REVIEW: Chirality and Handedness of Protein Structures
A. V. Efimov
Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; E-mail: efimov@protres.ru
Received May 25, 2017; Revision received June 19, 2017
In proteins, the polypeptide chain forms a number of right- and left-handed helices and superhelices, right- and left-turned hairpins, and some other structures that are nonsuperimposable, although they are not mirror images of each other as the L-amino acids are not converted to the D-amino acids. This property of protein structures will be referred to here as pseudo-chirality – or handedness. It has been shown that there are two kinds of handedness in proteins – helical handedness and handedness of arrangement. Some protein structures exhibit both the kinds of handedness. Handedness is observed at all levels of protein structural organization – from α-helices, β-strands, hairpins, βαβ-units up to complex structural motifs, superhelices, and supramolecular structures in fibrous and polymer proteins. There are several structures that have unique handedness in proteins, for example, α-helices, αα-corners, βαβ-units, abcd-units, and so on. This property of the polypeptide chain is of particular value in protein folding and protein modeling, because it drastically reduces the number of possible folds.
KEY WORDS: α-helix, β-hairpin, superhelix, structural motif, protein foldingDOI: 10.1134/S0006297918140092
In chemistry, chirality is a property of some molecules or their fragments of existing in two mirror-image forms. A chiral molecule is non-superposable on its mirror image. These two mirror images of a chiral molecules are referred to as configurations (or stereoisomers) of the molecule [1] whose interconversion requires the formal breaking and making of covalent bonds. On the other hand, polypeptide chains of proteins can form different types of left- and right-handed helices [2, 3] and superhelices [4] that are nonsuperimposable, although they are not mirror images of each other as the L-amino acids are not converted to the D-amino acids. In proteins, there are also right- and left-turned α-hairpins and β-hairpins [5, 6], right- and left-handed φ- and ψ-motifs [7-9], S-like and Z-like β-sheets [10], etc. The two forms of these structures are not real mirror images, and the left-handed forms can be converted to the right-handed ones and vice versa without breaking of chemical bonds (other than hydrogen bonds) because these forms are different conformations of the polypeptide chain. Thus, this property of protein structures will be referred to here as pseudo-chirality – or handedness.
In this review, two kinds of handedness of protein structures and several examples are considered and described. It should be noted that many of these protein structures have unique handedness. This property of the polypeptide chain is of particular value in protein folding and protein modeling, since it drastically reduces the number of possible folds.
RIGHT- AND LEFT-HANDED FORMS OF SECONDARY STRUCTURAL
ELEMENTS
At the level of secondary structure, the right-handed α-helix is the most energetically favorable and frequently found structural element in proteins [2]. In the ideal α-helix, there are 3.6 amino acid residues (a.a.) per turn, and every residue has torsion angles φ = –57, ψ = –47. The backbone of the α-helix can be represented as a square-sectioned parallelepiped with a right-handed twist. Each edge of the parallelepiped passes through Cα-atoms of amino acid residues in position i, i ± 4, i ± 8, …, while the faces are occupied by peptide groups. In theory, left-handed α-helix having the backbone twisted anticlockwise and with residue torsion angles φ = 57, ψ = 47 can be formed. The left-handed α-helix formed by any amino acid residues except glycine is energetically unfavorable [11] and occurs in proteins rather rarely. It is usually formed by 3-5 a.a. However, in an irregular region, one or two residues in the sterically constrained αL-conformations occur very often. Statistical analysis shows that approximately 80% of residues having αL-conformations in proteins are glycines, and others are residues with flexible side chains [12]. Right-handed 310 and π-helices are energetically less favorable than the α-helix and rarely occur as helical structures. However, in irregular regions of proteins, residues having 310 and/or π-conformations occur rather often.
Description of handedness of β-structure is more complex. In 1973, Chothia showed that β-sheets in proteins are almost invariably twisted in a right-handed sense when viewed along the polypeptide chain direction [13]. To maintain a large contact area without disturbing hydrogen bonds, β-strands must be coiled as well as twisted in strongly twisted β-sheets. A strongly twisted and coiled β-hairpin can form a right-handed double-helical structure [6], while flat and moderately twisted β-hairpins and triple-stranded β-sheets can form right- and left-handed forms depending on the arrangement of β-strands [6, 10] (see also below).
