High Level of Gene Transcription at the Embryonic Stage Leads to the Suppression of Heterochromatic Trans-Inactivation in Drosophila melanogaster Adults
A. A. Solodovnikov1, V. A. Gvozdev1, and S. A. Lavrov1,a*
1Institute of Molecular Genetics of the Russian Academy of Sciences, 123182 Moscow, Russia* To whom correspondence should be addressed.
Received December 30, 2019; Revised February 24, 2020; Accepted February 24, 2020
In some cases, gene transfer from euchromatin to constitutive heterochromatin as a result of chromosomal rearrangement is accompanied by epigenetic inactivation of this gene (cis-inactivation). In the case of trans-inactivation, transgenes in the normal chromosome are repressed by the cis-inactivation-causing rearranged homologous chromosome. Trans-inactivation is a result of the somatic pairing of homologs and the transfer of the normal chromosomal segment to the heterochromatic compartment of the nucleus. Previously, we have shown that the degree of trans-inactivation of the UAS-eGFP reporter gene in adult flies depends on its transcription level that can be regulated by temperature using the GAL4 transcription activator and its temperature-sensitive inhibitor GAL80ts. In this paper, we investigated the epigenetic inheritance of the active/repressed state of the trans-inactivated reporter gene at different expression levels by measuring eGFP fluorescence in the individual cells of Malpighian tubules in adult flies. High expression levels at the embryonic stage protected the eGFP gene from trans-inactivation in adult flies. The activated state was inherited over the entire period of development and differentiation, while the activating effect of GAL4 was turned off.
KEY WORDS: heterochromatin, Drosophila, position effect variegation, trans-inactivation, expression, GAL4, GAL80tsDOI: 10.1134/S0006297920040070
Abbreviations: A4, chromosomal inversion In(2)A4; PE, position effect; PEV, position effect variegation; UAS-eGFP, eGFP gene under the control of the UAS regulatory element.
The position effect (PE) is an alteration in the expression of a gene
when its location in the genome is changed without changes in
nucleotide sequence. A particular case of PE is heterochromatin-induced
position effect variegation (PEV), at which the expression of
euchromatin genes is repressed as they are translocated to constitutive
heterochromatin. As a result of chromosomal rearrangement, euchromatin
juxtaposed close to the new euchromatin/heterochromatin boundary
acquires the properties of heterochromatin (heterochromatinization),
and this process can occur over long distances (hundreds kbp to
millions bp). Heterochromatinization is accompanied by the recruitment
of heterochromatin-specific proteins (HP1a and others) and the
appearance of histone modifications (H3K9me2/3) in euchromatic regions
[1-3], leading to the
impairments in gene functioning. In heterochromatin-induced PEV, the
genes are inactivated in some cells at the embryonic stage, and the
resulting active/repressed state can be epigenetically inherited in the
cell line, eventually yielding tissues with mosaic gene expression.
In some cases, the rearranged chromosome with the new euchromatin/heterochromatin boundaries inactivates genes in normal (non- rearranged) chromosome [4-6]. This phenomenon (termed trans-inactivation) cannot be explained by heterochromatin spreading into euchromatin from the new boundary and, according to current concepts, is associated with the translocation of euchromatin regions to the heterochromatic compartment of the nucleus during somatic pairing of the normal and homologous rearranged chromosomes. Trans-inactivation is accompanied by de novo heterochromatin formation in the normal chromosome [7-10].
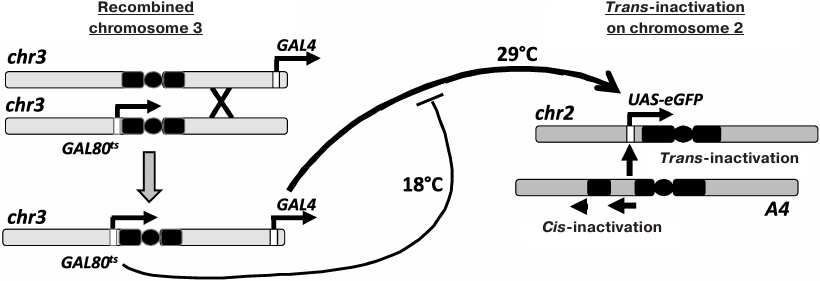
Chromosomal inversion In(2)A4 (A4; fig. 1) causes cis-inactivation of the genes located near the new euchromatin/heterochromatin boundaries and trans-inactivation of reporter constructs in the homologous chromosome [6, 9-12]. To investigate the inheritance of the epigenetic states of trans-inactivated genes during development, Drosophila strain containing reporter eGFP gene under the control of the UAS regulatory element (UAS-eGFP) in normal chromosome 2, the A4 inversion in the homologous chromosome, and the GAL4 trans-activator together with a thermosensitive form of GAL80 repressor (GAL80ts) in chromosome 3 (for the regulation of the reporter gene expression by varying the temperature) have been constructed. In the present work, this genetic system was used to study the interconnection between the reporter gene trans-inactivation in adult flies and the level of its expression at different developmental stages. It was shown that the high expression level at the embryonic stage resulted in the resistance of the reporter gene to trans-inactivation, which is epigenetically inherited throughout development.
