Differential MicroRNA Expression Profiles as Potential Biomarkers for Pancreatic Ductal Adenocarcinoma
Y. Zhu1,a#, J. Wang2,b#, F. Wang3, Z. Yan3, G. Liu2, Y. Ma4,c, W. Zhu5,d, Y. Li3, L. Xie3, A. V. Bazhin6,e*, and X. Guo3,f*
1Department of Oncology, International Joint Laboratory for Cell Medical Engineering of Henan Province, Henan University Huaihe Hospital, Kaifeng, 475000 Henan, People’s Republic of China2Department of Oncology, The Second Affiliated Hospital of Zhengzhou University, Zhengzhou, 450014 Henan, People’s Republic of China
3Department of Preventive Medicine, Cell Signal Transduction Laboratory, Joint National Laboratory for Antibody Drug Engineering, Institute of Biomedical Informatics, Medical School, Henan University, Kaifeng, 475004 Henan, People’s Republic of China
4College of Pharmacy and Tianjin Key Laboratory of Molecular Drug Research, Nankai University, Haihe Education Park, 300353 Tianjin, People’s Republic of China
5Department of Anesthesia, Stanford University, CA 94305, USA
6Department of General, Visceral, and Transplantation Surgery, Ludwig-Maximilians-University Munich, 81377 Munich, Germany
# These authors contributed equally to this work.
* To whom correspondence should be addressed.
Received December 4, 2018; Revised February 2, 2019; Accepted February 4, 2019
Pancreatic ductal adenocarcinoma (PDAC) remains a clinical challenge due to its poor prognosis. Therefore, the early diagnosis of PDAC is extremely important for achieving a cure. MicroRNAs (miRNAs) could serve as a potential biomarker for the early detection and prognosis of PDAC. In this work we analyzed plasma samples from healthy persons and PDAC patients to assess differential miRNA expression profiles by next generation sequencing technology and bioinformatics analysis. In this way, 165 mature miRNAs were found to be significantly deregulated in the patient group, of which 75 and 90 mature miRNAs were up- and down-regulated compared with healthy individuals, respectively. Furthermore, 1029 novel miRNAs were identified. In conclusion, plasma miRNA expression profiles are different between healthy individuals and patients with PDAC. These data provide a possibility for use of miRNA as diagnostic and prognostic biomarkers of PDAC.
KEY WORDS: pancreatic ductal adenocarcinoma, plasma miRNA, expression profilingDOI: 10.1134/S0006297919050122
Abbreviations: miRNA, microRNA; PDAC, pancreatic ductal adenocarcinoma.
Pancreatic ductal adenocarcinoma (PDAC) is a highly lethal disease with
overall 5-year survival rate of less than 5% [1].
Due to its high metastasis rate and absence of early clinical symptoms,
approximately 80% of patients are diagnosed at a late stage and lose
the optimal surgical therapy time [2]. Therefore,
novel approaches for early detection of PDAC are urgently required.
MicroRNAs (miRNAs), single-stranded RNAs with 19-25 nucleotides (nt) generated from endogenous hairpin-shaped transcripts, were discovered in 1993 [3]. MiRNAs do not encode protein but regulate approximately 50% of protein-coding genes [4]. These regulatory elements are first to be transcribed and then processed by Dicer and Drosha complexes into 21-23-nt mature miRNAs [5]. The miRNA is incorporated into the RNA-induced silencing complex (RISC) containing Dicer and other associated proteins [6]. RISC regulates posttranscriptional mRNA expression typically by binding to the 3′ untranslated region (3′-UTR) of the complementary mRNA sequence, preventing the recognition of cap by eIF4E and subsequent binding of translational factors [7]. Previous studies have demonstrated that miRNAs are not only highly correlated with tumorigenesis and progression [4, 8, 9], but also related to drug resistance, tumor metastasis, angiogenesis, cancer relapse, and poor clinical outcomes [10-13]. MiRNAs can be detected by a fully automated, high throughput procedure. MiRNAs produced from disrupted cells are transported in exosomes and released into blood [14, 15]. Being protected by exosomes, circulating miRNAs are stable biomarkers and can potentially be used for cancer detection, prognosis, and therapeutic evaluation [14, 16].
In PDAC, miRNAs have been reported to be responsible for apoptosis escape, proliferation, epithelial mesenchymal transition (EMT), metastasis, invasion, and drug resistance [17, 18]. Besides, it has been shown that they might be used as potential biomarkers for PDAC diagnosis [19]. Although some blood-sample based miRNA expression profiles of PDAC have been reported [20-23], a commonly used miRNAs panel for PDAC detection has not been determined yet.
In this study, we identified miRNA profiles of plasma samples of healthy individuals and PDAC patients by small RNA sequencing and found 165 mature miRNAs that are deregulated in PDAC.
