REVIEW: Search for Intracellular Sensors Involved in the Functioning of Monovalent Cations as Secondary Messengers
E. A. Klimanova1,a*, S. V. Sidorenko1, A. M. Tverskoi1, A. A. Shiyan1, L. V. Smolyaninova1, L. V. Kapilevich2, S. V. Gusakova3, G. V. Maksimov1, O. D. Lopina1, and S. N. Orlov1,2,3
1Lomonosov Moscow State University, Department of Biology, 119234 Moscow, Russia2National Research Tomsk State University, 634050 Tomsk, Russia
3Siberian State Medical University, Ministry of Health of Russian Federation, 634050 Tomsk, Russia
* To whom correspondence should be addressed.
Received June 3, 2019; Revised July 15, 2019; Accepted July 16, 2019
Maintenance of non-equilibrium Na+ and K+ distribution between cytoplasm and extracellular medium suggests existence of sensors responding with conformational transitions to the changes of these monovalent cations’ intracellular concentration. Molecular nature of monovalent cation sensors has been established in Na,K-ATPase, G-protein-coupled receptors, and heat shock proteins structural studies. Recently, it was found that changes in Na+ and K+ intracellular concentration are the key factors in the transcription and translation control, respectively. In this review, we summarize results of these studies and discuss physiological and pathophysiological significance of Na+i,K+i-dependent gene expression regulation mechanism.
KEY WORDS: sodium, potassium, transcription, translationDOI: 10.1134/S0006297919110063
Abbreviations: AMPK, AMP-activated protein kinase; CaMK, Ca2+/calmodulin-dependent protein kinase; COX-2, cyclooxygenase type 2; CREB, cyclic AMP response element-binding protein; CTS, cardiotonic steroids; EPS, electrical pulse stimulation; ERG, early response gene; GPCR, G-protein-coupled receptor; HIF-1, hypoxia-inducible factor 1; HRE, HIF-response element; HSP, heat shock protein; NaRE, identified Na+ response element, gene transcription regulator; NaREB, unidentified [Na+]i sensor, NaRE activator; NBD, nucleotide-binding domain; NFAT, nuclear factor of activated T cells; NFκB, nuclear factor kappa-light chain enhancer of activated B cells; NMDA, N-methyl-D-aspartate; SBD, substrate-binding domain; SERCA, skeletal muscle sarcoplasmic/endoplasmic-reticulum Ca-ATPase; SMC, smooth muscle cell; UTR, untranslated mRNA region.
In his works, Nobel prize winner Earl Wilbur Sutherland, Jr. has framed
a concept, according to which any molecule could be considered as an
intracellular signaling messenger if it satisfied following three major
requirements. (i) Changes in this molecule intracellular concentration
in response to the external stimuli occur prior to the generation of
cell functional response, and this compound concentration remains the
same after the stimulus ceases. (ii) In the absence of external
stimuli, changes in the messenger intracellular concentration are
sufficient for the generation of cell response. (iii) Cell functional
response is mediated by interaction between the messenger and its
highly selective intracellular sensors [1]. Further
works demonstrated that low-molecular-weight compounds, such as cyclic
nucleotides (cAMP, cGMP), products of phospholipid catabolism
(diacylglycerol, inositol-1,4,5-triphosphate), and Ca2+
ions, meet these requirements [2-5].
Similar to above mentioned canonic messengers, monovalent ions intracellular concentrations change temporarily in response to various stimuli. For example, Na+ex/H+i exchange activation by growth factors and subsequent [Na+]i increase have been observed in all investigated cell types [6-11]. Activation of Ca2+-dependent K+ channels in mammalian erythrocytes with membrane potential close to chloride equilibrium potential [Em = ECl ~ –(8-12) mV] resulted in 5- to 8-fold [K+]i decrease [12], which, in turn, was accompanied by cell shrinkage and eryptosis – special form of erythrocyte programmed death [13]. Short-term synaptic activity in neurons and astrocytes resulted in local [Na+]i increase from ~10 to 50-100 mM, which can be explained by Na+ influx through ionotropic glutamate receptors or Na+ex-coupled neuromediator transporters [14, 15]. Hypoxia of smooth muscle cells (SMCs) resulted in 2- to 3-fold [Na+]i increase,which together with hypoxia-induced transcription factor 1 (HIF-1) expression upregulation causes changes in gene expression profile [16, 17]. Extensive exercise was found to trigger dissipation of Na+ and K+ transmembrane gradients in skeletal muscles, leading to 5- to 10-fold [Na+]i/[K+]i ratio increase [18]. These data, as well as the results of studies on Na+, K+, Cl–, and HCO3– selective effects on the enzyme activity and gene expression, allowed Orlov and Hamet [19] to formulate a hypothesis that proposed monovalent ions as secondary messengers. Purpose of this review is to summarize new studies data on this topic with special emphasis on identification of monovalent cation intracellular sensors. Data on monovalent anions involvement in intracellular signaling has been reviewed in recent publications [20, 21].
IDENTIFIED SENSORS OF MONOVALENT CATIONS
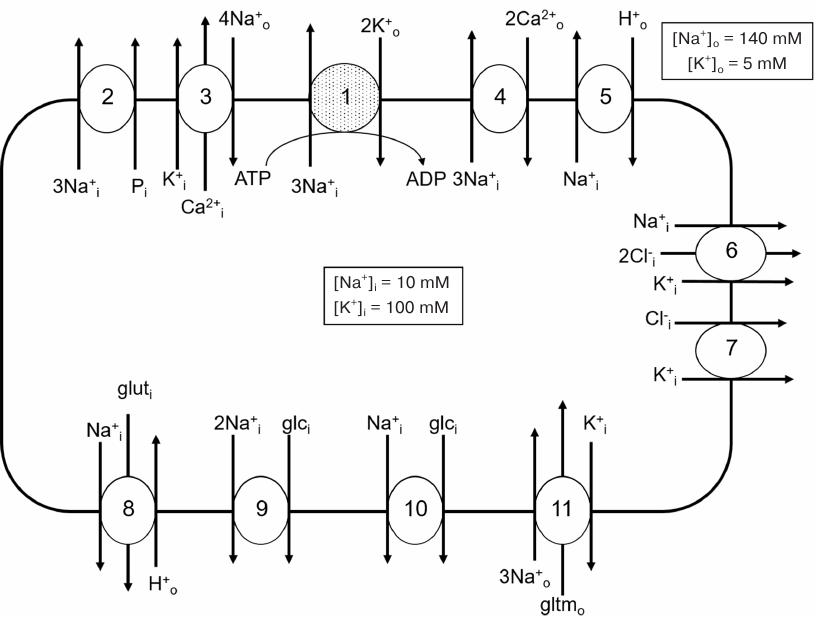
The most studied membrane transporters, which activity is controlled by Na+ and/or K+ ions in the concentrations comparable to their intracellular quantity (the first indication that these proteins might contain highly specific binding sites of these cations), are shown in Fig. 1. Other proteins exhibiting activity changes in response to monovalent cation concentration alteration have been also identified [22, 23]. In this section of our review, we summarize data of monovalent cation-binding sites structural studies.
Fig. 1. Membrane transporters whose activity is controlled by intracellular sodium (1, 2, 4-6, 8-10) and potassium (3, 6, 7, 11): 1) Na,K-ATPase; 2) 3Na+,phosphate cotransporter; 3) K+-dependent 4Na+/Ca2+ exchanger (KNCX); 4) 3Na+/Ca2+ exchanger (NCX); 5) Na+/H+ exchanger; 6) Na+,K+,2Cl– cotransporter (NKCC); 7) K+,Cl– cotransporter (KCC); 8) Н+-dependent Na+,glutamine cotransporter (SNAT3); 9) 2Na+,glucose cotransporter (SLC5A1); 10) Na+,glucose cotransporter (SLC5A2); 11) K+-dependent 3Na+,glutamate cotransporter. Shown only ion transport directions activated by the increase in Na+ and K+ intracellular concentrations. Values of sodium and potassium concentrations in extracellular fluids and cytoplasm of resting cells are shown on the example of blood plasma and human erythrocytes.
Na,K-ATPase. Although the first Na+ and K+ selective transporters were identified 70 years ago, structure of these cations binding sites has been investigated in detail only for Na,K-ATPase.
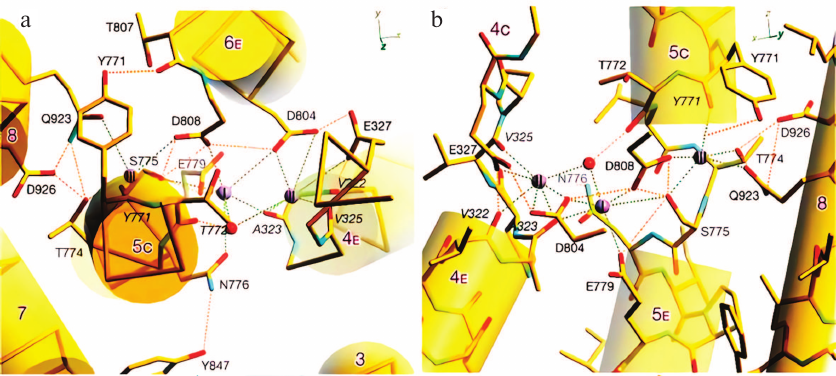
During its catalytic cycle, Na,K-ATPase exists alternatively in two conformational states; one of them displays high affinity for Na+ ions (E1) and the other – for K+ ions (E2). These two conformations’ spatial structure comparison made it possible to decipher the enzyme cation-binding structure (Fig. 2). According to conformation E1 X-ray diffraction data [24], three Na+-binding sites (I, II, and III) are located within the transmembrane part of the α-subunit (between transmembrane α-helices M4, M5, M6, and M8) at ~1/3 depth of membrane thickness from the cytoplasmic surface. Ala323, Glu779, Asp804, Asp808, Thr772, Ser775, Asn776 (site I); Val322, Ala323, Val325, Glu327, Asp804 (site II); and Tyr771, Thr772, Thr774, Ser775, Asp808, Gln923, Asp926 (site III) are the main amino acid residues of enzyme α-subunit that are involved in Na+-binding sites development. Site I is located at the same position as one of the K+-binding sites, while site II is 5 Å closer to the membrane cytoplasmic surface. Both sites I and II are situated in vicinity of the cytoplasmic part of the M5 transmembrane helix and separated by the Ser775 side chain. Site III is sterically limited (along M5) and too small to accommodate K+ ion.
