REVIEW: Interactions between the Translation Machinery and Microtubules
E. M. Chudinova1,2* and E. S. Nadezhdina1,3
1Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia; E-mail: chudiel@mail.ru2Peoples’ Friendship University of Russia, 117198 Moscow, Russia
3Lomonosov Moscow State University, 119991 Moscow, Russia
* To whom correspondence should be addressed.
Received June 24, 2017; Revision received July 17, 2017
Microtubules are components of eukaryotic cytoskeleton that are involved in the transport of various components from the nucleus to the cell periphery and back. They also act as a platform for assembly of complex molecular ensembles. Ribonucleoprotein (RNP) complexes, such as ribosomes and mRNPs, are transported over significant distances (e.g. to neuronal processes) along microtubules. The association of RNPs with microtubules and their transport along these structures are essential for compartmentalization of protein biosynthesis in cells. Microtubules greatly facilitate assembly of stress RNP granules formed by accumulation of translation machinery components during cell stress response. Microtubules are necessary for the cytoplasm-to-nucleus transport of proteins, including ribosomal proteins. At the same time, ribosomal proteins and RNA-binding proteins can influence cell mobility and cytoplasm organization by regulating microtubule dynamics. The molecular mechanisms underlying the association between the translation machinery components and microtubules have not been studied systematically; the results of such studies are mostly fragmentary. In this review, we attempt to fill this gap by summarizing and discussing the data on protein and RNA components of the translation machinery that directly interact with microtubules or microtubule motor proteins.
KEY WORDS: ribosome, polysome, mRNP, mRNA, kinesin, dynein, stress granuleDOI: 10.1134/S0006297918140146
Abbreviations: APC, adenomatous polyposis coli; eEF, eukaryotic translation elongation factor; Egl, Egalitarian; eIF, eukaryotic translation initiation factor; Hu antigen, antigen in human paraneoplastic neurological syndromes; JAKMIP1, Janus kinase and microtubule-interacting protein 1; KIF, kinesin family member; MAP1B LC1, microtubule-associated protein 1B light chain 1; mRNP, messenger ribonucleoprotein; PABP, poly(A)-binding protein; RpL, large subunit ribosomal protein; RpS, small subunit ribosomal protein; UTR, mRNA untranslated region; YB1, Y box-binding protein 1; ZBP1, zipcode-binding protein 1.
Protein synthesis (translation) is a complex process that requires the
contribution of multiple cell components. It involves formation of
macromolecular complexes of mRNAs, tRNAs, RNA-binding proteins,
ribosomes, and regulatory protein factors. For example, translation
initiation proceeds via successive formation of the 43S preinitiation
complex, 48S and 80S initiation complexes, and polysomes [1, 2]. The rate of translation in
cells is much higher than in cell-free systems. Disruption of the cell
membrane with saponin does not affect the rate of protein synthesis,
which stays 40 times higher than the rate of protein synthesis in an
in vitro preparation of polysomes isolated from the same cells
[3]. This suggests that efficient protein synthesis
requires some spatial organization of the translation machinery. It is
reasonable to assume that such spatial and temporal organization could
be provided by the cytoskeleton, i.e. an assembly of
detergent-resistant fibrillar structures in the cytoplasm. The
cytoskeleton is formed by actin and myosin filaments, microtubules, and
intermediate filaments, all of which can support cell organization and
cytoplasm transport. Indeed, experimentally induced depolymerization of
actin cytoskeleton significantly decreases the rate of protein
biosynthesis in cells [4, 5].
Similarly, microtubule disruption slows the processes of proteins
synthesis by approximately 20% (our data); microtubule depolymerization
targets only certain mRNAs for translational repression, i.e. it acts
selectively [6].
The cytoskeleton is essential for compartmentalization of protein biosynthesis in the cell. This phenomenon can be clearly observed in oocytes and early embryos of Drosophila melanogaster and Xenopus laevis, neurons, and budding yeast [7-12]. The concentration of β-actin mRNA increases in the regions of focal adhesion in fibroblasts [13]. Compartmentalization of protein biosynthesis is important for correct segmentation of insect embryos, axon growth, and directed fibroblast motility on substrates.
What are the mechanisms of interactions between the translation machinery and the cytoskeleton? Further in this article, the components of the translation machinery are attributed to ribosomes, ribosomal proteins, translation factors, and various mRNPs. It has been found that at least some of these elements can be transported across the cell along the cytoskeleton, or they can use the cytoskeleton for anchoring in the cytoplasm. The necessity for such transport arises from the fact that ribosomal proteins are synthesized in the cytoplasm and should be transported to the nucleus, while assembled ribosomal subunits and mRNPs should then be transported back to the cytoplasm and, in some cases, to cytoplasmic regions very distant from the nucleus. Relatively short actin filaments form a dense cytoplasmic network that serves for polysome anchoring [14] and regulation of transcription factors through binding [15-17]. Intracellular transport over long distances (e.g. from the nucleus to the cell periphery) is provided mostly by microtubules that form relatively long structures.
In this review, we focus on the interactions of the translation machinery with microtubules. Microtubules are an essential part of the cytoskeleton structural network. They are long (up to tens of microns) tubular structures 25 nm in diameter with walls composed of uniformly polymerized tubulin molecule dimers. The dimers in the microtubule can be exchanged with soluble tubulin dimers only at the microtubule ends. The character of tubulin organization in the microtubule provides its polarity: microtubules have (+) and (–) ends that differ in their dynamics. Microtubules are dynamic structures: they constantly polymerize and depolymerize in the cell. The half-life of a microtubule is within minutes or, in rare cases, tens of minutes. Microtubule depolymerization is accompanied by GTP hydrolysis by tubulin molecules. Numerous compounds can affect the ability of tubulin molecules to polymerize or cause microtubule disruption; in contrast, some compounds stabilize microtubules. Microtubules associate with many structural proteins that regulate microtubule dynamics. Changes in microtubule dynamics play an important regulatory role in the cell [18], although the mechanism of this process has not been completely elucidated. The major role of microtubules is intracellular transport – various cell structures are transported along the microtubules by attaching themselves to the motor proteins that are activated by microtubules and travel along them. Microtubule-associated motor proteins include kinesins and dyneins, as well as some myosins that can also bind to the microtubules. Motor proteins can travel either to the (+) (kinesins) or (–) (dyneins and some kinesins) ends of the microtubules. RNPs are transported over large distances (e.g. to neuronal processes) along microtubules [19]; experimental microtubule disruption arrests RNP transport.
RNP transport along microtubules is poorly studied; in most cases, molecular mechanisms of the association of RNPs with motor or microtubule proteins remain obscure. Cryoelectronic micrographs of the cytoskeleton and ribosomes show that some of the ribosomes localize very close to microtubules, but their orientation toward these microtubules can be different [20]. Therefore, it remains unclear which of the ribosomal proteins or ribosome-associated factors participate in microtubule binding and how specific this binding is. To understand better the interactions between the translation machinery and microtubules, it is necessary to elucidate which protein–protein or protein–RNA interactions are involved in the binding.
ASSOCIATION OF RIBOSOMAL PROTEINS, TRANSLATION FACTORS, AND
RNA-BINDING PROTEINS WITH MICROTUBULES
Ribosomal proteins and translation factors in preparations of microtubule-associated proteins. To identify microtubule-binding proteins, stabilized exogenous microtubules are added to a clarified tissue or cell homogenate. After incubation, the mixture is layered onto 40-50% glycerol, and microtubules together with the associated proteins are pelleted by centrifugation at 100,000g (microtubule cosedimentation test). As an alternative procedure, endogenous microtubules are polymerized in a homogenate by adding taxol. The classical method for microtubule isolation is purification by several cycles of tubulin polymerization–depolymerization [21]. This method is based on the ability of microtubules to depolymerize at low temperatures and reassemble upon heating or in the presence of 4 M glycerol. Highly purified tubulin preparations can be obtained using several centrifugation cycles in a specific temperature regime. Proteins bound to the microtubules can withstand several polymerization–depolymerization cycles. The resulting pellet is analyzed by 1D or 2D electrophoresis, immunoblotting, and mass spectrometry. The number of individual proteins that coprecipitate with microtubules is relatively high (200-600). Interestingly, these proteins always include translation machinery components, such as ribosomal proteins, translation initiation and elongation factors, and transcription regulators [22-29]. In plant homogenates, 14-19% of all microtubule-bound proteins are involved in translation [27, 28]; in homogenates of Drosophila embryos, the content of such proteins is 8%, i.e. more than of any other functional group [29]. It remains unclear if the translation machinery proteins bind directly to tubulin or to some other microtubule-associated proteins (e.g. motor proteins). Since in most studies the procedure for lysate preparation allows the presence of ribosomes or, at least of translation factors, in the content of a large common complex, it is also unknown which proteins of numerous RNPs or multisubunit factors bind directly to the microtubules, and which proteins associate with microtubules indirectly as components of microtubule-interacting protein complexes. Thus, when microtubules were pelleted from mitotic cell lysate clarified at 19,000g, the precipitate contained ribosomal proteins RpL6, RpL13, RpL19, RpL22, RpL23, RpS27A, P0, P1, P2, and some subunits of the eEF1, eEF2, eIF1, eIF2, eIF3, eIF4, and eIF5 translation factors [26]. Perhaps the pellet was contaminated with whole ribosomes that could be associated with translation factors. Similar arrays of proteins (translation factor subunits and ribosomal proteins) were found in other experiments on cosedimentation of microtubules, although the exact lists of proteins differed in different studies. More than 20 (out of approximately 80) ribosomal proteins were identified by mass spectrometry in isolated mitotic spindles (a variant of precipitated microtubules) [30].
