Molecular Pathway Activation Markers Are Associated with Efficacy of Trastuzumab Therapy in Metastatic HER2-Positive Breast Cancer Better than Individual Gene Expression Levels
M. Sorokin1,2,3, K. Ignatev4, V. Barbara4, U. Vladimirova1,3, A. Muraveva3, M. Suntsova1, N. Gaifullin5, I. Vorotnikov6, D. Kamashev1,3, A. Bondarenko7, M. Baranova7,8, E. Poddubskaya3,7, and A. Buzdin1,2,3,9,a*
1Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, 117997 Moscow, Russia2OmicsWay Corp., Walnut, 91789 CA, USA
3Sechenov First Moscow State Medical University (Sechenov University), 119991 Moscow, Russia
4Republican Oncological Dispensary, 185002 Petrozavodsk, Karelia Republic, Russia
5Faculty of Fundamental Medicine, Lomonosov Moscow State University, 119991 Moscow, Russia
6Blokhin Russian Cancer Research Center, 115478 Moscow, Russia
7Vitamed Oncological Clinics, Moscow, Russia
8Petrovsky Russian Research Center of Surgery, 119991 Moscow, Russia
9Moscow Institute of Physics and Technology, 141701 Dolgoprudny, Moscow Region, Russia
* To whom correspondence should be addressed.
Received March 31, 2020; Revised May 29, 2020; Accepted May 31, 2020
Increased expression or amplification of HER2 receptor tyrosine kinase gene ERBB2 is well-known and widely used as a prognostic biomarker of breast cancer (BC) response to the targeted treatment with trastuzumab and its analogs. Considering that part of the BC patients overexpressing HER2 does not respond to trastuzumab, clinical trial NCT03521245 was initiated to identify additional gene expression and molecular pathway activation response biomarkers to trastuzumab treatment in HER2-positive BC. Using RNA sequencing gene expression in 23 formalin-fixed, paraffin embedded HER2 positive BC tissue blocks from patients who either responded or not responded to trastuzumab treatment was profiled. Differentially regulated genes and molecular pathways were identified in the groups of trastuzumab responders and non-responders. These results were next compared with the 42 previously published BC trastuzumab responder and non-responder RNA sequencing profiles from the clinical trials NCT00513292 and NCT00353483. No correlation was observed between the response status and the expression levels of ERBB2 gene in the HER2 positive BC samples. Analysis of the differentially expressed genes and molecular pathways in the combined dataset revealed 15/27 commonly up/down regulated genes and 15/25 pathways, respectively. However, only the intersection of molecular pathways upregulated in trastuzumab responders vs non-responders was statistically significantly enriched compared to the random expectation model. A classifier built using the most significantly upregulated molecular pathway – cAMP Pathway Protein Retention – demonstrated the best performance for prediction of the HER2 positive BC response to trastuzumab for both our experimental and previously reported data. This pathway also predicted time to recurrence in the combined dataset with Log-rank p-value 0.041.
KEY WORDS: breast cancer, HER2, ERBB2, transcriptomics, RNA sequencing, trastuzumab, targeted therapy, personalized medicine, NCT03521245DOI: 10.1134/S0006297920070044
Abbreviations: AUC, area under ROC-curve; BC, breast cancer; ER, estrogen receptor; PAL, pathway activation level; PR, progesterone receptor.
INTRODUCTION
Breast cancer (BC) research and treatment strategies have been largely impacted by the discovery of potent prognostic biomarkers such as overexpression or amplification of HER2 tyrosine kinase receptor, expression of estrogen (ER) and progesterone (PR) receptors. Breast tumors lacking significant ER and PR expression with basal HER2 level are referred as triple-negative BC. ERBB2 gene amplification and pathologically increased expression of its encoded protein HER2 is observed in 15-35% of all invasive BC tumors [1-3]. Amplification/overexpression of ERBB2/HER2 referred to as HER2 positive tumor phenotype strongly positively correlates with the patient response to trastuzumab, a HER2-specific monoclonal antibody used for targeted therapy of BC and gastric cancer [4]. The HER2-targeted trastuzumab therapy is usually recommended for HER2 positive cancers independently on ER and PR status, and important gains in survival has been reported in recent years for this treatment strategy. HER2-targeted therapy is often combined with conventional chemotherapy or with hormone therapy. Most of the HER2-negative tumors are resistant to HER2-specific drugs and their exclusion greatly decreases failure of the targeted therapy [5, 6].
However, clinical practice demonstrated that only 25-80% of women with HER2 positive BC (determined by immunohistochemistry and in situ hybridization) respond to trastuzumab [7-9]. In general, response rate was higher when trastuzumab was administered in combination with chemotherapy or other targeted therapeutics and lower when the patients had metastatic cancer [10].
Therefore, finding additional molecular biomarkers associated with trastuzumab response in HER2 positive BC is of great fundamental and practical interest. Molecular biomarkers can include diagnostically relevant mutations or gene expression levels measured at the mRNA or protein levels [11]. In addition, molecular pathway activation levels calculated based on high throughput gene expression data [5, 12, 13] can serve as a new generation biomarkers with enhanced diagnostic values, when compared with the single gene expression levels or mutations [14-17].
This improved biomarker quality of the molecular pathways is due to their fundamental property of aggregating individual gene expression data into a resulting metric termed pathway activation level (PAL). Functional association of multiple gene products in molecular pathways provides a theoretical basis for measuring an overall pathway activation metric instead of the single gene expression levels [5, 18]. In addition, it has been shown for many cancers that PALs can efficiently suppress artifacts or batch effects in gene expression levels by decreasing technical errors of major experimental platforms [19, 20]. Knowledge of molecular pathway activation can be converted into a clinically relevant estimation of individual tumor response to targeted therapeutics, e.g., cetuximab [21] and tyrosine kinase inhibitors [22-24].
Clinical trial NCT03521245 was initiated to identify additional gene expression and molecular pathway activation biomarkers of trastuzumab response in HER2 positive BC. RNA sequencing was used to establish gene expression levels as a gold standard approach in cancer transcriptomics [11].
MATERIALS AND METHODS
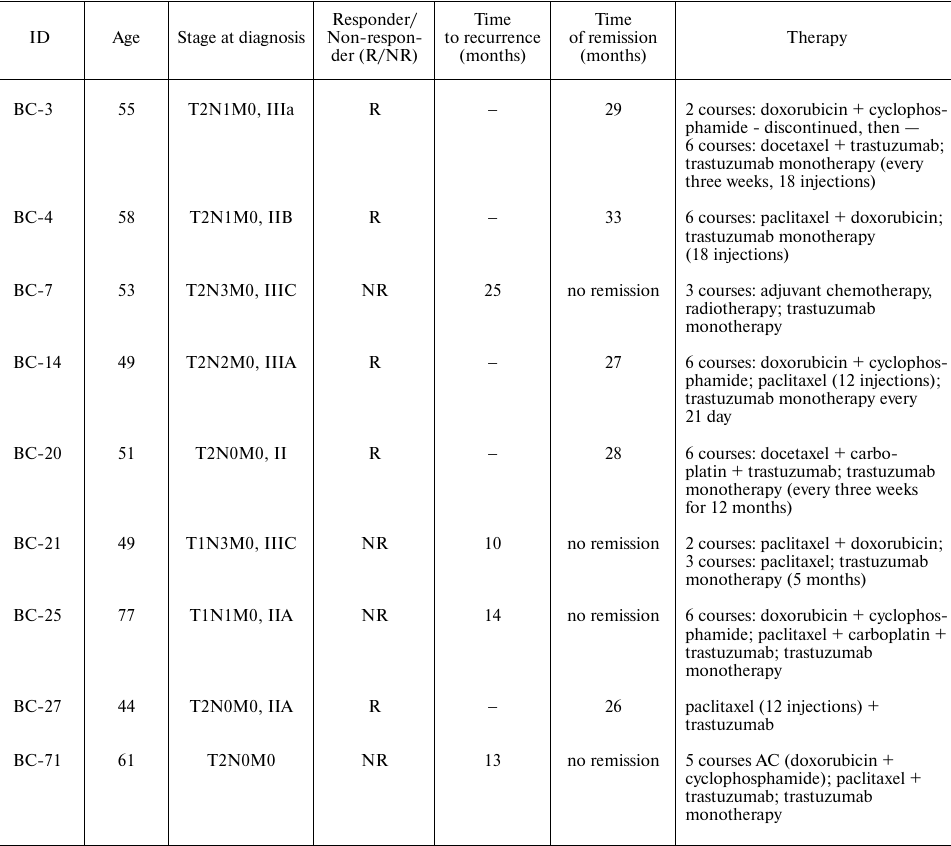
Biological samples from 23 women patients with mean age of 53.3 years (range 33-77 years old), who were diagnosed with HER2-positive BC and received either trastuzumab monotherapy or trastuzumab in combination with other anticancer drugs: docetaxel/paclitaxel + carboplatin/paclitaxel/docetaxel + carboplatin/aromatase inhibitor/pertuzumab/capecitabine/vinorelbine/gemcitabine, were included in the study. Clinical annotations of the investigated BC biosamples are summarized in Table S1 of the Supplement. Totally, nineteen patients received adjuvant therapy and four patients received neoadjuvant therapy.
