Characterization of the Altai Maral Chymosin Gene, Production of a Chymosin Recombinant Analog in the Prokaryotic Expression System, and Analysis of Its Several Biochemical Properties
S. V. Belenkaya1,2,a*, A. A. Bondar3, T. A. Kurgina2,3, V. V. Elchaninov4, A. Yu. Bakulina1,2, E. A. Rukhlova2, O. I. Lavrik3, A. A. Ilyichev1, and D. N. Shcherbakov1,5
1State Research Center of Virology and Biotechnology “Vector”, 630559 Koltsovo, Novosibirsk Region, Russia2Novosibirsk State University, 630090 Novosibirsk, Russia
3Institute of Chemical Biology and Fundamental Medicine, Siberian Branch of the Russian Academy of Sciences, 630090 Novosibirsk, Russia
4Federal Altai Scientific Center of Agrobiotechnologies, Siberian Research Institute of Cheese Making, 656910 Barnaul, Russia
5Altai State University, 656049 Barnaul, Russia
* To whom correspondence should be addressed.
Received May 5, 2020; Revised May 5, 2020; Accepted May 19, 2020
For the first time, the chymosin gene (CYM) of a maral was characterized. Its exon/intron organization was established using comparative analysis of the nucleotide sequence. The CYM mRNA sequence encoding a maral preprochymosin was reconstructed. Nucleotide sequence of the CYM maral mRNA allowed developing an expression vector to ensure production of a recombinant enzyme. Recombinant maral prochymosin was obtained in the expression system of Escherichia coli [strain BL21 (DE3)]. Total milk-coagulation activity (MCA) of the recombinant maral chymosin was 2330 AU/ml. The recombinant maral prochymosin relative activity was 52955 AU/mg. The recombinant maral chymosin showed 100-81% MCA in the temperature range 30-50°C, thermal stability (TS) threshold was 50°C, and the enzyme was completely inactivated at 70°C. Preparations of the recombinant chymosin of a single-humped camel and recombinant bovine chymosin were used as reference samples. Michaelis–Menten constant (Km), turnover number (kcat), and catalytic efficiency (kcat/Km) of the recombinant maral chymosin, were 1.18 ± 0.1 µM, 2.68 ± 0.08 s−1 and 2.27± 0.10 µm M−1·s−1, respectively.
KEY WORDS: recombinant chymosin, milk-clotting activity, thermal stability, Michaelis–Menten kinetics parametersDOI: 10.1134/S0006297920070068
Abbreviations: Chn, rChn, chymosin and recombinant chymosin; CYM, chymosin gene; κ-CS, κ-casein; ME, milk-clotting enzyme; MEP, milk-clotting enzyme preparation; rProChn, recombinant prochymosin; rChn Bos, recombinant bovine chymosin; rChn Cam, recombinant single-humped camel chymosin; TS, thermal stability.
INTRODUCTION
Chymosin (EC 3.4.23.4) is a pepsin-like aspartic endopeptidase [1]. It is synthesized in gastric mucosa cells as preprochymosin with 16-residue signal peptide necessary for chymosin intracellular transport. Next zymogen, termed prochymosin, is secreted into the stomach lumen. Zymogen activation takes place in an acidic environment involving cleavage of the N-terminal 42-residue propeptide that blocks the enzyme active site. As a result, prochymosin is transformed into the active enzyme – chymosin (Chn). Unlike pepsins, most of the known chymosins display narrow substrate specificity. Their primary natural substrate is κ-casein (κ-CS), milk protein located on the surface of casein micelles that provides micelle-stabilizing effect. Chymosin effectively hydrolyzes the specific peptide bond in a casein molecule (F105-M106 for the bovine casein), leading to destabilization and aggregation of casein micelles – milk-clotting. As a result, milk-clotting takes place.
The ability of chymosin to aggregate casein micelles is used in food industry to produce milk clot that is essential for producing rennet-added cheeses. Until recently, bovine (Bos taurus taurus L) chymosin was considered as the standard and universal milk-clotting enzyme (MCE) in cheese-making industry [2, 3]. Other natural proteases with milk-clotting properties have limited usage, mainly due to the excessive unspecific proteolytic activity and overly high thermal stability [2-6].