Fig. 2. Na,K-ATPase Na+-binding sites atomic model is presented in orientation (a) perpendicular to the membrane (view from the cytoplasmic side) and (b) parallel to the membrane (view from the M6 helix side). Amino acid residues shown in italic indicate that the carbonyl groups of the main chain facilitate Na+ coordination. Green dashed lines, possible coordination of Na+; orange lines, potential hydrogen bonds (provided that carboxyl groups are protonated) (data from Kanai et al. [24]).
Distance between I and II Na+-binding sites is 3.2-3.6 Å, which is less than the distance between the two K+-binding sites. It is likely sufficient for adjacent positioning of two sodium ions, but not for two potassium ions (ionic radii, 0.95 and 1.33 Å, respectively). It was suggested that these sites selectively bind Na+, because the cavity with 2.4 Å radius would be sufficient for Na+ binding, while K+ binding requires 2.8 Å radius cavity. Precisely this size cavity was found in Na,K-ATPase E2 conformation crystal (i.e., conformation with high K+ affinity). Site I is the least sterically limited; hence, it likely acts as the K+-binding site, because Ser775 and Asn776 side chains participate in the coordination of both Na+ and K+ ions. Sites II and III in the E1 conformation are not sufficiently large for K+ binding. However, it is possible that site II cavity enlarges during E1 to E2 transition, so this site becomes capable of K+ binding.
The following scenario is possible to explain sequential and cooperative binding of three Na+ ions: E1 conformation cavity size is sufficient for accepting the first cytoplasmic Na+ ion (but not K+ ion) in the site III only if Ser775 side chain shifts its position. Based on X-ray diffraction analysis data, it can be assumed that the first Na+ ion is transported to the site III and binds to it, but the sites I and II should not be completely formed at the moment of Na+ transport (otherwise Na+ would bind to them). Only after the first Na+ ion binds to site III and the second Na+ ion to the site I, the binding site II is formed.
It is known that Ca2+-binding sites in another P-type ATPase, specifically skeletal-muscle sarcoplasmic-reticulum/endoplasmic-reticulum Ca-ATPase (SERCA), are formed mostly by the same amino acid residues as in Na,K-ATPase [25]. Transmembrane M5 helix cytoplasmic portion orientation plays an important role in Na,K-ATPase selectivity toward Na+. According to the X-ray diffraction data, straightening of M5 helix (as in Ca-ATPase) makes K+ binding in the site III possible. Indeed, removal of C-terminal amino acid residues in Na,K-ATPase α-subunit increases M5 helix mobility and results in decrease of Na+ enzyme affinity and K+ binding in the site III.
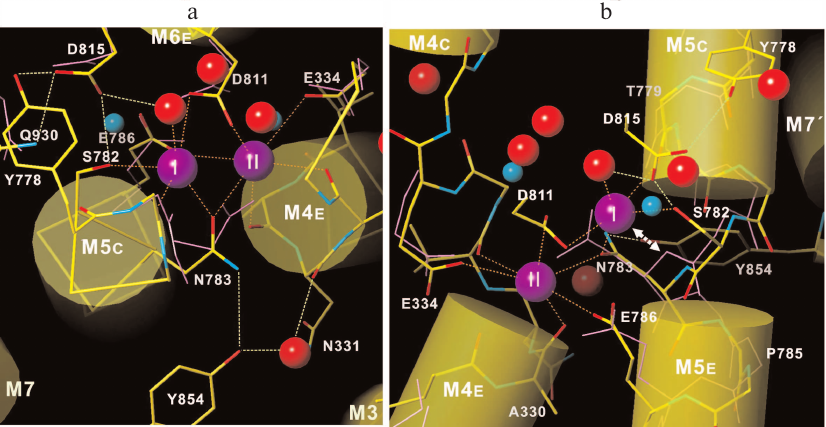
Na,K-ATPase belongs to the P-type ATPase family, whose members display significant homology in the amino acid sequences and similarity in catalytic subunit three-dimensional structure, as well as in ATP hydrolysis and ion transport mechanisms. This was the reason for Toyoshima et al. [26] to discuss in detail why Na,K-ATPase is capable of K+ and some other monovalent cations binding, while SERCA can bind H+ in addition to Ca2+. Based on data from Na,K-ATPase E2 conformation X-ray diffraction analysis with 2.4 Å resolution, the following conclusions can be made. First, unlike Ca2+-binding sites in SERCA, K+-binding sites are located at extremely close distances from each other, despite the fact that K+ ionic radius is much bigger than that of Ca2+ (1.353 and 0.99 Å, respectively) (Fig. 3). It was suggested that such close positioning is possible only for monovalent cation-binding sites, because in this case, oxygen atoms of carbonyl groups in the polypeptide chain coordinate cations in both enzymes. Second, enzyme structure comparison revealed that coordination geometry of bound cations differs significantly in Na,K-ATPase and SERCA, which suggests potentially low ion-binding sites affinity of SERCA to monovalent cations. Moreover, calculated Na+ and K+ valence values are slightly different, thus suggesting that these binding sites are not very K+ selective. Altogether, these data support the idea that Na,K-ATPase primary function is Na+ pumping. Third, amino acid side chains’ location and conformation in Na,K-ATPase are very similar to those in SERCA Ca2+-binding sites with the Asn783 exception. In Na,K-ATPase, this amino acid residue forms hydrogen bond with Tyr854 in M7 helix kink; M5 helix is also partially unfolded forming a kink due to Pro785 presence, which plays a crucial role in space and K+ coordination creation by interacting with Thr799 backbone carbonyl oxygen. In SERCA, Pro785 is replaced with Gly770, which results in less M5 helix bending and, respectively, smaller size of empty cavity. As a result, SERCA Asn768 amide (Na,K-ATPase Asn783 analogue) is very close to the first K+-binding site and to site II side chain, which prevents K+ binding to these SERCA sites. All these findings demonstrate that although formation of monovalent cation binding sites involves similar amino acid residues, sites emerge as a result of protein three-dimensional structure formation, and even slight alterations in the protein spatial structure significantly affect cation-binding sites’ structure.
Fig. 3. Na,K-ATPase K+-binding sites atomic model shown (a) from the cytoplasmic side along the M5-helix and (b) parallel to the membrane. Solid purple lines denote side and main chains of SERCA Ca2+-coordinating amino acid residues. Purple spheres, K+ ions; red spheres, water molecules; blue spheres, Ca2+ ions bound to SERCA. Double arrow in panel (b) depicts steric collision between K+-binding site I and Asn768 amide in SERCA (Na,K-ATPase Asn783 analogue) (data from Toyshima et al. [26]).
G-protein-coupled receptors. G-protein-coupled receptors (GPCRs) is the largest membrane proteins superfamily representing one third of all pharmacological targets. GPCRs found in the human genome (826 in total) are subdivided into four families (A, B, C, and F) varying in the amino acid sequences. In the middle of 1970s, Pert et al. [27] discovered that [Na+]i increase within 10-100 mM in brain homogenates decreased agonists binding to the family A opioid receptors without pronounced antagonist binding effect. Later, the same phenomenon was observed for adrenergic receptors, as well as adenosine, dopamine, bradykinin, and other GPCRs [28, 29]. GPCR mutant studies revealed
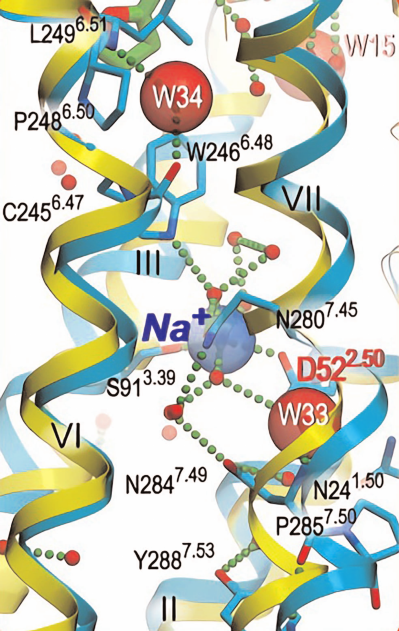
that Na+-dependent regulation of the receptor requires the aspartic acid presence in the second transmembrane GPCR domain. Together with high-resolution crystallography, these studies have established highly conservative Na+-binding site structure located in central cluster transmembrane domain composed of seven helices (Fig. 4). This cluster is a part of the water-filled channel located between ligand-binding and G-protein-binding sites. This channel has two narrow “necks” where its diameter decreases to the size slightly smaller than water molecule diameter (2.4 and 2.0 Å) due to the Trp246 and Tyr288 residue presence, respectively. Ligand binding changes receptor conformation, which results in the channel closing at these sites.
Fig. 4. GPCR transmembrane domain central cluster composed of seven helices (marked with Roman numerals) in antagonist-bound (cyan) and agonist-bound (yellow) states. Red spheres, water molecules; green dotted lines, salt bridges and hydrogen bonds; transparent blue sphere, Na+ ion; red semitransparent spheres, water molecules W33 and W34 that stabilize kinks in helices VI and VII (data from Liuetal et al. [30]).
Central part of the channel between Trp246 and Tyr288 (located 13 Å from each other) is completely filled with Na+ ion and 10 water molecules. Na+ is coordinated by Asp52 and Ser91 five oxygen atoms and three water molecules. In addition, Na+ forms hydrogen bonds with highly conserved amino acids Asn24, Trp246, Asn280, Asn284, and Tyr288 [30].
Considering that Na+ can enter central cluster from both sides of the membrane, the question arises whether it is extracellular or intracellular Na+ that controls the receptor activity. Na+ allosteric effect on ligand binding is a well-known fact reported in numerous studies. However, in the majority of investigated GPCRs, Na+ inhibits agonist binding with IC50 ~ 50 mM [29, 31, 32]. Based on these observations, as well as [Na+]ex value of 140 mM, extracellular Na+ involvement in GPCR activity regulation seems unlikely. The issue of intracellular Na+ contribution to GPCR functioning remains unsolved.
