Characterization of Tobacco Mosaic Virus Virions and Repolymerized Coat Protein Aggregates in Solution by Small-Angle X-Ray Scattering
A. L. Ksenofontov1,a*, M. V. Petoukhov2,3,b, A. N. Prusov1, N. V. Fedorova1, and E. V. Shtykova2,c
1Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, 119992 Moscow, Russia2Shubnikov Institute of Crystallography, Federal Scientific Research Centre “Crystallography and Photonics”, Russian Academy of Sciences, 119333 Moscow, Russia
3Frumkin Institute of Physical Chemistry and Electrochemistry, Russian Academy of Sciences, 119071 Moscow, Russia
* To whom correspondence should be addressed.
Received November 7, 2019; Revised December 20, 2019; Accepted December 22, 2019
The structure of tobacco mosaic virus (TMV) virions and stacked disk aggregates of TMV coat protein (CP) in solution was analyzed by synchrotron-based small-angle X-ray scattering (SAXS) and negative contrast transmission electron microscopy (TEM). TMV CP aggregates had a unique stability but did not have helical symmetry. According to the TEM data, they were stacked disks associated into transversely striated rod-shaped structures 300 to 800 Å long. According to modeling based on the crystallographic model of the 4-layer TMV CP aggregate (PDB: 1EI7), the stacked disks represented hollow cylinders. The calculated SAXS pattern for the disks was compared to the experimental one over the entire measured range. The best correlation with the SAXS data was found for the model with the repeating central pair of discs; the SAXS curves for the stacked disks were virtually identical irrespectively of the protein isolation method. The positions of maxima on the scatter curves could be used as characteristic features of the studied samples; some of the peaks were assigned to the existing elements of the quaternary structure (periodicity of aggregate structure, virion helix pitch). Low-resolution structural data for the repolymerized TMV CP aggregates in solution under conditions similar to natural were produced for the first time. Analysis of such nano-size objects is essential for their application in biomedicine and biotechnology.
KEY WORDS: TMV, coat protein, stacked disk aggregates, virions, small-angle X-ray scatteringDOI: 10.1134/S0006297920030062
Abbreviations: CP, coat protein; SAXS, small-angle X-ray scattering; TEM, transmission electron microscopy; TMV, tobacco mosaic virus.
Tobacco mosaic virus (TMV) is a well-characterized plant virus
consisting of 2130 identical coat protein (CP) subunits arranged in a
helical structure around a single-stranded RNA molecule [1]. The TMV structure has been investigated in detail
by crystallography and electron microscopy. Because of the availability
and high stability of TMV virions, their diameter and helix pitch
(22.92 Å) were measured with a high accuracy by the oriented
fiber X-ray diffraction method [2] and used as size
standards in elecron microscopy. The CP subunits form a hollow cylinder
with a length of 3000 Å and diameter of 180 Å. Isolated TMV
CP can form a large number of ordered aggregates of different structure
in solution. Depending on pH, ionic strength, temperature, and
concentration, TMV CP aggregates can exist in two major states:
disk-shaped and helical [3]. The examples of
disk-shaped CP aggregates are the two-layer 20S disks with the
sedimentation coefficient of 20S and stacked pairs of disks that form
during CP incubation in a solution with pH 7-9. TMV particles
themselves and helical CP aggregates that form at pH ≤ 6.0, as well
as the two-turn helix fragments, are examples of helical aggregates [3]. The structure of the two-layer 20S disk was solved
by the X-ray diffraction analysis at a 2.8 Å resolution [4]. The 20S disk consists of 34 protein subunits (17
subunits per layer). The two layers form contacts at the greater
radius, but not at the smaller radius [4], so that
the layers open apart onto the central hole, which ensures RNA binding.
These X-ray diffraction data have been used for developing a model of
the TMV self-assembly in vitro [5, 6], according to which TMV self-assembly is initiated
by the binding of RNA from the central hole side to the specific
RNA-binding site in the 20S disk, which causes transformation of the
cylindrical disk into the helix [6].
Unusual diffraction patterns of polymerized TMV CP from infected plants produced by Franklin in 1955 [7] were the first indication of the paired disk assembly into stacks (stacked disks). For a long time, it had been believed that stacked disks and helical aggregates are at the equilibrium and can transform into each other. This implied that stacked disks are polar, i.e., have all subunits oriented in the same direction (similar to protein in the viral particle) [8]. The proof of the bipolar nature of the stacked disks was obtained using monoclonal antibodies that bound only to one end of the TMV rod-like virion but were able to associate with both ends of the repolymerized CP aggregates [9]. Further studies of the aggregate structure at a 9-12 Å resolution using cryoelectron microscopy [10, 11] demonstrated that a larger portion of the two-layer disks had the quaternary structure similar to that of the central pair of rings in the crystallized four-layer disk aggregates [12].