All biosamples were formalin fixed, paraffin embedded (FFPE) tumor tissue blocks. Prior to further analysis, all biosamples were examined by pathologist to confirm the tumor tissue origin, and only specimens with the content of tumor cells greater than 70% were investigated. Among them, 19 breast cancer (BC) tissue samples were obtained from the Karelia Republic Oncological Hospital, Petrozavodsk, and 4 sample from the Vitamed Oncological Clinical Center, Moscow. Majority (21) of the biosamples were fragments of primary tumors and 2 represented node metastases. The complete dataset included RNA sequencing profiles for 23 patients with breast cancer, of which only nine met the following inclusion criteria: adjuvant therapy, negative ER/PR-status, patient had metastasis. Luminal B breast cancer subtype was excluded from the further analysis, because it was reported to have better prognosis than the rest of the HER2-positive subtypes [25, 26]. Immunohistochemical assays of the BC samples for HER2, ER, and PR proteins were done at Oncological Dispensary of the Republic of Karelia clinical diagnostic laboratory using antibody kits (Roche Diagnostics, USA) to identify the respective tumor status. For HER2, the output statuses were: (i) baseline staining (0), (ii) “+” (1), (iii) “++” (2) and (iv) “+++” (3). The “++” and “+++” statuses were confirmed using ISH DNA Probe Cocktail assay (Roche Diagnostics). For ER and PR status, scale 0-8 was used.
Nine patients selected for further analysis were women with HER2-positive ER/PR-negative BC, the mean age was 55.2 years (range 44-77 y.o.), who received adjuvant trastuzumab monotherapy or trastuzumab in combination with other therapeutics: docetaxel/paclitaxel + carboplatin/paclitaxel/docetaxel + carboplatin/capecitabine/vinorelbine/gemcitabine (Table 1). Of them, eight BC tissue samples were obtained from the Karelia Republic Oncological Hospital, Petrozavodsk, Russia, and one sample from the Vitamed Oncological Clinical Center, Moscow, Russia. Eight samples corresponded to primary tumors and one to lymph node metastasis (BC-20). The metastatic sample was included into the analysis because we aimed at identification of robust trastuzumab resistance biomarkers, which could potentially be applicable not only for biopsies derived from the primary site but also for metastases. The patients were considered responders if remission was observed for at least 25 months after biopsy. Totally, five patients were classified as the responders and four as the non-responders.
Table 1. Clinical and molecular annotation
of nine HER2+ BC patients who received adjuvant therapy, were
ER/PR-negative, and had metastases
Preparation of libraries and RNA sequencing. To isolate RNA, 10 µm-thick slices were cut from each FFPE tissue block using a microtome. RNA was extracted from FFPE slices using a QIAGEN RNeasy FFPE Kit according to the manufacturer’s protocol. RNA 6000 Nano Assay kits were used to measure RNA concentration. RNA Integrity Number (RIN) was evaluated using an Agilent 2100 bio-Analyzer. For depletion of ribosomal RNA and library construction, KAPA RNA Hyper with rRNA erase kit (HMR only) was used. Different adapters were used for multiplexing samples in one sequencing run. Library concentrations and quality were measured using a Qubit ds DNA HS Assay kit (Life Technologies, USA) and Agilent Tapestation (Agilent, USA). Single-end RNA sequencing, 50 bp read length, for approximately 30 million raw reads per sample was performed at the Omicslab LLC, Moscow, and at the Department of Pathology and Laboratory Medicine, University of California Los Angeles, using an Illumina HiSeq 3000 equipment. Data quality check was done using an Illumina SAV and de-multiplexing was performed with an Illumina Bcl2fastq2 v 2.17 software. Sequencing data were deposited to the NCBI Sequencing Read Archive (SRA) under accession ID PRJNA565016 and PRJNA578290.
Processing of RNA sequencing data. RNA sequencing FASTQ files were processed with STAR aligner [27] in “GeneCounts” mode with the Ensembl human transcriptome annotation (Build version GRCh38 and transcript annotation GRCh38.89). Ensembl gene IDs were converted to HGNC gene symbols using the Complete HGNC dataset (https://www.genenames.org/, database version from July 13, 2017). In total, expression levels were established for 36,596 annotated genes with the corresponding HGNC identifiers. Minimum number of uniquely mapped reads was 1.40 million for studied biosamples, with the mean value of 7.35 mln (8.03 for all 23 samples). Differential gene expression analysis was done using the DESeq2 software [28] and DESeq2-normalized counts of differentially expressed genes between responders and non-responders were used for further analysis.
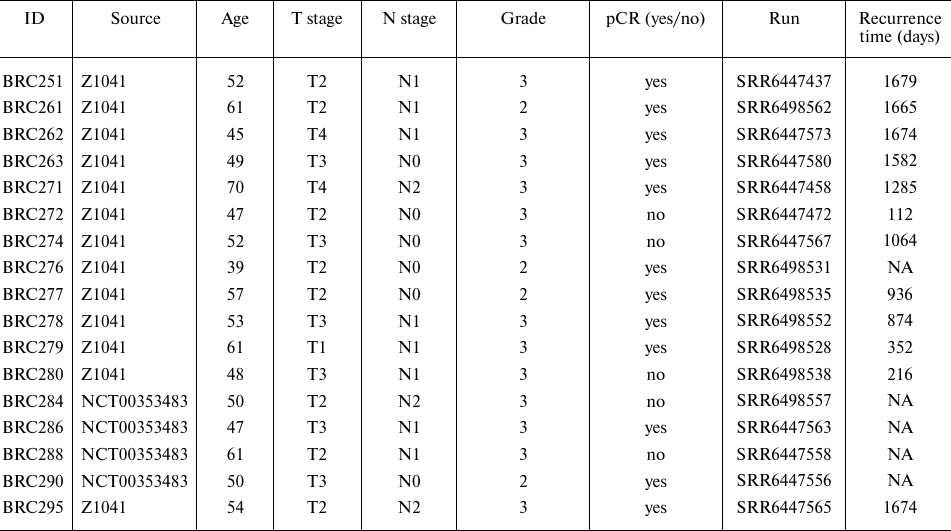
Literature gene expression data. The previously reported gene expression data for BC patients were extracted from the Database of Genotypes and Phenotypes (dbGAP), accession ID: phs001291.v1.p1, published in [4]. The patients were women with invasive breast cancer, not pregnant, mean age was 50.14 years (range 36-70 years old), who received neoadjuvant chemotherapy and trastuzumab. 31 cases were accessed from the American College of Surgeons Oncology Group Z1041 trial (NCT00513292) that compared the pCR rate of patients with HER2-positive breast cancer. Patients were randomly assigned to one of two arms: the first received fluorouracil 500 mg/m2, epirubicin 75 mg/m2, and cyclophosphamide 500 mg/m2 (FEC-75) on day 1 of a 21-day cycle for four cycles followed by paclitaxel 80 mg/m2 and trastuzumab 2 mg/kg (after a 4-mg/kg loading dose) once per week for 12 weeks, while the second group received paclitaxel and trastuzumab once a week for 12 weeks followed by four cycles of FEC-75 (on day 1 of each 21-day cycle) and once-weekly trastuzumab at the same doses as the first group. In addition to the patients enrolled in the Z1041 trial, eleven HER2-positive breast cancer cases included in the phs001291.v1.p1 were enrolled in a local trial at Washington University School of Medicine (NCT00353483) and received neoadjuvant treatment with trastuzumab in combination with other chemotherapeutics: trastuzumab + paclitaxel + carboplatin, or doxorubicin + carboplatin then trastuzumab + paclitaxel, or FEC then trastuzumab + paclitaxel [4].
All cases were identified as HER2-positive by immunohistochemistry and/or by fluorescence in situ hybridization (FISH). RNA samples were extracted from the pre-treatment snap-frozen tumor biopsies of BC patients and sequenced. The complete published dataset included RNA sequencing profiles for 42 patients with breast cancer (Table S2 in the Supplement), of which seventeen met the inclusion criteria of double negative hormonal ER and PR status.