Translation initiation factor eIF3 contains up to 11 subunits. The eIF3a (p170) subunit has been found in considerable amounts in microtubule preparation from adrenal medulla [31], bovine brain [32], and plant cells [28]. It coprecipitated with microtubules from rat brain homogenate [22] and cultured RAW264.7 macrophages [23], but not that from meiotic Xenopus oocytes [25]. eIF3a coprecipitated with microtubules from mitotic cells accumulated by the double thymidine block method [24], but not from cells synchronized at mitosis by 12-h incubation with Nocodazole followed by mitotic cells shake-off [26]. Perhaps the interactions between eIF3a and microtubules are determined by the functional state of the cell. It is unclear if eIF3 coprecipitates with microtubules as a whole multisubunit complex, or these are unbound free subunits that can interact and cosediment with microtubules. Identification of eIF3 complex subunits that associate with microtubules requires experiments utilizing individual purified proteins. Therefore, such “gross” analysis of proteins coprecipitating with microtubules is not a reliable method for elucidating interactions between the translation machinery and microtubules.
Immunofluorescence localization of proteins on microtubules. Microtubule-interacting proteins can be identified using various experimental approaches. One relatively simple method is immunofluorescence staining of a protein of interest followed by analysis of its intracellular location relative to the microtubules. Another approach is ectopic expression of GFP-labeled protein in cells. Fluorescence microscopy can show colocalization of the studied protein with microtubules only if the affinity of the protein to the microtubules is high (i.e. protein signal is higher than the background fluorescence). Thus, colocalization of the RPG1 protein (eIF3a ortholog in Saccharomyces cerevisiae yeast) with microtubules was shown by confocal microscopy [33]. eIF3a also exhibited weak colocalization with microtubules in cultured mammalian cells, as demonstrated by immunofluorescence microscopy [32].
Colocalization of microtubules with JAKMIP1 protein involved in regulation of mRNA expression in neurons was also shown by immunofluorescence staining [34]. Using expression of the JAKMIP1 fragments, it was found that the N-terminal region of JAKMIP1 interacts with the microtubules, while its C-terminal region binds Janus kinase (JAK). JAKMIP1 participates in the signal transduction from NMDA (N-methyl-D-aspartate) receptors and is essential for the normal development of neurons and synaptic plasticity. Mutations in JAKMIP1 are associated with autism and many neurological disorders due to the involvement of JAKMIP1 in the regulation of translation of the Grin2a, Grin2b, and Shank2 mRNAs coding the NMDA receptor subunits and the postsynaptic density structural protein, respectively. JAKMIP1 was found in transported RNP granules and polysomes in neurons [34].
Immunofluorescence staining does not distinguish between free proteins and proteins in the content of multisubunit complexes. Ribosomal proteins exist in the cell as both free molecules and components of ribosomes. Thus, expressed GFP–RpL22e was found along the microtubules and in the cytoplasm (Fig. 1), and it was impossible to tell apart protein molecules bound to the ribosomes and the pool of free GFP–RpL22e molecules.
The highest concentration of microtubules is in the mitotic spindle. If a protein has an affinity to microtubules, and this affinity is preserved during mitosis, such protein should accumulate in the mitotic spindle. Preparations of mitotic spindles contain many ribosomal proteins. It is reasonable to assume that ribosomes themselves bind to the mitotic spindles. However, antibodies against some of the ribosomal proteins (RpL11, RpS6) fail to stain mitotic spindles [35], i.e. it is more probable that interaction with mitotic spindles is characteristic of individual ribosomal proteins, but not entire ribosomes. Interestingly, RpL11 was identified in isolated mitotic spindles using mass spectrometry [28], but not by immunofluorescent staining. Immunofluorescence staining showed the presence of the ribosomal protein RpS3 in the spindle. Depletion of the RpS3 pool using RNA interference disturbed the shape of the mitotic spindle and resulted in partial arrest of cell division in metaphase, which indicates that RpS3 might be involved in regulation of microtubule dynamics in mitosis [35]. One of the smallest (25 a.a.) and most basic eukaryotic proteins – RpL41 – was also found in the mitotic spindle in anaphase and telophase. Depletion of the RpL41 pool by RNA interference leads to anomalies of the centrosomes, central spindle, and residual body in mitosis [36].
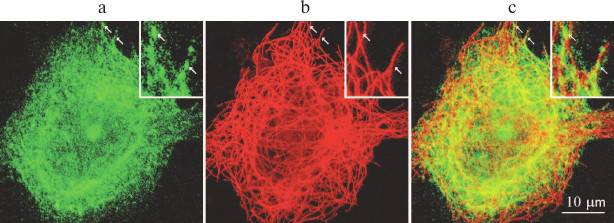
Fig. 1. GFP–RpL22 partially colocalizes with microtubules in cultured Vero cells: a) GFP–RpL22 (green fluorescence); b) immunostained microtubules (red fluorescence); c) merge of (a) and (b). Arrows indicate sites of colocalizations; inset, magnified cell fragment.
In vitro analysis of interactions between individual proteins and microtubules. The most reliable approach for elucidating molecular mechanisms underlying interaction between the translation machinery and microtubules is direct study of the binding of individual proteins to tubulin or other microtubule proteins. For example, association of the ribosomal RpL10 protein with tubulin was shown by immunoprecipitation [37]. However, these proteins were coprecipitated from a cell lysate clarified by centrifugation at 15,000g only, i.e. containing ribosomes and ribosomal subunits. Therefore, the RpL10 might not bind to the microtubules directly, but rather form a complex with some adaptor protein that, in its turn, binds to the microtubules. Direct unmediated interaction of RpL10 with microtubules could be demonstrated in experiment using purified RpL10 and tubulin proteins (or microtubule proteins). A few experiments of this kind have been reported. Thus, purified recombinant 6His-RpS3 was found to bind purified tubulin (as demonstrated using 6His-binding affinity resin) [35]. Purified RpS3 and RpL41 coprecipitated with taxol-stabilized microtubules [35, 36]. Purified recombinant RpL22 coprecipitated with microtubules polymerized from purified tubulin (Chudinova, et al., submitted for publication). The dissociation constant for the RpL22–microtubule complex was 1.3 µM, which is only slightly higher than dissociation constants for the “classical” structural proteins of microtubules. RpSA (also known as RpS40 or laminin receptor LamR) bound to tubulin immobilized on an ELISA plate. The similar bacterial RpS2 protein displayed no affinity for tubulin [38], thereby indicating that interaction between RpSA and tubulin is specific. According to our unpublished data, the N-terminal fragment of recombinant eIF3a protein coprecipitates with microtubules assembled from purified tubulin, which suggests that eIF3 binds to the microtubules through this subunit. The ability of other eIF3 subunits to bind tubulin has not been tested.
Binding of YB1 (Y box-binding protein 1) to microtubules has been convincingly shown and thoroughly studied. YB1 is a multifunctional protein involved in the regulation of transcription and translation [39]. It was found that YB1 facilitates polymerization of tubulin (both purified and containing associated proteins). It also coprecipitates with microtubules and binds tubulin in affinity chromatography. Tubulin could be eluted from YB1-Sepharose only with 600 mM NaCl (as compared to 150 mM NaCl required for tubulin elution from the control casein-Sepharose resin), which indicates that YB1 association with tubulin is specific. YB1 retains the ability to bind tubulin even after removal of the tubulin C-terminal fragment that contains numerous glutamate residues, i.e. after significant change in the tubulin molecule charge. Atomic-force microscopy showed that microtubules polymerized in the presence of YB1 have larger diameter, supposedly due to the coating of their outer surface with YB1, since the number of tubulin protofilaments in the microtubule wall does not change. YB1 forms aggregates with free tubulin dimers. In the presence of tubulin, the conformation of the mRNA–YB1 complexes changes, which could be seen from changes in the electrophoretic mobility of these complexes in native electrophoresis [40].