Here, we examined for the first time the structure of TMV rods and repolymerized TMV CP stacked disks in solution using the method of small angle X-ray scattering (SAXS). We also created a number of models of these structures using atomic coordinates of the four-layer aggregates [12] and compared their calculated scatter patterns with the experimental ones generated by SAXS.
Repolymerized TMV CP stacked disk are intriguing objects because of their amyloid-like stability: once assembled, the aggregates can be dissociated only under denaturing conditions [13, 14]. This type of nanoobjects (nanofibers and nanorods) could be used in nanoelectronics and various biomedical applications. Previously, we used SAXS to characterize and study dissociation of other virus-like particles, such as aggregates of the helical potato virus A potyvirus CP [15, 16].
MATERIALS AND METHODS
TMV CP isolation and repolymerization. TMV (strain U1) was kindly provided by V. V. Makarov. Repolymerized TMV CP aggregates were produced using two procedures: (i) dialysis of TMV CP isolated by the lithium method [17] against 10 mM Tris-HCl buffer (pH 8.0) containing 0.1 M NaCl; (ii) dialysis of TMV CP isolated by the acetate method [18] against 10 mM phosphate buffer (pH 8.0) containing 0.1 M NaCl. In both cases, dialysis was conducted for 24 h at 8°C, and the resulting protein preparations (1.5-5.0 mg/ml) were incubated for 5-7 days at 8°C for the formation of stacked disks followed by centrifugation for 10 min at 14,000 rpm (Benchtop Microfuge 22R; Beckman Coulter, USA). TMV CP yield was 50-70% of the calculated one.
Concentration of repolymerized CP aggregates and TMV particles was determined with a UV-2600 spectrophotometer (Hitachi, Japan) from the UV absorption spectra at 240-340 nm. Because of the significant contribution of scattering to the absorbance of repolymerized aggregates (40-50%), true absorption spectra (E) of protein suspensions were calculated in the 320-338 nm range using the extrapolation technique [19]. The absorption coefficients of CP and viral preparations were E0.1%280 = 1.3 and E0.1%260 = 2.3, respectively. The purity of the obtained samples was examined by Laemmli’s electrophoresis [20] in 15% polyacrylamide gel in a Mini-PROTEAN 3 Cell (Bio-Rad, USA). The gels were stained with 0.22% Coomassie Blue G-250 (Serva, Germany).
Transmission electron microscopy (TEM). The samples were applied onto carbon coated grids, incubated for 1 min at 20°C, washed with distilled water for 30 s, contrasted with 2% uranyl acetate for 10-30 s, washed again with distilled water, dried, and examined with a JEM-1400 transmission electron microscope (JEOL, Japan) at 60,000× magnification.
SAXS and analysis of experimental data. The structure of TMV particles and repolymerized CP was analyzed by SAXS of synchrotron radiation on a PetraIII synchrotron (DESY, Germany) on the P12 line. The P12 line was equipped with a device for automated sample change and a two-dimensional Pilatus 2M (DECTRIS, Switzerland) detector. The scattering intensity I(s) was recorded in the region of wave vector values 0.003 < s < 0.5 Å–1, where s = (4πsinθ)/λ ≤ 2π/d is scattering vector modulus; 2θ is scattering angle; λ = 1.24 Å is wavelength [21]; d is the size of scattering particles, structure periodicity. All measurements were carried out at 10°C; 20 individual scatter curves were recorded within 1 s for each sample in order to monitor possible radiation-inflicted damage. Initial data processing was carried out with the PRIMUS program [22]. Viral and protein preparation were examined at several concentrations (2-7 mg/ml) to account for the concentration dependence. MASSHA program [23] was used for modeling the structures of viral particles and repolymerized CP aggregates; the CRYSOL program [24] was used for the for calculation of scatter curves of the atomic resolution models and determination of gyration radii (Rg).
The distances in the crystallographic model of the four-layer disk aggregate (PDB: 1EI7) were measured with the tools provided in the RasMol program (http://www.rasmol.org/). The average value of relative deviation (Ō) of the scattering intensity vector maxima smi in the model from the experimental values siexp was used for comparing the modeled structures of repolymerized aggregates with the structures observed in solution: Ō = 1/n × ∑ (ǀsim – siexpǀ/siexp).