TWO KINDS OF HANDEDNESS OF SUPER-SECONDARY STRUCTURES
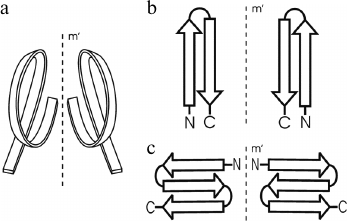
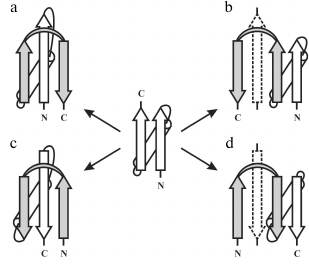
In globular proteins, the polypeptide chain folds over upon itself many times, and the elements of secondary structure that are close together in the chain have a strong tendency to be in close contact in three-dimensional structure. Very often, α-helices and/or β-strands that are adjacent along the chain form a few well-defined types of super-secondary structures or structural motifs, which frequently occur in both homologous and non-homologous proteins. Handedness of these protein structures can be of two kinds. The first kind is the helical handedness of 3D-structures (Fig. 1a) such as different helices (α-, π-, 310-helices) and superhelices (these are Rossmann folds, βαβ- and βαβαβ-units, etc.) [4]. The second kind is the handedness of relatively flat 2D-structures such as β-hairpins (Fig. 1b), S-like, and Z-like β-sheets (Fig. 1c). The β-hairpins can be right-turned or left-turned depending on whether the second β-strands runs on the right or left relative to the first when viewed from the same side (e.g., as viewed from the hydrophobic core). The polypeptide chain runs from the N-end to the C-end in the clockwise direction in the right-turned β-hairpin and in the anticlockwise direction in the left-turned β-hairpin (Fig. 1b). In the S-like β-sheet, the first β-hairpin is left-turned and the second is right-turned. In the Z-like β-sheet, the first β-hairpin is right-turned and the second is left-turned (Fig. 1c). As seen, the handedness of relatively flat structures is determined by mutual arrangement of the elements, so this kind of handedness will be referred to here as the handedness of arrangement.
Fig. 1. Two kinds of handedness of super-secondary structures. The polypeptide chain is shown as a ribbon in superhelices (a) and as arrows directed from the N- to C-ends in β-hairpins (b) and β-sheets (c).
HELICAL HANDEDNESS OF β-HAIRPINS AND β-SHEETS
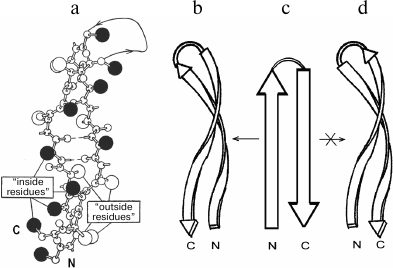
In proteins, β-hairpins and β-sheets are almost invariably twisted in a right-handed sense when viewed along the polypeptide chain direction [13]. The extent of the twist observed in proteins varies; the average dihedral angle between adjacent strands is close to –20° [13, 14]. In strongly twisted β-sheets, the β-strands must be coiled as well as twisted to maintain a large contact area without disturbing hydrogen bonds [14, 15]. A strongly twisted and coiled β-hairpin can be represented as a double-helical structure in which the strands are twisted and coiled in a right-handed sense (Fig. 2, a and b). Such a double superhelix has both concave and convex surfaces. A distinctive feature of this structure is that it is always formed by the right-turned β-hairpin when viewed from the concave side of the superhelix [6]. A similar superhelix is not formed by the left-turned β-hairpin (Fig. 2d), and such superhelices are not found in proteins. It should be noted that the strongly twisted and coiled β-hairpin can be arranged in a protein molecule so that its convex surface is directed into the hydrophobic core. In this case, the β-hairpin is left-turned if viewed from the hydrophobic core but remains right-turned when viewed from the concave side. Note that flat or moderately twisted β-hairpins can be both right- and left-turned, and they are widespread in proteins. Stereochemical analysis shows that in globular proteins there is the definite relationship between structure and amino acid sequence of strongly twisted and coiled β-hairpins [6, 16]. As shown, the “inside” positions of the β-hairpins are preferably occupied by hydrophobic residues and the “outside” positions by hydrophilic ones. Glycines and alanines have a strong tendency to occupy the “inside” positions in strongly twisted sites of β-strands, while prolines occupy the “outside” positions. There should be at least one glycine residue in the relatively short loop connecting the β-strands. Note that the interstrand S–S-bridges play an important role in folding and selection of the right- and left-turned β-hairpins [17].
Fig. 2. A ball-and-stick model (a) and a schematic representation (b) of a strongly twisted and coiled β-hairpin. Large black balls are “inside” side chains located on the concave surface of the double superhelix, and open balls are “outside” side chains located on the convex surface. A right-turned β-hairpin can be twisted and coiled to form right-handed double superhelix (b and c), but a left-turned β-hairpin (c and d) cannot.