Fig. 1. Control of the reporter gene expression in the Drosophila strain used in the study. Chromosome 2 with the A4 rearrangement causes trans-inactivation of the UAS-eGFP reporter gene in the homologous normal chromosome. Chromosome 3 was obtained through recombination and carries the genes for GAL4 (tubGal4) and GAL80ts (tubGal80ts). An increase in temperature results in the destruction of the GAL80ts repressor, an increase in the content of active GAL4, and activation of the UAS-eGFP transcription.
MATERIALS AND METHODS
Drosophila lines. The w67c23; In(2)A4/CyO line containing the trans-inactivation-inducing chromosome rearrangement In(2)A4 (hereinafter A4) has been obtained and studied in the Laboratory of Biochemical Genetics of Animals (Institute of Molecular Genetics, Russian Academy of Sciences) [6].
The following lines were provided by the Bloomington Drosophila Stock Center: line 7018 (w*; snaSc/CyO; P{tubP-GAL80ts}7), containing the gene for the GAL80ts protein under the control of the alphaTub84B promoter [13] on chromosome 3; line 5138 (y1 w*; P{tubP-GAL4}LL7/TM3 Sb1 Ser1), containing the gene for GAL4 under control of the alphaTub84B promoter [14].
Line w*; UAS-eGFP/CyO; P{tubP-GAL4}LL7; P{tubP-GAL80ts}7/TM3 Sb1 Ser1 contains the UAS-eGFP reporter gene on chromosome 2 [11] and the GAL4 and GAL80ts regulatory genes on chromosome 3. Chromosome 3 with GAL4 and GAL80ts was obtained by recombination between lines 7018 (w*; snaSc°/CyO; P{tubP-GAL80ts}7) and 5138 (y1 w*; P{tubP-GAL4}LL7/TM3 Sb1 Ser1). The crosses are described in the Supplement.
The effect of eGFP activation at different developmental stages on trans-inactivation in adult flies was studied by crossing w*; UAS-eGFP; GAL80tsGAL4/TM3 Sb1Ser1 and w*; In(2)A4/CyO; +/+ lines. The offspring from this cross contained flies of four genotypes. The w*; UAS-eGFP/In(2)A4; GAL80tsGAL4/+ (experiment) and w*; UAS-eGFP/CyO; GAL80tsGAL4/+ (control) flies carried GAL4 and GAL80ts genes. In these flies, an increase in growth temperature promotes eGFP expression. In the experimental flies, the UAS-eGFP gene undergoes trans-inactivation under the influence of In(2)A4; in the control flies, trans-inactivation is absent. The temperature of fly development was varied as shown in the table and Fig. 2a.
eGFP fluorescence in individual cells of Malpighian tubules was analyzed in 2-day-old adult females. Malpighian tubules were chosen due to the facts that mosaic PEV is well pronounced in these organs and that their tissue morphology (large isolated cells forming single-layer tubules) considerably facilitates quantitative measurements.
Temperature variations, T°C
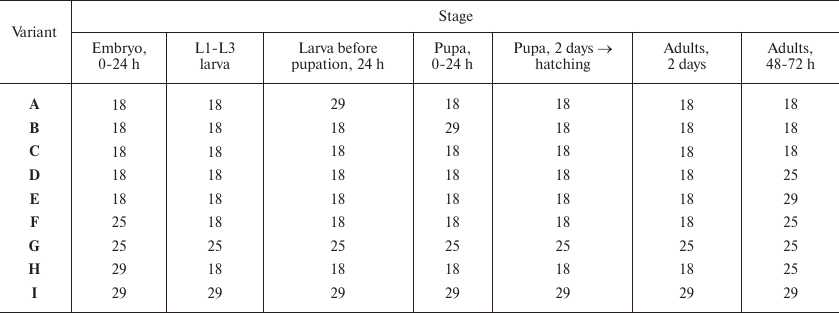
Notes: Variations of temperature at different stages of fly development
(see Fig. 1). For example (variant B), development
was carried out at 18°C until pupation; the pupae developed for 24
h at 29°C; then the development continued at 18°C. eGFP
fluorescence was evaluated in the cells of Malpighian tubules of adult
2-day-old flies. L1-L3 is the period of larval development (first-,
second-, and third-instar larvae, respectively).