RESULTS
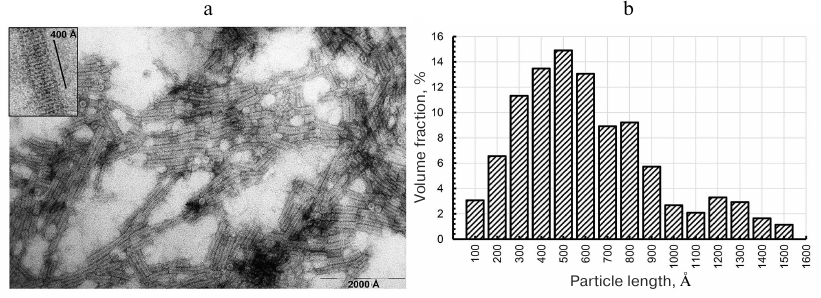
Electron microscopy of CP aggregates. Under certain conditions, purified TMV CP self-assembles into 20S disks, which transform into stable repolymerized aggregates (stacked disks) during further incubation [13, 14, 25]. First, we investigated the structure and length of the formed particles by negative contrast TEM. The images revealed the presence of transversely striated rod-shaped particles of varying lengths in all preparation of repolymerized CP aggregates produced by different methods (Fig. 1, a and b). The size distribution of the particles (Fig. 1b) showed that the main volume fraction (~70%) in the preparation were particles 300-800 Å in length. The 400 Å-long aggregates contained ~15-16 rings in the stack (15.5 ± 0.5) (Fig. 1, inset), which is in agreement with the previously reported TEM data [8, 9, 14]; the axial periodicity of the repolymerized protein aggregates was 26.0 ± 0.9 Å. Hence, the stacks contained 6 to 16 20S disks. The clusters of laterally connected repolymerized CP aggregates were also observed in the samples.
Fig. 1. Electron microscopy of repolymerized TMV CP aggregates (a) and particle size distribution (b). The length of ~250 particles was measured; the aggregates were produced from the TMV CP isolated by the lithium method [17] and contrasted with 2% uranyl acetate. Magnification, ×60,000 (a) and ×150,000 (inset).
Next, we characterized the structure of CP aggregates in solution by synchrotron-based SAXS and modeled the structure of virions and repolymerized TMV CP aggregates.
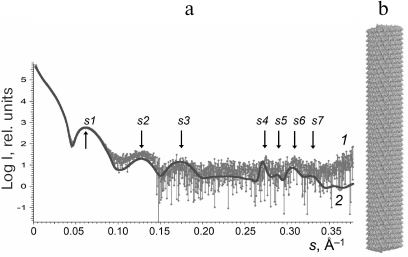
Structure modeling of TMV virions in solution. The scattering profiles of the TMV virions (3-6 mg/ml) were first obtained. The PDB: 2OM3 structure generated from the cryoelectron microscopy data (resolution, 4.4 Å) that contained a 3-turn fragment with the helical symmetry [26] was used for virion modeling. To model the virion structure (length, ~1200 Å; 784 subunits), this fragment was repeated 16 times along the helix axis. The calculated scattering pattern of the virion model produced with CRYSOL program [24] (Fig. 2, curve 2) was in a good agreement with the experimental data, including all peaks and minima of the scattering curve in the vector range 0.003 < s < 0.35 Å–1 (Fig. 2, curve 1).
Fig. 2. Experimental and calculated SAXS patterns (a) and structure model of TMV virion (b): 1) experimental data adjusted to zero concentration; 2) scattering pattern of the model virion structure calculated with the CRYSOL program and generated with the MASSHA program.
For more accurate comparison, we used the values of scattering vector s and the sizes of scattering particles d for all seven peaks (s1-s7) on the experimental and model curves (Table 1). Analysis of the data in Table 1 indicates that some maxima of the scattering vector intensity s are in good agreement with the known sizes in virions; e.g., d4 (linear size 22.9 Å) corresponds to the spiral pitch size of 22.9 Å evaluated from the X-ray diffraction of the oriented fibers [2].
Table 1. Comparison of the scattering
parameters of TMV virions in solution and model virion
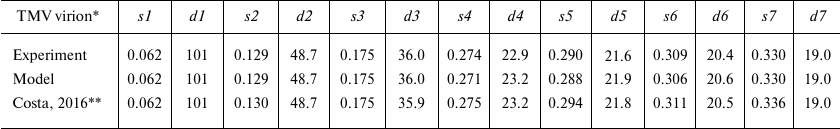
* PDB: 2OM3 model; s (Å–1), scattering
vector module; d = 2π/s (Å), structure
periodicity, size of scattering particles.