Heat shock proteins. Heat shock proteins with 70-kDa molecular mass (Hsp70) are conservative in almost all living organisms, where they are involved in numerous intracellular regulation processes, including folding of newly synthesized and/or denatured proteins to prevent their aggregation. Hsp70 family members display ATP-dependent chaperon activity. It was established that Hsp70 ATPase activity increases sharply in response to K+ concentration increase within 20-100 mM range, while Na+, Li+, and Cs+ produce almost no effect on the ATPase activity [33].
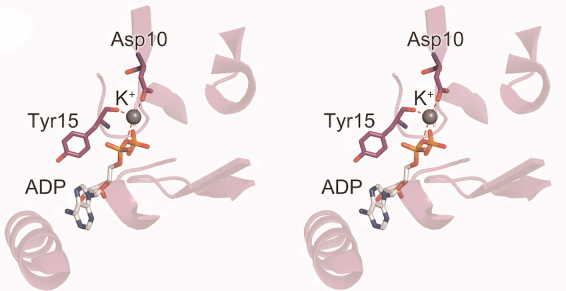
Hsp70 consists of N-terminal nucleotide-binding domain (NBD) and C-terminal substrate-binding domain (SBD). Hsp70 NBD in a complex with ADP crystal structure data from Escherichia coli revealed that this domain contains K+-binding site. When bound, K+ ion is coordinated by ADP β-phosphate group, Asp10 side chain, and Tyr15 main chain (Fig. 5). When K+ is replaced by Na+, the latter is also coordinated by ADP, Asp10, Tyr15 and located at the same position as K+ [34].
Fig. 5. Interaction between Hsp70 nucleotide-binding domain (NBD) from E. coli and ADP in the absence of Mg2+ ion. Grey sphere, K + ion; red dashed lines, coordination bonds (data from Arakawa et al. [34]).
Above data demonstrate fundamental difference in monovalent ions and Ca2+-binding sites structural organization. Indeed, Ca2+-binding sites in parvalbumin, calmodulin, troponin C, and a number of other proteins contain so-called EF-hand motif composed of 14-a.a. sequence [35, 36]. Examples given above show that monovalent cation binding sites are formed by amino acid residues located far apart from each other in the polypeptide chain with their geometry controlled by protein three-dimensional structure. This feature does not allow direct scanning of cDNA libraries for potential Na+ and K+ sensors, the strategy that has been successfully employed for Ca2+-binding proteins identification [37]. It must also be mentioned that unlike proteins with EF-hand motif and saturated at ~1 μM Ca2+ concentrations, monovalent cation binding sites affinity changes from 10 to 100 mM, which makes it challenging to use radioactive isotopes for protein identification in cell lysates.
SODIUM AS A TRANSCRIPTION REGULATOR
Cell shrinking and swelling are considered as universal markers of two different types of cell death – apoptosis and necrosis, respectively [38, 39]. In order to elucidate apoptosis mechanism, we compared effects of ion transport inhibitors on cell volume and viability. To our surprise, we discovered that prolonged rat aorta SMCs incubation with ouabain or in K+-free incubation medium (both inhibit Na,K-ATPase) attenuated apoptosis that has been induced by the lack of growth factors [40]. Na,K-ATPase inhibitors’ antiapoptotic effect was also demonstrated in renal epithelial proximal tubular cells culture [41], brain neurons [42], and vascular endothelial cells [43]. In further experiments we also found that SMCs Na,K-ATPase inhibition causes sharp activation of RNA synthesis [44] and generation of large amounts of newly synthesized proteins [45-47], which suggested expression of early response genes (ERGs). Indeed, cell treatment with ouabain resulted in significant c-Fos and c-Jun overexpression, the most studied ERG family representatives [47]. Taking into account that apoptosis prevention by Na,K-ATPase inhibitors was abolished by RNA (actinomycin D) and protein (cycloheximide) synthesis inhibitors [48], we suggested that this effect was mediated by transcription and/or translation regulation.
Data obtained during last 20 years have shown that ouabain and other cardiotonic steroids (CTSs) can generate intracellular signals independently from Na,K-ATPase inhibition [49, 50]. Considering this fact, we used Affymetrix technology and compared the effects of ouabain and potassium-depleted medium (as two independent methods for Na,K-ATPase inhibition) on SMCs, human umbilical vein endothelial cells (HUVECs), and HeLa cells transcriptomes [51]. Ouabain or the lack of K+ (Na,K-ATPase activator) altered expression of >500 genes with highly reliable positive correlation, which suggested dissipation of ion gradients and increase in [Na+]i/[K+]i ratio as the main mechanisms of transcription regulation. It was established that 80 of the Na+i/K+i-dependent gene transcripts were common for all three types of cells [51], as well as for rat brain neurons and mouse myoblast C2C12 cells [52]. Many ERGs, including Fos, FosB, Jun, JunB, Atf3, and Egr-1, were among these common Na+i/K+i-dependent genes.
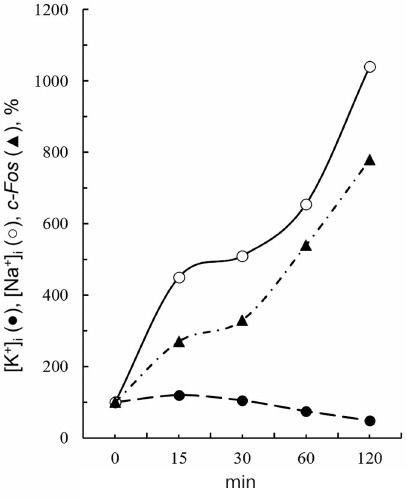
We have established in primary experiments that ouabain protected cells from apoptosis in a medium with elevated K+ content, this effect disappeared with Na+ transmembrane gradient dissipation [40, 53]. This indicates that ouabain antiapoptotic effect is due toNa+i increase rather than to K+i depletion. Conclusion on the leading role of Na+i increase is in agreement with the data comparing the kinetics of ouabain effects on ERG expression and Na+i and K+i content in SMCs [47]. Indeed, 4-fold decrease in c-Fos mRNA content was observed 30 min after ouabain addition, when intracellular Na+ content increased 5-fold, but K+i content decreased only by 10-15% (Fig. 6).
Fig. 6. Time course of the intracellular potassium content ([K+]i), sodium content ([Na+]i), and c-Fos mRNA in the aorta SMCs in response to 1 mM ouabain addition (data from Taurinetal et al. [47]).
Proof of Ca2+-independent signaling. Numerous studies have revealed that an increase in intracellular calcium concentration ([Ca2+]i) is accompanied by activation of several signaling systems involved in so-called excitation-transcription coupling [54-57]. First, Ca increase results in Ca2+/calmodulin-dependent protein kinase (CaMK) isoforms activation, IκB inhibitor phosphorylation, and NFκB (nuclear factor kappa-light-chain enhancer of activated B cells) translocation from the cytoplasm to the nucleus after phospho-IκB release. Second, [Ca2+]i increase is accompanied by nuclear factor of activated T cells (NFAT) dephosphorylation by Ca2+/calmodulin-dependent phosphatase calcineurin and translocation of the former to the nucleus. Third, an increase in [Ca2+]i leads to transcription regulator CREB (cyclic AMP response element-binding protein) phosphorylation. NFκB, NFAT, and CREB translocated into the nucleus interact with the corresponding binding sites in the 5′-untranslated regions (5′-UTRs) in gene promoters.
In some cell types, prolonged Na,K-ATPase inhibition is accompanied by [Ca2+]i increase due to the activation of Na+i/Ca2+o exchange and/or membrane depolarization followed by Ca2+ influx through the voltage-gated Ca2+ channels. In particular, we have shown that ouabain causes activation of Na+i/Ca2+o exchange in human lung fibroblasts [58] and of voltage-gated L-type Ca-channels in rat aorta SMCs [59]. It could be suggested that transcriptome changes in response to [Na+]i/[K+]i ratio increase are associated with [Ca2+]i increase and activation of above mentioned Ca2+-dependent transcription regulation mechanisms. This hypothesis, however, contradicts the data obtained with calcium chelators. Indeed, extracellular and intracellular Ca2+ chelators (EGTA and BAPTA-AM, respectively) did not affect ouabain-induced c-Fos upregulation in SMCs [47]. Moreover, loading cells with calcium chelators increased rather than decreased number of genes whose expression was altered in response to Na,K-ATPase inhibition [51]. Among universal Na+i/K+i-dependent genes, whose expressed changed more than 3-fold in the presence of Ca2+ chelators in all investigated cell types, were identified transcription factors (Fos, Jun, Hes1, Nfkbia), interleukin-6 (Il-6), and cyclooxygenase type 2 (COX-2 or Ptgs2).
While analyzing this data it should be kept in mind that calcium chelators can affect cell functions independently from calcium depletion. In particular, we observed that EGTA addition to SMCs caused an increase in sarcolemma permeability for monovalent cations [60] and ~10-fold [Na+]i/[K+]i ratio increase [59]. It is then not surprising that the content of 2071 transcripts was altered with high reliability in response to Na,K-ATPase inhibition (R2 > 0.51; p < 10–12) and addition of Ca2+ chelators [61]. Beside changing plasma membrane permeability, Ca2+ chelators can affect gene transcription via binding polyvalent cations, because EGTA and BAPTA affinity to Mn2+, Zn2+, Cu2+, Co2+, and Fe2+/3+ is several orders of magnitude higher than to Ca2+. This is the reason why calcium ions chelation is accompanied by changes in number of proteins (including numerous transcription regulation factors containing Zn2+-binding domains) regulated by polyvalent cations activity [62]. In 1981, Krishtal et al. [63] reported a sharp increase in calcium channel conductance in the presence of Ca2+ chelators, which also indirectly indicated Ca2+ chelators influence on cell metabolism.