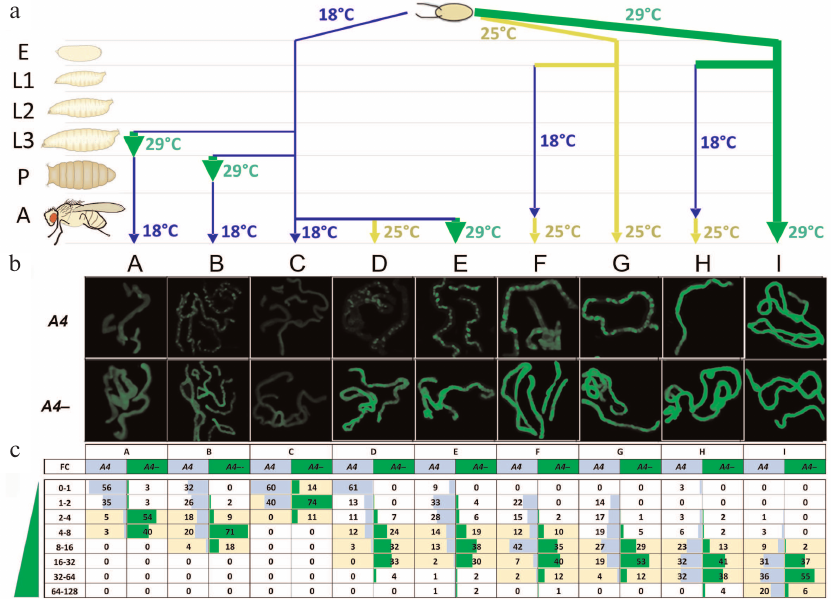
Fig. 2. Trans-inactivation of the UAS-eGFP reporter gene in adult flies after modulation of its expression at different stages of development. a) Variants of temperature treatment (A to I) (see also the table) in the development of eGFP/A4; GAL80ts GAL4/+ (A4, trans-inactivation) and eGFP/+; GAL80tsGAL4/+ (A4–, control) flies. E, embryos; L1-L3, 1-3 instar larvae; P, pupae; A, adult flies. b) eGFP fluorescence in the Malpighian tubules of flies with and without (A4–, control) the A4 chromosome. Variants B, D, E, F, and G (A4) demonstrate non-uniform fluorescence distribution representing heterochromatin trans-inactivation. c) Distribution of eGFP fluorescence in the cells of Malpighian tubules in the flies with trans-inactivation (A4) and the control flies (A4–). Fluorescence intensity was measured in each Malpighian tubule cell and normalized to the modal (most frequently occurring) level of fluorescence in the Malpighian tubules of A4– flies at 18°C that was taken as a unit (column C, A4–). The resulting values (FC, fold change) show how many times the level of fluorescence is higher than the level taken as a unit. The entire range of fluorescence values was divided into subranges (indicated in the leftmost column); the percentage of cells falling into a subrange was counted for each variant of temperature treatment and indicated in the table cells. Yellow color denotes the fluorescence range for the majority of the cells of Malpighian tubules in the flies without trans-inactivation (A4–), i.e., the normal value of fluorescence intensity for a given variant of temperature treatment. The significance of differences between the fluorescence levels in each variant (p-values) is given in Table S1 in the Supplement; standard deviations are given in Table S2 and Fig. S2 in the Supplement.
The w*; UAS-eGFP/In(2)A4; TM3 Sb1Ser1/+ and w*; UAS-eGFP/CyO; TM3 Sb1Ser1/+ flies lack the GAL4 activator. In these flies, eGFP was expressed at the background level. Comparison of eGFP expression levels with (UAS-eGFP/In(2)A4) and without (UAS-eGFP/CyO) trans-inactivation at different temperatures allowed us to assess the direct effect of temperature on trans-inactivation.