** Experimental data by Costa et al. [29].
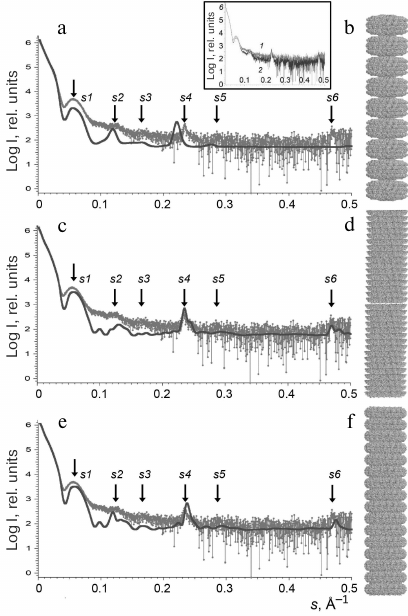
Structure modeling of repolymerized TMV CP aggregates in solution. According to the TEM data, repolymerized TMV CP aggregates represented a mixture of particles of varying length composed of repeating structural elements. The scatter profiles of the stacked disks consisting of TMV CP isolated by different methods [17, 18] were virtually identical (< 0.2% deviation of the scattering vectors maxima s1-s6) over the entire measured range (Fig. 3, inset). For this reason, further in the paper, we present SAXS data only for the repolymerized CP aggregates produced by the lithium method.
Originally, we used the PDB: 1EI7 crystallographic structure representing two attached paired rings (BA) with 17 protein subunits in each [12] for the model construction. The lower and the upper rings were denoted as B rings, and the central rings were denoted as A rings. The BAAB structural element was multiplied via spatial transformation. However, the scatter profile for the (BAAB)8 structure significantly differed from the experimentally obtained one (Figs. 3a and 3b and Table 2).
Fig. 3. Experimentally obtained SAXS profile of repolymerized TMV CP stacked disks (a, c, e) and calculated SAXS data for the corresponding model structures (b, d, f). Scattering was calculated with the CRYSOL program. The structures (BAAB)8 (b), (BA)16 (d), and (AA)16 (f) were modeled with the MASSHA program. Dots, experimental data; solid lines, calculated scattering of model structures; arrows, scattering vector intensity maxima. Inset, SAXS curves for the repolymerized CP aggregates produced by the lithium (1) and acetate (2) methods.
Table 2. Comparison of scattering parameters
of repolymerized TMV CP aggregates and scattering profiles of model
aggregates
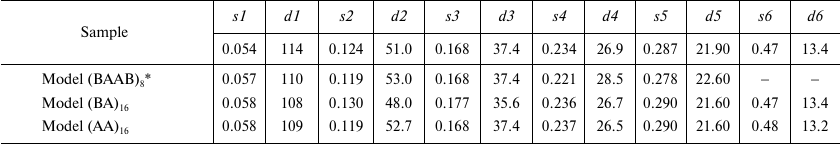
* PDB: 1EI7 crystal structure was used; values s
(Å–1) and d (Å) are the same as in
Table 1. Repolymerized CP aggregates was produced
by the lithium method.
Further in our work, we used the two other models based on the pair of rings from the PDB: 1EI7 structure: (i) lower pair of rings (BA; Fig. 3d) and (ii) central pair of rings (AA; Fig. 3f) that contained 544 subunits and had the axis length of ~820 Å. The ring elements were multiplied via spatial transformation with rotation of each element by 6.35° in accordance with the PDB: 1EI7 structure. The resulting models of stacked disks were hollow cylinders containing 16 two-layer disks each. Both models – (BA)16 and (AA)16 – provided adequate correlation between the experimental Iexp(s) values and calculated scatter curves. The positions of the intensity maxima on the scattering curves (s1-s6) could be used as specific parameters of the samples. To quantitatively evaluate the similarity between the generated models and repolymerized CP aggregates in solution, we used the averaged values of relative deviations of the scattering vector intensity maxima smi for the models from the experimental values: Ō = 1/n × ∑ (ǀsim – siexpǀ/siexp). The lowest Ō was found for the (AA)16 model (Ō = 0.02 ± 0.01), which indicated the closest similarity with the CP aggregate structure in solution in comparison to the (BA)16 (Ō = 0.03 ± 0.01) and (BAAB)8 (Ō = 0.04 ± 0.01) models. However, none of the model curves completely coincided with the experimental one.
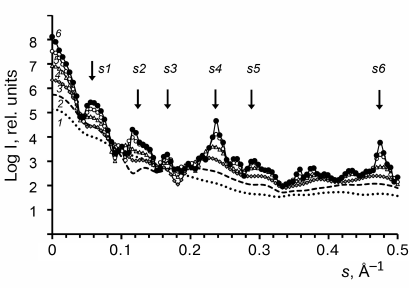
Effect of the repolymerized CP aggregate model length on the calculated SAXS curves. To evaluate the effect of the repolymerized CP aggregate length on the scatter curves, we used the CRYSOL program to calculate the scattering parameters for a series of model structures [24]: A, (AA)1, (AA)2, (AA)4, (AA)8, (AA)16 of the PDB: 1EI7 crystal [12], where A is a ring consisting of 17 protein subunits (Fig. 4).