MATERIALS AND METHODS
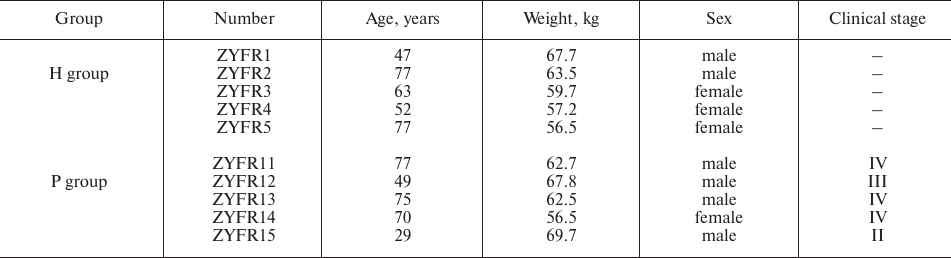
Patient samples. Five patients were recruited with the diagnosis of PDAC (P group) at the Department of General Surgery in the Second Affiliated Hospital of Zhengzhou University from October 2015 to March 2016. PDAC diagnosis was confirmed by histopathologic examination. Meanwhile, five healthy volunteers (H group) were randomly recruited from the Physical Examination Department or from the Department of Digestive Internal Medicine. The healthy individuals of the H group did not have diagnosis for any type of cancer or prior cancer history. The clinical-epidemiological characteristics of the participants are summarized in Table 1.
All blood samples were collected before surgical operation and gathered into polyethylene tubes that were pre-rinsed with EDTA. The samples were centrifuged at 2500 rpm for 5 min at 4°C. Plasma samples were collected by centrifugation. The plasma samples from each group were immediately collected and stored at −80°C.
Table 1. Clinical-epidemiological
characteristics of participants
RNA extraction and sequencing. Total RNA was extracted using a Norgen Plasma/Serum Circulating RNA Purification Mini Kit (Norgen, Canada) following the manufacturer’s procedure. The quantity and purity of total RNA were analyzed by an Agilent Bioanalyzer 2100 and RNA 6000 Nano LabChip Kit (Agilent, USA). The cutoff for RNA sequencing is RIN (RNA Integrity Number) number >7.0. Approximately 0.2-0.7 μg of total RNA was used to prepare a small RNA library according to the manufacturer’s protocol of TruSeq Small RNA Sample Prep Kits (Illumina, USA). Briefly, for small RNA library construction, the total RNA was size-fractionated on gel, and the 18-30-nt fraction was excised and purified. The gel-purified small RNAs were ligated to the 5′ RNA adapter and then the 3′ RNA adapter. The ligation products were purified, reverse transcribed, and amplified by RT-PCR. After the PCR products were purified, single-end sequencing (36 bp) was performed on an Illumina Hiseq2500 instrument at LC-BIO (Hangzhou, China) following the vendor’s protocol.
Bioinformatics and data processing. In this study, the CAP-miRSeq algorithm was used for a comprehensive analysis of small RNA sequencing reads to identify, annotate, and quantify miRNAs according to the user guide with some modifications [24]. In brief, the pre-processing of sequencing reads was performed by FastQC v. 0.10.1 to clean low-quality sequences and to remove adapter sequences from sequencing reads. Clean reads of more than 17 nt in length were aligned against the human Genome (hg19) and miRBase 20 using Bowtie v. 0.12.7 [25]. Subsequently, identification and quantification of known and predicted potential miRNAs were finished using miRDeep v. 2.0.0.5 [26, 27]. The significance of differential expression of miRNAs was determined using edgeR between healthy volunteers (H group) and PDAC patients (P group) [28]. The small RNA expression profiling data used for this study is publicly accessible through GEO (GSE144778).
Statistical analysis. All results are expressed as mean ± SD. Differences between groups were evaluated by analysis of variance, and post hoc analysis was performed by the Tukey–Kramer test. p value less than 0.05 was considered statistically significant.
RESULTS
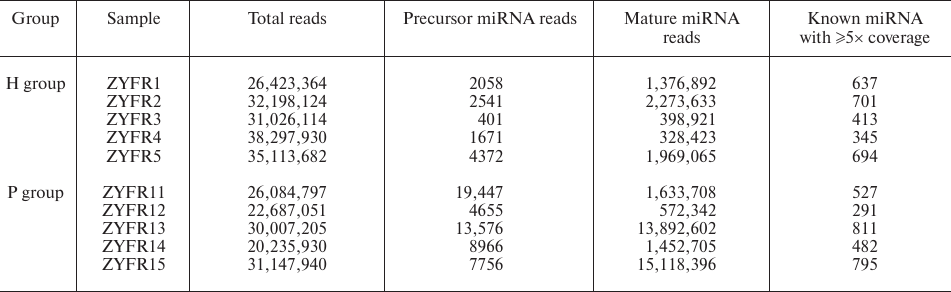
MiRNA expression profiles in blood plasma. To determine whether miRNAs could serve as potential biomarkers for PDAC diagnosis, five PDAC patients and five healthy volunteers were recruited (named as P group and H group, respectively). Blood samples were collected and the miRNAs from the plasma samples were sequenced by an Illumina Hiseq2500 instrument at LC-BIO. We found that a total of 130,162,923 and 163,059,214 reads are produced for the following analysis in P group (samples ZYFR11-15) and H group (samples ZYFR1-5), respectively. Among them, 581 of the known mature miRNAs with ≥5× coverage are detected in the P group and 558 of those in the H group (Table 2).