Currently recombinant chymosin preparations have been taking an increasing share of milk-clotting agents’ market [7, 8]. The first results on recombinant bovine chymosin (rChn Bos) expression in Escherichia coli were reported in 1980 [9]. After about ten years, rChn Bos was used in the cheese-making industry for the first time [10]. In fact, since the late 1990s and up to now the rChn Bos has been used as a technologically adequate replacement of the natural enzyme [1].
From 1980 to 2019, recombinant chymosins of several mammals were produced and characterized: sheep [11, 12], goat [13, 14], camel (dromedary) [15, 16], yak [17, 18], buffalo [19], and alpaca [20]. Examination of biochemical and technological properties of the produced enzymes revealed that the recombinant camel chymosin had higher milk-clotting activity and specificity than the bovine chymosin (milk-clotting and non-specific proteolytic activity ratio), but demonstrated higher thermal stability (TS) [16, 21, 22]. It is worth noting that the primary structures of camel and bovine κ-CS differ in several regions, including the region of contact with chymosin.
Tertiary structures of the bovine and camel chymosins were experimentally determined [16]. Chymosins, as other pepsin-like enzymes, consist of two predominantly beta-stranded domains, with each domain having a catalytic residue of asparagine amino acid. The substrate-binding cleft is located between the domains.
In addition to solving an applied problem of searching for new MCEs, the study of chymosins can provide insight into the relationship between enzyme structure, specificity, and TS. Chymosin is an exciting research target since there are many different chymosins in nature adapted to different κ-CS substrates.
The aim of the research was to characterize the chymosin gene of Altai maral [Cervus elaphus sibiricus (Severtzov, 1873)], to produce its recombinant form in prokaryotic expression system, and to examine several biochemical properties of this MCE. Achieving this goal is expected to expand the spectrum of known chymosins and open possibilities for comparative analyses of this class of enzymes.
MATERIALS AND METHODS
Nucleotide sequence of prochymosin from Altai maral. Total DNA was isolated from blood lymphocytes of a healthy 3-year-old male Altai maral (hereinafter − maral). Blood was collected by the staff of the veterinary service of the enterprise FSUE Novotalitskoye in the Charyshsky District of Altai Territory during a routine examination of animals. For blood sampling, vacutainers containing disodium EDTA as a coagulation inhibitor were used.
For the lymphocyte isolation, blood was layered on an LCM medium (Thermo Fisher Scientific, USA), centrifuged for 10 min at 900g (Eppendorf, Germany), and lymphocyte ring was collected. Genomic DNA was isolated from the lymphocyte suspension using a TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s recommendations. Total DNA concentration in the resulting preparation was 10 µg/ml.
The estimated length of the preprochymosin gene is ~11,000 bp; therefore, two pairs of oligonucleotide primers (Biosset, Russia) were designed to obtain amplicons of this sequence (Table 1). Two pairs, CYM_GL_9840F + CYM_GL_16024R and CYM_GR_15342F + CYM_Gene_21717R, are required to obtain left and right overlapping fragments (approximately 6000 bp). Amplification was performed according to the manufacturer’s recommendations using a Q5 polymerase (New England Biolabs, USA) with low error rates.
Table 1. Primers used to obtain DNA
fragments of the maral preprochymosin gene
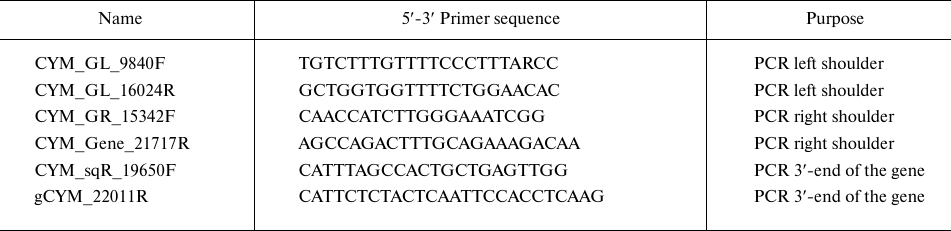
Subsequently, the DNA of both gene fragments was produced in preparative amounts, purified from PCR components by sorption on magnetic particles using an AMPure XP system (Beckman Coulter, USA), sequenced directly on both sides using the BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific) followed by analysis on an ABI3130XL automated gene analyzer (Applied Biosystems, USA). The sequenograms of the maral chymosin gene (CYM) with length of 850-950 bp were analyzed and next aligned with the CYM gene structures known for other Cetartiodactyla animals from the GenBank database to confirm belonging to the chymosin gene family and design primers for the next sequencing step. By repeating these steps, we finally obtained a set of overlapping sequenograms and assembled a contig of overlapping gene fragments. To control the correct assembly, central region of the gene was additionally amplified and sequenced directly. Additionally, to clarify the sequence of the 3′-end of the gene, we designed and used primers CYM_sqR_19650F and gCYM_22011R for PCR and Sanger sequencing (Table 1). The primer structures used for step sequencing are available upon request.