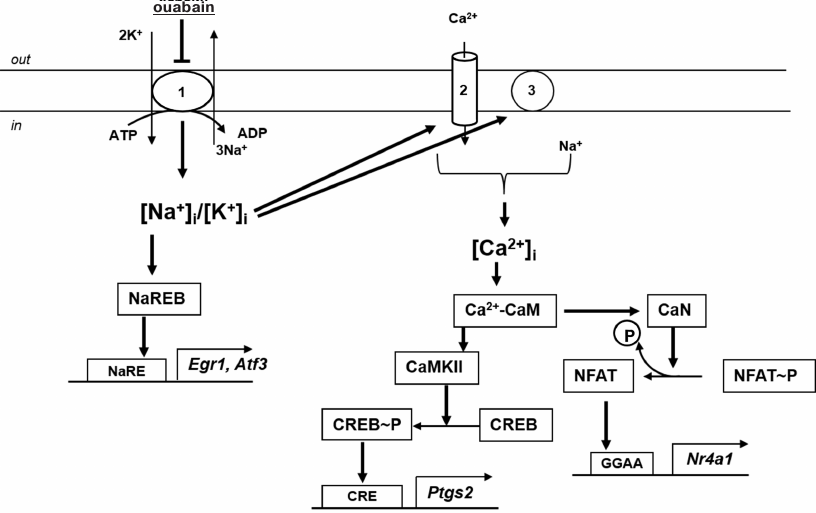
While searching for alternative approaches to study Na-sensitive gene transcription regulation mechanisms, we used Ca2+-dependent signaling systems inhibitors. We found that increase in Egr1 and Atf3 expression in ouabain-treated SMCs was independent from the presence of voltage-gated Ca2+ channels and Na+/Ca2+ exchange inhibitors, as well from calmodulin antagonists and Ca2+/calmodulin-dependent protein kinases and phosphatases inhibitors. At the same time, Ptgs2 and Nr4a1 expression upregulation was attenuated in the presence of Ca2+-dependent protein kinase (CaMKII) and calcineurin inhibitors, respectively (Fig. 7) [59]. All data presented in this section indicate Ca2+i-independent mechanisms involvement in the transcriptional regulation of genes, whose expression is altered by Na,K-ATPase and other stimuli causing an increase in [Na+]i inhibition.
Fig. 7. Involvement of monovalent cations and Ca2+ in the regulation of Egr1, Atf3, Ptgs2, and Nr4a1 transcription in rat aorta SMCs: 1) Na,K-ATPase; 2) voltage-gated Ca2+ channels; 3) Na+/Ca2+ exchanger; CaM, calmodulin; CaN, calcineurin; NaRE, identified Na+-dependent element regulating gene transcription; NaREB, non-identified sensor of [Na+]i changes activating NaRE.
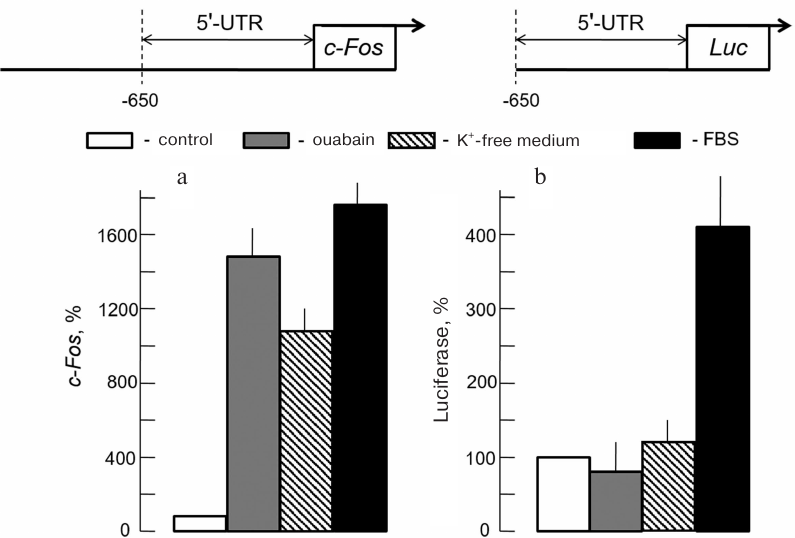
Canonical 5′-UTR promoters are not involved in the transcriptional regulation of Na+i-dependent genes. Mechanism of gene expression regulation mediated by interaction of ERG products and other transcription factors regulators with binding sites located in untranslated 5′-UTR sequence has been studied most comprehensively. Based on this fact, we have attempted to identify Na+-dependent transcription regulation element (Na+ response element, NaRE) in Na+-dependent gene c-Fos 5′-UTR sequence. This approach was justified by the absence of ouabain effect on the level of human c-Fos mRNA in NIH 3T3 mouse fibroblasts transfected with c-Fos gene with removed (–222)-(–70) sequence in the 5′-UTR [64]. Given the results of these studies HeLa cells were transfected with luciferase gene under control of 5′-UTR c-Fos long section ending at nucleotide (–650). This DNA fragment contained all studied promoter elements activated by growth factors and other canonical transcription regulators (Fig. 8). Cells treated with growth factors contained in the serum were used as a positive control. Similar to SMCs [47], Na,K-ATPase inhibition in HeLa cells with either ouabain or potassium-depleted medium was accompanied c-Fos expression upregulation (Fig. 8a). However, contrary to the endogenous c-Fos, luciferase expression was not regulated by ouabain, but instead was activated by addition of serum growth factors (Fig. 8b) [65]. These results indicated that increase in c-Fos mRNA content in response to Na,K-ATPase inhibition was not mediated by 5′-UTR site interaction with classic transcription regulators. Other Ca2+-independent regulation mechanisms of the Na+i-dependent transcriptome are discussed in the following section of this review.
Fig. 8. Changes of endogenous c-Fos mRNA content (a) and luciferase expression under 5′-UTR c-Fos control (b) in HeLa cells induced by growth factors addition (FBS) and Na,K-ATPase inhibition by ouabain or K+-free medium. Values obtained in the absence of stimuli are accepted as 100% (data from Haloui et al. [65]).
Search for sensors involved in the transcription regulation of Na+i-dependent genes. Negative results presented above can be explained by the existence of the following alternative mechanisms. (i) Na+ interacts with Ca2+-binding sites of calpain at relatively low [Ca2+]i (~100 nM) causing half-maximum enzyme activation at [Na+]i = 15 mM [66]. Role of Ca2+-binding proteins in transcription regulation via interaction with monovalent cations remains poorly understood. (ii) 5′-UTR site responsible for Na+i-mediated c-Fos transcription regulation is located far from the canonical promoter. In fact, it was established that chromatin reorganization affects c-Fos through 5′-UTR adjacent nucleotide (–1900) [67]. Farther located tissue-specific alternative promoters have been identified in a number of genes (complex promoters consisting of two or more independently functioning parts located upstream of one or several exons and ensuring different size mRNA molecules development from the same gene). For example, alternative promoters for gene Ppargc1a encoding coactivator of peroxisome proliferation activator receptor were found in skeletal muscles and nervous tissue. These promoters were located 14 and 587 kb upstream of the canonical promoter, respectively [68, 69]. (iii) 5'-UTR transcription can be regulated by the sequences located in introns or 3′-UTR. All these possibilities have been established for c-Fos transcription [70, 71]. In this regard, our laboratory has been screening for genes, whose transcription is altered at initial stages of monovalent ion transmembrane gradient dissipation. We expect that this approach will allow us to identify their common Na+-dependent sites (NaRE) and, based on these data, to elucidate the nature of Na+-dependent proteins (NaREBs) interacting with these NaREs. (iv) By now, large body of evidence has been accumulated on the regulation of gene expression during gene translocation in the nucleus (so-called three-dimensional transcription regulation model) [72]. Indeed, in the interphase, the most condensed chromatin part (heterochromatin) is located at the nucleus periphery, while less condensed chromatin (euchromatin) localizes at nucleus center [73]. Transcription factories visualized via incorporation of fluorescently labeled uridine-5′-triphosphate have been mainly observed precisely in the euchromatin [74]. Rearrangement of hetero- and euchromatin is controlled by such epigenetic factors as DNA methylation and acetylation of chromatin. Role of [Na+]i/[K+]i ratio increase in the spatial organization regulation of hetero- and euchromatin remains elusive. However, recently reported data indicate that K+ and Na+ ions stabilize four-helix guanidine-rich structures (G-quadruplexes) differently. G4-quadruplexes specifically bind mono- and divalent metal ions inside the cavity formed by guanine carbonyl groups. Cations lose their hydrate shells during this process. Maximum stabilization has been observed for K+, while Na+ displays less pronounced stabilizing effect. Interestingly enough, G-quadruplexes are often found in promoters. Further studies will allow to establish whether changes in the stability of G-quadruplexes caused by the increase in the [Na+]i/[K+]i ratio are associated with the changes in gene expression [75].
POTASSIUM AS A TRANSLATIONAL REGULATOR
Fifty years ago, it was found that protein synthesis in prokaryotic cell extracts decreases along with K+ concentration [76]. Later, this phenomenon has been also observed in animal and plant origin eukaryotic cells (see review by Orlov and Hamet [19]). In this view, it is not surprising that in the vast majority of investigated cell types, including fibroblasts [77], epithelial cells [78], and reticulocytes [79], Na,K-ATPase inhibition is accompanied by protein synthesis reduction without affecting transmembrane transport of labeled amino acids, intracellular ATP content, and ribosome assembly. In case of reticulocyte lysates, half-maximum increase in globin synthesis in media containing 60, 90, and 125 mM Na+ was observed at [K+] = 15, 25, and 40 mM, respectively [79]. This led to the suggestion that Na+ (unlike K+) does not activate protein synthesis, but rather competes with K+ for the sites involved in the translation regulation. Indeed, in contrast to K+ and Rb+ ions, Na+, Li+, and Cs+ did not produce any reliably detected effect on the activity of purified human tyrosyl-tRNA synthetase [80].