For twelve of these profiles, the patients demonstrated a pathological complete response (pCR) after treatment, while five patients showed residual disease as the primary outcome. Clinical annotation of the biosamples and outcomes are summarized in Table 2.
Table 2. Clinical and molecular annotation
of seventeen HER2+, hormone-negative BC biosamples from
phs001291.v1.p1
Molecular pathway analysis. Pathway activation levels (PALs) were calculated using Oncobox method and software [29]. For the analysis 1754 molecular pathways containing each at least 10 gene products were extracted from the public databases Reactome [30], NCI Pathway Interaction Database [31], Kyoto Encyclopedia of Genes and Genomes [32], HumanCyc [33], Biocarta [34], and Qiagen Pathway Central and listed in Table S3 in the Supplement.
For calculating PALs, all the experimental BC gene expression profiles for the responders were normalized to the non-responders profiles. Similarly, all the literature BC gene expression profiles for the responders were normalized to the non-responders profiles. Molecular pathways were visualized using the Oncobox pathway visualization/reconstruction tool [16, 29, 35-37].
RESULTS
In this study we compared differential gene expression and molecular pathways activation levels between the experimental and literature BC samples with better and worse responses to trastuzumab treatment.
Clinical data. We used experimental data obtained during observational clinical investigation NCT03521245 “Molecular pathway activation markers predicting efficacy of trastuzumab therapy for HER2-positive breast cancer”. The patients were residents of north-western and central parts of the Russian Federation who underwent treatment in Vitamed Oncological Clinic, Moscow, or in Oncological Dispensary of Karelia Republic, Petrozavodsk, Russia. The inclusion criteria were: adult females with histologically confirmed HER2 positive breast cancer and available formalin fixed paraffin embedded (FFPE) samples of BC tissue; patients treated with Trastuzumab alone or in combination(s) with other chemotherapy regimens with known outcome according to RECIST 1.1; patients with BC stage II or higher who have signed an informed consent to participate in this clinical trial. The exclusion criterion was presence of less than 70% of intact tumor cells in the available FFPE BC tissue samples (Fig. 1). Totally, 23 patients were enrolled, and the respective tumor tissue specimens were profiled by RNA sequencing (Table S1 in the Supplement). The mean age of the patients was 53.3 years. The tumor response criteria used was remission for 25 months since the biopsy. The patients were then classified accordingly as either responders or non-responders to treatment with trastuzumab containing therapies. However, we could not directly compare trastuzumab response statuses for all the patients included because they had different types of HER2+ breast cancer (HER2+ luminal B type, or HER2+ ER–/PR– type). These two types of BC have different prognosis [25, 26] and it seems inadequate to compare them directly. For further analysis we selected a cohort of patients who received adjuvant therapy, had ER/PR-negative BC, and had metastases; in total five treatment responders and four non-responders were selected. All patients received trastuzumab in various combinations. The most common combinations included taxanes (paclitaxel or docetaxel, n = 7) and doxorubicin (n = 6, Table 1).
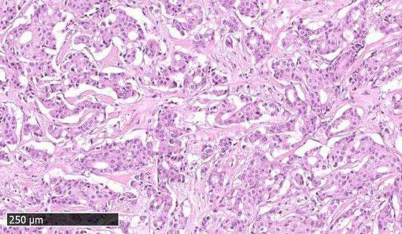
Fig. 1. Representative histological image of breast cancer tissue under investigation. Microphotograph of hematoxylin and eosin stained sample showed moderately differentiated infiltrative ductal cancer (BC71 HER2+/ER–/PR–).
The previously published [4] collection of 42 RNA sequencing BC gene expression profiles available in the Database of Genotypes and Phenotypes (dbGAP) with accession ID phs001291.v1.p1 (Table S2 in the Supplement) was used for comparison.
The patients were females with invasive BC, mean age of the patients was 50.14 years. Only samples corresponding to the patients with ER/PR negative BC were selected for further analysis. Based on the outcomes, the patients were also classified as trastuzumab responders (pathological complete response) and non-responders (residual disease). In total, seventeen patient profiles were included corresponding to twelve responders and five non-responders (Table 2).
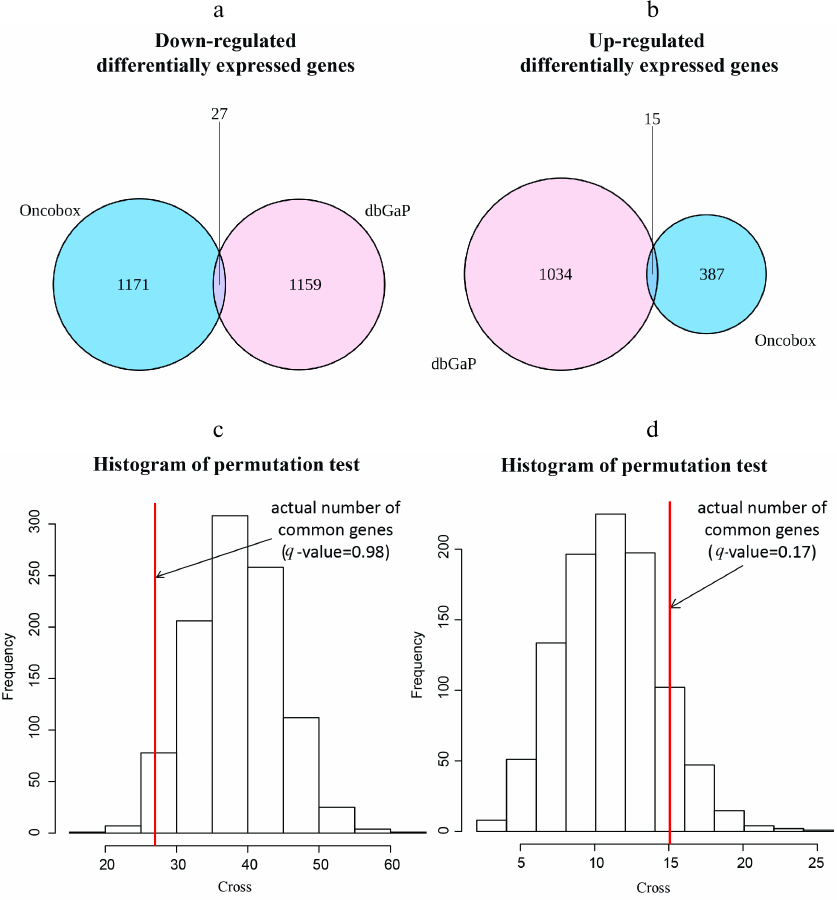
Differentially regulated genes and molecular pathways. In both experimental and literature datasets, we identified differentially expressed genes between the trastuzumab responder and non-responder tumors using the DESeq2 software. In total, 1600 genes passed threshold p-value < 0.05 for the experimental dataset (402 genes were upregulated and 1198 downregulated in the responders group). In the literature dataset, we found 2235 differentially expressed genes (p < 0.05), where 1049 genes were upregulated and 1186 downregulated in the non-responders group), Table S4 in the Supplement. Further intersection of differentially regulated gene sets for both experimental and literature data identified in total 15 commonly upregulated and 27 downregulated genes in the trastuzumab responders group (Fig. 2, a and b). Next statistical significance of this intersection by randomly permutating gene names and by intersecting random gene groups of the same sizes as above for the actual data was examined. It was found that there was no statistically significant enrichment in the number of intersecting genes for both upregulated and downregulated genes, which implied that commonly regulated genes could appear due to a random coincidence (Fig. 2, c and d). Expression of the ERBB2 gene encoding HER2 protein, a molecular target of trastuzumab, was also compared between the trastuzumab responders and non-responders; no significant difference between these groups in both datasets was observed (Fig. S1 in the Supplement).
Fig. 2. Intersection of differentially expressed genes between the experimental and literature datasets. Venn diagrams show numbers of downregulated (a) and upregulated (b) differentially expressed genes between the experimental dataset (NCT03521245) and the literature dataset (dbGAP accession: phs001291.v1.p1). Histograms (c, d) show actual numbers of differentially regulated genes and random permutation models, number of cycles = 1000. c) Commonly downregulated differentially expressed genes, q-value of observing higher number of intersecting genes is 0.98.d) Commonly upregulated differentially expressed genes, q-value of observing higher number of intersecting genes is 0.17.