Table 2. General information of small RNA
sequencing from plasma samples in the H and P groups
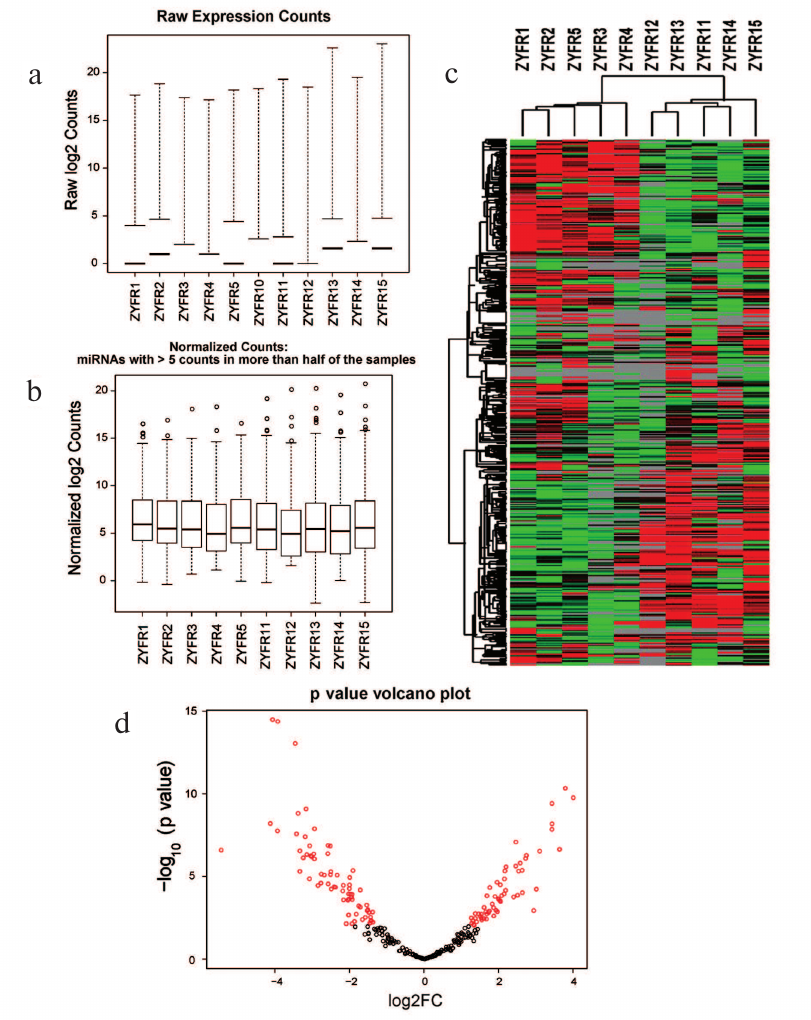
After that, the miRNA gene expression profiles were normalized (Fig. 1, a and b). For this purpose, the unsupervised hierarchical clustering of miRNA expression data was performed and presented as a heatmap (Fig. 1c). The p value of comparison of differential expression between H and P groups was calculated and visualized as shown in the volcano plot (Fig. 1d).
Fig. 1. Statistics and heatmap of sequencing data: a) raw expression counts of each sample; b) normalized counts of each sample; c) heatmap of miRNA expression in H and P groups; d) volcano plot.