Positive charge of most RNA-binding proteins promotes their binding to negatively charged RNA molecules and microtubules. The question arises if there is a competition between RNA and microtubules for the protein binding. Atomic-force microscopy showed that YB1 in saturating concentrations can act as adaptor molecules, i.e. bind to both RNA and microtubules. Large RNP aggregates were found to form on microtubules in the presence of YB1 (Fig. 2). A similar phenomenon was observed for poly(A)-binding protein (PABP) [41]. These observations suggest the importance of microtubules and tubulin in transcription regulation and pose a question of the possible involvement of translation-associated proteins in the regulation of microtubule dynamics. So far, this issue has not been investigated.
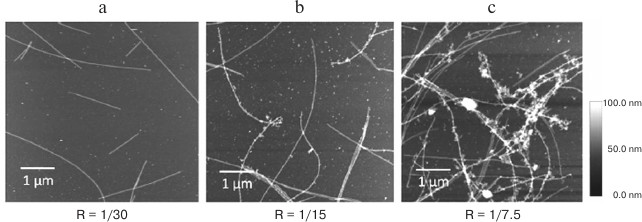
Fig. 2. Atomic force microscopy imaging of microtubule preparations with luciferase mRNA and YB1 at different YB1/mRNA molar ratios (R). Interaction between microtubules and RNPs occurs only at high YB1/mRNA ratio (from [41] with kind permission from John Willey & Sons, Inc.).
Translation machinery components and cytoskeleton dynamics. Another microtubule-associated protein that was found to interact with mRNA is the structural oncosuppressor APC (adenomatous polyposis coli) [42]. This protein binds to the (+) end of the microtubule, as well as associating with cell edges. APC is involved in the Wnt signaling pathway. It also interacts with multiple proteins, and at least with 260 different mRNAs, as demonstrated by the HITS-CLIP method (a technique that uses UV cross-linking to identify RNAs directly bound to a protein of interest). One of these RNAs encodes β2B-tubulin (Tubb2b), an isoform of β-tubulin found in dynamic microtubules in the axon growth cone. Inhibition of the APC binding to the Tubb2b mRNA inhibits microtubule dynamics and impairs neuron migration in vivo. Therefore, APC-mediated anchoring of Tubb2b mRNA in the axon growth cone regulates microtubule dynamics [42]. APC is also important for anchoring the transcript at the leading edge of fibroblasts [43] and local translation of Lis1 (dynein cofactor) mRNA in axons in response to stimulation by nerve growth factor [44].
Elongation factor 1α (EF-1α) also binds to microtubules. When added to tubulin at a 1 : 20 molar ratio, EF-1α stabilizes microtubules, lessens the frequency of their depolymerization, and decreases 2-3-fold the shortening velocity of microtubules polymerized from plant tubulin or tubulin from bovine brain homogenate in vitro [45]. Interestingly, at the EF-1α/tubulin ratio of 1 : 3, EF-1α causes fragmentation of taxol-stabilized microtubules in analogous in vitro experimental systems. Microinjection of EF-1α into fibroblasts results in microtubule fragmentation [46]. Based on the results of the two above-mentioned studies, we assume that microtubule dynamics depend on the concentration of free EF-1α. EF-1α also modulates the cytoskeleton in yeast. Overexpression of EF-1α in yeast cells increases cell sensitivity to the cytoskeleton-disrupting agents cytochalasin D and thiabendazole and causes aberrant cell morphology typical for cells with defective cytoskeleton [47]. Perhaps EF-1α mediates the relationship between the intensity of translation and microtubule cytoskeleton dynamics. The concentration of free EF-1α during active translation is low and favors microtubule stabilization; when the translation rate slows (e.g. because of cell transition to mitosis), an increase in free EF-1α concentration increases dynamic instability of the microtubules. Unfortunately, this hypothesis has not been tested by contemporary experimental methods, and the mechanism of EF-1α action on microtubules remains obscure.
There are indirect data suggesting that microtubules bind translation initiation factor eIF4G. Overexpression of the eIF4G conserved domain in Saccharomyces pombe causes destruction of both actin and microtubule cytoskeletal structures [48]. Since the authors used a fragment of eIF4G, it is difficult to estimate the importance of the full-size eIF4G in the regulation of cytoskeleton dynamics.
A yeast two-hybrid system was used to demonstrate interactions between P0 protein of the ribosome lateral protrusion (P-stalk) and the kinesin-like protein KIF4 and to map the interaction site in KIF4, which turned out to be the ezrin-radixin-moesin-like domain. Suppression of the KIF4 synthesis by RNA interference or removal of the ezrin-radixin-moesin-like domain prevented ribosome translocation to axons [49].
Depolymerization of microtubules impairs the nucleus-to-cytoplasm translocation of some proteins, although no direct interactions between these proteins and microtubules have been demonstrated [50, 51]. One of them is the regulatory RNA-binding protein HuR. Hu family proteins (antigens in human paraneoplastic neurologic disorders) are closely related to the RNA-binding ELAV (embryonic lethal abnormal visual system) protein in Drosophila; they are responsible for the posttranscriptional regulation of gene expression. The neuron-specific HuD stimulates cap-dependent translation by interacting with eIF4a and mRNA poly(A)-tail [52]. Using a yeast two-hybrid system, it was shown that HuD binds to the microtubule-associated protein 1B light chain 1 (MAP1B LC1). Association of HuD with microtubules through MAP1B LC1 has been demonstrated in pull-down experiments and by immunofluorescence microscopy. MAP1B LC1 also binds other neuron-specific proteins of the Hu family, such as HuB and HuC, while ubiquitously expressed HuR does not interact with MAP1B LC1 [53]. Even though no direct binding between HuR and microtubules has been demonstrated, disruption of the microtubule cytoskeleton by the compound MPT0B098 impairs nucleus-to-cytoplasm transport of HuR and causes HuR accumulation in the nucleus [54]. The mechanism of this phenomenon remains unclear, although it might be related to either disturbances in the protein transport along the microtubules or cell stress response.
MICROTUBULES AS A PLATFORM FOR ASSEMBLY AND DISASSEMBLY OF RNP
COMPLEXES
Microtubules can act as a scaffold for the formation of micrometric granules from translation machinery components. As mentioned above, in the presence of YB1 and/or PABP proteins, in vitro mRNAs form large granules on microtubules, probably because of the ability of YB1 and PABP to serve as adaptor molecules in the binding of mRNA molecules to each other and to the microtubules [41].
Chierchia et al. [37] suggested that microtubules might also play a role as a scaffold for the disassembly of protein complexes. They found that microtubules facilitate dissociation of the anti-association eIF6 protein from the ribosomal 60S subunit. Translation of some mRNAs requires their association with microtubules. For example, translation of the hypoxia inducible factor 1 (HIF-1) is microtubule-dependent. Disruption of microtubules with Nocodazole or their stabilization with taxol arrest HIF-1 translation; its mRNA is deposited into P-bodies and degraded [6, 55]. P-bodies are dense subcellular cytoplasmic RNP structures in which mRNA is decapped and 5′-3′ degraded. Microtubule destabilization enhances P-body formation [55], supposedly because translation by the microtubule-associated polysomes stops and mRNA gets transferred to the P-bodies.
When translation initiation is inhibited (e.g. by oxidative stress), preinitiation complexes that include mRNAs, mRNA-associated proteins, small ribosomal subunits, and some initiation factors aggregate into dense formations up to several microns in size, so-called stress granules (Fig. 3). Stress granules differ from P-bodies: components of the translation machinery in the stress granules are present in the “frozen” inactive state, while mRNAs gradually leave stress granules and are either degraded or used for protein synthesis. This process is accompanied by mRNA sorting: mRNAs coding for the proteins that counteract the stress are retained; the rest of the mRNAs are degraded. After the stress is eliminated, stress granules dissociate, and protein synthesis resumes with the remaining mRNAs [56-58].
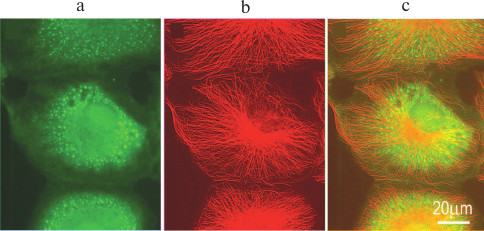
Fig. 3. Stress granules in cultured CV-1 cell after treatment with sodium arsenite: a) eIF3a (green fluorescence), stress granule marker; b) microtubules (red fluorescence); c) merge of (a) and (b).