eGFP fluorescence in the cells of Malpighian tubules. The images of Malpighian tubules (Fig. 2b) were obtained on LeicaDM6000 fluorescence microscope equipped with a Hamamatsu ORCA ER camera and a set of filters for fluorescence detection. Malpighian tubules were dissected in PBT (phosphate buffer with 0.1% Tween 20), then incubated in PBT with DAPI for 15 min at room temperature and photographed. Images were captured at a 5× magnification and with the same acquisition parameters (exposure, gain, resolution); the exposure was selected to avoid image saturation in the brightest samples. The samples were photographed in the GFP (488 nm) and DAPI (405 nm) channels; the images were saved as 16-bit TIFF files and processed with ImageJ. The DAPI channel was used for image segmentation (Threshold). The resulting set of regions of interest (ROIs) was transferred to the eGFP image; the region without the objects was chosen for background subtraction. The following parameters were measured for each ROI: area, modal optical density, a standard deviation of the pixel intensity and ROI sphericity (MultiMeasure in ImageJ). The data were transferred to an Excel datasheet and the records were filtered by the area (30-300 pixels), standard deviation (no more than the ROI intensity) and sphericity (>0.2). As a result, ~100 records of eGFP fluorescence measurements corresponding to individual cells were obtained for each image. These data were used to construct histograms of the eGFP fluorescence intensity distribution. The results were obtained from two independent experiments and processed statistically in MS Excel and R (https://cran.r-project.org/). Five to ten flies of each genotype were analyzed.
Quantitative PCR. RNA was isolated from adult flies of the w*; UAS-eGFP/In(2)A4; GAL80tsGAL4/+ (experiment); w*; UAS-eGFP/CyO; GAL80tsGAL4/+ (control); and w*; UAS-eGFP/In(2)A4 (the study of the direct effect of temperature increase on trans-inactivation) lines. Reverse transcription and quantitative PCR with the eGFP gene-specific primers were carried out as described in [11]. The amount of eGFP mRNA was normalized to the amount of mRNA for the housekeeping gene Rpl32.
RESULTS
The reporter construct used in the work did not contain any functional elements except for UAS; in the absence of GAL4, eGFP expressed at a low level at all stages of fly development. The UAS-eGFP expression can be regulated by GAL4 transcription activator and its thermosensitive repressor GAL80ts [11]. The genes for GAL4 transactivator and GAL80ts repressor are placed under the control of the promoter of the ubiquitously active alpha-tubulin gene and expressed at similarly high levels at all developmental stages. GAL80ts blocks the GAL4 activator domain at 18°C; however, it undergoes degradation at higher temperatures with the release of the GAL4 activator, leading to the upregulation of eGFP expression [13]. The temperature decrease results in the accumulation of the active form of GAL80ts, GAL4 inactivation, and reduction in the reporter expression level. The activation of the reporter gene does not require GAL4 expression but is determined by the protein-protein interactions between GAL4 and GAL80ts already present in the cells. Thus, the combined effect of GAL4 and GAL80ts makes it possible to predictably regulate the level of gene expression by incubation temperature adjustment [11]. The use of eGFP as a reporter gene allows us to observe PEV in vivo and to assess the level of eGFP expression by measuring eGFP fluorescence (which correlates with the amount of eGFP mRNA) (Fig. S1 in the Supplement). Previous measurements of the eGFP mRNA level by PCR have shown that it increased ~3-fold at 18°C, ~50-fold at 25°C and ~100-fold at 29°C in whole adult flies compared to flies lacking the GAL4 gene (background level) [11]. The degrees of the eGFP transcription activation at different temperatures were similar in the adult flies and fly embryos (unpublished data). Hereinafter, the expression of eGFP denoted as weak (~3-fold) at 18°C, moderate (~50-fold) at 25°C, and strong (~100-fold) at 29°C.
Figure 1 shows the genetic system with recombinant chromosome 3 carrying GAL4 and GAL80ts sources used in this study. In the flies of UAS-eGFP/A4; GAL4 GAL80ts/+ (eGFP/A4) genotype, the reporter gene was trans-inactivated under the influence of the A4 inversion. The UAS-eGFP/CyO; GAL4 GAL80ts/+ (eGFP/+) flies lacking chromosome A4 were used as a control. Trans-inactivation was manifested as the appearance of cells with the low level or absence of eGFP fluorescence in the Malpighian tubules of adult flies (Fig. 2b, variants B, D-G).
One of our goals was to examine the stability of epigenetic inheritance of the expression level of a gene undergoing heterochromatinization induced by homologous rearranged chromosome A4. We investigated the dependence of the degree of trans-inactivation of reporter gene eGFP in adult flies on the level of its expression (weak, moderate, and strong) at the earlier developmental stages, beginning from the embryo. The degree of trans-inactivation was estimated by measuring the level of eGFP fluorescence in the cells of Malpighian tubules in adult flies A4 and A4– (Fig. 2, b and c). The temperature was varied during the development as shown in Fig. 2a and the table (variants A-I).