Obtaining nucleotide sequence of the Altai maral cytochrome b. Primers (MTCB_F 5′-CCHCCATAAATAGGNGAAGG-3′ and MTCB_R 5′-WAGAAYTTCAGCTTTGGG-3′) were used to amplify a fragment of the mitochondrial genome that overlaps the MT-CYB gene. DNA of the fragment was amplified in preparative amounts, purified from PCR components by sorption on AMPure XP magnetic particles, and subjected to direct sequencing on both sides by the Sanger method on an ABI3130XL automatic gene analyzer. We compared nucleotide sequence of the MT-CYB C.e. sibiricus (registration number GenBank MT331597) with the structures of the corresponding mitochondrial DNA sections of 12 subspecies of red deer Cervus elaphus, available in the GenBank database.
Cloning of maral CYM gene fragments into pJET vector. Cloning of maral CYM gene fragments into the pJET vector was carried out using a Thermo Scientific CloneJET PCR Cloning Kit (Thermo Fisher Scientific), according to the manufacturer’s recommendations. For this purpose, PCR products were separated on an agarose gel and isolated using a QIAquick Gel Extraction Kit (Thermo Fisher Scientific). The produced DNA fragment was mixed with the vector, and ligation was carried out for 5 min at 22°C. Next, E. coli cells (NEB Stable strain, New England Biolabs, USA) were transformed with the ligation products.
Producing of recombinant maral prochymosin. After establishing the intron-exon organization of the gene, coding sequence of the prochymosin (ProChn) gene was determined. Codon composition of the gene was optimized for the selected expression system using the online service Integrated DNA Technologies (https://eu.idtdna.com/CodonOpt). The synthesized nucleotide sequence was integrated into the expression vector pET21a (Novagen, Germany) at the unique restriction sites BamHI and HindIII.
Chemical transformation of E. coli strain BL21 (DE3) with the resulting construct was carried out to obtain the target protein. Individual E. coli colonies containing recombinant plasmids were cultured overnight on an orbital shaker (180 rpm, Biosan, Latvia) in LB medium (AppliChem, USA) containing 100 µg/ml ampicillin at 37°C. The inoculum was transferred to an Erlenmeyer flask containing LB medium at a ratio of 1/100 and cultivated to optical density 0.8 (at λ = 600 nm). Isopropyl-β-D-1-thiogalactopyranoside (IPTG) (Anatrace Products, USA) was added to a final concentration of 1 mM to induce the culture followed by additional cultivation on a shaker for 12 h at 25°C and 180 rpm. To isolate inclusion bodies the produced biomass was first centrifuged (Beckman Coulter, USA) for 20 min at 5000g and 4°C. Next the resulting precipitate was dissolved in STET buffer (AppliChem, USA) (8% sucrose; 50 mM Tris-HCl; 20 mM EDTA; 5% (w/v) Triton X-100 pH 8.0) at a ratio 20 ml per 1 gram of biomass and incubated overnight at 4°C. After incubation, the cells were destroyed using a Soniprep 150 Plus ultrasonic homogenizer (MSE, PRC), inclusion bodies were precipitated by centrifugation (Beckman Coulter, USA) at 20,000g for 20 min at 4°C [23]. The precipitated inclusion bodies were solubilized in buffer A (50 mM KH2PO4, 150 mM NaCl, pH 10.7) containing 8 M urea, incubated for 24 h at 15°C and centrifuged at 20,000g for 20 min. Further work was carried out with a supernatant containing recombinant prochymosin (rProChn). The target protein was renatured using the procedure recommended by Wei et al. [23]. Supernatant was diluted 3-fold with buffer A and incubated for 12 h at 15°C. Following incubation, the supernatant diluted with alkaline buffer was adjusted to pH 8.0 with 1.0 M HCl, kept at 15°C for 1 h, and dialyzed against buffer B (50 mM Tris, 150 mM NaCl, pH 8.0) overnight at 4°C [23]. As a result, preparation of the maral rProChn was obtained.