When microtubules are experimentally depolymerized, stress granules form much more slowly, and they are smaller in size [59-64]. Under moderate stress conditions, microtubule disruption might completely inhibit stress granule formation [59]. Disintegration of stress granules after elimination of the stress slows in the absence of microtubules [64]. Interestingly, microtubule stabilization with taxol also inhibits stress granule formation [63]. It was suggested [63] that microtubules push stress granule components with the growing (+) end or drag them along during disassembly. This “pushing or pulling” mechanism facilitates association of stress granule components with each other.
The exact role of microtubules in stress granule dynamics remains unclear. Many experiments have shown that simple diffusion of stress granule components across the cytoplasm cannot ensure rapid assembly of such large particles, since it would be hindered by the high viscosity of the cytoplasm, actin filaments, membranes, etc. However, simple diffusion of RNPs is possible in cells with disrupted microtubules [62]. Stress granule assembly requires facilitation of the transfer of the component across the cytoplasm by either increasing the transfer velocity or decreasing the transfer distance. Microtubules can fulfill both these conditions. There are several possible mechanisms of the involvement of microtubules in stress granule assembly. Stress granule components might: (i) be moved along the microtubules by the active transport mechanism by motor proteins; (ii) move along the microtubules by the one-dimensional diffusion mechanism; (iii) attach to the microtubules, while the microtubules move along each other using motor proteins; (iv) be pushed and pulled by dynamically unstable microtubules, as suggested by Chernov et al. [63]. The last mechanism has not been experimentally confirmed; there are no published data on specific interactions between dynamic microtubule ends and RNPs.
Analysis of stress granule translocation in cells showed that they move in a diffuse manner, i.e. chaotically and slowly (up to 0.15 µm/s). However, sometimes stress granules make fast (over 0.5 µm/s), although rare, movements along the microtubules, which might be cases of active transport [64]. Data on the involvement of motor proteins (kinesins and dynein) in stress granule assembly are contradictory. Some authors reported that inhibition of dynein (suppression of retrograde transport) disturbs stress granule assembly [65, 66], while inhibition of kinesin 1 (suppression of anterograde transport) impairs stress granule disassembly [66]. Other researchers failed to find the influence of dynein inhibition on stress granule assembly [61, 63]. Acidification of the culture medium prevents formation of stress granules in sodium arsenite-treated HeLa cells [67], probably due to inhibition of the motor proteins. We believe that involvement of motor proteins in stress granule assembly requires further study of certain components using methods for specific inhibition of motor proteins.
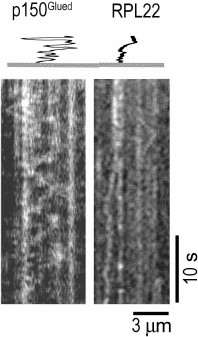
One-dimensional diffusion of stress granule components and large RNP complexes along the microtubules might play an important role in the assembly of these particles in the cell. Association with microtubules considerably increases the probability of interactions between the stress granule components due to the local increase in their concentration, as well as one-dimensional diffusion. At the same time, diffusion, i.e. multidimensional ATP-independent translocation, might be preferable to directed transport by motor proteins, since the latter will rather cause separation of stress granule components. Translocation of molecules along microtubules by one-dimensional diffusion presumably occurs due to the interaction of the negatively charged C-terminal region of tubulin and positively charged translocated proteins [68]. High ionic strength, changes in pH, or removal of the tubulin C-terminus with subtilisin block one-dimensional diffusion along microtubules [69, 70]. One-dimensional diffusion along microtubules has been demonstrated for many microtubule-associated proteins [68-72]. Unfortunately, no diffusion along the microtubule has been so far observed for any RNA-binding protein. Thus, ribosomal RpL22 protein can move chaotically along the microtubules only over small distances (Chudinova et al., submitted for publication) as compared to the dynactin p150Glued subunit that is capable of high-amplitude one-dimensional diffusion (Fig. 4).
Fig. 4. Diffusion of GFP–RpL22 and GFP–p150Glued proteins along the microtubule in vitro. Kymograms were registered with a TIRF microscope; the tracks are presented using the altered time scale.
RNP TRANSPORT ALONG MICROTUBULES
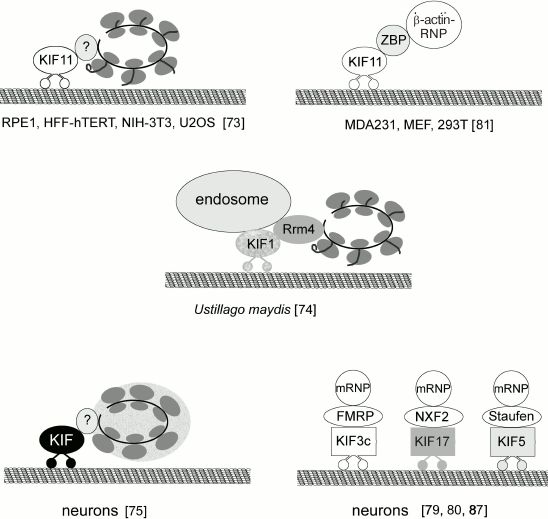
Translocation of polysomes and transport granules. Cells have both general and specific (i.e. related to the synthesis of a particular protein) mechanisms of polysome transport. In both cases, ribosomes should be translocated from the nucleus to the cell periphery (e.g. neuron processes). Polysomes can bind to the microtubules through the kinesin Eg5 (KIF11). Inhibition of Eg5 by monastrol or downregulation of its biosynthesis by RNA interference decrease total translation activity by 40% [73]. Analysis of the polysome profile confirmed the association of Eg5 with the polysomes. It remains unclear if Eg5 binds to ribosomes directly or through some adaptor protein. Since similar results were obtained for several cell lines tested, it is reasonable to assume that polysome association with microtubules through Eg5 is a common mechanism for polysome transport from the perinuclear area to the cell periphery in mammalian cells (Fig. 5).
Fig. 5. Interactions of polyribosomes and various mRNPs with kinesins.
Another mechanism of polysome translocation was found in basidiomycetes. Higuchi et al. [74] convincingly demonstrated that ribosomes could be transported along microtubules as components of early endosomes. In elongated cells of the Ustilago maydis fungus, ribosomes accumulated in the apical regions of the hyphae due to the transport of their complexes with early endosomes by the motor proteins dynein and kinesin. Translocation of ribosomes also requires involvement of the RNA-binding Rrm4 (RNA recognition motif 4) protein. In fungal cells, Rrm4 binds over 50% of mRNAs. Whether Rrm4 associates with motor proteins directly or through elements of early endosomes still needs to be elucidated. Deletion of Rrm4, as well as mutations in kinesin 3 (KIF1) and dynein, disrupt polysome transport and cause ribosomes to accumulate in the perinuclear region. Transport usually involves association of several mRNAs, probably in the content of polysomes, into one transported granule. It is important to note that only actively translating polysomes are transported; inhibition of protein synthesis and polysome dissociation block ribosome transport.
Graber et al. [75] suggested that in neurons, mRNAs are transported to axons and dendrites as components of translationally repressed polysomes. They suggested that translation initiation and the first elongation cycle take place in the neuron soma; inactive polysomes are then transported to neurites, where translation is rapidly reactivated by external stimuli. Inactive polysomes form so-called transport granules that differ in their content from both stress granules and P-bodies [19]. Proteomic analysis of isolated transport granules and high-resolution electron microscopy also provide evidence for the possibility of ribosome transport in the content of neuronal RNP granules [75, 76].
Selective transport of mRNAs. As mentioned above, mRNPs and other components of the translation machinery are subjects of intracellular transport [9-13, 19]; moreover, some mRNPs are transported selectively. Pichon et al. [77] developed an elegant method for visualizing the translation process in living cells. They used cells that constitutively expressed a GFP-labeled fragment of antibody against tandem repeat sequence from the yeast Gcn4 protein. The N-termini of the proteins of interest were modified by fusing with 56 tandem repeats to ensure their interaction with the GFP-labeled antibodies, so that the newly synthesized protein molecule could be visualized as a bright green dot in the cell. The mRNAs of interest were modified with an array of MS2 stem-loops in the 3′-ends to visualize single mRNAs as red dots using the fluorescent RFP version of the MS2 coat protein. This system allowed observing translation in a living cell at a single-molecule level. It was found that polysomes that translated RNA polymerase II (POLR2A) or Ki67 nuclear protein diffused through the cytoplasm, whereas polysomes that translated dynein heavy chain displayed rapid microtubule-dependent rectilinear movements with median speed of 1 µm/s. This suggests that translocation of some polysomes in the cytoplasm involves microtubule-associated motor proteins, whereas other polysomes do not associate with motor proteins. It is reasonable to assume that microtubule-associated motor proteins selectively bind certain mRNAs.