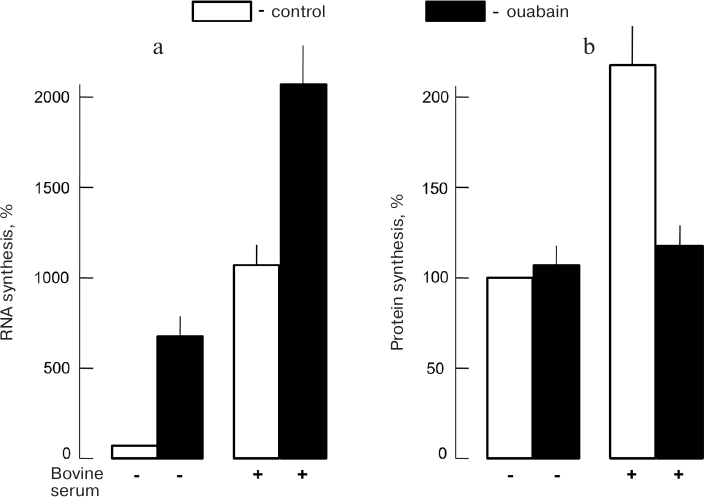
We must emphasize observed protein synthesis suppression in intact cells in response to [K+]i decrease can be partially compensated by [Na+]i increase and activation of previously mentioned Na+-dependent mechanism of gene transcription regulation, including those involved in elongation [81, 82]. In particular, we established that Na,K-ATPase inhibition with CTSs in human endothelial cells was accompanied by 3-fold decrease in mRNA content for the translation elongation factor EIF5 [83]. It should also be mentioned that K+-dependent translation relative contribution to gene expression regulation is tissue-specific. Indeed, no 24-h ouabain Na,K-ATPase inhibition reliable effect on [3H]leucine incorporation into protein fraction of the rat aorta SMCs was observed in the absence of growth factors, which was contrary to 7-8-fold RNA synthesis activation measured by [3H]uridine incorporation (Fig. 9). In the presence of growth factors, RNA synthesis increased 10-fold and protein synthesis increased 2-fold. In this case ouabain led to 2-fold protein synthesis decrease while RNA synthesis increased 2-fold [44].
Fig. 9. Effect of ouabain (1 mM, 24 h) on RNA (a) and protein (b) synthesis in rat aorta SMCs; CS, 1% bovine serum in the medium (data from Orlov et al. [44]).
Three hypotheses can be suggested to explain this phenomenon. (i) K+i-dependent sensor content involved in translation regulation is tissue-specific and reduced in SMCs. Indeed, unlike mammalian tyrosyl-tRNA synthetase, K+ does not affect bacterial enzyme activity [84, 85]. (ii) K+i-sensitive translation regulation mechanism is limited to certain genes [86]. (iii) Gene translation inhibition caused by [K+]i decrease can be compensated by above-described mechanism of Na+i-dependent transcription activation of both studied gene and ERGs involved in the transcription regulation. At present, these hypotheses validation is underway in our laboratory.
PHYSIOLOGICAL AND PATHOPHYSIOLOGICAL SIGNIFICANCE OF
Na+i,K+i-DEPENDENT GENE EXPRESSION REGULATION MECHANISM
In this section, we will limit ourselves to few examples demonstrating significance of Na+i,K+i-dependent gene expression regulation mechanism and functional cell response generation.
Myokines secretion. It was established in early 2000s that skeletal muscles can function as an endocrine organ producing cytokines and other low-molecular-weight proteins (later termed myokines) in response to increasing physical activity. Three mechanisms have been suggested that might explain this phenomenon [57]. (i) Excitation of skeletal muscles is accompanied by L-type Ca2+ channels activation, [Ca2+]i increase, and gene transcription changes by the mechanisms discussed in the “Proof of Ca2+-independent signaling” section. (ii) Physical activity causes increase in AMP/ATP ratio and activation of AMPK involved in gene transcription regulation. (iii) Intense physical activity is accompanied by local decrease in partial oxygen pressure (pO2), increase in hypoxia-induced factor HIF-1α synthesis, and activation by HIF-1α/HIF-1β heterodimer of HREs (HIF-1 response elements) found in 5′-UTRs promoters of some pO2-depending genes [18, 87]. In our study, we have obtained evidence for alternative signaling mechanism existence proceeding via transcription activation in response to [Na+]i increase.
Cultured SMCs subjected to electrical pulse stimulation (EPS) are considered the most adequate contracting sceletal muscule in vitro model [88]. We have established that similarly to the in vivo experiments [89], 4-h mouse myoblasts C2C12 cells EPS resulted in [Na+]i/[K+]i 5-fold increase [90]. Considering the absence of hypoxia in this type of experiments, we investigated role of Ca2+- and AMPK-dependent signaling in C2C12 cells transcriptome change [91]. It was shown that in addition to Na+ accumulation and K+ loss, EPS was accompanied by rhythmic [Ca2+]i oscillations and changes in 3215 transcripts content. Nicardipine addition (L-type Ca2+ channel blocker) abolished [Ca2+]i oscillations without affecting [Na+]i/[K+]i ratio. It was found that EPS-induced alterations in transcription of 1018 genes were maintained in nicardipine presence. Contrary to the changes in ion balance, EPS did not affect acetyl-CoA carboxylase and Unc-51 factor recognized as markers of AMPK activation phosphorylation. Around 300 transcripts, whose content was also altered by the addition of ouabain at concentrations causing [Na+]i/[K+]i increase corresponding to ones observed in EPS subjected cells, were identified among EPS-dependent genes. Altogether, these data indicate an involvement of Na+i-dependent, Ca2+i- and AMPK-independent excitation-transcription coupling mechanism in the regulation of gene expression in SMCs.
Cell memory formation. Between the 20th and 21st centuries, it was demonstrated that excitation highly increases Erg1 and c-Fos mRNA content in brain neurons [92-94]. Comparison of ouabain and potassium-free medium action on SMCs and cells of electrically non-excitable tissues allowed to classify these genes as Na+i,K+i-dependent ERGs [51, 52]. Ability for learning in Egr1+/– mice deteriorated sharply [95], which indicated the key role of changes in the neuronal transcriptome mediated by these genes in the development of long-term memory (see review by Thiel et al. [96]). It was established that Egr1 and other ERGs transcription upregulation induced by neuron excitation was at least partly via activation of N-methyl-D-aspartate (NMDA) receptors. Considering that activation of NMDA receptors is accompanied by [Ca2+]i increase, it was suggested that Ca2+i-mediated signaling plays an essential role in long-term memory development [97].
However, NMDA receptors permeability to monovalent cations is higher than to Ca2+ (PNa ~ PK >> PCa), and their activation can result in [Na+]i/[K+]i ratio increase. Indeed, [Na+]i increase was observed upon both electrical [98] and pharamacological [99] excitation of brain neurons. Using intracellular Na+ and Ca2+ selective fluorescent indicators, it was demonstrated that Na+ influx during pyramidal neurons excitation occurs prior to [Ca2+]i increase. It was also established that unlike fast [Ca2+]i normalization, [Na+]i increase, due to the neuron excitation, returns to normal level within longer period of time [100].
In our latest studies, we investigated ouabain action on [Na+]i/[K+]i ratio and transcriptome of cultured rat brain neurons [101]. Increase in Npas4, Fos, Junb, Crem, Atf3, Klf4, and Crebzf expression was observed among [Na+]i/[K+]i-dependent transcription regulators. Previously, activation of these ERGs transcription was detected in neurons subjected to electrostimulation or treated with neurotransmitters [93, 94, 102-104]. Participation of these genes in learning and synaptic plasticity has been demonstrated in a number of studies [105, 106]. Currently, we examine the role of Ca2+i-mediated and Ca2+i-independent signaling activated upon dissipation of monovalent cations transmembrane gradients in brain neurons and involved in the alteration of transcriptome in response to electrical and pharmacological stimulation.
Hypoxia. As mentioned above, inhibition of HIF-1 degradation in response to pO2 decrease and HIF-1 interaction with hypoxia-dependent HREs are considered as certain genes transcriptional regulation main mechanisms in hypoxia. The most studied of these genes are Vegfa, Edn1, Klf10, and Nr4a1 [107, 108]. 5′-UTRs of pO2-dependent genes, such as Egr1, Atf3, Ptgs2, Il6, Ppp1r5, Hes1, Nfkbiz, Txnip, Adamts1, Egr3, Cxcl2, and Hsp70, do not contain HREs [51]. It has attracted out attention that transcription of Egr1, Atf3, Ptgs2, Il6, Ppp1r5, Hes1, Nfkbiz, and Txnip in SMCs is activated in response to the increase in the [Na+]i/[K+]i ratio [51]. In order to investigate the mechanism of this phenomenon, we compared the effects of Na,K-ATPase inhibition and hypoxia in combination with glucose starvation on the SMC transcriptome. Under these conditions, hypoxia resulted in 3-fold [Na+]i increase and 2-fold [K+]i increase. Both Na,K-ATPase and hypoxia inhibition caused unidirectional transcription changes in dozens of genes, including Cyp1a1, Fos, Atf3, Klf10, Ptgs2, Nr4a1, Per2, and Hes1. Elimination of monovalent cations transmembrane gradient using medium with reduced Na+ content and increased K+ content abolished ouabain effect on both [Na+]i/[K+]i ratio and expression of the above genes. Same procedure also completely prevented hypoxia effect on [Na+]i/[K+]i ratio and Fos, Atf3, Ptgs2, and Per2 transcription and strongly reduced Klf10, Edn1, Nr4a1, and Hes1 upregulation. SMCs transfection with Hif-1a siRNA reduced increase Vegfa, Edn1, Klf10, and Nr4a1 mRNAs content observed under hypoxia, but did not affect Fos, Atf3, Ptgs2, and Per2 expression [16, 17]. These results indicate that Na+i/K+i-mediated, Hif-1α-independent mechanism of transcription regulation contributes significantly to the regulation of gene expression under hypoxic conditions.