Two β-strands that are not connected by loops or formed by rather distant regions of the polypeptide chain can form an antiparallel β-sheet stitched by hydrogen bonds, which is very similar to the β-hairpin. Such two-stranded β-sheets will be referred to here as ββ-pairs. Like the β-hairpin, the ββ-pair can be right- or left-turned when viewed from the same side, e.g. from the hydrophobic core of a protein. Strongly twisted and coiled ββ-pairs that are similar to the β-hairpin shown in Fig. 2 (a and b), in which the loop is very long or absent, can also be formed. Such two-stranded right-handed superhelices are formed by the right-turned ββ-pairs when viewed from the concave side.
It is rather difficult to find preference for the S-like or Z-like β-sheets if they are compared as isolated flat or moderately twisted β-sheets. The preference is observed at the level of super-secondary structures of higher order that include β-sheets or if triple-stranded β-sheet folds into a three-dimensional structure itself. Thus, some higher order structures include S-like and others Z-like β-sheets.
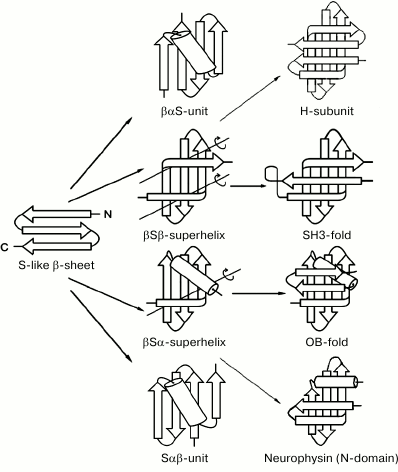
Figure 3 shows several intermediate and completed protein structures that include S-like β-sheets. The βαS- and Sαβ-units can be represented as combinations of S-like β-sheet and right-handed split βαβ-unit, which occur rather often in proteins [10, 18]. Note that the split βαβ-units in these combinations as well as most βαβ-units in α/β-proteins form right-handed superhelices [4, 19, 20]. In βSβ-superhelix, the S-like β-sheet is flanked by two right-handed β-bends. In accordance with the definition [21], in right-handed β-bends the strands bend through ~90° in the right-handed direction when passing from one β-sheet to the other. The overall fold of the resulting structure can be represented as right-handed βSβ-superhelix if the S-like β-sheet is replaced by one imaginary strand. In the right-handed βSα-superhelix, the S-like β-sheet is flanked by right-handed β-bend at the N-terminus and by an α-helix at the C-terminus. Some examples of proteins containing the βSβ- and βSα-superhelices are shown in Fig. 3 (see also [10, 22]).
Fig. 3. Schematic representation of some intermediate and complete protein structures that include S-like β-sheets. The β-strands are shown as arrows directed from the N- to C-ends and α-helices as cylinders. In the βSβ- and βSα-superhelices, imaginary polypeptide chain rotation axes at the sites where strands bend and pass from one layer to another are shown by lines with circular arrows.
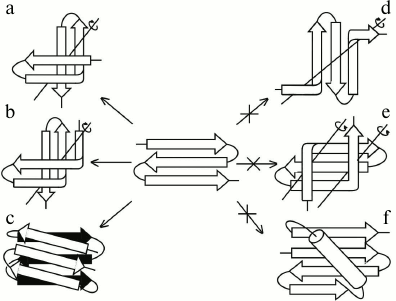
The main difference between S-like and Z-like β-sheets is that the Z-like β-sheet can fold into a compact structure of higher order itself. The 3β-corner is a structural motif that can be represented as Z-like β-sheet folded upon itself so that its two β-hairpins are packed approximately orthogonally in different layers and the central β-strand is bent by ~90° when passing from one layer to the other to form a half-turn of the right-handed superhelix (Fig. 4, a and b) [23], similar to that in βSβ- and βSα-superhelices (see Fig. 3) and β-bends [21]. Note that S-like β-sheet is not able to fold upon itself into a similar compact structure. It could fold into a compact structure if the central β-strand forms a left-handed β-bend when passing from one layer to the other, but this is prohibited, and such structures are not found in proteins [10]. On the other hand, Z-like β-sheet cannot be included into structures similar to those that include S-like β-sheet. For example, a compact βZβ-superhelix (Fig. 4e) is left-handed, contains two prohibited left-handed β-bends, and does not occur in proteins [10]. If the Z-like β-sheet is flanked by two right-handed β-bends (Fig. 4d), the resulting unfolded structure does not seem to be stable, and it also does not occur in proteins. The Zαβ-unit shown in Fig. 4f contains the prohibited left-handed βαβ-unit and does not occur in proteins [10, 18]. Thus, it can be concluded that selection of allowed structures of higher order, which include S-like and Z-like β-sheets, depends on their handedness and compactness.