Binding of mRNAs to motor proteins requires involvement of adaptor proteins that are capable of recognition of a specific sequence in the mRNAs as well as interactions with motor proteins. So far, about 45 genes for kinesin group proteins have been found; the actual number of protein isoforms is even higher because of alternative splicing. According to Hirokawa and Tanaka [78], there are 15 kinesin families. Proteins that can act as adaptor molecules to provide interactions between kinesins and mRNAs are poorly studied and have been identified for only a few kinesin types (Fig. 5). Neurospecific kinesin KIF3C binds mRNA through FMRP (fragile X mental retardation protein) [79]. KIF17 interacts with RNP granules through NXF2 (nuclear RNA export factor) that exports mRNA from the nucleus and also acts as an adaptor in RNP granule transport across the cytoplasm [80]. Kinesin KIF11 (Eg5) binds to the β-actin mRNA through ZBP1 (zipcode-binding protein 1) [81]. ZBP1 recognizes the so-called zipcode sequence – 54 nucleotides at the 3′-UTR of the β-actin mRNA [82]. ZBP1 is one of the proteins that inhibit premature translation. In NG108-15 neuroblastoma cells, β-actin mRNA binds ZBP1 cotranscriptionally and is transported by kinesin to the cell periphery, where ZBP1 dissociates from the mRNA after phosphorylation of Tyr396 by Src kinase, thereby initiating β-actin synthesis [83]. The zipcode sequence has been identified in some other mRNAs; other signal sequences have been found in 5′- and 3′-UTRs of some mRNAs that presumably regulate the transport of these mRNAs and translation loci [84, 85]. Interactions between these regulatory elements and cytoskeleton have not been investigated yet.
Kinesin KIF5 (kinesin-1) has a conserved 59-a.a. sequence in the tail domain of its heavy chain that is essential and sufficient for the binding of transported RNP granules. Pull-down experiments identified 42 proteins in the RNP granules from mouse brain homogenate that bind this conserved sequence [86]. The most probable candidate for the adaptor molecule to provide binding between mRNAs and the KIF5 family kinesins is Staufen 1 [87].
Many studies of the RNA transport mechanism have been conducted in Drosophila oocytes. Asymmetric localization of some mRNAs in the oocyte is required for correct segmentation of the developing fly larva. Drosophila oocytes are located in the egg chamber and surrounded by 15 nurse cells. Nurse cells and the oocyte are connected via a system of intercellular bridges called ring canals. Each group of cells has one common microtubule-organizing center in the oocyte, from which microtubules protrude to the nurse cells, with the microtubule (–) ends in the oocytes and (+) ends in the nurse cells. mRNA is synthesized in the nurse cells and then transported to the oocyte through the ring canals.
Using an extensive collection of transgenic Drosophila flies, Gaspar et al. [88] showed that most of the oskar mRNA molecules bind to kinesin-1 immediately after mRNA export to the cytoplasm in the perinuclear area of the nurse cells. However, kinesin-1 is inactive in nurse cells, and RNPs are translocated to the oocyte, i.e. to the microtubule (–) ends, driven by dynein [88, 89]. Kinesin is activated during mid-oogenesis; oskar mRNA is translocated to the oocyte posterior, where it anchors to actin filaments. Perhaps an important role in targeted mRNA delivery in Drosophila oocytes is played by cytoplasmic movement (streaming) resulting from the sliding of microtubules along each other driven by kinesin-1 (KIF5a) [90]. Efficient cytoplasm mixing in Drosophila oocytes is possible because of the presence of two microtubule populations: stable microtubules attached to the cortex, and actively moving cytoplasmic microtubules. In Drosophila kinesin-1 mutants deficient in microtubule sliding but able to transport most cargos, oskar mRNA distribution in the oocytes is more diffuse [91]. Therefore, correct localization of oskar mRNA involves two processes: active transport along the microtubules and oocyte cytoplasmic streaming provided by microtubule–microtubule sliding [91].
Recently, two research groups independently demonstrated that an adaptor molecule that binds kinesin 1 and oskar mRNA is the RNA-binding atypical tropomyosin (tropomyosin 1-I/C) [88, 92]. Both kinesin 1 and Staufen 1 were identified as tropomyosin-binding partners in coimmunoprecipitation experiments, although it is still unknown whether Staufen 1 interacts with tropomyosin directly or through kinesin. UV-induced (254 nm) crosslinking showed that oskar mRNA binds directly to tropomyosin [86].
The association of mRNA with dynein is mediated by the Egalitarian (Egl) and Bicaudal D proteins [93]. Bicaudal D is a dynein cofactor involved in the binding of various types of cargos [94]. The N-terminal and central fragments of Bicaudal D are responsible for the interaction with the dynein–dynactin complex; the C-terminal fragment binds adaptor molecules, including Egl. In vitro studies showed that Egl directly associates with mRNAs. Dienstbier et al. [95] used localizing and nonlocalizing RNA sequences that were appended to streptavidin-binding aptamers and then immobilized on a streptavidin-agarose matrix. The localizing sequences contained signals for mRNA apical localization (TLS, ILS, GLS); the nonlocalizing sequences (TLSas, TLSΔbub, TLSU6C, ILSas) did not. In vitro translated Egl bound all RNAs tested, but the binding efficiency for localizing RNA was more than two times higher than for the nonlocalizing controls [95]. Egl does not contain classical RNA-binding motifs; interactions with RNA are determined by an extended fragment of the protein sequence (a.a. 1-814) [95]. Formation of the RNA–Egl complexes occurs with the involvement of dynein light chains [96].
Dynein is required for anterior localization of the bicoid mRNA in Drosophila oocytes. This RNA is essential for correct segmentation of the larval body. Dynein binding of the bicoid mRNA involves the ESCRT-II protein complex, whose other function is sorting of endosomal proteins into internal vesicles. One of the ESCRT-II complex proteins, VPS36, recognizes specific sequence in the bicoid mRNA 3′-UTR, binds dynein, and localizes to the anterior of the oocyte, where it recruits Staufen 1 to the bicoid complex [97]. No adaptor molecules that provide mRNA binding to dynein in other types of cells have been studied.
There is no doubt that translation machinery components interact with the microtubule portion of the cytoskeleton, a structure that forms transportation routes and scaffold for multimolecular cellular complexes. The functional role of these interactions might be the transport of proteins and RNPs, as well as assembly or disintegration of protein and RNP complexes.
Direct binding of free ribosomal proteins RpS3, RpSA (RpS40, LamR), RpL11, RpL22, and RpL41 to tubulin microtubules has been unambiguously demonstrated; however, it remains unclear whether these proteins can interact with microtubules as components of ribosomes. Other proteins that associate with microtubules are transcriptional factors eEF-1α and eIF3; the latter binds to microtubules through the eIF3a subunit. Microtubule-associated mRNA-binding protein YB1 provides nonspecific binding of mRNAs to microtubules. Another microtubule-associated protein, APC, interacts with many mRNAs; MAP1B LC1 interacts with RNA-binding proteins of the Hu family. All these interactions result in the formation of static complexes, as well as providing the possibility for one-dimensional diffusion of proteins and RNPs along the microtubules.
A significant portion of mRNPs and/or polysomes interact with microtubules through motor proteins to ensure mRNP and polysome transport across the cytoplasm, often followed by local translation. Local translation of selected mRNAs is essential in various cell processes such as segmentation of insect larva body or directed motility of mammalian fibroblasts on substrates. Functioning of cells with long protrusions (e.g. neurons) requires active transport of all translation machinery component to the cytoplasm periphery. Many motors proteins (kinesins, dyneins) are involved in the transport of the translation machinery components. Thus, kinesin KIF4 binds to ribosomal protein P0; kinesin KIF11 (Eg5) binds to polysomes, although to a still unknown interaction partner. Fungal kinesin KIF1 interacts with polysomes through the Rrm4 protein. In neurons, KIF3C binds the RNP protein FMRP; kinesin KIF17 binds NXF2. Another adaptor protein is ZBP, which associates with a specific nucleotide sequence (zipcode) at the mRNA 3′-end and with kinesin KIF11. “Common” kinesin 1 (KIF5) participates in mRNA transport in Drosophila oocytes by binding to Staufen 1 and atypical tropomyosin, which in turn recognizes oskar mRNA. The multiprotein dynein complex interacts with the RNA-binding protein Egalitarian (Egl) and VPS36 (component of ESCRT-II) through its component Bicaudal to provide correct localization of certain mRNAs in the oocyte.