Data presented in this review demonstrate that activity of some enzymes, as well as transcription and translation, are regulated via changes in Na+ and K+ intracellular concentrations within physiological range, which allows to consider monovalent cations as secondary messengers. However, it must be emphasized that unlike Ca2+ and other secondary messengers, molecular structures of Na+- and K+-binding sites formed by protein tertiary structure have been investigated only for a limited number of sensors (see the section “Identified sensors of monovalent cations”). In addition to these works obvious fundamental significance, continuation of these studies might offer substantial practical applications. For example, it was found that myofibroblasts differentiation activated by tumor transforming growth factor (TGFb1) was suppressed by CTSs [58]. CTS injections inhibited myofibroblasts accumulation in the lungs of laboratory animals (model of idiopathic pulmonary fibrosis in humans) [109]. It was established that CTSs action is mediated by Na,K-ATPase inhibition, [Na+]i increase, and following activation of transcription of Ptgs2 gene encoding COX-2 [58] accompanied by transcription suppression of TGFBR2 subunit of TGFβ1 receptor [58]. It was also shown both in in vitro and in vivo that CTSs inhibit influenza virus replication through [K+]i decrease and suppression of K+i-dependent translation stages [78]. However, CTSs pharmacological application in doses that increase [Na+]i/[K+]i ratio is problematic because of their toxic effect [110]. In this connection, it seems promising to develop drugs that provide tissue-specific effects on the activity of monovalent cation sensors involved in the transcription and translation regulation.
Funding. This work was supported by the Russian Science Foundation (project 16-15-10026-p, section “Physiological and pathophysiological significance of Na+i,K+i-dependent mechanism of gene expression regulation”) and Russian Foundation for Basic Research (project 18-04-00063, sections “Identified sensors of monovalent cations”, “Sodium as transcription regulator”, and “Potassium as translation regulator”).
Acknowledgements. The authors express their gratitude to Prof. A. A. Mongin (Albany Medical College, USA) for reading this review and providing critical comments.
Conflict of interest. The authors declare no conflict of interest in financial or any other sphere.
Ethical approval. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
REFERENCES
1.Sutherland, E. W. (1972) Studies on the mechanism
of hormone action, Science, 177, 401-408.
2.Robison, G. A., Butcher, R. W., and Sutherland, E.
W. (1971) Cyclic AMP, Academic Press, New York.
3.Lincoln, T. M., and Cornwell, T. L. (1993)
Intracellular cyclic GMP receptor proteins, FASEB J., 7,
328-338.
4.Carafoli, E. (2002) Calcium signaling: a tale for
all seasons, Proc. Natl. Acad. Sci. USA, 99,
1115-1122.
5.Berridge, M. J. (1993) Inositol triphosphate and
calcium signalling, Nature, 361, 315-325.
6.Grinstein, S., Smith, J. D., Benedict, S. H., and
Gelfand, E. W. (1989) Activation of sodium-hydrogen exchange by
mitogens, Curr. Topics Membr. Transport, 34, 331-343.
7Prasad, K. V. S., Severini, A., and Kaplan, J. G. (1987) Sodium ion
fluxes in proliferating lymphocytes: an early component of mitogenic
signal, Arch. Biochem. Biophys., 252, 515-525.
8.Wakabayashi, S., Shigekawa, M., and Poyssegur, J.
(1997) Molecular physiology of vertebrate Na+/H+
exchanger, Physiol. Rev., 77, 51-74.
9.Marakhova, I. I., Vereninov, A. A., Toropova, F.
V., and Vinogradova, T. A. (1998) Na,K-ATPase pump in activated human
lymphocytes: on the mechanisms of rapid and long-term increase in K
influxes during the initiation of phytohemagglutinin-induced
proliferation, Biochim. Biophys. Acta, 1368, 61-72.
10.Burns, C. P., and Rozengurt, E. (1984)
Extracellular Na+ and initiation of DNA synthesis: role of
intracellular pH and K+, J. Cell Biol., 98,
1082-1089.
11.Hamet, P., Orlov, S. N., DeBlois, D., Sun, Y.,
Kren, V., and Tremblay, J. (2004) Angiotensin As a Cytokine
Implicated in Accelerated Cellular Turnover, Springer Verlag, N.
Y., pp. 71-98.
12.Bennekou, P., and Christophersen, P. (2003)
Ion Channels, Springer, Berlin, pp. 139-152.
13.Schneider, J., Nicolay, J. P., Foller, M.,
Wieder, T., and Lang, F. (2007) Suicidal erythrocyte death following
cellular K+ loss, Cell Physiol. Biochem., 20,
35-44.
14.Rose, C. R., and Konnerth, A. (2001)
NMDA-receptor-mediated Na+ signals in spines and dendrites,
J. Neurosci., 21, 4207-4214.
15.Verkhratsky, A., Noda, M., Parpura, V., and
Kirischuk, S. (2013) Sodium fluxes and astroglial function, Adv.
Exp. Med. Biol., 961, 295-305.
16.Koltsova, S. V., Shilov, B., Burulina, J. G.,
Akimova, O. A., Haloui, M., Kapilevich, L. V., Gusakova, S. V.,
Tremblay, J., Hamet, P., and Orlov, S. N. (2014) Transcriptomic changes
triggered by hypoxia: evidence for HIF-1a -independent,
[Na+]i/[K+]i-mediated
excitation-transcription coupling, PLoS One, 9,
e110597.
17.Orlov, S. N., Birulina, Y. G., Smaglii, L. V.,
and Gusakova, S. V. (2017) Vascular smooth muscle as an oxygen sensor:
role of elevation of the
[Na+]i/[K+]i, in Hypoxia
and Human Diseases (Zheng, J., and Zhou, C., eds.) IntechOpen, Vol.
4, pp. 73-90; doi: 10.5772/65384.
18.Kapilevich, L. V., Kironenko, T. A., Zaharova, A.
N., Kotelevtsev, Yu., Dulin, N. O., and Orlov, S. N. (2015) Skeletal
muscle as an endicrine organ: role of
[Na+]i/[K+]i-mediated
excitation-transcription coupling, Genes Dis., 2,
328-336.
19.Orlov, S. N., and Hamet, P. (2006) Intracellular
monovalent ions as second messengers, J. Membr. Biol.,
210, 161-172.
20.Shekarabi, M., Zhang, J., Khanna, A. R., Ellison,
D. H., Delpire, E., and Kahle, K. T. (2017) WNK kinase signaling in ion
homeostasis and human disease, Cell Metab., 25,
285-299.
21.Wilson, C. S., and Mongin, A. A. (2019) The
signaling role for chloride in the bidirectional communication between
neurons and astrocytes, Neurosci. Lett., 689, 33-44.
22.Page, M. J., and Di Cera, E. (2006) Role of
Na+ and K+ in enzyme function, Physiol.
Rev., 86, 1049-1092.
23.Linden, D. J., Smeyne, M., and Connor, J. A.
(1993) Induction of cerebellar long-term depression in culture requires
postsynaptic action of sodium ions, Neuron, 11,
1093-1100.
24.Kanai, R., Ogawa, H., Vilsen, B., Cornelius, F.,
and Toyoshima, C. (2013) Crystal structure of Na+-bound
Na+,K+-ATPase preceding the E1P state,
Nature, 502, 201-206.
25.Sweadner, K. J., and Donnet, C. (2001) Structural
similarities of Na,K-ATPase and SERCA, the Ca2+-ATPase of
the sarcoplasmic reticulum, Biochem. J., 356, 685-704;
doi: 10.1042/0264-6021:3560685.
26.Toyoshima, C., Kanai, R., and Cornelius, F.
(2011) First crystal structure of Na+,K+-ATPase:
new light on the oldest ion pump, Structure, 19,
1732-1738; doi: 10.1016/j.str.2011.10.016.
27.Pert, C. B., Pasternak, G., and Snyder, S. H.
(1973) Opiate agonists and antagonists discriminated by receptor
binding in brain, Science, 182, 1359-1361.
28.Katritch, V., Fenalti, G., Abola, E. E., Roth, B.
L., Cherezov, V., and Stevens, R. C. (2014) Allosteric sodium in class
A GPCR signaling, Trends Biochem. Sci., 39, 233-244.
29.Strasser, A., Wittmann, H.-J., Schneider, E. H.,
and Seifert, R. (2015) Modulation of GPCRs by mnovalent cations and
anions, Naunyn Schmied. Arch. Pharmacol., 388, 363-380;
doi: 10.1007/s00210-014-1073-2.
30.Liu, W., Chun, E., Thompson, A. A., Chubukov, P.,
Xu, F., Katritch, V., Han, G. W., Roth, C. B., Heitman, L. H.,
Ijzerman, A. P., Cherezov, V., and Stevens, R. C. (2012) Structural
basis for allosteric regulation of GPCRs by sodium ions,
Science, 337, 232-236.
31.Fenalti, G., Giguere, P. M., Katritch, V., Huang,
X. P., Thompson, A. A., Cherezov, V., Roth, B. L., and Stevens, R. C.
(2014) Molecular control of d-opioid receptor signalling,
Nature, 506, 191-196.
32.Gutierrez-de-Teran, H., Massink, A., Rodriguez,
D., Liu, W., Han, G. W., Joseph, J. S., Katritch, I., Heitman, L. H.,
Xia, L., Ijzerman, A. P., Cherezov, V., Katritch, V., and Stevens, R.
C. (2013) The role of sodium ion-binding site in the allosteric
modulation of the A2A adenosine G protein-coupled receptor,
Structure, 21, 2175-2185.
33.O'Brien, M.-C., and McKay, D. B. (1995) How
potassium affects the activity of the molecular chaperone Hsc70. I.
Potassium is required for optimal ATPase activity, J. Biol.
Chem., 270, 2247-2250.
34.Arakawa, A., Handa, N., Shirouzu, M., and
Yokoyama, S. (2011) Biochemical and structural studies on the high
affinity of Hsp70 for ADP, Protein Sci., 20,
1367-1379.
35.Orlov, S. N. (1987) Clamodulin: General
Problems of Physocochemical Biology [in Russian], VINITI, Moscow,
Vol. 8, pp. 5-212.
36.Permyakov, E. A., Uversky, V. N., and Permyakov,
S. E. (2017) Parvalbumin as a pleomorphic protein, Curr. Protein
Pept. Sci., 18, 780-794.
37.Heizmann, C. W., and Hunziker, W. (1991)
Intracellular calcium-binding proteins: more sites than insights,
Trends Biochem. Sci., 16, 98-103.