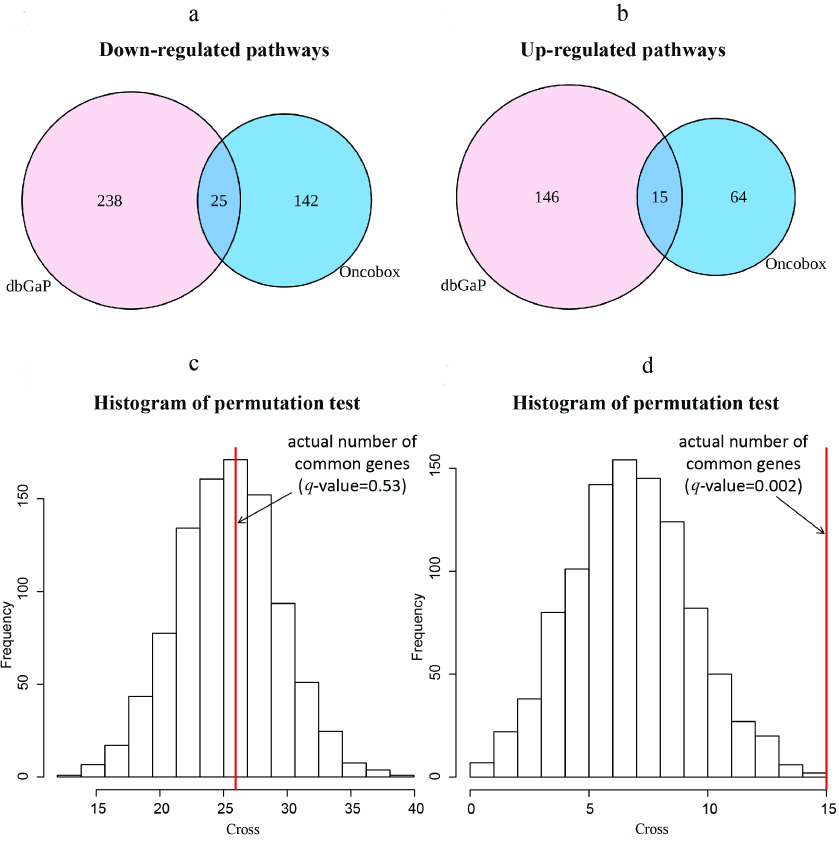
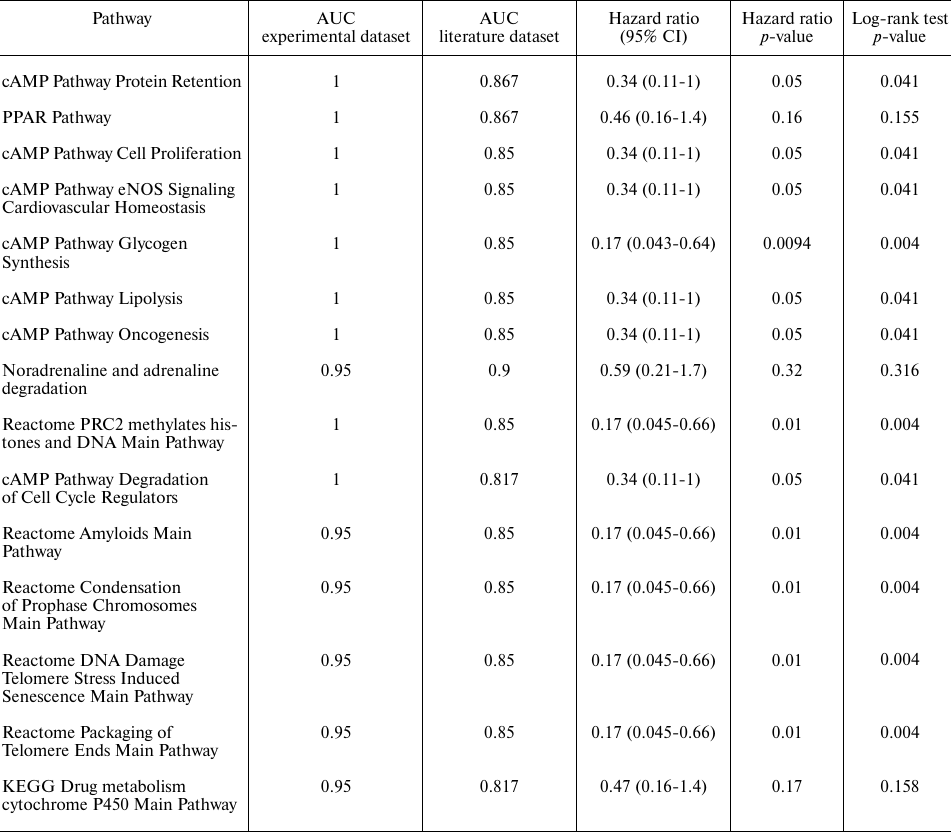
Pathway activation level (PAL) values for 1754 intracellular molecular pathways containing each at least 10 gene products was calculated for the differentially regulated gene sets using the Oncobox software [29]. Statistically significant differentially regulated molecular pathways (p < 0.05) were identified including 246 pathways for the experimental dataset (79 upregulated and 167 downregulated in the trastuzumab responders group) and 424 pathways for the literature dataset (161 upregulated and 263 downregulated in the trastuzumab responders group), Table S5 in the Supplement. Next, we intersected commonly regulated differential pathways and observed a highly likely random intersection for the 25 downregulated pathways, but non-random, significantly enriched intersection for the 15 upregulated pathways in the responders group, p = 0.002 (Fig. 3).
Fig. 3. Intersection of differentially regulated molecular pathways between the experimental and literature datasets. Venn diagrams show numbers of downregulated (a) and upregulated (b) differential pathways between the experimental dataset (NCT03521245) and the literature dataset (dbGAP accession: phs001291.v1.p1). Histograms (c, d) show actual numbers of differentially regulated pathways and random permutation models, number of cycles = 1000. c) Commonly downregulated differentially expressed pathways, q-value of observing higher number of intersecting pathways is 0.53. d) Commonly upregulated differentially expressed pathways, q-value of observing higher number of intersecting pathways is 0.002
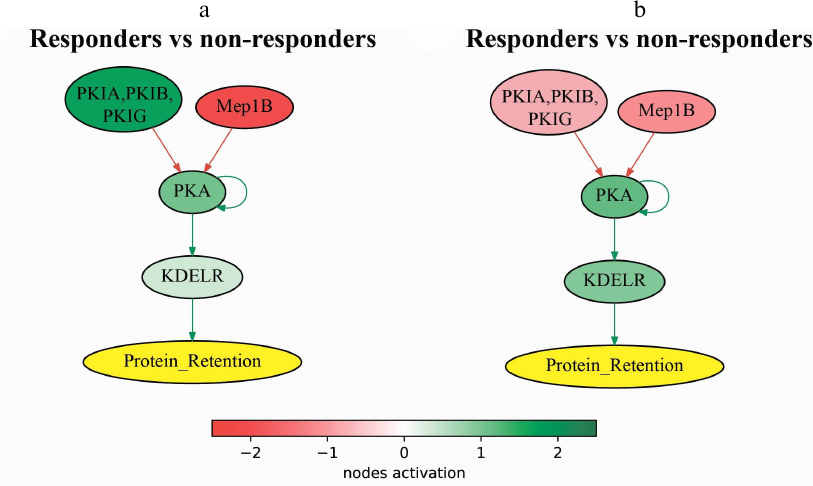
Among the latter fifteen molecular pathways (Table 3), the following two were the most statistically significantly (Wilcoxon rank sum test) upregulated in the trastuzumab responders group: PPAR Pathway and cAMP Protein Retention Pathway. Pathway activation profile for the cAMP Protein Retention Pathway, which is also associated with the progression-free survival in further analysis, is provided for trastuzumab responders and non-responders in Fig. 4.
Fig. 4. Pathway activation profile of cAMP Pathway Protein Retention in the trastuzumab responders normalized to the non-responders data in (a) experimental dataset (NCT03521245) and (b) literature dataset (dbGAP accession: phs001291.v1.p1).