In conclusion, interactions between translation machinery and microtubules include nonspecific ubiquitous binding of ribosomal proteins, translation factors, and other components with microtubules, as well as selective association of some mRNAs with microtubule motor proteins through adaptor proteins capable of recognizing specific sequences in mRNAs (Fig. 5). The existence of selective transport of mRNAs supports the idea of microtubule cytoskeleton involvement in the regulation of gene expression via local translation of mRNAs. Nonspecific interactions might facilitate this process; besides, they might regulate translation in the cell at a general level and contribute to microtubule dynamics.
Acknowledgments
This work was supported by the State Project No. 01201362159.
REFERENCES
1.Jackson, R. J., Hellen, C. U., and Pestova, T. V.
(2010) The mechanism of eukaryotic translation initiation and
principles of its regulation, Rev. Mol. Cell Biol., 11,
113-127.
2.Afonina, Zh. A., and Shirokov, V. A. (2018)
Three-dimensional organization of polyribosomes – a modern
approach, Biochemistry (Moscow), 83, Suppl. 1,
S48-S55.
3.Negrutskii, B. S., Stapulionis, R., and Deutscher,
M. P. (1994) Supramolecular organization of the mammalian translation
system, Proc. Natl. Acad. Sci. USA, 91, 964-968.
4.Gross, S. R., and Kinzy, T. G. (2007) Improper
organization of the actin cytoskeleton affects protein synthesis at
initiation, Mol. Cell Biol., 27, 1974-1989.
5.Morelli, J. K., Zhou, W., Yu, J., Lu, C., and
Vayda, M. E. (1998) Actin depolymerization affects stress-induced
translational activity of potato tuber tissue, Plant Physiol.,
116, 1227-1237.
6.Carbonaro, M., O’Brate, A., and Giannakakou,
P. (2011) Microtubule disruption targets HIF-1alpha mRNA to cytoplasmic
P-bodies for translational repression, J. Cell Biol.,
192, 83-99.
7.King, M. L., Messitt, T. J., and Mowry, K. L.
(2005) Putting RNAs in the right place at the right time: RNA
localization in the frog oocyte, Biol. Cell, 97,
19-33.
8.Lecuyer, E., Yoshida, H., Parthasarathy, N., Alm,
C., Babak, T., Cerovina, T., Hughes, T. R., Tomancak, P., and Krause,
H. M. (2007) Global analysis of mRNA localization reveals a prominent
role in organizing cellular architecture and function, Cell,
131, 174-187.
9.Jansen, R. P., and Niessing, D. (2012) Assembly of
mRNA–protein complexes for directional mRNA transport in
eukaryotes – an overview, Curr. Protein Pept. Sci.,
13, 284-293.
10.Parton, R. M., Davidson, A., Davis, I., and Weil,
T. T. (2014) Subcellular mRNA localisation at a glance, J. Cell
Sci., 127, 2127-2133.
11.Buxbaum, A. R., Yoon, Y. J., Singer, R. H., and
Park, H. Y. (2015) Single-molecule insights into mRNA dynamics in
neurons, Trends Cell Biol., 25, 468-475.
12.Piper, M., Lee, A. C., van Horck, F. P.,
McNeilly, H., Lu, T. B., Harris, W. A., and Holt, C. E. (2015)
Differential requirement of F-actin and microtubule cytoskeleton in
cue-induced local protein synthesis in axonal growth cones, Neural
Dev., 10, 3.
13.Katz, Z. B., Wells, A. L., Park, H. Y., Wu, B.,
Shenoy, S. M., and Singer, R. H. (2012) β-Actin mRNA
compartmentalization enhances focal adhesion stability and directs cell
migration, Genes Dev., 26, 1885-1890.
14.Lenk, R., Ransom, L., Kaufmann, Y., and Penman,
S. (1977) A cytoskeletal structure with associated polyribosomes
obtained from HeLa cells, Cell, 10, 67-78.
15.Shestakova, E. A., Motuz, L. P., and Gavrilova,
L. P. (1993) Co-localization of components of the protein-synthesizing
machinery with the cytoskeleton in G0-arrested cells, Cell Biol.
Int., 17, 417-424.
16.Edmonds, B. T., Bell, A., Wyckoff, J., Condeelis,
J., and Leyh, T. S. (1998) The effect of F-actin on the binding and
hydrolysis of guanine nucleotide by Dictyostelium elongation
factor 1A, J. Biol. Chem., 273, 10288-10295.
17.Perez, W. B., and Kinzy, T. G. (2014) Translation
elongation factor 1A mutants with altered actin bundling activity show
reduced aminoacyl-tRNA binding and alter initiation via eIF2α
phosphorylation, J. Biol. Chem., 289, 20928-20938.
18.Kaverina, I., and Straube, A. (2011) Regulation
of cell migration by dynamic microtubules, Semin. Cell Dev.
Biol., 22, 968-974.
19.Carson, J. H., and Barbarese, E. (2005) Systems
analysis of RNA trafficking in neural cells, Biol. Cell,
97, 51-62.
20.Mahamid, J., Pfeffer, S., Schaffer, M., Villa,
E., Danev, R., Cuellar, L. K., Forster, F., Hyman, A. A., Plitzko, J.
M., and Baumeister, W. (2016) Visualizing the molecular sociology at
the HeLa cell nuclear periphery, Science, 351,
1969-1972.
21.Castoldi, M., and Popov, A. V. (2003)
Purification of brain tubulin through two cycles of
polymerization-depolymerization in a high-molarity buffer, Protein
Expr. Purif., 32, 83-88.
22.Sakamoto, T., Uezu, A., Kawauchi, S., Kuramoto,
T., Makino, K., Umeda, K., Araki, N., Baba, H., and Nakanishi, H.
(2008) Mass spectrometric analysis of microtubule co-sedimented
proteins from rat brain, Genes Cells, 13, 3295-3312.
23.Patel, P. C., Fisher, K. H., Yang, E. C., Deane,
C. M., and Harrison, R. E. (2009) Proteomic analysis of
microtubule-associated proteins during macrophage activation, Mol.
Cell Proteomics, 8, 2500-2514.
24.Ozlu, N., Monigatti, F., Renard, B. Y., Field, C.
M., Steen, H., Mitchison, T. J., and Steen, J. J. (2010) Binding
partner switching on microtubules and aurora-B in the mitosis to
cytokinesis transition, Mol. Cell Proteomics, 9,
336-350.
25.Gache, V., Waridel, P., Winter, C., Juhem, A.,
Schroeder, M., Shevchenko, A., and Popov, A. V. (2010) Xenopus
meiotic microtubule-associated interactome, PLoS One, 17,
e9248.
26.Volkov, V. A., Grissom, P. M., Arzhanik, V. K.,
Zaytsev, A. V., Renganathan, K., McClure-Begley, T., Old, W. M., Ahn,
N., and McIntosh, J. R. (2015) Centromere protein F includes two sites
that couple efficiently to depolymerizing microtubules, J. Cell
Biol., 209, 813-828.
27.Chuong, S. D., Good, A. G., Taylor, G. J.,
Freeman, M. C., Moorhead, G. B., and Muench, D. G. (2004) Large-scale
identification of tubulin-binding proteins provides insight on
subcellular trafficking, metabolic channeling, and signaling in plant
cells, Mol. Cell Proteomics, 3, 970-983.
28.Derbyshire, P., Menard, D., Green, P., Saalbach,
G., Buschmann, H., Lloyd, C. W., and Pesquet, E. (2015) Proteomic
analysis of microtubule interacting proteins over the course of xylem
tracheary element formation in Arabidopsis, Plant Cell,
27, 2709-2726.
29.Hughes, J. R., Meireles, A. M., Fisher, K. H.,
Garcia, A., Antrobus, P. R., Wainman, A., Zitzmann, N., Deane, C.,
Ohkura, H., and Wakefield, J. G. (2008) A microtubule interactome:
complexes with roles in cell cycle and mitosis, PLoS Biol.,
22, e98.
30.Sauer, G., Korner, R., Hanisch, A., Ries, A.,
Nigg, E. A., and Sillje, H. H. (2005) Proteome analysis of the human
mitotic spindle, Mol. Cell Proteomics, 4, 35-43.
31.Severin, F. F., Shanina, N. A., Shevchenko, A.,
Solovyanova, O. B., Koretsky, V. V., and Nadezhdina, E. S. (1997) A
major 170 kDa protein associated with bovine adrenal medulla
microtubules: a member of the centrosomin family? FEBS Lett.,
420, 125-128.
32.Shanina, N. A., Ivanov, P. A., Chudinova, E. M.,
Severin, F. F., and Nadezhdina, E. S. (2001) Translation initiation
factor eIF3 is able to bind with microtubules in mammalian cells,
Mol. Biol. (Moscow), 35, 638-646.
33.Hasek, J., Kovarik, P., Valasek, L., Malinska,
K., Schneider, J., Kohlwein, S. D., and Ruis, H. (2000) Rpg1p, the
subunit of the Saccharomyces cerevisiae eIF3 core complex, is a
microtubule-interacting protein, Cell Motil. Cytoskeleton,
45, 235-246.