38.Okada, Y., Maeno, E., Shimizu, T., Dezaki, K.,
Wang, J., and Morishima, S. (2001) Receptor-mediated control of
regulatory volume decrease (RVD) and apoptotic volume decrease (AVD),
J. Physiol., 532, 3-16.
39.Orlov, S. N., Platonova, A. A., Hamet, P., and
Grygorczyk, R. (2013) Cell volume and monovalent ion transporters:
their role in the triggereing and progression of the cell death
machinery, Am. J. Physiol. Cell Physiol., 305,
C361-C372.
40.Orlov, S. N., Thorin-Trescases, N., Kotelevtsev,
S. V., Tremblay, J., and Hamet, P. (1999) Inversion of the
intracellular Na+/K+ ratio blocks apoptosis in
vascular smooth muscle at a site upstream of caspase-3, J. Biol.
Chem., 274, 16545-16552.
41.Zhou, X., Jiang, G., Zhao, A., Bondeva, T.,
Hirzel, P., and Balla, T. (2001) Inhibition of Na,K-ATPase activates
PI3 kinase and inhibits apoptosis in LLC-PK1 cells, Biochem.
Biophys. Res. Commun., 285, 46-51.
42.Isaev, N. K., Stelmashook, E. V., Halle, A.,
Harms, C., Lautenschlager, M., Weih, M., Dirnagl, U., Victorov, I. V.,
and Zorov, D. B. (2000) Inhibition of
Na+,K+-ATPase activity in cultured cerebellar
granule cells prevents the onset of apoptosis induced by low potassium,
Neurosci. Lett., 283, 41-44.
43.Trevisi, L., Visentin, B., Cusinato, F., Pighin,
I., and Luciani, S. (2004) Antiapoptotic effect of ouabain on human
umbilical endothelial cells, Biochem. Biophys. Res. Commun.,
321, 716-721.
44.Orlov, S. N., Taurin, S., Tremblay, J., and
Hamet, P. (2001) Inhibition of Na+,K+ pump
affects nucleic acid synthesis and smooth muscle cell proliferation via
elevation of the
[Na+]i/[K+]i ratio:
possible implication in vascular remodeling, J. Hypertens.,
19, 1559-1565.
45.Taurin, S., Seyrantepe, V., Orlov, S. N.,
Tremblay, T.-L., Thibaut, P., Bennett, M. R., Hamet, P., and
Pshezhetsky, A. V. (2002) Proteome analysis and functional expression
identify mortalin as an anti-apoptotic gene induced by elevation of
[Na+]i/[K+]i ratio in
cultured vascular smooth muscle cells, Circ. Res., 91,
915-922.
46.Orlov, S. N., and Hamet, P. (2004) Apoptosis vs
oncosis: role of cell volume and intracellular monovalent cations,
Adv. Exp. Med. Biol., 559, 219-233.
47.Taurin, S., Dulin, N. O., Pchejetski, D.,
Grygorczyk, R., Tremblay, J., Hamet, P., and Orlov, S. N. (2002) c-Fos
expression in ouabain-treated vascular smooth muscle cells from rat
aorta: evidence for an intracellular-sodium-mediated,
calcium-independent mechanism, J. Physiol., 543,
835-847.
48.Orlov, S. N., Taurin, S., Thorin-Trescases, N.,
Dulin, N. O., Tremblay, J., and Hamet, P. (2000) Inversion of the
intracellular Na+/K+ ratio blocks apoptosis in
vascular smooth muscle cells by induction of RNA synthesis,
Hypertension, 35, 1062-1068.
49.Schoner, W., and Scheiner-Bobis, G. (2007)
Endogenous and exogenous cardiac glycosides: their role in
hypertension, salt metabolism, and cell growth, Am. J. Physiol. Cell
Physiol., 293, C509-C536.
50.Orlov, S. N., Klimanova, E. A., Tverskoi, A. M.,
Vladychenskaya, E. A., Smolyaninova, L. V., and Lopina, O. D. (2017)
Na+i,K+i-dependent and
-independent signaling triggered by cardiotonic steroids: facts and
artifacts, Molecules, 22, E635; doi:
10.3390/molecules22040635.
51.Koltsova, S. V., Trushina, Y., Haloui, M.,
Akimova, O. A., Tremblay, J., Hamet, P., and Orlov, S. N. (2012)
Ubiquitous
[Na+]i/[K+]i-sensitive
transcriptome in mammalian cells: evidence for
Ca2+i-independent excitation-transcription
coupling, PLoS One, 7, e38032.
52.Klimanova, E. A., Sidorenko, S. V., Smolyaninova,
L. V., Kapilevich, L. V., Gusakova, S. V., Lopina, O. D., and Orlov, S.
N. (2019) Ubiquitous and cell type-specific transciptomic changes
triggered by dissipation of monovalent cation gradients in rodent
cells: physiological and pathophysiological implications, Curr. Top.
Membr., 83, 107-149; doi: 10.1016/bs.ctm.2019.01.006.
53.Orlov, S. N., Thorin-Trescases, N., Pchejetski,
D., Taurin, S., Farhat, N., Tremblay, J., Thorin, E., and Hamet, P.
(2004) Na+/K+ pump and endothelial cell survival:
[Na+]i/[K+]i-independent
necrosis triggered by ouabain, and protection against apoptosis
mediated by elevation of [Na+]i, Pflugers
Arch., 448, 335-345.
54.Alonso, M. T., and Garcia-Sancho, J. (2011)
Nuclear Ca2+ signalling, Cell Calcium, 49,
280-289.
55.Taurin, S., Hamet, P., and Orlov, S. N. (2003)
Na/K pump and intracellular monovalent cations: novel mechanism of
excitation-transcription coupling involved in inhibition of apoptosis,
Mol. Biol., 37, 371-381.
56.Santana, L. F. (2008) NFAT-dependent
excitation-transcription coupling in heart, Circ. Res.,
103, 681-683.
57.Gundersen, K. (2011) Excitation-transcription
coupling in skeletal muscle: the molecular pathways of exercise,
Biol. Rev., 86, 564-600.
58.La, J., Reed, E. B., Koltsova, S. V., Akimova, O.
A., Hamanaka, R. B., Mutlu, R. B., Orlov, S. N., and Dulin, N. O.
(2016) Regulation of myofibroblast differentiation by cardiac
glycosides, Am. J. Physiol. Lung Cell. Mol. Physiol.,
310, L815-L823.
59.Smolyaninova, L. V., Koltsova, S. V., Sidorenko,
S. V., and Orlov, S. N. (2017) Augemented gene expression triggered by
Na+,K+-ATPase inhibition: role of
Ca2+-mediated and -independent excitation-transcription
coupling, Cell Calcium, 68, 5-13.
60.Orlov, S. N., Aksentsev, S. L., and Kotelevtsev,
S. V. (2005) Extracellular calcium is required for the maintenance of
plasma membrane integrity in nucleated cells, Cell Calcium,
38, 53-57.
61.Koltsova, S. V., Tremblay, J., Hamet, P., and
Orlov, S. N. (2015) Transcriptomic changes in Ca2+-depleted
cells: role of elevated intracellular [Na+]/[K+]
ratio, Cell Calcium, 58, 317-324.
62.Matt, T., Martinez-Yamout, M. A., Dyson, H. J.,
and Wright, P. E. (2004) The CBP/p300 XAZ1 domain in its native state
is not a binding patner of MDM2, Biochem. J., 381,
685-691.
63.Krishtal, O. A., Pidoplichko, V. I., and
Shakhovalov, Y. A. (1981) Conductance of the calcium channel in the
membrane of snail neurones, J. Physiol., 301,
423-434.
64.Nakagawa, Y., Rivera, V., and Larner, A. C.
(1992) A role for Na/K-ATPase in the control of human c-fos and c-jun
transcription, J. Biol. Chem., 267, 8785-8788.
65.Haloui, M., Taurin, S., Akimova, O. A., Guo,
D.-F., Tremblay, J., Dulin, N. O., Hamet, P., and Orlov, S. N. (2007)
Na+i-induced c-Fos expression is not
mediated by activation of the 5′-promoter containing known
transcriptional elements, FEBS J., 274, 3257-3267.
66.Ono, Y., Ojimam, K., Torii, F., Takaya, E., Doi,
N., Nakagawa, K., Hata, S., Abe, K., and Sorimachi, H. (2010) Skeletal
muscle-specific calpain is an intracellular Na+-dependent
protease, J. Biol. Chem., 285, 22986-22998.
67.Herrera, R. E., Nordheim, A., and Stewart, A. F.
(1997) Chromatin structure analysis of the human c-Fos promoter reveals
a centrally positioned nucleosome, Chromosoma, 106,
284-292.
68.Chinsomboon, J., Ruas, J., Gupta, R. K., Thom,
R., Shoag, J., Rowe, G. C., Sawada, N., Raghuram, S., and Arany, Z.
(2009) The transcriptional coactivator PGC-1alpha mediates
exercise-induced angiogenesis in skeletal muscle, Proc. Natl. Acad.
Sci. USA, 106, 21401-21405.
69.Soyal, S. M., Felder, T. K., Auer, S., Hahne, P.,
Oberkofler, H., Witting, A., Paulmichl, M., Landwehrmeyer, G. B.,
Weydt, P., and Patsch, W. (2012) A greatly extended PPARGC1A genomic
locus encodes several new brain-specific isoforms and influences
Huntington disease age of onset, Hum. Mol. Genet., 21,
3461-3470.
70.Mechti, N., Piechaczyk, M., Blanchard, J. M.,
Jeanteur, P., and Lebleu, B. (1991) Sequence requirements for premature
transcription arrest within the first intron of the mouse c-fos
gene, Mol. Cell. Biol., 11, 2832-2841.
71.Coulon, V., Veyrune, J.-L., Tourkine, N., Vie,
A., Hipskind, R. A., and Blanchard, J.-M. (1999) A novel calcium
signaling pathway targets the c-fos intragenic transcriptional
pausing site, J. Biol. Chem., 274, 30439-30446.
72.Lanctot, C., Cheutin, T., Cremer, M., Cavalli,
G., and Cremer, T. (2007) Dynamic genome architecture in the nuclear
space: regulation of gene expression in three dimensions, Nature
Rev. Genet., 8, 104-115.