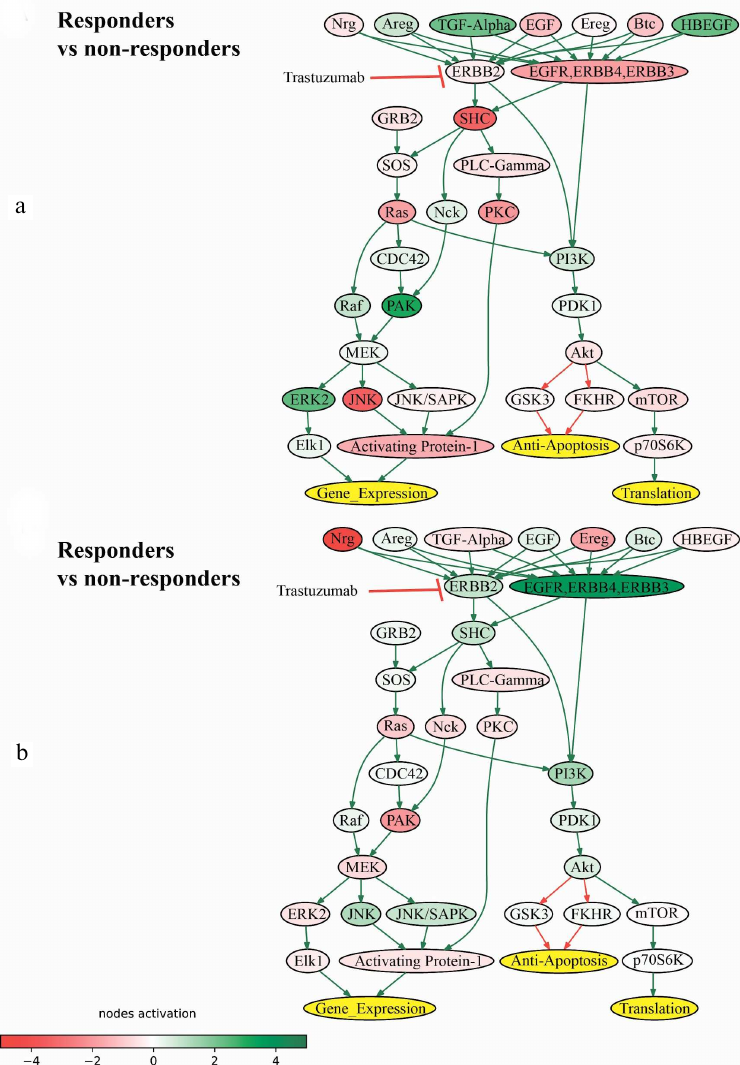
In addition, we identified the ErbB Family Pathway that included ERBB2 gene product among the intersected molecular pathways that were downregulated in the trastuzumab responders group (Fig. 5). However, as was shown previously, the intersection of the downregulated pathways between the datasets could be due to coincidence (Fig. 3c).
Fig. 5. Pathway activation profile of ErbB Family Pathway in the trastuzumab responders normalized to the non-responders data in (a) experimental dataset (NCT03521245) and (b) literature dataset (dbGAP accession: phs001291.v1.p1).
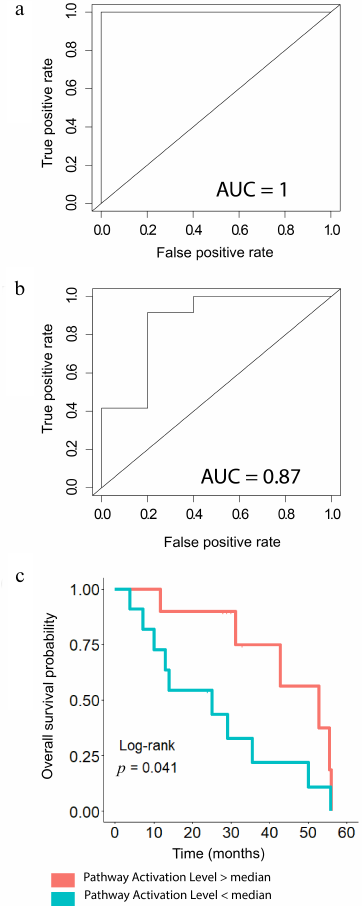
Hazard Ratio and Log-rank test p-values for each of the fifteen pathways upregulated in the trastuzumab responders group was calculated next to analyze their abilities to discriminate between the trastuzumab treatment responders and non-responders. To perform this analysis, we pooled patients with the time to progression information available from the both datasets. The patients were then divided into two groups that had the pathway activation levels higher and lower than the median value, respectively. It was established that all the pathways tested could be predictive of the progression-free survival, except the PPAR, noradrenaline and adrenaline degradation and Drug metabolism cytochrome P450 pathways, for which Log-rank p-value exceeded 0.05 (Table 3). The cAMP Protein Retention pathway showed the best performance for both discriminating trastuzumab responders from non-responders and for predicting time to tumor recurrence (Fig. 6). To evaluate that we calculated area under the receiver-operating characteristic curve (AUC) value, which is the universal characteristic of biomarker robustness depending on its sensitivity and specificity [38]. This statistical approach is broadly applicable to different types of biomarkers in oncology [14, 16, 17, 39-41]. AUC positively correlates with the quality of a biomarker and varies from 0.5 to 1. The standard discrimination threshold is 0.7 and the parameters with greater AUC are considered high-quality biomarkers, and vice versa [42]. The top ranked molecular pathway was the cAMP Pathway Protein Retention, which showed the highest AUC and the lowest hazard ratio observed [AUC 1 for the experimental dataset and 0.867 for the literature dataset; hazard ratio 0.34 (95% CI: 0.11-1, p-value = 0.05)], Table 3. Total number of genes per pathway, as well as the number of differentially expressed genes in both sets are provided in Table S6 in the Supplement.
Fig. 6. Predictive power of the cAMP Pathway Protein Retention for BC tumor response and time to progression after trastuzumab therapy. Area under the ROC curve for separation of the responders and non-responders using the cAMP Pathway Protein Retention pathway activation level for (a) experimental patients and (b) patients from the literature dataset, dbGAP accession: phs001291.v1.p1. (c) Predictive power of the cAMP Pathway Protein Retention pathway activation level for the progression-free survival in BC patients from both experimental and literature datasets. Cohorts of patients with pathway activation level lower or higher than the median are shown separately. Hazard ratio = 0.34 (95%CI: 0.11-1, p-value = 0.05).
Table 3. Characteristics of biomarker
quality for fifteen intersected non-random differentially upregulated
pathways in the trastuzumab responder groups in both groups of
patients
DISCUSSION
Gene expression in 23 HER2 positive BC tissue samples from patients who either responded or not responded to trastuzumab treatment was examined. Differentially regulated genes and molecular pathways were identified in both groups and compared with the 42 published BC trastuzumab responder and non-responder RNA sequencing profiles from the previous clinical trials NCT00513292 and NCT00353483 [4]. No correlation between the response status and the expression levels of ERBB2 gene in the HER2 positive BC samples was observed. 15/27 commonly up/down regulated genes and 15/26 pathways, respectively, were identified in the combined dataset. However, only the intersection of molecular pathways upregulated in the trastuzumab responders vs non-responders was statistically significantly enriched compared to the random expectation model. A classifier built using the most significant downregulated molecular pathway – cAMP Pathway Protein Retention – showed quality of AUC = 1 and 0.87 for prediction of HER2 positive BC response to trastuzumab in our experimental and literature data, respectively. This pathway also predicted time to recurrence in a combined dataset with Log-rank p-value 0.041.
While numerous studies examining gene expression in the BC samples from patients treated with trastuzumab have been published [40, 43, 44], only a minor fraction of them contained therapy response status linked to the publicly available expression profile for the clinical cases reported and the longitudinal survival data. Moreover, most of the studies on trastuzumab resistant phenotype used microarray technology for tumor profiling [45-47]. Here we used RNA sequencing, which is now recognized as a gold standard method for profiling gene expression [48, 49]. In addition, we used total RNA sequencing without poly(A) enrichment, thus our dataset could be used also to investigate non-coding RNAs association with trastuzumab resistance in future studies. Finally, this is the only dataset of BC samples associated with the trastuzumab response, which is fully compatible with the database of healthy normal tissues, because we used the same experimental protocol as for the previous ANTE database [50].
Expression of genes and pathways that distinguished trastuzumab responders and non-responders in our experimental data and in the previously published clinically annotated RNA sequencing dataset were compared [4]. The earlier trial aimed to identify biomarkers of trastuzumab response on the genomic and transcriptomic level. The authors did not find any gene or gene signature associated with the trastuzumab response based on the RNA sequencing data [4]. This is probably due to the fact that the authors did not stratify patients according to the ER and PR hormone receptor status, as we did in the present study. While the number of common differentially expressed genes in both datasets was probably random, the actual number of significantly upregulated pathways was significantly higher than expected from the analysis of random permutations. In particular, the best association with trastuzumab sensitivity was observed for a branch of cyclic AMP pathway that activates protein retention (Table 3).
Cyclic AMP is the first identified secondary messenger, which has a fundamental role in the cellular response to various extracellular stimuli [51]. One function of cAMP molecular pathway is activation of KDELR (KDEL (Lys-Asp-Glu-Leu) Endoplasmic Reticulum Protein Retention Receptor) to promote retrieval of proteins (protein retention) from Golgi complex to endoplasmic reticulum, thereby maintaining steady state of the cell [52]. It was shown previously that alterations in such retrograde trafficking could drive breast cancer metastasis through receptor mislocalization [53]. In addition, intrinsic defects in the endocytic machinery responsible for HER2 downregulation correlate with unresponsiveness to trastuzumab therapy [54]. However, to the best of our knowledge the present study is the first to support the plausible link between the retrograde trafficking and resistance to trastuzumab.