Fig. 4. Allowed (a, b, c) and prohibited (d, e, f) structures that include Z-like β-sheets. Designations are as in Fig. 3.
STRUCTURAL MOTIFS CONTAINING βαβ- AND
βββ-SUPERHELICES
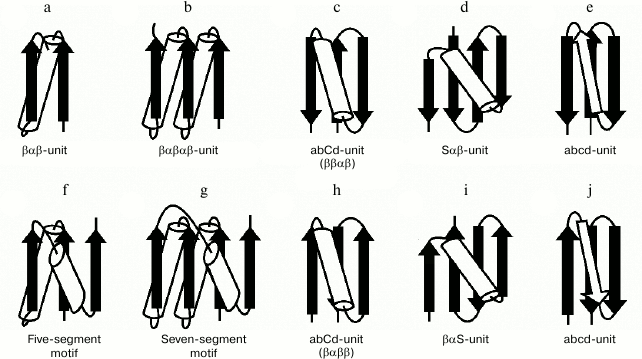
The βαβ-unit or βαβ-superhelix is a two-layer structure in which the β-strands form a parallel β-sheet and the α-helix is located in the other layer, so that the overall fold of the polypeptide chain represents a large cycle of a right-handed superhelix (Fig. 5a). Rao and Rossmann [4] were the first to observe and describe the βαβ-unit and analogous structures later called Rossmann folds. The βαβ-unit usually forms right-handed superhelix in homologous and non-homologous α/β-proteins [4, 19, 20] although there are some left-handed βαβ-superhelices (see below). The βαβαβ-unit (Fig. 5b), which is also a commonly occurring folding unit in α/β-proteins, has two turns of right-handed superhelix. There are seven βαβ-units and consequently seven turns of the right-handed superhelix in the so-called (α/β)8-barrels.
Fig. 5. Structural motifs containing βαβ- and βββ-superhelices. See also the text.
In three-layer α/β-proteins, there are two more complex structural motifs composed of five (Fig. 5f) or seven (Fig. 5g) elements of secondary structure folded into βαβ- or βαβαβ-units [24]. It should be noted that all the α/β-proteins and domains in which α-helices and β-strands alternate along the polypeptide chain can be represented as combinations of βαβ-units that differ in the number of βαβ-units and their arrangement in three-dimensional space. In some subclasses of (α+β)- and α/β-proteins, the βαβ-units form combinations with other elements of secondary structure. For example, addition of one more β-strand to the right-handed βαβ-unit results in formation of the commonly occurring structural motif called abCd-unit (Fig. 5, c and h) [24, 25]. Addition of a β-hairpin, S-like, or Z-like β-sheet to the βαβ-unit results in formation of structural motifs analogous to the abCd-unit (see, e.g. Fig. 5, d and i) that are also widespread in proteins [18, 25]. The βαβ-units and ψ-motifs can also form some combinations [26]. A distinctive feature of these combinations is that the frequency of occurrence of left-handed βαβ-units in them (about 11%) is much higher than average in α/β-proteins (see also below).
In most two-layer β-proteins with aligned β-sheet packings, there is a four-stranded super-secondary structure referred to as the abcd-unit [24, 25, 27]. In the abcd-unit, the β-hairpin formed by strands a and b is closed into a large circle by superhelix bcd (Fig. 5, e and j). The superhelix bcd is analogous to the βαβ-superhelix in the abCd-unit (Fig. 5, c and h) and always occurs in right-handed form in proteins. As seen, the abcd- and abCd-units have the same overall fold of the chain but different conformations of regions c and C [25]. The abcd- and abCd-units having the direct chain orientation include right-turned β-hairpins (Fig. 5, c and e), and those with the reverse chain orientation include left-turned β-hairpins (Fig. 5, h and j). A complex variant of the abcd-unit can include a Z-like β-sheet instead of strand c (Fig. 4c).
Apparently, the so-called parallel β-helices are the largest superhelices formed by β-strands [28-30]. There are two kinds of parallel β-helices, the right-handed β-helix (RβH) and left-handed β-helix (LβH). Each rung (or coil) of the canonical β-helix consists of three β-strands connected by standard β-turns or loop regions. The β-strands of the adjacent rungs form flat parallel β-sheets whose close packing results in formation of triangular β-helix. There are several types of β-helices that differ in number of β-strands and/or residues per rung. In the right-handed β-helices (RβH), a rung can consist of 2-4 β-strands, and the overall fold of these β-helices depends of the number of β-sheets. It is of interest that there is a fold, which was named right-handed quadrilateral β-helix, that exhibits size, shape, and electrostatic similarity to the B-form DNA (Mfp, PDB ID 2BM4) [31].
