997) CspA, the
major cold-shock protein of Escherichia coli, is an RNA
chaperone, J. Biol. Chem., 272, 196-202.
4.Phadtare, S., and Severinov, K. (2010) RNA
remodeling and gene regulation by cold shock proteins, RNA
Biol., 7, 788-795.
5.Mihailovich, M., Militti, C., Gabaldon, T., and
Gebauer, F. (2010) Eukaryotic cold shock domain proteins: highly
versatile regulators of gene expression, Bioessays, 32,
109-118.
6.Chaikam, V., and Karlson, D. T. (2010) Comparison
of structure, function and regulation of plant cold shock domain
proteins to bacterial and animal cold shock domain proteins,
Biochem. Mol. Biol. Rep., 43, 1-8.
7.Lasham, A., Moloney, S., Hale, T., Homer, C.,
Zhang, Y. F., Murison, J. G., Braithwaite, A. W., and Watson, J. (2003)
The Y-box-binding protein, YB1, is a potential negative regulator of
the p53 tumor suppressor, J. Biol. Chem., 278,
35516-35523.
8.Moss, E. G., Lee, R. C., and Ambros, V. (1997) The
cold shock domain protein LIN-28 controls developmental timing in C.
elegans and is regulated by the lin-4 RNA, Cell, 88,
637-646.
9.Newman, M. A., Thomson, J. M., and Hammond, S. M.
(2008) Lin-28 interaction with the Let-7 precursor loop mediates
regulated microRNA processing, RNA, 14,
1539-1549.
10.Mayr, F., Schutz, A., Doge, N., and Heinemann, U.
(2012) The Lin28 cold-shock domain remodels pre-let-7 microRNA,
Nucleic Acids Res., 40, 7492-7506.
11.Sasaki, K., and Imai, R. (2012) Pleiotropic roles
of cold shock domain proteins in plants, Front. Plant Sci.,
2, 116.
12.Nakaminami, K., Hill, K., Perry, S. E., Sentoku,
N., Long, J. A., and Karlson, D. T. (2009) Arabidopsis cold
shock domain proteins: relationships to floral and silique development,
J. Exp. Bot., 60, 1047-1062.
13.Chaikam, V., and Karlson, D. T. (2010) Comparison
of structure, function and regulation of plant cold shock domain
proteins to bacterial and animal cold shock domain proteins, BMB
Rep., 43, 1-8.
14.Karlson, D., Nakaminami, K., Toyomasu, T., and
Imai, R. (2002) A cold regulated nucleic acid binding protein of winter
wheat shares a domain with bacterial cold shock proteins, J. Biol.
Chem., 277, 35248-35356.
15.Radkova, M., Vitamvas, P., Sasaki, K., and Imai,
R. (2014) Development- and cold-regulated accumulation of cold shock
domain proteins in wheat, Plant Physiol. Biochem., 77,
44-48.
16.Chaikman, V., and Karlson, D. (2008) Functional
characterization of two cold shock domain proteins from Oryza
sativa, Plant Cell Env., 31, 995-1006.
17.Karlson, D., and Imai, R. (2003) Conservation of
the cold shock domain protein family in plants, Plant Physiol.,
131, 12-15.
18.Taranov, V. V., Berdnikova, M. V., Nosov, A. V.,
Galkin, A. V., and Babakov, A. V. (2010) Cold shock domain proteins in
the extremophyte Thellungiella salsuginea (salt cress):
gene structure and differential response to cold, Mol. Biol.
(Moscow), 44, 787-794.
19.Choi, M. J., Park, Y. R., Park, S. J., and Kang,
H. (2015) Stress-responsive expression patterns and functional
characterization of cold shock domain proteins in cabbage (Brassica
rapa) under abiotic stress conditions, Plant Physiol.
Biochem., 96, 132-140.
20.Sasaki, K., Kim, M. H., and Imai, R. (2007)
Arabidopsis cold shock domain protein 2 is an RNA chaperone that
is regulated by cold and developmental signals, Biochem. Biophys.