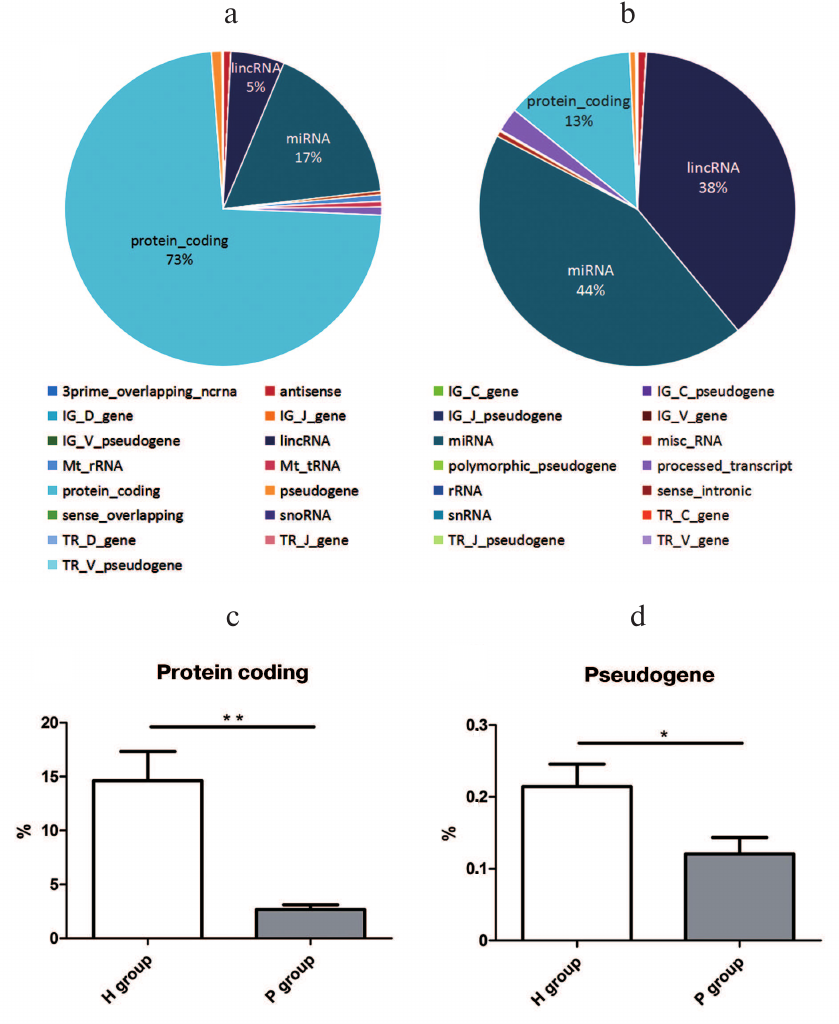
Small RNA distribution by genomic annotation. When compared with the human genome annotation (hg19), the small RNAs distributed in 27 types of genomic loci, including the protein coding region, miRNA loci, lncRNA loci, etc. Among them, the top three small RNA mapping regions with the most proportions in the H group were protein-coding ones (73%), miRNA (17%), and lncRNA (5%) loci (Fig. 2a), while the P group consisted of miRNA (44%), lncRNA (38%), and protein-coding (13%) loci (Fig. 2b). Among the 27 genomic loci with small RNAs mapped, proportions of small RNAs from protein-coding (15%) and pseudogene loci (0.2%) in the H group were significantly higher than the ones in the P group (2 and 0.1%) with p value 0.0024 and 0.0419, respectively (Fig. 2, c and d). The data indicated that the miRNAs are significantly differential expressed between PDAC patients and healthy persons.
Fig. 2. Small RNA gene loci in the H (a) and P (b) groups; c) proportional difference of protein-coding gene loci between H and P groups, ** p = 0.0024; d) proportional difference of pseudogene gene loci between H and P groups, * p = 0.0422.
Differential expression of mature miRNAs and novel miRNA in P versus H groups. To assess potential diagnostic value of the identified small RNAs, differential analysis by edgeR was performed to determine the expression differences between the P and H groups. The analysis revealed that the expression of 165 miRNAs were significantly (p < 0.05) different between the P and H groups. From these, mature miRNAs were significantly up- (75) and down-regulated (90) in the P group rather than in the H group (p < 0.05; Table S1, see Supplement to this paper on the website of the journal (http://protein.bio.msu.ru/biokhimiya) and Springer site (Link.springer.com)).
In addition, a total of 279 novel miRNAs in the H group and 785 in the P group were found (Tables S2 and S3, see Supplement), of which 35 novel miRNAs were shared by the H and P groups.
DISCUSSION
As mentioned in the introductory part, early diagnosis is extremely important for PDAC patients. Plasma miRNAs have been reported as a type of reliable biomarkers for PDAC early detection due to its sensitivity, specificity, and stability. Although miRNA studies based on blood samples have been performed for clinical utilization, there is no reproducible miRNA marker panel shared by all studies [19-23]. In our study, plasma miRNA profiles from healthy individuals (H group) and PDAC patients (P group) in a Chinese Han population were investigated with genome-wide small RNA expression profiling. A total of 165 mature miRNAs are significantly differentially expressed between the P group and H group, among which 75 are up-regulated and 90 are down-regulated in the P group rather than in H group, respectively. The top two significantly up-regulated miRNAs in the P group are hsa-miR-182-5p (logFold-Change = 4.00, p = 1.77E-10) and hsa-miR-4732-5p (logFold-Change = 3.78, p = 4.76E-11), while the two significantly down-regulated miRNAs in P group are hsa-miR-139-5p (logFold-Change = –4.07, p = 3.35E-15) and hsa-miR-23b-3p (logFold-Change = –3.93, p = 4.31E-15).
It has been reported that miR-182-5p (previous ID: miR-182, MiRBase) is overexpressed in PDAC cell lines and tumor tissues and promotes tumor cell proliferation and migration via direct targeting of the β-TrCP/β-catenin pathway [29]. In addition, the overexpression of miR-182-5p could depress FOXO3a (Forkhead box O3) and lead to the BIM/Bax-dependent mitochondrial apoptosis signaling pathway by directly inhibiting FOXO3a expression [30, 31]. Consistent with the above-mentioned studies, plasma miR-182-5p was also found in our study. In addition, Chen et al. found that circulating miR-182 in pancreatic cancer patients was significantly higher than that in healthy controls, which may become useful as a non-invasive tumor marker for diagnosis and prognosis of PDAC [32] after careful further validation in a larger cohort.