COMBINATIONS OF Π-LIKE MODULES AND βαβ-UNITS,
β-HAIRPINS, S- AND Z-LIKE β-SHEETS
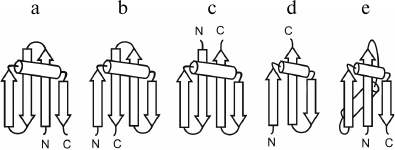
The overall fold of the Π-module can be represented as β-strand–crossover loop–β-strand, which are arranged so that they resemble the Greek letter Π or a clip. In contrast to β-hairpin, the β-strands of the Π-module do not form H-bonds between each other, but then interact with another one or two additional β-strands packed in between them in the β-sheet. Thus, the Π-module can be represented as a split β-hairpin. The crossover loop connects two β-strands of the Π-module, crosses over the central β-strands of the β-sheet, and provides a reserve turn of the polypeptide chain. There are right- and left-turned Π-modules. When viewed from the crossover loop, the polypeptide chain runs from the N- to the C-end in the clockwise direction in the right-turned Π-module (Fig. 6a) and in the anticlockwise direction in the left-turned Π-module (Fig. 6c).
Fig. 6. Possible pathways of formation of combinations between the right-handed βαβ-unit and the right-turned (a, d) and left-turned (b, c) Π-module. Π-modules are highlighted in gray.
Analysis of our database shows that in φ-motifs the Π-modules are predominantly right-turned (>95%) [7]. In combinations of the Π-module with S- and Z-like β-sheets (denoted as the SΠ- and ZΠ-motifs), all the Π-modules are right-turned [32], but in combinations of Π-modules and βαβ-units 34% of the Π-modules are left-turned and 66% are right-turned [26]. A more detailed analysis of this problem shows that selection of the right- or left-turned Π-module is determined by mutual arrangement of the Π-module and the βαβ-unit as well as by compactness and stability of the resulting combinations. If the Π-module follows the right-handed βαβ-unit along the chain and forms a compact closed structure as shown in Fig. 6a, the right-turned Π-module is selected. This combination denoted βαβΠ-motif occurs most often. In the Πβαβ-motif, the right-handed βαβ-unit follows the Π-module along the chain, so the left-turned Π-module is selected (Fig. 6c). Note that in the βαβΠ- and Πβαβ-motifs the Π-module clips together the β-strands of the βαβ-units, making the resulting structural motifs more cooperative and stable. Addition of the left-turned Π-module to the C-end of the right-handed βαβ-unit results in formation of an unfolded and unfavorable structure in which the second β-strand of the Π-module does not interact with the βαβ-unit (Fig. 6b). An analogous unstable structure results if the right-turned Π-module is added to the N-end of the right-handed βαβ-unit (Fig. 6d). For stabilization of these combinations, it is possible to add β-strands shown by dotted lines or to transform the Π-modules shown in Fig. 6, b and d, into β-hairpins, but this is another story. Thus, there is a definite relationship between mutual arrangement of the Π-module and the βαβ-unit and their handedness in resulting combinations.
The structures of the crossover loops are rather different in right- and left-turned Π-modules. Analysis shows that 73% of right-turned Π-modules contain α-helices (up to 2-3 turns) in the crossover loops, while only 7% of left-turned Π-modules have α-helices in the loops. As a rule, in the right-turned Π-modules the α-helix and the second β-strand is connected by a small standard arch. Thus, the overall structure of most right-turned Π-modules can be represented as β-strand–α-helix–arch–β-strand. Such Π-modules are also widespread in other structural motifs, for example, in combinations of Π-modules and β-hairpins and in S-like and Z-like β-sheets (Fig. 7) [7, 22, 26, 32, 33]. Structural alignment and comparative analysis of their amino acid sequences have shown that the Π-modules found in unrelated proteins and in different combinations have very similar sequence patterns of the key hydrophobic, hydrophilic, glycine, and proline residues [32, 34]. Apparently, the isolated Π-module is not stable because its β-strands do not interact with each other and its overall fold is not compact. However, in combination with the S-like and Z-like β-sheets, β-hairpins, and βαβ-units, the Π-module fastens their β-strand like a clip, which results in formation of compact and stable structural motifs that can act as nuclei and/or “ready-made” building blocks in protein folding.