Fig. 4. Model SAXS curves calculated with the CRYSOL program [24] for a series of models (curves 1-6): A, (AA)1, (AA)2, (AA)4, (AA)8, and (AA)16, respectively. Arrows indicate the scattering vector intensity maxima (see Fig. 3).
The Bragg’s scattering maxima were completely absent in the scatter profiles of the ring A and disk (AA)1 models (Fig. 4, curves 1 and 2); the scattering intensity increased at the positions of angle vectors s1, s4, and s5 (curves 3 and 4) for the (AA)2 and (AA)4 aggregates. For the (AA)8 and (AA)16 models, the scattering intensity significantly increased at the position of angle vectors s2, s4, and s6 (curves 5 and 6), which corresponded to the following scattering sizes of the repolymerized aggregates: d2 = 51 Å, d4 = 26.9 Å, and d6 = 13.4 Å (Table 2). The d4 value was close to the value of axial periodicity of the repolymerized ring-shaped aggregates (26.0 ± 0.9 Å) that was determined using TEM. The double value (52 Å) corresponded to the periodicity of disks in the repolymerized aggregates and was close to the scattering size d2, which was estimated as 53 Å by Franklin and Commoner based on the data of X-ray diffraction of oriented fibers [7]. The calculated invariants (molecular mass Mr and gyration radius Rg) for these models and repolymerized aggregates are shown in Table 3. The Rg values for the models of repolymerized aggregates consisting of 1 to 32 rings were in the range of 64.2 to 253.4 Å. The invariants for the (AA)16 model were the closest to the experimentally determined values for the repolymerized CP aggregates in solution.
Table 3. Comparison of invariants for model
structures and repolymerized TMV CP aggregates
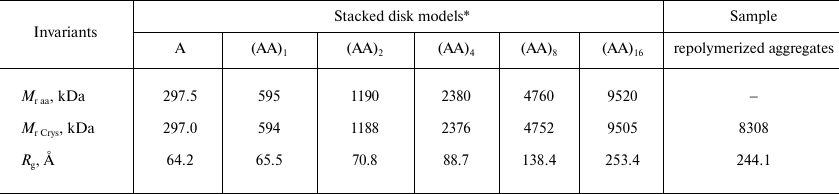
* Molecular masses (Mr Crys) and gyration
radii (Rg) for the modeled structures A,
(AA)1, (AA)2, (AA)4, (AA)8,
and (AA)16 were calculated with the CRYSOL program [24] using PDB: 1EI7 structure; Mr
aa values were calculated from the protein amino acid
sequences.
DISCUSSION
By combining modern methods for interpretation of synchrotron-based SAXS data and results of electron microscopy, we were able to analyze in detail the structure of repolymerized TMV CP aggregates (stacked disks). SAXS data allow to determine molecular architecture of proteins in multicomponent solutions [27] and proteins with flexible unstructured fragments [28]. New approaches to the analysis of SAXS data make it possible to obtain information on the quaternary structure of biological objects. SAXS can be used for visualizing the structures of investigated objects of various sizes (~10-2100 Å), depending on the energy of X-ray radiation and angle of measurement. Considering that the longer axis of the TMV virion is ~3000 Å, its length cannot be measured within the scattering angle range typical for SAXS (0.003 < s < 0.5 Å–1). Experimental and calculated curves diverged after s = 0.35 Å–1 (Fig. 2), which can be explained by the low accuracy of SAXS measurement of objects smaller than 15 Å or by the difference in the structures of the 3-turn fragment (PDB: 2OM3) and corresponding element of the virion in solution. Recently, TMV virions were examined by SAXS on the small-angle line BM29 (ESRF, Grenoble, λ = 0.99 Å) by Costa et al. [29]. The authors were able to produce the scattering curve at the virus concentration, which was 4 times higher than in our study (26 mg/ml). It can be seen in Table 1 that the s and d values obtained by Costa et al. are in good agreement with the data produced in our work (relative deviation below 1.5%). The fitting of geometric models was done with the Genfit program [30]. By using the three-shell cylinder model of the virion, the authors managed to reproduce the maxima of two peaks (s1 and s2) from the seven peaks observed on the experimental scattering curve [29].