MiR-4732-5p was reported to be expressed in breast tissue [33]. MiR-4732-5p may directly bind the 5′-UTR region of Wrap53 mRNA in breast cancer and prohibit the binding of p53 mRNA [34], indicating a possible linkage between miR-4732-5p and tumor suppressive protein p53. As recently shown, p53 plays an important role in the late-stage progression of PDAC, indicating that miR-4732-5p may also play an important role in PDAC genesis [35].
The functions of miR-139-5p (previous ID: miR-139, MiRBase) in cancer are still controversial between different studies. Li et al. reported miR-139 is significantly overexpressed in endothelial cells of human pancreatic tumor and may be positively correlated with migration and angiogenesis [36] and could also promote the progression of colorectal cancer and oral tongue squamous cell carcinoma [37, 38]. However, some studies proposed that miR-139 is a tumor suppresser and is significantly down-regulated in non-small cell lung cancer (NSCLC), hepatocellular carcinoma, basal cell carcinoma, and adult acute myeloid leukemia [39-43].
The second most down-regulated miRNAs in the P group is hsa-miR-23b-3p (previous ID miR-23b, MiRBase). However, the function of this miRNA is still to be determined. Previous studies showed that miR-23b-3p was down-regulated in the tissue of NSCLC patients [44], but other researchers reported that miR-23b-3p was an up-regulated factor in circulating exosomes of NSCLC patients [45, 46]. Wang et al. found that miR-23b inhibits autophagy in human pancreatic cancer cell line BxPC3 and Panc-1 by direct targeting of ATG12 (autophagy-related protein 12), and further sensitized pancreatic cancer cells to radiation treatment [47]. An inversely correlated expression pattern of miR-23b and ATG12 was found in human PDAC. In our study, the expression level of miR-23b-3p was significantly decreased in the plasma of PDAC patients, indicating its potential diagnostic role in PDAC detection.
In conclusion, a genome wide small RNA expression profiling was performed for plasma samples from PDAC patients and healthy volunteers, a total of 165 mature miRNAs were differentially expressed between PDAC and healthy groups, and these miRNAs could be potential early and non-invasive diagnostic biomarkers for PDAC when further validated in a large cohort.
Acknowledgements
The authors would like to thank Prof. Deling Yin from the Department of Internal Medicine and Biomedical Sciences, East Tennessee State University Quillen College of Medicine, for his kind instruction and help.
Funding
This work was supported in part by following grants: The Project of Department of Science and Technology of Henan Province (No. 162102410006); The Health and Family Planning Commission of Henan Province (No. 2017049); The National Natural Science Foundation of China (No. 81602362); The program for Science and Technology Development in Henan Province (No. 162102310391); The program for Young Key Teacher of Henan Province (2016GGJS-214); The supporting grants of Henan University (No. 2015YBZR048; No. B2015151); Yellow River Scholar Program (No. H2016012); The program for Innovative Talents of Science and Technology in Henan Province (No. 18HASTIT048).
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
This study was approved (Approval number LL201508001) by the Ethics Committee of the Second Affiliated Hospital of Zhengzhou University, Zhengzhou, China. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants involved in the study.
REFERENCES
1.Raimondi, S., Maisonneuve, P., and Lowenfels, A. B.
(2009) Epidemiology of pancreatic cancer: an overview, Nat. Rev.
Gastroenterol. Hepatol., 6, 699-708; doi:
10.1038/nrgastro.2009.177.
2.Hartwig, W., Werner, J., Jager, D., Debus, J., and
Buchler, M. W. (2013) Improvement of surgical results for pancreatic
cancer, Lancet Oncol., 14, 476-485; doi:
10.1016/S1470-2045(13)70172-4.
3.Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993)
The C. elegans heterochronic gene lin-4 encodes small
RNAs with antisense complementarity to lin-14, Cell,
75, 843-854; doi: 10.1016/0092-8674(93)90529-Y.
4.Krol, J., Loedige, I., and Filipowicz, W. (2010)
The widespread regulation of microRNA biogenesis, function and decay,
Nat. Rev. Genet., 11, 597-610; doi: 10.1038/nrg2843.
5.Van Kouwenhove, M., Kedde, M., and Agami, R. (2011)
MicroRNA regulation by RNA-binding proteins and its implications for
cancer, Nat. Rev. Cancer, 11, 644-656; doi:
10.1038/nrc3107.
6.Rana, T. M. (2007) Illuminating the silence:
understanding the structure and function of small RNAs, Nat. Rev.
Mol. Cell Biol., 8, 23-36; doi: 10.1038/nrm2085.
7.Stefani, G., and Slack, F. J. (2008) Small
non-coding RNAs in animal development, Nat. Rev. Mol. Cell
Biol., 9, 219-230; doi: 10.1038/nrm2347.
8.Suzuki, H. I., Arase, M., Matsuyama, H., Choi, Y.