Fig. 7. Schematic representation of combinations of the Π-module and the S-like β-sheet (a), the Z-like β-sheet (b), two β-hairpins that are the part of the cleaved φ-motif (c), a β-hairpin that is a part of the φ-motif (d), and the βαβ-unit (e).
HANDEDNESS OF α-HELICAL STRUCTURES
In proteins, the packing of α-helices is restricted by both the principle of close packing and the chemical nature of the side chains. As a rule, α-helices are amphipathic and pack so that their hydrophobic faces are buried in a hydrophobic core and polar side chains are accessible to water molecules or form hydrogen bonds and ion pairs between themselves or with cofactors. The first detailed model for helix-to-helix packing was proposed by Crick in 1953 [35]. In this model, a side chain from one α-helix (knob) packs into a space surrounded by four side chains of the opposite α-helix (hole), and vice versa. This model gives two preferred orientations of closely packed α-helices, Ω = 20° and Ω = –70° (Ω denotes the torsion angle between the helix axes). If α-helices are long enough, while packed at Ω = 20° they form a left-handed superhelix having helical pitch of about 140 Å. There are such coiled-coil structures in tropomyosin, α-keratin, etc. Note that in these coiled-coil structures the right-handed α-helices form left-handed superhelix. It was shown that amino acid sequences encoding coiled-coil structures of this type have so-called heptad repeats [36]. The seven positions of repeats are conventionally referred to as a-b-c-d-e-f-g, where a and d positions are occupied by hydrophobic and e and g positions by hydrophilic or charged amino acid residues [36] (see also reviews [37, 38]). On the surface of α-helix having heptad repeats, a hydrophobic stripe twisted anticlockwise around the α-helical axis is formed. Close packing of two or more α-helices having such hydrophobic stripes to form a closely packed hydrophobic core results in formation of a left-handed superhelix (or a coiled-coil) in which α-helices are packed at Ω = 20°. The other widespread repeat in α-helices is the so-called undecatad repeat, the eleven positions of which are conventionally referred to as letters a to k. In the undecatad repeat, positions a, d, and h are occupied by hydrophobic residues, which produce a right-handed hydrophobic stripe on the surface of the α-helix. Close packing of α-helices having undecatad repeat gives rise to a right-handed superhelix or supercoil (see, e.g. [39, 40]).
In globular proteins, both kinds of handedness are observed in structures formed by α-helices. For example, α-helical hairpins can be right- or left-turned when viewed from one side, e.g. from the hydrophobic core. On the other hand, two connected α-helices can be packed approximately crosswise so that, in three dimensions, the polypeptide chain passes through almost one complete turn of a left-handed superhelix [41]. Variants of this structure were initially found in two homologous protein families, “EF-hands” in calcium-binding proteins [42] and “helix-turn-helix” motifs in DNA-binding proteins [43]. It was shown later that two-α-helical structures of this kind referred to as αα-corners are widespread in both homologous and nonhomologous proteins and are found practically always in one form, as the left-handed superhelix [24, 41]. Addition of other α-helices to the αα-corner gives rise to many higher-order structures, some of which are superhelices having definite handedness (for details, see [24, 41, 44]).
The three-α-helix bundle occurs in proteins in two forms, as left-handed and right-handed superhelix. Below a schematic representation of the right- and left-handed superhelices viewed end-on is shown, where A, B, and C are three α-helices, double lines are near connections, and single lines are distant connections.
Four-α-helical super-secondary structures in proteins can be of two types – type I and type II [45]. When viewed from the N-end of helix A, the arrangement of four α-helices A, B, C, and D in these bundles is as shown below:
Moreover, four α-helices adjacent along the polypeptide chain can fold into a superhelix. For example, this can be obtained by addition of a fourth α-helix to a right-handed three-α-helical superhelix (see above). Such two-layer α/α-superhelices, also called α-solenoids, can be formed by 3-4 up to 17-18 α-helices as, e.g. in lipovitellin [46]. The arrangement of α-helices in the α/α-superhelix can be schematically represented as follows:
Such α/α-superhelices have now been found in some dozens of proteins, and most of them are right-handed. The predominance of the right-handed form of α/α-superhelices as well as the handedness of other structural motifs are determined by several factors, but essentially it is a result of the homochirality of L-amino acid residues in proteins. It should be noted that the problem of selection of right- or left-handed three-α-helical bundles, superhelices, α-hairpins, and four-α-helix bundles of type I or II can be solved if their α-helices are connected by short loops (see, e.g. [5, 41]).