73.Trinkle-Mulchany, L., and Lamond, A. I. (2008)
Nuclear functions in space and time: gene expression in a dynamic,
constrained environment, FEBS Lett., 582, 1960-1970.
74.Maharana, S., Sharma, D., Shi, X., and
Shivashankar, G. V. (2012) Dynamic organization of transcription
comparments is dependent in functional nuclear architecture,
Biophys. J., 103, 851-859.
75.Dolinnaya, N. G., Ogloblina, A. M., and
Yakubovskaya, M. G. (2016) Structure, properties, and biological
relevance of the DNA and RNA G-quadruplexes: overview 50 years after
their discovery, Biochemistry (Moscow), 81,
1602-1649.
76.Lubin, M., and Ennis, H. L. (1964) On the role of
intracellular potassium in protein synthesis, Biochim. Biophys.
Acta, 80, 614-631.
77.Ledbetter, M. L. S., and Lubin, M. (1977) Control
of protein synthesis in human fibroblasts by intracellular potassium,
Exp. Cell Res., 105, 223-236.
78.Amarelle, L., Katzen, J., Shigemura, M., Welch,
L. C., Cajigas, H., Peteranderl, C., Celli, D., Herold, S., Lecuona,
E., and Sznajder, J. L. (2019) Cardiac glycosides decrease influenza
virus replication by inhibiting cell protein translation machinery,
Am. J. Physiol. Lung Cell. Mol. Physiol., 316,
L1094-L1106; doi: 10.1152/ajplung.00173.2018.
79.Cahn, F., and Lubin, M. (1978) Inhibition of
elongation steps of protein synthesis at reduced potassium
concentrations in reticulocytes and reticulocyte lysate, J. Biol.
Chem., 253, 7798-7803.
80.Austin, J., and First, E. A. (2002) Potassium
functionally replaced the second lysine of the KMSKS signaure sequence
in human tyrosyl-tRNA synthetase, J. Biol. Chem., 277,
20243-20248.
81.Jennings, M. D., and Pavitt, G. D. (2010) eIF5 is
a dual function GAP and GDI for eukariotic translational control,
Small GTPases, 1, 118-123.
82.Cao, J., He, L., Lin, G., Hu, C., Dong, R.,
Zhang, J., Zhu, H., Hu, Y., Wagner, C. R., He, Q., and Yang, B. (2014)
Cap-dependent translation initiation factor, eIF4E, is the target for
ouabain-mediated inhibition of HIF-1a, Biochem. Pharmacol.,
89, 20-30.
83.Klimanova, E. A., Tverskoi, A. M., Koltsova, S.
V., Sidorenko, S. V., Lopina, O. D., Tremblay, J., Hamet, P.,
Kapilevich, L. V., and Orlov, S. N. (2017) Time- and dose-dependent
actions of cardiotonic steroids on transcriptome and intracellular
content of Na+ and K+: a comparative analysis,
Sci. Rep., 7, 45403; doi: 10.1038/srep45403.
84.Fisher, P. A., Goodman, J. G., and Kull, F. L.
(1976) Thyroid ribonucelic acid-iodopeptides. Comparison of
thyrosyl-complex II and thyrosyl-tRNA, Biochemistry, 15,
4065-4070.
85.Quivy, J. P., and Chroboczek, J. (1988)
Tyrpsyl-tRNA synthetase from wheat germ, J. Biol. Chem.,
263, 15277-15281.
86.Dever, T. E. (2002) Gene-specific regulation by
general translation factors, Cell, 108, 545-556.
87.Pedersen, B. K., and Febbraio, M. A. (2008)
Muscle as an endocrine organ: focus on muscle-derived interleukin-6,
Physiol. Rev., 88, 1379-1406.
88.Nikolic, N., Gordens, S. W., Thoresen, G. H.,
Aas, V., Eckel, J., and Eckardt, K. (2017) Electrical pulse stimulation
of cultured skeletal muscle cells as a model for in vitro
exercise – possibilities and limitations, Acta Physiol.,
220, 310-331.
89.Murphy, K. T., Nielsen, O. B., and Clausen, T.
(2008) Analysis of exercise-induced Na+-K+
exchange in rat skeletal muscle, Exp. Physiol., 93,
1249-1262.
90.Danilov, K., Sidorenko, S. V., Milovanova, K.,
Klimanova, E. A., Kapilevich, L. V., and Orlov, S. N. (2017) Electrical
pulse stimulation decreases electrochemical Na+ and
K+ gradients in C2C12 myotubes, Biochem. Biophys. Res.
Commun., 493, 875-878.
91.Sidorenko, S. V., Klimanova, E. A., Milovanova,
K., Lopina, O. D., Kapilevich, L. V., Chibalin, A. V., and Orlov, S. N.
(2018) Transciptomic changes in C2C12 myotubes triggered by electrical
stimulation: role of Ca2+i-mediated and
Ca2+i-independent signaling and elevated
[Na+]i/[K+]i ratio, Cell
Calcium, 76, 72-86.
92.Bakowski, D., and Parekh, A. B. (2002) Monovalent
cation permeability and Ca2+ block of the store-operated
Ca2+ current ICRAC in rat basophylic lleukemia cells,
Pfluger Arch. Eur. J. Physiol., 443, 892-902.
93.Hunt, S. P., Pini, A., and Evan, G. (1987)
Induction of c-Fos-like protein in spinal cord neurons following
sensory stimulation, Nature, 328, 632-634.
94.Cole, A. J., Saffen, D. W., Baraban, J. M., and
Worley, P. F. (1989) Rapid increase of an immediate early gene
meseenger RNA in hippocampal neurons by synaptic NMDA receptor
activation, Nature, 340, 474-476.
95.Jones, M. W., Errington, M. L., French, P. J.,
Fine, P. J., Bliss, T. V. P., Garel, S., Charney, P., Bozon, B.,
Laroche, S., and Davis, S. (2001) A requirement for the immediate early
response gene Zif268 in the expression of late LTP and long-term
memories, Nat. Neurosci., 4, 289-296.
96.Thiel, G., Mayer, S. I., Muller, I., Stefano, L.,
and Rossler, O. G. (2010) Egr-1 – a Ca2+-regulated
transcription factor, Cell Calcium, 47, 397-403.
97.Alberini, C. A. (2009) Transcription factors in
long-term memory and synaptic plasticity, Physiol. Rev.,
89, 121-145.
98.Bennay, M., Langer, J., Meier, S. D., Kafitz, K.
W., and Rose, C. R. (2008) Sodium signals in cerebellar Purkinje
neurons and bergmann glial cells evoked by glutamatergic synaptic
transmission, Glia, 56, 1138-1149.
99.Jaffe, D. B., Johnston, D., Lasser-Ross, N.,
Lisman, J. E., Miyakawa, H., and Ross, W. N. (1992) The spread of
Na+ spikes determines the pattern of dendritic
Ca2+ entry into hippocampal neurones, Nature,
357, 244-246.
100.Baeza-Lehnert, F., Saab, A. S., Gutierrez, R.,
Larenas, V., Diaz, E., Horn, M., Vargas, M., Hosli, L., Stobart, J.,
Hirrlinger, J., Weber, B., and Barros, L. F. (2019) Non-canonical
control of neuronal energy status by the Na+ pump, Cell
Metab., 29, 1-13.
101.Smolyaninova, L. V., Shiyan, A. A., Kapilevich,
L. V., Lopachev, A. V., Fedorova, T. N., Klementieva, T. S.,
Moskovtsev, A. A., Kubatiev, A. A., and Orlov, S. N. (2019)
Transcriptomic changes triggered by ouabain in rat cerebellum granule
cells: role of a3- and a1-mediated signaling, PLoS One, in
press.
102.Coba, M. P., Valor, L. M., Kopanitsa, M. V.,
Afinowi, N. O., and Grant, S. G. (2008) Kinase networks integrate
profiles of N-methyl-D-aspartate receptor-mediated gene expression in
hippocampus, J. Biol. Chem., 283, 34101-34107.
103.Barry, D. N., and Commins, S. (2017) Temporal
dynamics of emmediate early gene expression during cellular
consolidation of spatial memory, Behav. Brain Res., 327,
44-53.
104.Zhu, S., Tai, C., MacVicar, B. A., Jia, W., and
Cynader, M. S. (2009) Glutamatergic stimulation triggers rapid
Krupple-like factor 4 expression in neurons and the overexpression of
KLF4 sensitizes neurons to NMDA-induced caspase-3 activity, Brain
Res., 1250, 49-62.
105.Flavell, S. W., and Greenberg, M. E. (2008)
Signaling mechanisms linking neuronal activity to gene expressoin and
plasticity of the nervous system, Annu. Rev. Neurosci.,
31, 563-590.
106.Malik, A. N., Vierbuchen, T., Hemberg, M.,
Rubin, A. A., Ling, E., Couch, C. H., Stroud, H., Spiegel, I., Kai-How
Farth, K., Harmin, D. A., and Greenberg, M. E. (2014) Genome-wide
identification and characterization of functional activity-dependent
enhancers, Nat. Neurosci., 17, 1330-1339.
107.Semenza, G. L. (2003) Targeting HIF-1 for
cancer therapy, Nat. Rev. Cancer, 3, 721-732.
108.Wood, I. S., Perez de Heredia, F., Wang, B.,
and Trayhurn, P. (2009) Cellular hypoxia and adipose tisue dysfunction
in obesity, Proc. Nutr. Soc., 68, 370-377; doi:
10.1017/S0029665109990206.
109.Orlov, S. N., La, J., Smolyaninova, L. V., and
Dulin, N. O. (2019) Na+,K+-ATPase as a target for
treatment of tissue fibrosis, Curr. Med. Chem., 26,
564-575; doi: 10.2174/0929867324666170619105407.
110.Akimova, O. A., Tverskoi, A. M., Smolyaninova,
L. V., Mongin, A. A., Lopina, O. D., La, J., Dulin, N. O., and Orlov,
S. N. (2015) Critical role of the
α1-Na+,K+-ATPase subunit in insensitivity
of rodent cells to cytotoxic action of ouabain, Apoptosis,
20, 1200-1210.