Previous studies showed several other potential mechanisms of trastuzumab resistance in breast cancer [55]. First, other HER family receptors could be associated with such resistance; for example, HER2 positive patients that are negative for HER3 have mostly metastasis-free survival [56]. Non-HER family receptors such as FGFR and IGF-1R can also play a significant role in trastuzumab resistance [57, 58]. Androgen receptor (AR) could be also involved, because AR expression negatively correlates with the immune cells infiltration, which, in turn, is associated with trastuzumab resistance [59, 60].
Based on the data generated in this study, activation level of the “cAMP Pathway Protein Retention” pathway appeared to be a robust biomarker that separates trastuzumab responders from non-responders in both our experimental and previously published datasets. Moreover, activation of this pathway is associated with favorable prognosis in terms of predicting time to tumor recurrence. Thus, activation level of this pathway could be potentially used for further development of companion diagnostic tools for early detection of non-responders to HER2-targeted therapies to avoid ineffective treatment. This pathway contains 14 genes; hence, such diagnostic tool may be based on rt-PCR, targeted RNA sequencing, NanoString assay, or any other platform for gene expression profiling. However, further clinical investigations of this approach are needed to validate findings presented in this manuscript.
Funding. This study was financially supported by the OmicsWay research program in oncology and by the Russian Science Foundation (project No. 18-15-00061, M. Su., M. So., U. V., and A. B.).
Ethics declarations. For all the biosamples, informed written consents were collected from the patients to participate in the clinical trial NCT03521245 “Molecular pathway activation markers predicting efficacy of trastuzumab therapy for HER2-positive breast cancer” and to communicate the results in this report. All procedures performed in studies involving human participants were in accordance with the ethical standards of the Karelia Republic Oncological Hospital, Petrozavodsk, and the Vitamed Oncological Clinical Center, Moscow.
The authors M. Sorokin and A. Buzdin were employed by the company OmicsWay Corp. The authors declare that this study received funding from OmicsWay Corp. The funder had the following involvement in the study: data analysis and interpretation as well as the writing of this manuscript. The funder was not involved in the study design, collection of data, decision to submit this article for publication. The remaining authors declare no conflict of interest in financial or any other sphere.
Electronic supplementary material. Supplementary materials are available in the electronic version on the journal website (http://protein.bio.msu.ru/biokhimiya) and Springer site (Link.springer.com).
REFERENCES
1.Press, M. F., Bernstein, L., Thomas, P. A.,
Meisner, L. F., Zhou, J. Y., Ma, Y., Hung, G., Robinson, R. A., Harris,
C., El-Naggar, A., Slamon, D. J., Phillips, R. N., Ross, J. S., Wolman,
S. R., and Flom, K. J. (1997) HER-2/neu gene amplification
characterized by fluorescence in situ hybridization: poor
prognosis in node-negative breast carcinomas, J. Clin. Oncol.,
15, 2894-2904, doi: 10.1200/JCO.1997.15.8.2894.
2.Slamon, D. J., Clark, G. M., Wong, S. G., Levin, W.
J., Ullrich, A., and McGuire, W. L. (1987) Human breast cancer:
correlation of relapse and survival with amplification of the HER-2/neu
oncogene, Science, 235, 177-182,
doi: 10.1126/science.3798106.
3.Hammond, M. E. H., Hayes, D. F., Dowsett, M.,
Allred, D. C., Hagerty, K. L., et al. (2010) American Society of
Clinical Oncology/College of American Pathologists guideline
recommendations for immunohistochemical testing of estrogen and
progesterone receptors in breast cancer, Arch. Pathol. Lab.
Med., 134, 907-922,
doi: 10.1043/1543-2165-134.6.907.
4.Lesurf, R., Griffith, O. L., Griffith, M., Hundal,
J., Trani, L., Watson, M. A., Aft, R., Ellis, M. J., Ota, D., Suman, V.
J., Meric-Bernstam, F., Leitch, A. M., Boughey, J. C., Unzeitig, G.,
Buzdar, A. U., Hunt, K. K., and Mardis, E. R. (2017) Genomic
characterization of HER2-positive breast cancer and response to
neoadjuvant trastuzumab and chemotherapy-results from the ACOSOG Z1041
(Alliance) trial, Ann. Oncol., 28, 1070-1077,
doi: 10.1093/annonc/mdx048.
5.Buzdin, A., Sorokin, M., Garazha, A., Sekacheva,
M., Kim, E., Zhukov, N., Wang, Y., Li, X., Kar, S., Hartmann, C.,
Samii, A., Giese, A., and Borisov, N. (2018) Molecular pathway
activation – new type of biomarkers for tumor morphology and
personalized selection of target drugs, Semin. Cancer Biol.,
53, 110-124, doi: 10.1016/j.semcancer.2018.06.003.
6.Artemov, A., Aliper, A., Korzinkin, M., Lezhnina,
K., Jellen, L., Zhukov, N., Roumiantsev, S., Gaifullin, N.,
Zhavoronkov, A., Borisov, N., and Buzdin, A. (2015) A method for
predicting target drug efficiency in cancer based on the analysis of
signaling pathway activation, Oncotarget, 6, 29347-29356,
doi: 10.18632/oncotarget.5119.
7.Pagani, O., Klingbiel, D., Ruhstaller, T.,
Nolè, F., Eppenberger, S., et al. (2017) Do all patients with
advanced HER2 positive breast cancer need upfront-chemo when receiving
trastuzumab? Randomized phase III trial SAKK 22/99, Ann. Oncol.,
28, 305-312, doi: 10.1093/annonc/mdw622.
8.Schmid, S., Klingbiel, D., Aebi, S., Goldhirsch,
A., Mamot, C., Munzone, E., Nolè, F., Oehlschlegel, C., Pagani,
O., Pestalozzi, B., Rochlitz, C., Thürlimann, B., von Moos, R.,
Weder, P., Zaman, K., and Ruhstaller, T. (2019) Long-term responders to
trastuzumab monotherapy in first-line HER-2+ advanced breast cancer:
characteristics and survival data, BMC Cancer, 19, 902,
doi: 10.1186/s12885-019-6105-3.
9.Zhu, X., and Verma, S. (2015) Targeted therapy in
HER2-positive metastatic breast cancer: a review of the literature,
Curr. Oncol., 22, S19-28,
doi: 10.3747/co.22.2363.
10.Slamon, D. J., Leyland-Jones, B., Shak, S.,
Fuchs, H., Paton, V., Bajamonde, A., Fleming, T., Eiermann, W., Wolter,
J., Pegram, M., Baselga, J., and Norton, L. (2001) Use of chemotherapy
plus a monoclonal antibody against HER2 for metastatic breast cancer
that overexpresses HER2, N. Engl. J. Med., 344, 783-792,
doi: 10.1056/NEJM200103153441101.
11.Buzdin, A., Sorokin, M., Garazha, A., Glusker,
A., Aleshin, A., Poddubskaya, E., Sekacheva, M., Kim, E., Gaifullin,
N., Giese, A., Seryakov, A., Rumiantsev, P., Moshkovskii, S., and
Moiseev, A. (2019) RNA sequencing for research and diagnostics in
clinical oncology, Semin. Cancer Biol., doi:
10.1016/j.semcancer.2019.07.010.
12.Artcibasova, A. V., Korzinkin, M. B., Sorokin, M.
I., Shegay, P. V., Zhavoronkov, A. A., Gaifullin, N., Alekseev, B. Y.,
Vorobyev, N. V., Kuzmin, D. V., Kaprin, A. D., Borisov, N. M., and
Buzdin, A. A. (2016) MiRImpact, a new bioinformatic method using
complete microRNA expression profiles to assess their overall influence
on the activity of intracellular molecular pathways, Cell Cycle,
15, 689-698, doi: 10.1080/15384101.2016.1147633.
13.Buzdin, A. A., Prassolov, V., Zhavoronkov, A. A.,
and Borisov, N. M. (2017) Bioinformatics meets biomedicine: OncoFinder,
a quantitative approach for interrogating molecular pathways using gene
expression data, Methods Mol. Biol., 1613, 53-83,
doi: 10.1007/978-1-4939-7027-8_4.
14.Borisov, N. M., Terekhanova, N. V., Aliper, A.
M., Venkova, L. S., Smirnov, P. Y., Roumiantsev, S., Korzinkin, M. B.,
Zhavoronkov, A. A., and Buzdin, A. A. (2014) Signaling pathways
activation profiles make better markers of cancer than expression of
individual genes, Oncotarget, 5, 10198-10205,
doi: 10.18632/oncotarget.2548.