The main conclusion of this work is that protein molecules have two very important properties, chirality and handedness, which should be distinguishable. As described above, there are two kinds of handedness in proteins – helical handedness and handedness of arrangement. Some protein structures exhibit both the kinds of handedness. Handedness is observed at all the levels of protein structural organization, from α-helices, β-strands, hairpins, and βαβ-units up to complex structural motifs, superhelices, and supramolecular structures in fibrous and polymer proteins. Some super-secondary structures such as β-hairpins, Π-modules, and triple-stranded β-sheets can exist in two forms, but at the level of higher order structures that include them, only one form is selected. There are many protein structures that have a unique handedness, for example α-helices, αα-corners, βαβ-units, abcd-units, and so on. This property of polypeptide chains is of particular value in protein folding and protein modeling since it drastically reduces the number of possible folds.
Acknowledgments
I thank Ms. E. A. Boshkova and Dr. A. M. Kargatov for help in drawing the figures and preparation of the manuscript.
This work was supported by the Russian Foundation for Basic Research (projects Nos. 13-04-00150 and 17-04-00242).
REFERENCES
1.IUPAC-IUB Commission on Biochemical Nomenclature
(1970) J. Mol. Biol., 52, 1-17.
2.Pauling, L., and Corey, R. B. (1951) Atomic
coordinates and structure factors for two helical configurations of
polypeptide chains, Proc. Natl. Acad. Sci. USA, 37,
235-240.
3.Watson, J. D., and Crick, F. H. C. (1953) Molecular
structure of nucleic acids. A structure for deoxyribose nucleic acid,
Nature, 171, 737-738.
4.Rao, S. T., and Rossmann, M. G. (1973) Comparison
of super-secondary structures in proteins, J. Mol. Biol.,
76, 241-256.
5.Efimov, A. V. (1991) Structure of
α-α-hairpins with short connections, Protein Eng.,
4, 245-250.
6.Efimov, A. V. (1991) Structure of coiled
β-β-hairpins and β-β-corners, FEBS Lett.,
284, 288-292.
7.Efimov, A. V. (2008) Structural trees for proteins
containing φ-motifs, Biochemistry (Moscow), 73,
23-28.
8.Suguna, K., Bott, R. R., Padlan, E. A.,
Subramanian, E., Sheriff, S., Cohen, G. H., and Davies, D. R. (1987)
Structure and refinement at 1.8 Å resolution of the aspartic
proteinase from Rhizopus chinensis, J. Mol. Biol.,
196, 877-900.
9.Castillo, R. M., Mizuguchi, K., Dhanaraj, V.,
Albert, A., Blundell, T. L., and Murzin, A. G. (1999) A six-stranded
double-psi beta barrel is shared by several protein superfamilies,
Structure, 7, 227-236.
10.Efimov, A. V. (1993) Super-secondary structures
involving triple-strand β-sheets, FEBS Lett., 334,
253-256.
11.Ramachandran, G. N., and Sasisekharan, V. (1968)
Conformation of polypeptides and proteins, Adv. Protein Chem.,
7, 283-438.
12.Efimov, A. V. (1986) Standard conformations of
polypeptide chain in irregular regions of proteins, Mol. Biol.
(Moscow), 20, 208-216.
13.Chothia, C. (1973) Conformation of twisted
β-pleated sheets in proteins, Proc. Natl. Acad. Sci. USA,
75, 295-302.
14.Chothia, C. (1983) Coiling of β-pleated
sheets, J. Mol. Biol., 163, 107-117.
15.Nishikawa, K., and Scheraga, H. A. (1976)
Geometrical criteria for formation of coiled-coil structure of
polypeptide chains, Macromolecules, 9, 395-407.
16.Boshkova, E. A., Brazhnikov, E. V., and Efimov,
A. V. (2016) Relationship between structure and amino acid sequence of
strongly twisted and coiled β-hairpins in globular proteins,
Mol. Biol. (Moscow), 50, 777-782.
17.Brazhnikov, E. V., and Efimov, A. V. (2010)
Structure of β-β-hairpins closed into cycles by
S–S-bridges, Mol. Biol. (Moscow), 44, 466-472.
18.Gordeev, A. B., and Efimov, A. V. (2013) Modeling
of folds and folding pathways for some protein families of
(α+β)- and (α/β)-classes, J. Biomol. Struct.
Dyn., 31, 4-16.
19.Sternberg, M. J. E., and Thornton, J. M. (1976)
On the conformation of proteins: the handedness of the
β-strand-α-helix-β-strand unit, J. Mol. Biol.,
105, 367-382.