The eGFP fluorescence in Malpighian tubule cells in the experimental (UAS-eGFP/A4, A4 in Fig. 2, b and c) and control (UAS-eGFP/CyO, A4– in Fig. 2, b and c) flies was measures and normalized to the modal (most frequently occurring) level of fluorescence in the Malpighian tubules of control flies developing at 18°C (variant C, A4–) that was taken as a unit. The data obtained for all variants of temperature treatment of the experimental and control flies were used to construct histograms showing the percentage of cells falling within the given ranges of fluorescence intensity. For example, in the case D (Fig. 2c), both control and experimental flies developed to the adult stage at 18°C (weak eGFP activation); then, the reporter gene was moderately activated at 25°C before fluorescence measurements. In 61% of cells, the level of fluorescence in case of trans-inactivation (D, column A4) was below or equal to the value taken as a unit, while in the control flies without trans-inactivation (D, column A4–), the level of expression exceeded this value 4-32 times in 89% of cells. The distribution of fluorescence intensity in the experimental and control flies was different, demonstrating strong trans-inactivation under the influence of the A4 inversion.
Data analysis has led us to the following conclusions. The low level of reporter gene expression at the early stages of development was accompanied by its strong trans-inactivation in adult flies (variants A-D); in all variants, a population of cells with the background level of eGFP fluorescence was observed. In the case of moderate and strong expression at the embryonic stage, the cells with the background level of eGFP fluorescence were not observed in adult flies (variants F-I). In the case of moderate expression throughout the development (25°C, variant G), ~45% of cells demonstrated trans-inactivation of eGFP and their fluorescence was below the minimum level of fluorescence in the control. Strong expression of eGFP throughout the development prevented trans-inactivation in most cells; the distribution of fluorescence was not significantly different in the experimental and control flies (variant I). However, in ~10% of cells, the reporter gene was trans-inactivated. Unexpectedly, the fluorescence level in ~10% of cells of UAS-eGFP/A4 flies exceeds the maximum level of fluorescence in the control. The repressed state of the reporter gene established at the early stages of development could be overcome by strong transcriptional activation. For example, in variant D, eGFP was weakly expressed over the entire period of development (18°C) and then moderately activated in adult flies (25°C), while in variant E, its expression was strongly activated in adult flies (29°C). As a result, only 9% of the cells in the variant E demonstrated complete repression of the reporter gene, compared to variant D, where the number of such cells was up to 60%. In other words, up to 80% of cells with trans-inactivated eGFP (Fig. 2c, variant E) resumed its active expression upon strong transcriptional activation in adult flies. At the same time, the level of fluorescence in variant E in the flies with trans-inactivation was generally lower than in the control (A4–).
Comparison of variants D, F, and H shows that activation of the reporter gene at the embryonic stage in the presence of A4 chromosome leads to the formation of stable active epigenetic imprinting. eGFP was weakly activated (18°C) in variant D, moderately activated in variant F (25°C), and strongly activated in variant H (29°C) at the embryonic stage. Further fly development in all cases occurred at 18°C (weak expression of the reporter gene) until hatching; then, expression of the reporter gene was moderately activated (25°C) to analyze the degree of trans-inactivation. The fraction of the Malpighian tubule cells with repressed eGFP substantially varied between the variants. In variant D, we observed strong PEV (85% cells with impaired reporter activation); repression was found in ~40% cells in variant F and only in 12% cells in variant H. Comparison of variants D, F, and H shows that the maintenance of the active state of chromatin (resistance to trans-inactivation) until the adult stage requires high expression levels at the embryonic stage, whereas at the post-embryonic stages, active transcription of the reporter gene is not obligatory.
It is well known that the manifestation of the heterochromatin PEV usually decreases at higher temperatures [1, 3]. Since the level of the reporter gene expression in the studied system was regulated by varying the temperature, such effect could influence the degree of trans-inactivation directly (not only through the activation of the reporter gene expression by GAL4). However, quantitative PCR showed no evident differences in the eGFP mRNA amount in the UAS-eGFP/A4 flies lacking the GAL4 gene (UAS-eGFP/A4; +/+) that developed at 18°C and 29°C (Fig. S3 in the Supplement). It is also known that temperature dependence of PEV varies for different rearrangements and, in some cases, the effect of temperature on repression is absent [3, 15]. It seems more probable that the observed influence of the reporter gene expression level on the degree of its trans-inactivation is determined by the activation of transcription with the involvement of GAL4.