Hiragi et al. [31] examined TMV CP aggregates formed in solution with pH 7.2 and ionic strength of 0.1-0.2 M at 15-25°C by SAXS on the small-angle line (Photon Factory, Japan; λ = 1.49 Å). The authors used cylindrical model of the 20S disk with outer and inner diameters of 180 and 40 Å, respectively, two 23-Å layers, and gyration radius of Rg 66.5 Å for approximation. The studied sample was found to be significantly disperse, with mostly two- and four-layer aggregates and small amount of longer aggregates. The gyration radius of the sample increased with the increase in ionic strength, temperature, and concentration, and reached 85.6 Å in 100 mM phosphate buffer at 25°C and 12 mg/ml, which was close to the suggested (AA)4 model of repolymerized aggregates with Rg of 88.7 Å (Table 3). In another study by the same research group, the kinetics of reassembly of TMV and cucumber green mottle mosaic virions from proteins and RNA was investigated by SAXS. It was demonstrated based on the changes in the Rg values, that the rate of TMV assembly was significantly higher [32]. The kinetics of formation of helical fragments (possible intermediates in the TMV assembly) and repolymerization of long helical protein aggregates in acidic medium was investigated by Potschka et al. [33] on the DORIS-I line (DESY, Hamburg; λ = 1.50 Å).
In this work, we characterized for the first time the structure of TMV virions and repolymerized CP aggregates in solution by SAXS and modeled the SAXS curves in the entire measured range. According to the TEM data, repolymerized CP aggregates represented an array of stacked disks varying in length (Fig. 1). The main fraction represented particles composed of 6-16 20S disks. The measured values of the average gyration radius (Rg = 244.1 Å) and molecular weight (Mr = 8308 kDa) were found to be close to the (AA)16 model (Rg = 253.4 Å and Mr = 9505 kDa, respectively) (Table 3). It was established previously by cryoelectron microscopy that the structure of the TMV CP stacked disks is similar to that of the two central rings in the crystallized four-layer aggregate, and the thickness of the individual ring is 26.8 Å [10, 11], which is close to the periodicity of the repolymerized CP aggregates determined in our study (d4 = 26.9 Å; Table 2).
The reason for low correlation between the model and experimental SAXS curves could be the difference between the lengths of the used (AA)16 model (~820 Å) and repolymerized CP aggregates (300-800 Å). As can be seen in Fig. 4, the increase in the particle length results in the increase in intensity of peaks s2, s4, and s6 corresponding to the disk packaging in the aggregate structure. Different hydration degree of 20S disks in the crystal and in solution could also result in the data discrepancies. Franklin and Commoner [7] discovered significant hydration of disks in solution and decrease in the size of axial periodicity in the disk stacks from 53 to 43 Å upon water removal from the preparations. It is also possible that the protein atomic structure in the PDB: 1EI7 crystal model is slightly different from the structure of repolymerized CP aggregates in solution, which might be the reason for unique stability of repolymerized TMV CP aggregates in solution and can explain the binding of certain dyes characteristic for amyloids [13, 14].
One of the advantages of SAXS is the possibility to characterize all particles in solution under conditions close to physiological ones. More labor-intensive method of X-ray diffraction allows to determine the crystal structure, while cryoelectron microscopy examines the frozen particles. Here, we used SAXS to characterize for the first time the structure of TMV virions and repolymerized CP aggregates in solution and modeled the structure of CP aggregates based on the SAXS data in the entire measured range. Some of the intensity maxima on the scatter curves were assigned to the elements of the aggregate quaternary structure (structure periodicity, helix pitch in virion). The best agreement with the experimental data was obtained for the model of repolymerized CP aggregates consisting of repeated 20S disks composed of the central pair of rings from the PDB: 1EI7 crystal model.
Funding. This work supported by the Russian Foundation for Basic Research (grant 18-04-00525a) and Ministry of Science and Higher Education of the Russian Federation (State Budget Project “Crystallography and Photonics”, SAXS experiments).
Conflict of interest. The authors declare no conflict of interest in financial or any other sphere.
Ethical approval. This article does not contain description of studies with human participants or animals performed by any of the authors.
REFERENCES
1.Caspar, D. L., and Namba, K. (1990) Switching in
the self-assembly of tobacco mosaic virus, Adv. Biophys.,
26, 157-185; doi: 10.1016/0065-227x(90)90011-h.
2.Kendall, A., McDonald, M., and Stubbs, G. (2007)
Precise determination of the helical repeat of tobacco mosaic virus,
Virology, 369, 226-227; doi:
10.1016/j.virol.2007.08.013.
3.Butler, P. J. (1999) Self-assembly of tobacco
mosaic virus: the role of an intermediate aggregate in generating both
specificity and speed, Philos. Trans. R. Soc. Lond. B Biol.
Sci., 354, 537-550; doi: 10.1098/rstb.1999.0405.