L., Ueno, T., Mano, H., Sugimoto, K., and Miyazono, K. (2011) MCPIP1
ribonuclease antagonizes dicer and terminates microRNA biogenesis
through precursor microRNA degradation, Mol. Cell, 44,
424-436; doi: 10.1016/j.molcel.2011.09.012.
9.Esquela-Kerscher, A., and Slack, F. J. (2006)
Oncomirs – microRNAs with a role in cancer, Nat. Rev.
Cancer, 6, 259-269; doi: 10.1038/nrc1840.
10.An, X., Sarmiento, C., Tan, T., and Zhu, H.
(2017) Regulation of multidrug resistance by microRNAs in anti-cancer
therapy, Acta Pharm. Sin. B, 7, 38-51; doi:
10.1016/j.apsb.2016.09.002.
11.Jafri, M. A., Al-Qahtani, M. H., and Shay, J. W.
(2017) Role of miRNAs in human cancer metastasis: implications for
therapeutic intervention, Semin. Cancer Biol., 44,
117-131; doi: 10.1016/j.semcancer.2017.02.004.
12.Fattore, L., Costantini, S., Malpicci, D.,
Ruggiero, C. F., Ascierto, P. A., Croce, C. M., Mancini, R., and
Ciliberto, G. (2017) MicroRNAs in melanoma development and resistance
to target therapy, Oncotarget, 8, 22262-22278; doi:
10.18632/oncotarget.14763.
13.Fabbri, M., Paone, A., Calore, F., Galli, R.,
Gaudio, E., Santhanam, R., Lovat, F., Fadda, P., Mao, C., Nuovo, G. J.,
Zanesi, N., Crawford, M., Ozer, G. H., Wernicke, D., Alder, H.,
Caligiuri, M. A., Nana-Sinkam, P., Perrotti, D., and Croce, C. M.
(2012) MicroRNAs bind to Toll-like receptors to induce prometastatic
inflammatory response, Proc. Natl. Acad. Sci. USA, 109,
2110-2116; doi: 10.1073/pnas.1209414109.
14.Cortez, M. A., Bueso-Ramos, C., Ferdin, J.,
Lopez-Berestein, G., Sood, A. K., and Calin, G. A. (2011) MicroRNAs in
body fluids – the mix of hormones and biomarkers, Nat. Rev.
Clin. Oncol., 8, 467-477; doi:
10.1038/nrclinonc.2011.76.
15.Stroun, M., Lyautey, J., Lederrey, C.,
Olson-Sand, A., and Anker, P. (2001) About the possible origin and
mechanism of circulating DNA apoptosis and active DNA release, Clin.
Chim. Acta, 313, 139-142; doi:
10.1016/S0009-8981(01)00665-9.
16.Mitchell, P. S., Parkin, R. K., Kroh, E. M.,
Fritz, B. R., Wyman, S. K., Pogosova-Agadjanyan, E. L., Peterson, A.,
Noteboom, J., O'Briant, K. C., Allen, A., Lin, D. W., Urban, N.,
Drescher, C. W., Knudsen, B. S., Stirewalt, D. L., Gentleman, R.,
Vessella, R. L., Nelson, P. S., Martin, D. B., and Tewari, M. (2008)
Circulating microRNAs as stable blood-based markers for cancer
detection, Proc. Natl. Acad. Sci. USA, 105, 10513-10518;
doi: 10.1073/pnas.0804549105.
17.Karius, T., Schnekenburger, M., Dicato, M., and
Diederich, M. (2012) MicroRNAs in cancer management and their
modulation by dietary agents, Biochem. Pharmacol., 83,
1591-1601; doi: 10.1016/j.bcp.2012.02.004.
18.Brunetti, O., Russo, A., Scarpa, A., Santini, D.,
Reni, M., Bittoni, A., Azzariti, A., Aprile, G., Delcuratolo, S.,
Signorile, M., Gnoni, A., Palermo, L., Lorusso, V., Cascinu, S., and
Silvestris, N. (2015) MicroRNA in pancreatic adenocarcinoma:
predictive/prognostic biomarkers or therapeutic targets?
Oncotarget, 6, 23323-23341; doi:
10.18632/oncotarget.4492.
19.Liu, R., Chen, X., Du, Y., Yao, W., Shen, L.,
Wang, C., Hu, Z., Zhuang, R., Ning, G., Zhang, C., Yuan, Y., Li, Z.,
Zen, K., Ba, Y., and Zhang, C. Y. (2012) Serum microRNA expression
profile as a biomarker in the diagnosis and prognosis of pancreatic
cancer, Clin. Chem., 58, 610-618; doi:
10.1373/clinchem.2011.172767.
20.Cote, G. A., Gore, A. J., McElyea, S. D.,
Heathers, L. E., Xu, H., Sherman, S., and Korc, M. (2014) A pilot study
to develop a diagnostic test for pancreatic ductal adenocarcinoma based
on differential expression of select miRNA in plasma and bile, Am.
J. Gastroenterol., 109, 1942-1952; doi:
10.1038/ajg.2014.331.