DISCUSSION
Gene repression in the case of heterochromatin PEV during individual development has been previously investigated in [16-19]. In studies [18, 19], the behavior of a reporter construct carrying UAS-eGFP (as well as other functional elements) under the influence of heterochromatin was investigated. Sage et al. [19] studied the trans-acting PEV [19].
Ahmad and Henikoff [18] used a reporter construct containing the GFP gene (under control of UAS) and mini-white gene. The construct was affected by PEV and the GFP expression was triggered at different stages of development using the GAL4 gene placed under the control of tissue- or stage-specific promoters. The authors analyzed imaginal discs of larvae and eyes of adult flies and showed that the high level of activation of GFP reporter gene at the embryonic stage (GAL4 driver under the actin promoter) led to the PEV suppression; however, PEV was observable at a lower level of expression activation (GAL4 under the control of the armadillo [arm] gene promoter, weak but active in most tissues and at all stages of development). PEV of the reporter gene was also observed if its transcriptional activation occurred at the post-embryonic stages. Derepression of the reporter gene (emergence of GFP-expressing cells) was observed during differentiation of imaginal discs in larvae; derepression occurred in both mitotically active and already differentiated cells. This observation implies that the cell cycle progression is not necessary for the reactivation of genes repressed in PEV.
Trans-inactivation of the reporter construct carrying UAS-eGFP and mini-white under the influence of the BwD allele on the homologous chromosome was studied in [19]. BwD is an insertion of satellite DNA in the Brown gene. UAS-eGFP expression was activated using the GAL4 gene placed under the control of promoters of various strength and specificity (including actin and arm, like in [18]); then the degree of the reporter gene trans-inactivation was analyzed in the tissues of the third-instar larvae and adults. The trans-inactivation of UAS-eGFP in this work demonstrated significant variability in different tissues and was observed in the case of the low level of the reporter gene expression at the developmental stages preceding the studied ones (third-instar larvae and adult flies).
Analysis of data from earlier studies revealed the following properties of gene inactivation, common to cis- and trans-acting PEV during development. PEV – caused repression begins at the gastrulation stage in the embryos. If a gene is expressed at a high level starting from the embryonic stage (e.g., under control of actin promoter), it is resistant to PEV. If a gene is expressed at a low/medium level (arm promoter) starting from the embryonic stage, it can be repressed. Genes whose expression is turned on at the post-embryonic stages are also repressed. A gene repressed at the early stages of development can be reactivated at the later stages (usually during metamorphosis). Reactivation does not require cell cycle progression [18, 19].
Our system allows us to predictably and quantitatively regulate the reporter gene expression level [11], while in the previous works, the expression profiles of PEV-affected reporter genes in different types of tissues during development were characterized only qualitatively. In our study, the trans-inactivation was investigated in only one type of tissue (Malpighian tubules of adult flies). Malpighian tubules originate from the ectoderm and mesoderm at stages 5-13 of embryonic development. Cell proliferation is completed at stage 13, after which the number of cells in these organs does not change. Malpighian tubules do not undergo histolysis during metamorphosis [20]. Polytene chromosomes in the nuclei of Malpighian tubule cells pass through up to eight cycles of DNA endoreplication without cytokinesis during the development and can contain up to 256 sister chromatids [21]. Malpighian tubules consist of terminally differentiated cells like salivary glands cell; but in contrast to the latter, they are preserved in adult flies.
According to our data, trans-inactivation of the reporter gene under the influence of the A4 inversion behaves similarly to PEV cases in the above-mentioned works, presumably reflecting the characteristic features of heterochromatin repression in general. For example, high-level expression of UAS-eGFP at the embryonic stage suppressed trans-inactivation in adult flies. No trans-inactivation in adult flies was observed at a 100-fold increase in the reporter expression at the embryonic stage; however, well-pronounced repression was found at a 30-50-fold activation. At a 100-fold activation, the level of eGFP expression was about 17% of the expression level of the housekeeping gene Rpl32 [11]. This fact suggests that genes expressed at similar or higher levels at the embryonic stage will not be subjects to heterochromatin repression. The studies of gene transcription levels in the case of cis-acting PEV have shown that only a few of dozens of genes existing in the region of PEV undergo heterochromatin repression [9, 22], which might be explained by low transcription levels of these genes at the embryonic stage.