15.Buzdin, A., Sorokin, M., Poddubskaya, E., and
Borisov, N. (2019) High-throughput mutation data now complement
transcriptomic profiling: advances in molecular pathway activation
analysis approach in cancer biology, Cancer Inform., 18,
1176935119838844, doi: 10.1177/1176935119838844.
16.Zolotovskaia, M. A., Sorokin, M. I., Roumiantsev,
S. A., Borisov, N. M., and Buzdin, A. A. (2018) Pathway instability is
an effective new mutation-based type of cancer biomarkers, Front.
Oncol., 8, 658, doi: 10.3389/fonc.2018.00658.
17.Lezhnina, K., Kovalchuk, O., Zhavoronkov, A. A.,
Korzinkin, M. B., Zabolotneva, A. A., Shegay, P. V., Sokov, D. G.,
Gaifullin, N. M., Rusakov, I. G., Aliper, A. M., Roumiantsev, S. A.,
Alekseev, B. Y., Borisov, N. M., and Buzdin, A. A. (2014) Novel robust
biomarkers for human bladder cancer based on activation of
intracellular signaling pathways, Oncotarget, 5,
9022-9032, doi: 10.18632/oncotarget.2493.
18.Ozerov, I. V., Lezhnina, K. V., Izumchenko, E.,
Artemov, A. V., Medintsev, S., Vanhaelen, Q., Aliper, A., Vijg, J.,
Osipov, A. N., Labat, I., West, M. D., Buzdin, A., Cantor, C. R.,
Nikolsky, Y., Borisov, N., Irincheeva, I., Khokhlovich, E., Sidransky,
D., Camargo, M. L., and Zhavoronkov, A. (2016) In silico Pathway
Activation Network Decomposition Analysis (iPANDA) as a method for
biomarker development, Nat. Commun., 7, 13427,
doi: 10.1038/ncomms13427.
19.Buzdin, A. A., Zhavoronkov, A. A., Korzinkin, M.
B., Roumiantsev, S. A., Aliper, A. M., Venkova, L. S., Smirnov, P. Y.,
and Borisov, N. M. (2014) The OncoFinder algorithm for minimizing the
errors introduced by the high-throughput methods of transcriptome
analysis, Front. Mol. Biosci., 1, 8,
doi: 10.3389/fmolb.2014.00008.
20.Borisov, N., Suntsova, M., Sorokin, M., Garazha,
A., Kovalchuk, O., et al. (2017) Data aggregation at the level of
molecular pathways improves stability of experimental transcriptomic
and proteomic data, Cell Cycle, 16, 1810-1823,
doi: 10.1080/15384101.2017.1361068.
21.Zhu, Q., Izumchenko, E., Aliper, A. M., Makarev,
E., Paz, K., Buzdin, A. A., Zhavoronkov, A. A., and Sidransky, D.
(2015) Pathway activation strength is a novel independent prognostic
biomarker for cetuximab sensitivity in colorectal cancer patients,
Hum. Genome Var., 2, 15009,
doi: 10.1038/hgv.2015.9.
22.Poddubskaya, E. V., Baranova, M. P., Allina, D.
O., Sekacheva, M. I., Makovskaia, L. A., Kamashev, D. E., Suntsova, M.
V., Barbara, V. S., Kochergina-Nikitskaya, I. N., and Aleshin, A. A.
(2019) Personalized prescription of imatinib in recurrent granulosa
cell tumor of the ovary: case report, Cold Spring Harb. Mol. Case
Stud., 5, doi: 10.1101/mcs.a003434.
23.Poddubskaya, E. V., Baranova, M. P., Allina, D.
O., Smirnov, P. Y., Albert, E. A., Kirilchev, A. P., Aleshin, A. A.,
Sekacheva, M. I., and Suntsova, M. V. (2018) Personalized prescription
of tyrosine kinase inhibitors in unresectable metastatic
cholangiocarcinoma, Exp. Hematol. Oncol., 7, 21,
doi: 10.1186/s40164-018-0113-x.
24.Poddubskaya, E., Bondarenko, A., Boroda, A.,
Zotova, E., Glusker, A., Sletina, S., Makovskaia, L., Kopylov, P.,
Sekacheva, M., Moisseev, A., and Baranova, M. (2019)
Transcriptomics-guided personalized prescription of targeted
therapeutics for metastatic ALK-positive lung cancer case following
recurrence on ALK inhibitors, Front. Oncol., 9, 1026,
doi: 10.3389/fonc.2019.01026.
25.Howlader, N., Cronin, K. A., Kurian, A. W., and
Andridge, R. (2018) Differences in breast cancer survival by molecular
subtypes in the United States, Cancer Epidemiol. Biomarkers
Prev., 27, 619-626,
doi: 10.1158/1055-9965.EPI-17-0627.
26.Prat, A., Carey, L. A., Adamo, B., Vidal, M.,
Tabernero, J., Cortés, J., Parker, J. S., Perou, C. M., and
Baselga, J. (2014) Molecular features and survival outcomes of the
intrinsic subtypes within HER2-positive breast cancer, J. Natl.
Cancer Inst., 106, doi: 10.1093/jnci/dju152.
27.Dobin, A., Davis, C. A., Schlesinger, F.,
Drenkow, J., Zaleski, C., Jha, S., Batut, P., Chaisson, M., and
Gingeras, T. R. (2013) STAR: ultrafast universal RNA-seq aligner,
Bioinformatics, 29, 15-21,
doi: 10.1093/bioinformatics/bts635.
28.Love, M. I., Huber, W., and Anders, S. (2014)
Moderated estimation of fold change and dispersion for RNA-seq data
with DESeq2, Genome Biol., 15, 550,
doi: 10.1186/s13059-014-0550-8.
29.Sorokin, M., Kholodenko, R., Suntsova, M.,
Malakhova, G., Garazha, A., Kholodenko, I., Poddubskaya, E., Lantsov,
D., Stilidi, I., Arhiri, P., Osipov, A., and Buzdin, A. (2018) Oncobox
bioinformatical platform for selecting potentially effective
combinations of target cancer drugs using high-throughput gene
expression data, Cancers, 10,
doi: 10.3390/cancers10100365.
30.Croft, D., Mundo, A. F., Haw, R., Milacic, M.,
Weiser, J., et al. (2014) The reactome pathway knowledgebase,
Nucleic Acids Res., 42, D472-D477,
doi: 10.1093/nar/gkt1102.
31.Schaefer, C. F., Anthony, K., Krupa, S., Buchoff,
J., Day, M., Hannay, T., and Buetow, K. H. (2009) PID: the Pathway
Interaction Database, Nucleic Acids Res., 37, D674-D679,
doi: 10.1093/nar/gkn653.
32.Kanehisa, M. (2000) KEGG: Kyoto Encyclopedia of
Genes and Genomes, Nucleic Acids Res., 28, 27-30,
doi: 10.1093/nar/28.1.27.
33.Romero, P., Wagg, J., Green, M. L., Kaiser, D.,
Krummenacker, M., and Karp, P. D. (2005) Computational prediction of
human metabolic pathways from the complete human genome, Genome
Biol., 6, R2, doi: 10.1186/gb-2004-6-1-r2.
34.Nishimura, D. (2001) BioCarta, Biotech
Software & Internet Report, 2, 117-120,
doi: 10.1089/152791601750294344.
35.Igolkina, A. A., Zinkevich, A., Karandasheva, K.
O., Popov, A. A., Selifanova, M. V., Nikolaeva, D., Tkachev, V.,
Penzar, D., Nikitin, D. M., and Buzdin, A. (2019) H3K4me3, H3K9ac,
H3K27ac, H3K27me3 and H3K9me3 histone tags suggest distinct regulatory
evolution of open and condensed chromatin landmarks, Cells,
8, doi: 10.3390/cells8091034.
36.Marggraf, M. B., Panteleev, P. V., Emelianova, A.
A., Sorokin, M. I., Bolosov, I. A., Buzdin, A. A., Kuzmin, D. V., and
Ovchinnikova, T. V. (2018) Cytotoxic potential of the novel horseshoe
crab peptide polyphemusin III, Mar. Drugs, 16,
doi: 10.3390/md16120466.
37.Buzdin, A., Garazha, A., Sorokin, M., Glusker,
A., Aleshin, A., Allina, D., Suntsova, M., Tkachev, V., Borger, P.,
Borisov, N., and Gaifullin, N. (2019) RNA sequencing analysis for
profiling activation of cancer-associated molecular pathways, J.
Clin. Oncol., 37, e13032-e13032,
doi: 10.1200/jco.2019.37.15_suppl.e13032.