4.Bloomer, A. C., Champness, J. N., Bricogne, G.,
Staden, R., and Klug, A. (1978) Protein disk of tobacco mosaic virus at
2.8 Å resolution showing the interactions within and between
subunits, Nature, 276, 362-368; doi:
10.1038/276362a0.
5.Klug, A. (1999) The tobacco mosaic virus particle:
structure and assembly, Philos. Trans. R. Soc. Lond. B Biol.
Sci., 354, 531-535.
6.Butler, P. J. (1984) The current picture of the
structure and assembly of tobacco mosaic virus, J. Gen. Virol.,
65 (Pt. 2), 253-279; doi: 10.1099/0022-1317-65-2-253.
7.Franklin, R. E., and Commoner, B. (1955) Abnormal
protein associated with tobacco mosaic virus; X-ray diffraction by an
abnormal protein (B8) associated with tobacco mosaic virus,
Nature, 175, 1076-1077; doi: 10.1038/1751076a0.
8.Unwin, P. N., and Klug, A. (1974) Electron
microscopy of the stacked disk aggregate of tobacco mosaic virus
protein. I. Three-dimensional image reconstruction, J. Mol.
Biol., 87, 641-656; doi: 10.1016/0022-2836(74)90075-8.
9.Dore, I., Ruhlmann, C., Oudet, P., Cahoon, M.,
Caspar, D. L., and Van Regenmortel, M. H. (1990) Polarity of binding of
monoclonal antibodies to tobacco mosaic virus rods and stacked disks,
Virology, 176, 25-29; doi:
10.1016/0042-6822(90)90226-h.
10.Diaz-Avalos, R., and Caspar, D. L. (1998)
Structure of the stacked disk aggregate of tobacco mosaic virus
protein, Biophys. J., 74, 595-603; doi:
10.1016/S0006-3495(98)77818-X.
11.Diaz-Avalos, R., and Caspar, D. L. (2000)
Hyperstable stacked-disk structure of tobacco mosaic virus protein:
electron cryomicroscopy image reconstruction related to atomic models,
J. Mol. Biol., 297, 67-72; doi:
10.1006/jmbi.1999.3481.
12.Bhyravbhatla, B., Watowich, S. J., and Caspar, D.
L. (1998) Refined atomic model of the four-layer aggregate of the
tobacco mosaic virus coat protein at 2.4-Å resolution,
Biophys. J., 74, 604-615; doi:
10.1016/S0006-3495(98)77819-1.
13.Raghavendra, K., Adams, M. L., and Schuster, T.
M. (1985) Tobacco mosaic virus protein aggregates in solution:
structural comparison of 20S aggregates with those near conditions for
disk crystallization, Biochemistry, 24, 3298-3304; doi:
10.1021/bi00334a034.
14.Raghavendra, K., Salunke, D. M., Caspar, D. L.,
and Schuster, T. M. (1986) Disk aggregates of tobacco mosaic virus
protein in solution: electron microscopy observations,
Biochemistry, 25, 6276-6279; doi:
10.1021/bi00368a066.
15.Ksenofontov, A. L., Dobrov, E. N., Fedorova, N.
V., Arutyunyan, A. M., Golanikov, A. E., Jarvekulg, L., and Shtykova,
E. V. (2018) Structure of potato virus A coat protein particles and
their dissociation, Mol. Biol., 52, 913-921; doi:
10.1134/S0026893318060109.
16.Ksenofontov, A. L., Dobrov, E. N., Fedorova, N.
V., Serebryakova, M. V., Prusov, A. N., Baratova, L. A., Paalme, V.,
Jarvekulg, L., and Shtykova, E. V. (2018) Isolated potato virus A coat
protein possesses unusual properties and forms different short
virus-like particles, J. Biomol. Struct. Dyn., 36,
1728-1738; doi: 10.1080/07391102.2017.1333457.
17.Goodman, R. M. (1975) Reconstitution of potato
virus X in vitro. I. Properties of the dissociated protein
structural subunits, Virology, 68, 287-298; doi:
10.1016/0042-6822(75)90272-x.
18.Fraenkel-Conrat, H. (1957) Degradation of tobacco
mosaic virus with acetic acid, Virology, 4, 1-4; doi:
10.1016/0042-6822(57)90038-7.
19.Ksenofontov, A. L., Kozlovskii, V. S.,
Kordiukova, L. V., Radiukhin, V. A., Timofeeva, A. V., and Dobrov, E.
N. (2006) Determination of concentration and aggregate size in
influenza virus preparations from true UV absorption spectra, Mol.
Biol., 40, 152-158.