In the case of trans-inactivation, the epigenetic effect is associated with changes in the intranuclear location of the reporter gene. It has been shown previously [7-9, 23] that repression is accompanied by gene transfer to the heterochromatic compartment of the nucleus, while the maintenance of the active state, on the contrary, leads the gene localization to the euchromatin. Local activation of UAS-eGFP can result in the disturbed pairing of the trans-inactivated region of the normal chromosome and the inversion and the transfer of the region containing the reporter gene to the euchromatic compartment of the nucleus. It would be interesting to use FISH hybridization to compare the location of low-active and 100-fold activated reporter genes in the Malpighian tube cells with different degree of trans-inactivation.
We have shown that trans-inactivation of the reporter gene established at the embryonic stage can be overcome by the high-level activation of transcription in young adult flies (Fig. 2b, variant E, A4). The temperature for the expression activation corresponded to that necessary for preventing trans-inactivation at the embryonic stage. It is possible that the threshold level of the reporter gene expression, at which the heterochromatinized chromatin state in the region of the gene becomes unstable, was achieved. Heterochromatin on the reporter gene, in this case, does not form in the embryos and gets disturbed in adult flies. Since Malpighian tubules are a non-proliferating terminally differentiated tissue, changes in the chromatin organization/nuclear location of the reporter gene during reactivation is achieved in the absence of cell division and in polytenized nuclei.
When the reporter gene was strongly activated throughout the development (29°C) in the presence of A4 inversion, up to ~10% Malpighian tubule cells demonstrate higher fluorescence compared to the control (Fig. 2, variant I). It indicates that the heterochromatic environment can maintain a high level of expression and enhance transcription. It appears that heterochromatin is not a nonspecific repressor of euchromatin genes – some of them can behave similarly to heterochromatin genes demonstrating high levels of transcription in the heterochromatic environment. Hyperactivated reporter genes may be located in the heterochromatin compartment of the nucleus and, concerning chromatin organization, should be similar to highly expressed heterochromatin genes, i.e., include both heterochromatin markers (HP1a and H3K9me2/3) and the markers of active transcription, e.g., H3K4me3 [24].
In this work, we demonstrated that epigenetic changes in gene expression upon heterochromatin-induced trans-inactivation can be investigated in a genetic system with the adjustable expression of the reporter gene during development. The system allows monitoring of the inheritance of epigenetic state up to the adult stage in the absence of transcription inducer. We found that the high-level activation of gene expression at the embryonic stage switches the repressed state to the active state in adult flies. The genetic system proposed in this work can be used for further studies of heterochromatin-induced PEV, including identification of new genes involved in epigenetic inheritance, as well as the mechanisms of heterochromatic repression.
Funding. The work was supported by the Russian Science Foundation (project No. 19-74-20178) and by the Russian Foundation for Basic Research (projects Nos. 17-04-01984 A and 17-00-00282 KOMFI).
Conflict of interest. The authors declare no conflict of interest.
Compliance with ethical standards. This article does not contain any studies involving animals or human participants performed by any of the authors.
REFERENCES
1.Elgin, S. C., and Reuter, G. (2013) Position-effect
variegation, heterochromatin formation, and gene silencing in
Drosophila, Cold Spring Harb. Perspect. Biol., 5,
a017780, doi: 10.1101/cshperspect.a017780.
2.Weiler, K. S., and Wakimoto, B. T. (1995)
Heterochromatin and gene expression in Drosophila, Annu. Rev.
Genet., 29, 577-605,
doi: 10.1146/annurev.ge.29.120195.003045.
3.Spofford, J. B. (1976) Position-effect variegation
in Drosophila, in The Genetics and Biology of Drosophila,
Academic press, p. 955.
4.Csink, A. K., Bounoutas, A., Griffith, M. L., Sabl,
J. F., and Sage, B. T. (2002) Differential gene silencing by
trans-heterochromatin in Drosophila melanogaster,
Genetics, 160, 257-269.
5.Martin-Morris, L. E., Csink, A. K., Dorer, D. R.,
Talbert, P. B., and Henikoff, S. (1997) Heterochromatic
trans-inactivation of Drosophila white transgenes,
Genetics, 147, 671-677.
6.Abramov, Y. A., Kibanov, M. V., Gvozdev, V. A., and
Lavrov, S. A. (2011) Genetic and molecular analysis of the phenomenon
of gene trans-inactivation in Drosophila
melanogaster determined by eu-heterochromatic rearrangement of
the homologous chromosome, Dokl. Akad. Nauk, 437,
261-265.
7.Thakar, R., and Csink, A. K. (2005) Changing
chromatin dynamics and nuclear organization during differentiation in
Drosophila larval tissue, J. Cell Sci., 118,
951-960, doi: 10.1242/jcs.01684.