Res. Commun., 364, 633-638.
21.Nakaminami, K., Sasaki, K., Kajita, S., Takeda,
H., Karlson, D., Ohgi, K., and Imai, R. (2005) Heat stable
ssDNA/RNA-binding activity of a wheat cold shock domain protein,
FEBS Lett., 579, 4887-4891.
22.Nakaminami, K., Karlson, D. T., and Imai, R.
(2006) Functional conservation of cold shock domains in bacteria and
higher plants, Proc. Natl. Acad. Sci. USA, 103,
10122-10127.
23.Zlobin, N., Evlakov, K., Alekseev, Y.,
Blagodatskikh, K., Babakov, A., and Taranov, V. (2016) High DNA melting
activity of extremophyte Eutrema salsugineum cold shock domain
proteins EsCSDP1 and EsCSDP3, Biochem. Biophys. Rep., 5,
502-508.
24.Juntawong, P., Sorenson, R., and Bailey-Serres,
J. (2013) Cold shock protein 1 chaperones mRNAs during translation in
Arabidopsis thaliana, Plant J., 74, 1016-1028.
25.Stefani, G., Chen, X., Zhao, H., and Slack, F. J.
(2015) A novel mechanism of LIN-28 regulation of let-7 microRNA
expression revealed by in vivo HITS-CLIP in C. elegans,
RNA, 21, 985-996.
26.Kudryashova, E. V., Gladilin, A. K., and
Levashov, A. V. (2002) Proteins in supramolecular assemblies: structure
study by time-resolved fluorescence anisotropy, Uspekhi Biol.
Khim., 42, 257-294.
27.Xia, B., Ke, H., and Inouye, M. (2001)
Acquirement of cold sensitivity by quadruple deletion of the cspA
family and its suppression by PNPase S1 domain in Escherichia
coli, Mol. Microbiol., 40, 179-188.
28.Kim, M. H., and Imai, R. (2015) Determination of
RNA chaperone activity using an Escherichia coli mutant, in
RNA Remodeling Proteins, Humana Press, New York, NY, pp.
117-123.
29.Nam, Y., Chen, C., Gregory, R. I., Chou, J. J.,
and Sliz, P. (2011) Molecular basis for interaction of let-7 microRNAs
with Lin28, Cell, 147, 1080-1091.
30.Loughlin, F. E., Gebert, L. F., Towbin, H.,
Brunschweiger, A., Hall, J., and Allain, F. H. (2012) Structural basis
of pre-let-7 miRNA recognition by the zinc knuckles of pluripotency
factor Lin28, Nat. Struct. Mol. Biol., 19, 84-89.
31.Kim, M. H., Sonoda, Y., Sasaki, K., Kaminaka, H.,
and Imai, R. (2013) Interactome analysis reveals versatile functions of
Arabidopsis cold shock domain protein 3 in RNA processing within
the nucleus and cytoplasm, Cell Stress Chaperones, 18,
517-525.
32.Li, C., Sako, Y., Imai, A., Nishiyama, T.,
Thompson, K., Kubo, M., Hiwatashi, Y., Kabeya, Y., Karlson, D., Wu,
S.-H., Ishikawa, M., Murata, T., Benfey, P. N., Sato, Y., Tamada, Y.,
and Hasebel, M. (2017) A Lin28 homologue reprograms differentiated
cells to stem cells in the moss Physcomitrella patens, Nat.
Commun., 8, 14242.
33.Yu, J., Vodyanik, M. A., Smuga-Otto, K.,
Antosiewicz-Bourget, J., Frane, J. L., Tian, S., Nie, J., Jonsdottir,
G. A., Ruotti, V., Stewart, R., Slukvin, I. I., and Thomson, A. (2007)
Induced pluripotent stem cell lines derived from human somatic cells,
Science, 318, 1917-1920.
34.Mayr, F., and Heinemann, U. (2013) Mechanisms of
Lin28-mediated miRNA and mRNA regulation – a structural and
functional perspective, Int. J. Mol. Sci., 14,
16532-16553.