20.Laemmli, U. K. (1970) Cleavage of structural
proteins during the assembly of the head of bacteriophage T4,
Nature, 227, 680-685; doi: 10.1038/227680a0.
21.Blanchet, C. E., Spilotros, A., Schwemmer, F.,
Graewert, M. A., Kikhney, A., Jeffries, C. M., Franke, D., Mark, D.,
Zengerle, R., Cipriani, F., Fiedler, S., Roessle, M., and Svergun, D.
I. (2015) Versatile sample environments and automation for biological
solution X-ray scattering experiments at the P12 beamline (PETRA III,
DESY), J. Appl. Crystallogr., 48, 431-443; doi:
10.1107/S160057671500254X.
22.Konarev, P. V., Volkov, V. V., Sokolova, A. V.,
Koch, M. H. J., and Svergun, D. I. (2003) PRIMUS: a Windows PC-based
system for small-angle scattering data analysis, J. Appl.
Crystallogr., 36, 1277-1282, doi:
10.1107/S0021889803012779.
23.Konarev, P. V., Petoukhov, M. V., and Svergun, D.
I. (2001) MASSHA – a graphics system for rigid-body modelling of
macromolecular complexes against solution scattering data, J. Appl.
Crystallogr., 34, 527-532; doi:
10.1107/S0021889801006100.
24.Svergun, D., Barberato, C., and Koch, M. H.
(1995) CRYSOL – a program to evaluate X-ray solution scattering
of biological macromolecules from atomic coordinates, J. Appl.
Crystallogr., 28, 768-773; doi:
10.1107/S0021889895007047.
25.Durham, A. C., Finch, J. T., and Klug, A. (1971)
States of aggregation of tobacco mosaic virus protein, Nat. New
Biol., 229, 37-42; doi: 10.1038/newbio229037a0.
26.Sachse, C., Chen, J. Z., Coureux, P. D., Stroupe,
M. E., Fandrich, M., and Grigorieff, N. (2007) High-resolution electron
microscopy of helical specimens: a fresh look at tobacco mosaic virus,
J. Mol. Biol., 371, 812-835; doi:
10.1016/j.jmb.2007.05.088.
27.Blanchet, C. E., and Svergun, D. I. (2013)
Small-angle X-ray scattering on biological macromolecules and
nanocomposites in solution, Annu. Rev. Phys. Chem., 64,
37-54; doi: 10.1146/annurev-physchem-040412-110132.
28.Shtykova, E. V., Baratova, L. A., Fedorova, N.
V., Radyukhin, V. A., Ksenofontov, A. L., Volkov, V. V., Shishkov, A.
V., Dolgov, A. A., Shilova, L. A., Batishchev, O. V., Jeffries, C. M.,
and Svergun, D. I. (2013) Structural analysis of influenza A virus
matrix protein M1 and its self-assemblies at low pH, PLoS One,
8, e82431; doi: 10.1371/journal.pone.0082431.
29.Costa, L., Andriatis, A., Brennich, M., Teulon,
J. M., Chen, S. W., Pellequer, J. L., and Round, A. (2016) Combined
small angle X-ray solution scattering with atomic force microscopy for
characterizing radiation damage on biological macromolecules, BMC
Struct. Biol., 16, 18; doi: 10.1186/s12900-016-0068-2.
30.Spinozzi, F., Ferrero, C., Ortore, M. G.,
Antolinos, A. D., and Mariani, P. (2014) GENFIT: software for the
analysis of small-angle X-ray and neutron scattering data of
macromolecules in solution, J. Appl. Crystallogr., 47,
1132-1139; doi: 10.1107/S1600576714005147.
31.Hiragi, Y., Inoue, H., Sano, Y., Kajiwara, K.,
Ueki, T., Kataoka, M., Tagawa, H., Izumi, Y., Muroga, Y., and Amemiya,
Y. (1988) Temperature dependence of the structure of aggregates of
tobacco mosaic virus protein at pH 7.2. Static synchrotron small-angle
X-ray scattering, J. Mol. Biol., 204, 129-140; doi:
10.1016/0022-2836(88)90604-3.
32.Sano, Y., Inoue, H., and Hiragi, Y. (1999)
Differences of reconstitution process between tobacco mosaic virus and
cucumber green mottle mosaic virus by synchrotron small angle X-ray
scattering using low-temperature quenching, J. Protein Chem.,
18, 801-805; doi: 10.1023/a:1020689720082.
33.Potschka, M., Koch, M. H., Adams, M. L., and
Schuster, T. M. (1988) Time-resolved solution X-ray scattering of
tobacco mosaic virus coat protein: kinetics and structure of
intermediates, Biochemistry, 27, 8481-8491; doi:
10.1021/bi00422a028.