8.Thakar, R., Gordon, G., and Csink, A. K. (2006)
Dynamics and anchoring of heterochromatic loci during development,
J. Cell Sci., 119, 4165-4175,
doi: 10.1242/jcs.03183.
9.Abramov, Y. A., Shatskikh, A. S., Maksimenko, O.
G., Bonaccorsi, S., Gvozdev, V. A., and Lavrov, S. A. (2016) The
differences between cis- and trans-gene inactivation
caused by heterochromatin in Drosophila, Genetics,
202, 93-106, doi: 10.1534/genetics.115.181693.
10.Shatskikh, A. S., Abramov, Y. A., and Lavrov, S.
A. (2017) Trans-inactivation: repression in a wrong place,
Fly (Austin), 11, 96-103,
doi: 10.1080/19336934.2016.1225634.
11.Shatskikh, A. S., Olenkina, O. M., Solodovnikov,
A. A., and Lavrov, S. A. (2018) Regulated gene expression as a tool for
analysis of heterochromatin position effect in Drosophila,
Biochemistry (Moscow), 83, 542-551,
doi: 10.1134/S0006297918050073.
12.Lavrov, S. A., Shatskikh, A. S., Kibanov, M. V.,
and Gvozdev, V. A. (2013) Transcriptional inactivation of genes under
the effec of position in D. melanogaster correlates at
the level of individual cells with their transfer to the
heterochromatin compartment of the nucleus, Molekulyarnaya
Biologiya, 47, 1-6.
13.Mcguire, S. E., Le, P. T., Osborn, A. J.,
Matsumoto, K., and Davis, R. L. (2003) Spatiotemporal rescue of memory
dysfunction in Drosophila, Science, 302,
1765-1768, doi: 10.1126/science.1089035.
14.Lee, T., and Luo, L. (1999) Mosaic analysis with
a repressible cell marker for studies of gene function in neuronal
morphogenesis, Neuron, 22, 451-461,
doi: 10.1016/s0896-6273(00)80701-1.
15.Lloyd, V. K., Dyment, D., Sinclair, D. A., and
Grigliatti, T. A. (2003) Different patterns of gene silencing in
position-effect variegation, Genome, 46, 1104-1117,
doi: 10.1139/g03-070.
16.Lu, B. Y., Bishop, C. P., and Eissenberg, J. C.
(1996) Developmental timing and tissue specificity of
heterochromatin-mediated silencing, EMBO J., 15,
1323-1332.
17.Weiler, K. S., and Wakimoto, B. T. (1998)
Chromosome rearrangements induce both variegated and reduced, uniform
expression of heterochromatic genes in a development-specific manner,
Genetics, 149, 1451-1464.
18.Ahmad, K., and Henikoff, S. (2001) Modulation of
a transcription factor counteracts heterochromatic gene silencing in
Drosophila, Cell, 104, 839-847.
19.Sage, B. T., Wu, M. D., and Csink, A. K. (2008)
Interplay of developmentally regulated gene expression and
heterochromatic silencing in trans in Drosophila,
Genetics, 178, 749-759,
doi: 10.1534/genetics.107.083105.
20.Gautam, N. K., Verma, P., and Tapadia, M. G.,
(2017) Drosophila malpighian tubules: a model for understanding
kidney development, function, and disease, in Kidney Development and
Disease (Miller, R.K., ed.), Springer International Publishing,
Cham, pp. 3-25.
21.Lamb, M. J. (1982) The DNA content of polytene
nuclei in midgut and Malpighian tubule cells of adult Drosophila
melanogaster, Wilhelm Roux’s Arch. Dev. Biol.,
191, 381-384, doi: 10.1007/bf00879628.
22.Vogel, M. J., Pagie, L., Talhout, W., Nieuwland,
M., Kerkhoven, R. M., and Van Steensel, B. (2009) High-resolution
mapping of heterochromatin redistribution in a Drosophila
position-effect variegation model, Epigenetics Chromatin,
2, 1, doi: 10.1186/1756-8935-2-1.
23.Harmon, B., and Sedat, J. (2005) Cell-by-cell
dissection of gene expression and chromosomal interactions reveals
consequences of nuclear reorganization, PLoS Biol., 3,
e67, doi: 10.1371/journal.pbio.0030067.
24.Saha, P., Sowpati, D. T., and Mishra, R. K.
(2019) Epigenomic and genomic landscape of Drosophila
melanogaster heterochromatic genes, Genomics, 111,
177-185, doi: 10.1016/j.ygeno.2018.02.001.
Supplementary Figures S1, S2 and Tables S1-S3 (PDF)
[Download Reprint (PDF)]