34.Berg, J. M., Lee, C., Chen, L., Galvan, L.,
Cepeda, C., Chen, J. Y., Penagarikano, O., Stein, J. L., Li, A.,
Oguro-Ando, A., Miller, J. A., Vashisht, A. A., Starks, M. E., Kite, E.
P., Tam, E., Gdalyahu, A., Al-Sharif, N. B., Burkett, Z. D., White, S.
A., Fears, S. C., Levine, M. S., Wohlschlegel, J. A., and Geschwind, D.
H. (2015) JAKMIP1, a novel regulator of neuronal translation, modulates
synaptic function and autistic-like behaviors in mouse, Neuron,
88, 1173-1191.
35.Jang, C. Y., Kim, H. D., Zhang, X., Chang, J. S.,
and Kim, J. (2012) Ribosomal protein S3 localizes on the mitotic
spindle and functions as a microtubule associated protein in mitosis,
Biochem. Biophys. Res. Commun., 429, 57-62.
36.Wang, S., Huang, J., He, J., Wang, A., Xu, S.,
Huang, S. F., and Xiao, S. (2010) RPL41, a small ribosomal peptide
deregulated in tumors, is essential for mitosis and centrosome
integrity, Neoplasia, 12, 284-293.
37.Chierchia, L., Tussellino, M., Guarino, D.,
Carotenuto, R., DeMarco, N., Campanella, C., Biffo, S., and Vaccaro, M.
C. (2015) Cytoskeletal proteins associate with components of the
ribosomal maturation and translation apparatus in Xenopus stage
I oocytes, Zygote, 23, 669-682.
38.Venticinque, L., Jamieson, K. V., and Meruelo, D.
(2011) Interactions between laminin receptor and the cytoskeleton
during translation and cell motility, PLoS One, 6,
e15895.
39.Eliseeva, I. A., Kim, E. R., Guryanov, S. G.,
Ovchinnikov, L. P., and Lyabin, D. N. (2011) Y-box-binding protein 1
(YB-1) and its functions, Biochemistry (Moscow), 76,
1402-1433.
40.Chernov, K. G., Mechulam, A., Popova, N. V.,
Pastre, D., Nadezhdina, E. S., Skabkina, O. V., Shanina, N. A.,
Vasiliev, V. D., Tarrade, A., Melki, J., Joshi, V., Baconnais, S.,
Toma, F., Ovchinnikov, L. P., and Curmi, P. A. (2008) YB-1 promotes
microtubule assembly in vitro through interaction with tubulin
and microtubules, BMC Biochem., 9, e23.
41.Chernov, K. G., Curmi, P. A., Hamon, L.,
Mechulam, A., Ovchinnikov, L. P., and Pastré, D. (2008) Atomic
force microscopy reveals binding of mRNA to microtubules mediated by
two major mRNP proteins YB-1 and PABP, FEBS Lett., 582,
2875-2881.
42.Preitner, N., Quan, J., Nowakowski, D. W.,
Hancock, M. L., Shi, J., Tcherkezian, J., Young-Pearse, T. L., and
Flanagan, J. G. (2014) APC is an RNA-binding protein, and its
interactome provides a link to neural development and microtubule
assembly, Cell, 158, 368-382.
43.Mili, S., Moissoglu, K., and Macara, I. G. (2008)
Genome-wide screen reveals APC-associated RNAs enriched in cell
protrusions, Nature, 453, 115-119.
44.Villarin, J. M., McCurdy, E. P., Martinez, J. C.,
and Hengst, U. (2016) Local synthesis of dynein cofactors matches
retrograde transport to acutely changing demands, Nat. Commun.,
7, 13865.
45.Moore, R. C., Durso, N. A., and Cyr, R. J. (1998)
Elongation factor-1 alpha stabilizes microtubules in a
calcium/calmodulin-dependent manner, Cell Motil. Cytoskeleton,
41, 168-180.
46.Shiina, N., Gotoh, Y., Kubomura, N., Iwamatsu,
A., and Nishida, E. (1994) Microtubule severing by elongation factor 1
alpha, Science, 266, 282-285.
47.Suda, M., Fukui, M., Sogabe, Y., Sato, K.,
Morimatsu, A., Arai, R., Motegi, F., Miyakawa, T., Mabuchi, I., and
Hirata, D. (1999) Overproduction of elongation factor 1 alpha, an
essential translational component, causes aberrant cell morphology by
affecting the control of growth polarity in fission yeast, Genes
Cells, 4, 517-527.
48.Hashemzadeh-Bonehi, L., Curtis, P. S., Morley, S.
J., Thorpe, J. R., and Pain, V. M. (2003) Overproduction of a conserved
domain of fission yeast and mammalian translation initiation factor
eIF4G causes aberrant cell morphology and results in disruption of the
localization of F-actin and the organization of microtubules, Genes
Cells, 8, 163-178.
49.Bisbal, M., Wojnacki, J., Peretti, D., Ropolo,
A., Sesma, J., Jausoro, I., and Caceres, A. (2009) KIF4 mediates
anterograde translocation and positioning of ribosomal constituents to
axons, J. Biol. Chem., 284, 9489-9497.
50.Campbell, E. M., and Hope, T. J. (2003) Role of
the cytoskeleton in nuclear import, Adv. Drug Deliv. Rev.,
55, 761-771.
51.Roth, D. M., Moseley, G. W., Glover, D., Pouton,
C. W., and Jans, D. A. (2007) A microtubule-facilitated nuclear import
pathway for cancer regulatory proteins, Traffic, 8,
673-686.
52.Fukao, A., Sasano, Y., Imataka, H., Inoue, K.,
Sakamoto, H., Sonenberg, N., Thoma, C., and Fujiwara, T. (2009) The
ELAV protein HuD stimulates cap-dependent translation in a poly(A)- and
eIF4A-dependent manner, Mol. Cell, 36, 1007-1017.
53.Fujiwara, Y., Kasashima, K., Saito, K., Fukuda,
M., Fukao, A., Sasano, Y., Inoue, K., Fujiwara, T., and Sakamoto, H.
(2011) Microtubule association of a neuronal RNA-binding protein HuD
through its binding to the light chain of MAP1B, Biochimie,
93, 817-822.
54.Cheng, Y. C., Liou, J. P., Kuo, C. C., Lai, W.
Y., Shih, K. H., Chang, C. Y., Pan, W. Y., Tseng, J. T., and Chang, J.
Y. (2013) MPT0B098, a novel microtubule inhibitor that destabilizes the
hypoxia-inducible factor-1α mRNA through decreasing
nuclear-cytoplasmic translocation of RNA-binding protein HuR, Mol.
Cancer Ther., 12, 1202-1212.
55.Sweet, T. J., Boyer, B., Hu, W., Baker, K. E.,
and Coller, J. (2007) Microtubule disruption stimulates P-body
formation, RNA, 13, 493-502.
56.Kedersha, N., Ivanov, P., and Anderson, P. (2013)
Stress granules and cell signaling: more than just a passing phase?
Trends Biochem. Sci., 38, 494-506.
57.Buchan, J. R. (2014) mRNP granules: assembly,
function, and connections with disease, RNA Biol., 11,
1019-1030.
58.Anderson, P., Kedersha, N., and Ivanov, P. (2015)
Stress granules, P-bodies and cancer, Biochim. Biophys. Acta,
1849, 861-870.
59.Ivanov, P. A., Chudinova, E. M., and Nadezhdina,
E. S. (2003) Disruption of microtubules inhibits cytoplasmic
ribonucleoprotein stress granule formation, Exp. Cell Res.,
290, 227-233.
60.Kwon, S., Zhang, Y., and Matthias, P. (2007) The
deacetylase HDAC6 is a novel critical component of stress granules
involved in the stress response, Genes Dev., 21,
3381-3394.
61.Fujimura, K., Katahira, J., Kano, F., Yoneda, Y.,
and Murata, M. (2009) Microscopic dissection of the process of stress
granule assembly, Biochim. Biophys. Acta, 1793,
1728-1737.
62.Kolobova, E., Efimov, A., Kaverina, I., Rishi, A.
K., Schrader, J. W., Ham, A. J., Larocca, M. C., and Goldenring, J. R.
(2009) Microtubule-dependent association of AKAP350A and CCAR1 with RNA
stress granules, Exp. Cell Res., 315, 542-555.
63.Chernov, K. G., Barbet, A., Hamon, L.,
Ovchinnikov, L. P., Curmi, P. A., and Pastre, D. (2009) Role of
microtubules in stress granule assembly: microtubule dynamical
instability favors the formation of micrometric stress granules in
cells, J. Biol. Chem., 284, 36569-3680.