21.Wang, J., Chen, J., Chang, P., LeBlanc, A., Li,
D., Abbruzzesse, J. L., Frazier, M. L., Killary, A. M., and Sen, S.
(2009) MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients
as novel blood-based biomarkers of disease, Cancer Prev. Res.
(Phila), 2, 807-813; doi:
10.1158/1940-6207.CAPR-09-0094.
22.Liu, J., Gao, J., Du, Y., Li, Z., Ren, Y., Gu,
J., Wang, X., Gong, Y., Wang, W., and Kong, X. (2012) Combination of
plasma microRNAs with serum CA19-9 for early detection of pancreatic
cancer, Int. J. Cancer, 131, 683-691; doi:
10.1002/ijc.26422.
23.Schultz, N. A., Dehlendorff, C., Jensen, B. V.,
Bjerregaard, J. K., Nielsen, K. R., Bojesen, S. E., Calatayud, D.,
Nielsen, S. E., Yilmaz, M., Hollander, N. H., Andersen, K. K., and
Johansen, J. S. (2014) MicroRNA biomarkers in whole blood for detection
of pancreatic cancer, JAMA, 311, 392-404; doi:
10.1001/jama.2013.284664.
24.Sun, Z., Evans, J., Bhagwate, A., Middha, S.,
Bockol, M., Yan, H., and Kocher, J. P. (2014) CAP-miRSeq: a
comprehensive analysis pipeline for microRNA sequencing data, BMC
Genomics, 15, 1-10; doi: 10.1186/1471-2164-15-423.
25.Langmead, B., Trapnell, C., Pop, M., and
Salzberg, S. L. (2009) Ultrafast and memory-efficient alignment of
short DNA sequences to the human genome, Genome Biol.,
10, R25; doi: 10.1186/gb-2009-10-3-r25.
26.An, J., Lai, J., Lehman, M. L., and Nelson, C. C.
(2013) miRDeep*: an integrated application tool for miRNA
identification from RNA sequencing data, Nucleic Acids Res.,
41, 727-737; doi: 10.1093/nar/gks1187.
27.Friedlander, M. R., Mackowiak, S. D., Li, N.,
Chen, W., and Rajewsky, N. (2012) miRDeep2 accurately identifies known
and hundreds of novel microRNA genes in seven animal clades, Nucleic
Acids Res., 40, 37-52; doi: 10.1093/nar/gkr688.
28.Robinson, M. D., McCarthy, D. J., and Smyth, G.
K. (2010) edgeR: a bioconductor package for differential expression
analysis of digital gene expression data, Bioinformatics,
26, 139-140; doi: 10.1093/bioinformatics/btp616.
29.Chen, X., Su, Z., Wang, S., and Xu, H. (2016)
Clinical and prognostic significance of Arl4c expression in colorectal
cancer, Cancer Biomark, 16, 253-257; doi:
10.3233/cbm-150562.
30.Li, Y., Li, A., Wu, J., He, Y., Yu, H., Chai, R.,
and Li, H. (2016) MiR-182-5p protects inner ear hair cells from
cisplatin-induced apoptosis by inhibiting FOXO3a, Cell Death
Dis., 7, e2362; doi: 10.1038/cddis.2016.246.
31.Czabotar, P. E., Colman, P. M., and Huang, D. C.
(2009) Bax activation by Bim? Cell Death Differ., 16,
1187-1191; doi: 10.1038/cdd.2009.83.
32.Chen, Q., Yang, L., Xiao, Y., Zhu, J., and Li, Z.
(2014) Circulating microRNA-182 in plasma and its potential diagnostic
and prognostic value for pancreatic cancer, Med. Oncol.,
31, 225; doi: 10.1007/s12032-014-0225-z.
33.Persson, H., Kvist, A., Rego, N., Staaf, J.,
Vallon-Christersson, J., Luts, L., Loman, N., Jonsson, G., Naya, H.,
Hoglund, M., Borg, A., and Rovira, C. (2011) Identification of new
microRNAs in paired normal and tumor breast tissue suggests a dual role
for the ERBB2/Her2 gene, Cancer Res., 71, 78-86;
doi: 10.1158/0008-5472.CAN-10-1869.
34.Pouladi, N., Kouhsari, S. M., Feizi, M. H.,
Gavgani, R. R., and Azarfam, P. (2013) Overlapping region of p53/wrap53
transcripts: mutational analysis and sequence similarity with
microRNA-4732-5p, Asian Pac. J. Cancer Prev., 14,
3503-3507; doi: 10.7314/APJCP.2013.14.6.3503
35.Maitra, A., Adsay, N. V., Argani, P.,
Iacobuzio-Donahue, C., De Marzo, A., Cameron, J. L., Yeo, C. J., and
Hruban, R. H. (2003) Multicomponent analysis of the pancreatic
adenocarcinoma progression model using a pancreatic intraepithelial
neoplasia tissue microarray, Mod. Pathol., 16, 902-912;
doi: 10.1097/01.MP.0000086072.56290.FB.