The recombinant protein produced in E. coli cells was analyzed by electrophoresis in the presence of sodium dodecyl sulfate (SDS-PAGE) according to the Laemmli method [24]. To analyze electrophoretic mobility of the protein and determine its size, a molecular weight marker PageRuler Unstained Protein Ladder (Termo Fisher Scientific) was used. Protein concentration was determined by the Bradford assay [25].
Activation of maral recombinant prochymosin. Activation of the maral rProChn was carried out by a stepwise change of pH [26]. HCl (2.0 M) was added to the rProChn sample adjusting pH to 3.0 with continuous stirring. Then stirring was stopped, and the mixture was incubated at pH 3.0 for 2 h. After incubation pH of the sample was adjusted to 5.8 using 0.5 M NaOH. As a result, the rChn of maral was obtained.
Determination of milk-clotting activity. A 10% solution of standardized skimmed milk powder (MZSF OJSC, Russia) in 5 mM CaCl2, pH 6.5, was used as a substrate. A 1% aqueous solution of a dry control rennet sample (MZSF OJSC, Russia) with known MCA was used as a standard. Prior to determining MCA, the control sample and the liquid preparation of recombinant chymosin (rChn) were kept in a water bath at 35°C for 15 min and cooled to room temperature.
The procedure for determining MCA was carried out in a water bath at 35°C. Substrate solution (2.5 ml) was placed into a glass tube and heated at 35°C for 5 min. An aliquot (0.25 ml) of an enzyme was added to the substrate, a stopwatch was turned on, and the resulting reaction mixture was immediately thoroughly mixed. Time when first flakes of the coagulated substrate were observed in the drops of reaction mixture applied onto the tube wall was considered to be the clotting time. Milk-clotting activity was expressed in arbitrary units per 1 ml (AU/ml) and calculated using equation (1):
MCA(AU/ml) = 0,01 × A × T1/T2, (1)
where A – certified milk-clotting activity of the control sample of chymosin, in arbitrary units per 1 gram, T1 being coagulation time for the control sample of chymosin, and T2 – coagulation time for the test sample.
Determination of total MCA in each sample was performed in triplicate (n = 3). Relative MCA of maral rChn was calculated after determining the total MCA and protein concentration. For comparison, commercial preparations of bovine rChn (granular dry form with declared MCA – 2235 IMCU/g) and camel rChn (liquid form with declared MCA – 1000 IMCU/ml) (Chr. Hansen, Denmark) were used. To convert IMCU values (International Milk Clotting Units) into AU, multiplication coefficient of 130 was used. A 1.0% aqueous solution of bovine rChn was prepared to assess relative MCA of the commercial reference preparations, and the liquid camel rChn was diluted 10 times with distilled water. Protein concentration was determined in the resulting solutions with the Bradford assay. The MCA values declared by the manufacturer were used to calculate relative MCA of the commercial reference enzymes. Relative MCA was expressed in AU/mg.
Comparison of the structure of maral, bovine, and camel chymosins. For comparing the primary structures, protein sequences were aligned using the BioEdit program [27].
To compare tertiary structures, we used the structure of bovine Chn from the PDB base (identifier 4AA8) and the model of maral we constructed. The construction was performed by the homology simulation method on the Swiss-Model server [28]. The bovine Chn structure was used as a template for modeling, with all modeling parameters left to default values. Visual analysis of the structures was carried out with the Chimera program [29].