64.Nadezhdina, E. S., Lomakin, A. J., Shpilman, A.
A., Chudinova, E. M., and Ivanov, P. A. (2010) Microtubules govern
stress granule mobility and dynamics, Biochim. Biophys. Acta,
1803, 361-371.
65.Tsai, N. P., Tsui, Y. C., and Wei, L. N. (2009)
Dynein motor contributes to stress granule dynamics in primary neurons,
Neuroscience, 159, 647-656.
66.Loschi, M., Leishman, C. C., Berardone, N., and
Boccacio, G. L. (2009) Dynein and kinesin regulate stress-granule and
P-body dynamics, J. Cell Sci., 122, 3973-3982.
67.Chudinova, E. M., Nadezhdina, E. S., and Ivanov,
P. A. (2012) Cellular acidosis inhibits assembly, disassembly, and
motility of stress granules, Biochemistry (Moscow), 77,
1277-1284.
68.Cooper, J. R., and Wordeman, L. (2009) The
diffusive interaction of microtubule binding proteins, Curr. Opin.
Cell Biol., 21, 68-73.
69.Helenius, J., Brouhard, G., Kalaidzidis, Y.,
Diez, S., and Howard, J. (2006) The depolymerizing kinesin MCAK uses
lattice diffusion to rapidly target microtubule ends, Nature,
441, 115-119.
70.Hinrichs, M. H., Jalal, A., Brenner, B.,
Mandelkow, E., Kumar, S., and Scholz, T. (2012) Tau protein diffuses
along the microtubule lattice, J. Biol. Chem., 287,
38559-38568.
71.Ali, M. Y., Krementsova, E. B., Kennedy, G. G.,
Mahaffy, R., Pollard, T. D., Trybus, K. M., and Warshaw, D. M. (2007)
Myosin Va maneuvers through actin intersections and diffuses along
microtubules, Proc. Natl. Acad. Sci. USA, 13,
4332-4336.
72.Ayloo, S., Lazarus, J. E., Dodda, A., Tokito, M.,
Ostap, E. M., and Holzbaur, E. L. (2014) Dynactin functions as both a
dynamic tether and brake during dynein-driven motility, Nat.
Commun., 5, 4807.
73.Bartoli, K. M., Jakovljevic, J., Woolford, J. L.,
Jr., and Saunders, W. S. (2011) Kinesin molecular motor Eg5 functions
during polypeptide synthesis, Mol. Biol. Cell, 22,
3420-3430.
74.Higuchi, Y., Ashwin, P., Roger, Y., and
Steinberg, G. (2014) Early endosome motility spatially organizes
polysome distribution, J. Cell Biol., 204, 343-357.
75.Graber, T. E., Hebert-Seropian, S., Khoutorsky,
A., David, A., Yewdell, J. W., Lacaille, J. C., and Sossin, W. S.
(2013) Reactivation of stalled polyribosomes in synaptic plasticity,
Proc. Natl. Acad. Sci. USA, 110, 16205-16210.
76.El Fatimy, R., Davidovic, L., Tremblay, S.,
Jaglin, X., Dury, A., Robert, C., De Koninck, P., and Khandjian, E. W.
(2016) Tracking the fragile X mental retardation protein in a highly
ordered neuronal ribonucleoparticles population: a link between stalled
polyribosomes and RNA granules, PLoS Genet., 12,
e1006192.
77.Pichon, X., Bastide, A., Safieddine, A., Chouaib,
R., Samacoits, A., Basyuk, E., Peter, M., Mueller, F., and Bertrand, E.
(2016) Visualization of single endogenous polysomes reveals the
dynamics of translation in live human cells, J. Cell Biol.,
214, 769-781.
78.Hirokawa, N., and Tanaka, Y. (2015) Kinesin
superfamily proteins (KIFs): various functions and their relevance for
important phenomena in life and diseases, Exp. Cell Res.,
334, 16-25.
79.Davidovic, L., Jaglin, X. H., Lepagnol-Bestel, A.
M., Tremblay, S., Simonneau, M., Bardoni, B., and Khandjian, E. W.
(2007) The fragile X mental retardation protein is a molecular adaptor
between the neurospecific KIF3C kinesin and dendritic RNA granules,
Hum. Mol. Genet., 16, 3047-3058.
80.Takano, K., Miki, T., Katahira, J., and Yoneda,
Y. (2007) NXF2 is involved in cytoplasmic mRNA dynamics through
interactions with motor proteins, Nucleic Acids Res., 35,
2513-2521.
81.Song, T., Zheng, Y., Wang, Y., Katz, Z., Liu, X.,
Chen, S., Singer, R. H., and Gu, W. (2015) Specific interaction of
KIF11 with ZBP1 regulates the transport of β-actin mRNA and cell
motility, J. Cell Sci., 128, 1001-1010.
82.Kislauskis, E. H., Zhu, X., and Singer, R. H.
(1997) β-Actin messenger RNA localization and protein synthesis
augment cell motility, J. Cell Biol., 136, 1263-1270.
83.Huttelmaier, S., Zenklusen, D., Lederer, M.,
Dictenberg, J., Lorenz, M., Meng, X., Bassell, G. J., Condeelis, J.,
and Singer, R. H. (2005) Spatial regulation of beta-actin translation
by Src-dependent phosphorylation of ZBP1, Nature, 24,
438, 512-515.
84.Shigeoka, T., Jung, H., Jung, J., Turner-Bridger,
B., Ohk, J., Lin, J. Q., Amieux, P. S., and Holt, C. E. (2016) Dynamic
axonal translation in developing and mature visual circuits,
Cell, 166, 181-192.
85.Simon, B., Masiewicz, P., Ephrussi, A., and
Carlomagno, T. (2015) The structure of the SOLE element of oskar
mRNA, RNA, 21, 1444-1453.
86.Kanai, Y., Dohmae, N., and Hirokawa, N. (2004)
Kinesin transports RNA: isolation and characterization of an
RNA-transporting granule, Neuron, 43, 513-525.
87.Falley, K., Schutt, J., Iglauer, P., Menke, K.,
Maas, C., Kneussel, M., Kindler, S., Wouters, F. S., Richter, D., and
Kreienkamp, H. J. (2009) Shank1 mRNA: dendritic transport by kinesin
and translational control by the 5′-untranslated region,
Traffic, 10, 844-857.
88.Gaspar, I., Sysoev, V., Komissarov, A., and
Ephrussi, A. (2017) An RNA-binding atypical tropomyosin recruits
kinesin-1 dynamically to oskar mRNPs, EMBO J., 36,
319-333.
89.Jambor, H., Mueller, S., Bullock, S. L., and
Ephrussi, A. (2014) A stem-loop structure directs oskar mRNA to
microtubule minus ends, RNA, 20, 429-439.
90.Palacios, I. M., and St. Johnston, D. (2002)
Kinesin light chain-independent function of the kinesin heavy chain in
cytoplasmic streaming and posterior localization in the
Drosophila oocyte, Development, 129,
5473-5485.
91.Lu, W., Winding, M., Lakonishok, M., Wildonger,
J., and Gelfand, V. I. (2016) Microtubule-microtubule sliding by
kinesin-1 is essential for normal cytoplasmic streaming in
Drosophila oocytes, Proc. Natl. Acad. Sci. USA,
113, E4995-5004.
92.Veeranan-Karmegam, R., Boggupalli, D. P., Liu,
G., and Gonsalvez, G. B. (2016) A new isoform of Drosophila
non-muscle tropomyosin 1 interacts with kinesin-1 and functions in
oskar mRNA localization, J. Cell Sci., 129,
4252-4264.
93.Mach, J. M., and Lehmann, R. (1997) An
Egalitarian–Bicaudal D complex is essential for oocyte
specification and axis determination in Drosophila, Genes
Dev., 11, 423-435.
94.Hoogenraad, C. C., and Akhmanova, A. (2016)
Bicaudal D family of motor adaptors: linking dynein motility to cargo
binding, Trends Cell Biol., 26, 327-340.
95.Dienstbier, M., Boehl, F., Li, X., and Bullock,
S. L. (2009) Egalitarian is a selective RNA-binding protein linking
mRNA localization signals to the dynein motor, Genes Dev.,
23, 1546-1558.
96.Navarro, C., Puthalakath, H., Adams, J. M.,
Strasser, A., and Lehmann, R. (2004) Egalitarian binds dynein light
chain to establish oocyte polarity and maintain oocyte fate, Nat.
Cell Biol., 6, 427-435.
97.Irion, U., and St. Johnston, D. (2007) Bicoid RNA
localization requires specific binding of an endosomal sorting complex,
Nature, 445, 554-558.