36.Li, L., Li, B., Chen, D., Liu, L., Huang, C., Lu,
Z., Lun, L., and Wan, X. (2015) miR-139 and miR-200c regulate
pancreatic cancer endothelial cell migration and angiogenesis,
Oncol. Rep., 34, 51-58; doi: 10.3892/or.2015.3945.
37.Miyoshi, J., Toden, S., Yoshida, K., Toiyama, Y.,
Alberts, S. R., Kusunoki, M., Sinicrope, F. A., and Goel, A. (2017)
MiR-139-5p as a novel serum biomarker for recurrence and metastasis in
colorectal cancer, Sci. Rep., 7, 43393; doi:
10.1038/srep43393.
38.Chen, Z., Yu, T., Cabay, R. J., Jin, Y.,
Mahjabeen, I., Luan, X., Huang, L., Dai, Y., and Zhou, X. (2017)
miR-486-3p, miR-139-5p, and miR-21 as biomarkers for the detection of
oral tongue squamous cell carcinoma, Biomark. Cancer, 9,
1-8; doi: 10.4137/BIC.S40981.
39.Sun, C., Sang, M., Li, S., Sun, X., Yang, C., Xi,
Y., Wang, L., Zhang, F., Bi, Y., Fu, Y., and Li, D. (2015)
Hsa-miR-139-5p inhibits proliferation and causes apoptosis associated
with down-regulation of c-Met, Oncotarget, 6,
39756-39792; doi: 10.18632/oncotarget.5476.
40.Mo, Y., Lu, Y., Wang, P., Huang, S., He, L., Li,
D., Li, F., Huang, J., Lin, X., Li, X., Che, S., and Chen, Q. (2017)
Long non-coding RNA XIST promotes cell growth by regulating
miR-139-5p/PDK1/AKT axis in hepatocellular carcinoma, Tumour
Biol., 39, 1010428317690999; doi:
10.1177/1010428317690999.
41.Sand, M., Skrygan, M., Sand, D., Georgas, D.,
Hahn, S. A., Gambichler, T., Altmeyer, P., and Bechara, F. G. (2012)
Expression of microRNAs in basal cell carcinoma, Br. J.
Dermatol., 167, 847-855; doi:
10.1111/j.1365-2133.2012.11022.x.
42.Xu, K., Shen, K., Liang, X., Li, Y., Nagao, N.,
Li, J., Liu, J., and Yin, P. (2016) MiR-139-5p reverses
CD44+/CD133+-associated multidrug resistance by downregulating NOTCH1
in colorectal carcinoma cells, Oncotarget, 7,
75118-75129; doi: 10.18632/oncotarget.12611.
43.Chen, H., Xu, H., Meng, Y. G., Zhang, Y., Chen,
J. Y., and Wei, X. N. (2016) miR-139-5p regulates proliferation,
apoptosis, and cell cycle of uterine leiomyoma cells by targeting
TPD52, Onco. Targets Ther., 9, 6151-6160; doi:
10.2147/OTT.S108890.
44.Janikova, M., Zizkova, V., Skarda, J.,
Kharaishvili, G., Radova, L., and Kolar, Z. (2016) Prognostic
significance of miR-23b in combination with P-gp, MRP and LRP/MVP
expression in non-small cell lung cancer, Neoplasma, 63,
576-587; doi: 10.4149/neo_2016_411.
45.Cinegaglia, N. C., Andrade, S. C., Tokar, T.,
Pinheiro, M., Severino, F. E., Oliveira, R. A., Hasimoto, E. N.,
Cataneo, D. C., Cataneo, A. J., Defaveri, J., Souza, C. P., Marques, M.
M., Carvalho, R. F., Coutinho, L. L., Gross, J. L., Rogatto, S. R.,
Lam, W. L., Jurisica, I., and Reis, P. P. (2016) Integrative
transcriptome analysis identifies deregulated microRNA-transcription
factor networks in lung adenocarcinoma, Oncotarget, 7,
28920-28934; doi: 10.18632/oncotarget.8713.
46.Begum, S., Hayashi, M., Ogawa, T., Jabboure, F.
J., Brait, M., Izumchenko, E., Tabak, S., Ahrendt, S. A., Westra, W.
H., Koch, W., Sidransky, D., and Hoque, M. O. (2015) An integrated
genome-wide approach to discover deregulated microRNAs in non-small
cell lung cancer: clinical significance of miR-23b-3p deregulation,
Sci. Rep., 5, 13236; doi: 10.1038/srep13236.
47.Wang, P., Zhang, J., Zhang, L., Zhu, Z., Fan, J.,
Chen, L., Zhuang, L., Luo, J., Chen, H., Liu, L., Chen, Z., and Meng,
Z. (2013) MicroRNA 23b regulates autophagy associated with
radioresistance of pancreatic cancer cells, Gastroenterology,
145, 1133-1143; doi: 10.1053/j.gastro.2013.07.048.
Supplementary Tables S1-S3 (PDF)