38.Green, D. M., and Swets, J. A. (1966) Signal
Detection Theory and Psychophysics, Wiley, New York.
39.Chen, L., Zhou, Y., Tang, X., Yang, C., Tian, Y.,
Xie, R., Chen, T., Yang, J., Jing, M., Chen, F., Wang, C., Sun, H., and
Huang, Y. (2019) EGFR mutation decreases FDG uptake in non-small cell
lung cancer via the NOX4/ROS/GLUT1 axis, Int. J. Oncol.,
54, 370-380, doi: 10.3892/ijo.2018.4626.
40.Tanioka, M., Fan, C., Parker, J. S., Hoadley, K.
A., Hu, Z., Li, Y., et al. (2018) Integrated analysis of RNA and DNA
from the phase III trial CALGB 40601 identifies predictors of response
to trastuzumab-based neoadjuvant chemotherapy in HER2-positive breast
cancer, Clin. Cancer Res., 24, 5292-5304,
doi: 10.1158/1078-0432.CCR-17-3431.
41.Liu, T., Cheng, G., Kang, X., Xi, Y., Zhu, Y.,
Wang, K., Sun, C., Ye, J., Li, P., and Yin, H. (2018) Noninvasively
evaluating the grading and IDH1 mutation status of diffuse gliomas by
three-dimensional pseudo-continuous arterial spin labeling and
diffusion-weighted imaging, Neuroradiology, 60, 693-702,
doi: 10.1007/s00234-018-2021-5.
42.Boyd, J. C. (1997) Mathematical tools for
demonstrating the clinical usefulness of biochemical markers, Scand.
J. Clin. Lab. Invest. Suppl., 227, 46-63.
43.Hendricks, W. P. D., Briones, N., Halperin, R.
F., Facista, S., Heaton, P. R., Mahadevan, D., and Kim, S. (2019)
PD-1-associated gene expression signature of neoadjuvant
trastuzumab-treated tumors correlates with patient survival in
HER2-positive breast cancer, Cancers, 11,
doi: 10.3390/cancers11101566.
44.Okuma, H. S., Koizumi, F., Hirakawa, A.,
Nakatochi, M., Komori, O., Hashimoto, J., Kodaira, M., Yunokawa, M.,
Yamamoto, H., Yonemori, K., Shimizu, C., Fujiwara, Y., and Tamura, K.
(2016) Clinical and microarray analysis of breast cancers of all
subtypes from two prospective preoperative chemotherapy studies, Br.
J. Cancer, 115, 411-419, doi: 10.1038/bjc.2016.184.
45.Popovici, V., Chen, W., Gallas, B. G., Hatzis,
C., Shi, W., Samuelson, F. W., Nikolsky, Y., Tsyganova, M., Ishkin, A.,
Nikolskaya, T., Hess, K. R., Valero, V., Booser, D., Delorenzi, M.,
Hortobagyi, G. N., Shi, L., Symmans, W. F., and Pusztai, L. (2010)
Effect of training-sample size and classification difficulty on the
accuracy of genomic predictors, Breast Cancer Res., 12,
R5, doi: 10.1186/bcr2468.
46.Shi, L., Campbell, G., Jones, W. D., Campagne,
F., Wen, Z., et al. (2010) The MicroArray Quality Control (MAQC)-II
study of common practices for the development and validation of
microarray-based predictive models, Nat. Biotechnol., 28,
827-838, doi: 10.1038/nbt.1665.
47.Sonnenblick, A., Brohée, S., Fumagalli,
D., Rothé, F., Vincent, D., Ignatiadis, M., Desmedt, C.,
Salgado, R., Sirtaine, N., Loi, S., Neven, P., Loibl, S., Denkert, C.,
Joensuu, H., Piccart, M., and Sotiriou, C. (2015) Integrative proteomic
and gene expression analysis identify potential biomarkers for adjuvant
trastuzumab resistance: analysis from the Fin-her phase III randomized
trial, Oncotarget, 6, 30306-30316,
doi: 10.18632/oncotarget.5080.
48.Nault, R., Fader, K. A., and Zacharewski, T.
(2015) RNA-Seq versus oligonucleotide array assessment of
dose-dependent TCDD-elicited hepatic gene expression in mice, BMC
Genomics, 16, 373, doi: 10.1186/s12864-015-1527-z.
49.SEQC/MAQC-III Consortium (2014) A comprehensive
assessment of RNA-seq accuracy, reproducibility and information content
by the Sequencing Quality Control Consortium, Nat. Biotechnol.,
32, 903-914, doi: 10.1038/nbt.2957.
50.Suntsova, M., Gaifullin, N., Allina, D.,
Reshetun, A., Li, X., Mendeleeva, L., Surin, V., Sergeeva, A., Spirin,
P., Prassolov, V., Morgan, A., Garazha, A., Sorokin, M., and Buzdin, A.
(2019) Atlas of RNA sequencing profiles for normal human tissues,
Sci. Data, 6, 36,
doi: 10.1038/s41597-019-0043-4.
51.Kumar, N., Prasad, P., Jash, E., Saini, M.,
Husain, A., Goldman, A., and Sehrawat, S. (2018) Insights into exchange
factor directly activated by cAMP (EPAC) as potential target for cancer
treatment, Mol. Cell. Biochem., 447, 77-92,
doi: 10.1007/s11010-018-3294-z.
52.Capitani, M., and Sallese, M. (2009) The KDEL
receptor: new functions for an old protein, FEBS Lett.,
583, 3863-3871, doi: 10.1016/j.febslet.2009.10.053.
53.Maisel, S. A., and Schroeder, J. (2019) Wrong
place at the wrong time: how retrograde trafficking drives cancer
metastasis through receptor mislocalization, J. Cancer Metastasis
Treat., 5, 7, doi: 10.20517/2394-4722.2018.82.
54.Valabrega, G., Montemurro, F., Sarotto, I.,
Petrelli, A., Rubini, P., Tacchetti, C., Aglietta, M., Comoglio, P. M.,
and Giordano, S. (2005) TGFα expression impairs
Trastuzumab-induced HER2 downregulation, Oncogene, 24,
3002-3010, doi: 10.1038/sj.onc.1208478.
55.Ahmad, A. (2019) Current updates on Trastuzumab
resistance in HER2 overexpressing breast cancers, Adv. Exp. Med.
Biol., 1152, 217-228,
doi: 10.1007/978-3-030-20301-6_10.
56.Adamczyk, A., Kruczak, A., Harazin-Lechowska, A.,
Ambicka, A., Grela-Wojewoda, A., Domagala-Haduch, M., Janecka-Widla,
A., Majchrzyk, K., Cichocka, A., Rys, J., and Niemiec, J. (2018)
Relationship between HER2 gene status and selected potential biological
features related to trastuzumab resistance and its influence on
survival of breast cancer patients undergoing trastuzumab adjuvant
treatment, Onco Targets Ther., 11, 4525-4535,
doi: 10.2147/OTT.S166983.
57.Hanker, A. B., Garrett, J. T., Estrada, M. V.,
Moore, P. D., Ericsson, P. G., et al. (2017) HER2-overexpressing breast
cancers amplify FGFR signaling upon acquisition of resistance to dual
therapeutic blockade of HER2, Clin. Cancer Res., 23,
4323-4334, doi: 10.1158/1078-0432.CCR-16-2287.
58.Lenz, G., Hamilton, A., Geng, S., Hong, T.,
Kalkum, M., Momand, J., Kane, S. E., and Huss, J. M. (2018) t-Darpp
activates IGF-1R signaling to regulate glucose metabolism in
Trastuzumab-resistant breast cancer cells, Clin. Cancer Res.,
24, 1216-1226, doi: 10.1158/1078-0432.CCR-17-0824.
59.Martinez, V. G., O’Neill, S., Salimu, J.,
Breslin, S., Clayton, A., Crown, J., and O’Driscoll, L. (2017)
Resistance to HER2-targeted anti-cancer drugs is associated with immune
evasion in cancer cells and their derived extracellular vesicles,
Oncoimmunology, 6, e1362530,
doi: 10.1080/2162402X.2017.1362530.
60.Van Rooijen, J. M., Qiu, S. Q., Timmer-Bosscha,
H., van der Vegt, B., Boers, J. E., Schroder, C. P., and de Vries, E.
G. E. (2018) Androgen receptor expression inversely correlates with
immune cell infiltration in human epidermal growth factor receptor
2-positive breast cancer, Eur. J. Cancer, 103, 52-60,
doi: 10.1016/j.ejca.2018.08.001.
Supplementary Figure S1 (PDF)
Supplementary Tables S1-S6 (ZIP)