20.Richardson, J. S. (1976) Handedness of crossover
connections in β-sheets, Proc. Natl. Acad. Sci. USA,
73, 2619-2623.
21.Chothia, C., and Janin, J. (1982) Orthogonal
packing of β-pleated sheets in proteins, Biochemistry,
21, 3955-3965.
22.Efimov, A. V. (1998) A structural tree for
proteins containing S-like β-sheets, FEBS Lett.,
437, 246-250.
23.Efimov, A. V. (1992) A novel super-secondary
structure of β-proteins: a triple-strand corner, FEBS
Lett., 298, 261-265.
24.Efimov, A. V. (1997) Structural trees for protein
superfamilies, Proteins, 28, 241-260.
25.Efimov, A. V. (1995) Structural similarity
between two-layer α/β and β-proteins, J. Mol.
Biol., 245, 402-415.
26.Kargatov, A. M., and Efimov, A. V. (2010) A novel
structural motif and structural trees for proteins containing it,
Biochemistry (Moscow), 75, 249-256.
27.Efimov, A. V. (1982) Super-secondary structure of
β-proteins, Mol. Biol. (Moscow), 16, 635-641.
28.Jenkins, J., and Pickersgill, R. (2001) The
architecture of parallel β-helices and related folds, Prog.
Biophys. Mol. Biol., 77, 111-175.
29.Ienger, P., Joshi, N. V., and Balaram, P. (2006)
Conformational and sequence signatures in β-helix proteins,
Structure, 14, 529-542.
30.Kajava, A. V., and Steven, A. C. (2006)
Beta-rolls, beta-helices, and other beta-solenoid proteins, Adv.
Protein Chem., 73, 55-96.
31.Hegde, S. S., Vetting, M. W., Roderick, S. L.,
Mitchenall, L. A., Maxwell, A., Takiff, H. E., and Blanchard, J. S.
(2005) A fluoroquinolone resistance protein from Mycobacterium
tuberculosis that mimics DNA, Science, 308,
1480-1483.
32.Efimov, A. V. (2017) Structural motifs in which
β-strands are clipped together with the Π-like module,
Proteins, doi: 10.1002/prot.25346.
33.Murzin, A. G. (1993)
OB(oligonucleotide/oligosaccharide binding)-fold: common structural and
functional solution for non-homologous sequences, EMBO J.,
12, 861-867.
34.Kargatov, A. M., and Efimov, A. V. (2018) Unique
combinations of βαβ-units and Π-like modules in
proteins and features of their amino acid sequences, Mol. Biol.
(Moscow), 52, in press.
35.Crick, F. H. C. (1953) The packing of
α-helices: simple coiled-coils, Acta Cryst., 6,
689-697.
36.McLachlan, A. D., and Stewart, M. (1975)
Tropomyosin coiled-coil interactions: evidence for an unstaggered
structure, J. Mol. Biol., 98, 293-304.
37.Lupas, A., and Gruber, M. (2005) The structure of
α-helical coiled coils, Adv. Protein Chem., 70,
37-78.
38.Efimov, A. V. (1999) Complementary packing of
α-helices in proteins, FEBS Lett., 463, 3-6.
39.Plecs, J. J., Harbury, P. B., Kim, P. S., and
Alber, T. (2004) Structural test of the parameterized-backbone method
for protein design, J. Mol. Biol., 342, 289-297.
40.Stetefeld, J., Jenny, M., Schulthess, T.,
Landwehr, R., Engel, J., and Kammerer, R. A. (2000) Crystal structure
of a naturally occurring parallel right-handed coiled coil tetramer,
Nat. Struct. Biol., 7, 772-776.
41.Efimov, A. V. (1984) A novel super-secondary
structure of proteins and the relation between the structure and the
amino acid sequence, FEBS Lett., 166, 33-38.
42.Kretsinger, R. H., and Nockolds, C. E. (1973)
Carp muscle calcium-binding protein. II. Structure determination and
general description, J. Biol. Chem., 248, 3313-3326.
43.Pabo, C. O., and Sauer, R. T. (1984)
Protein–DNA recognition, Annu. Rev. Biochem., 53,
293-321.
44.Efimov, A. V. (1997) Novel structural motifs in
α-helical proteins, Russ. J. Bioorg. Chem., 23,
223-230.
45.Argos, P., Rossmann, M. G., and Johnson, J. E.
(1977) A four-helical super-secondary structure, Biochem. Biophys.
Res. Commun., 75, 83-86.
46.Anderson, T. A., Levitt, D. G., and Banaszak, L.
J. (1998) The structural basis of lipid interactions in lipovitellin, a
soluble lipoprotein, Structure, 6, 895-909.