Determination of Michaelis–Menten kinetics parameters of recombinant chymosins by fluorescence spectrometry. A synthetic peptide of the following composition was used as a substrate: Dabcyl-HPHPHLSFMAIPK (5-FAM) KK-NH2 (Dabcyl = 4-(dimethylaminoazo) benzene-4-carboxylic acid, 5-FAM = 5-carboxyfluorescein) (CPC Scientific, USA) containing the Chn-sensitive region of bovine κ-casein. A 5-FAM fluorophore was located at the C-terminal end of the substrate, and a Dabcyl quencher was located at the N-terminal end. The quencher and fluorophore were brought together in such a way that the fluorescence resonance energy transfer (FRET) from 5-FAM to Dabcyl made fluorescence of 5-carboxyfluorescein minimal. Hydrolysis of the substrate with chymosin led to the fluorescence intensity increase due to separation of the fluorophore from the quencher. The rate of fluorescence increase reflected the rate of enzymatic reaction. All measurements were performed with a CLARIOstar fluorescence plate reader (BMG LABTECH, Germany) using excitation and emission wavelengths 495 nm and 520 nm, respectively.
Reaction mixtures were prepared on ice in a 384-well plate with subsequent incubation at room temperature for 5 min. All measurements were carried out at 25°C. Each well contained one reaction mixture. The instrument was calibrated using a solution of a peptide with a concentration of 20 µM, subjected to complete hydrolysis. Fluorescence intensity of this mixture was taken as 80%. Measurements were carried out in the kinetic scan mode. Duration of the cycle and number of the cycles were determined individually, depending on the number of scanned wells. All experiments were repeated three times (n = 3). Commercial recombinant bovine and camel Chn (Chr. Hansen, Denmark) were used as reference samples.
Reaction mixtures (5 µl volume) contained 50 mM acetate buffer (pH 5.6) and peptide of various concentrations (0-20 µM). The reaction was initiated by adding of 5 µl of Chn to a reaction mixtures using a pump built into the plate reader. The final concentration of Chn was 5 nM.
Kinetic constants were calculated using the accompanying MARS Data Analysis software (BMG LABTECH, Germany). Michaelis constants (Km) were determined using the Michaelis–Menten equation [30], hydrolysis rate constants or enzyme turnover numbers (kcat) were calculated using equation (2):
kcat = V max/[E], (2)
where kcat is hydrolysis rate constant; Vmax is maximum reaction rate obtained from the Michaelis–Menten equation; [E] – concentration of the studied recombinant chymosin.
Catalytic efficiency (specificity constant) was determined as a kcat/Km ratio.
Determination of thermal stability. Solutions of MCEs were heated in a water bath in the temperature range 30-70°C for 30 min, and residual MCA was determined in them. The MCA values obtained in the samples heated at 30°C were taken as 100%. TS of maral rChn was compared to the dry commercial rChn Bos (Bos rChn) and to the liquid commercial rChn of one-humped camel (rChn Cam), which were produced in the eukaryotic expression system Aspergillus niger var. Awamori [1, 16]. Before the test, a 0.5% aqueous solution of the bovine rChn was prepared; the camel rChn was diluted 100 times with distilled water. Milk-clotting activity of the solutions of bovine rChn and rChn of a dromedary camel was normalized to the activity of maral rChn (Cer rChn). Temperature at which the studied MCE retained at least 80% of the maximum MCA was considered to be the TS threshold.
Statistical processing of the obtained data was carried out using Excel 2008 software (Microsoft Corporation, USA). The results are presented as an arithmetic mean with standard deviation (M ± SD).
RESULTS AND DISCUSSION
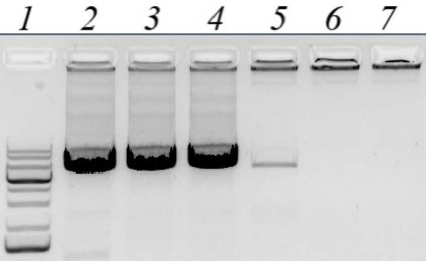
Obtaining the nucleotide sequence of the Altai maral preprochymosin gene. To design PCR primers for preprochymosin (CYM) gene amplification, BLAST resource was used to search for DNA sequences homologous to the bovine CYM mRNA in the NCBI Gene database (www.ncbi.nlm.nih.gov/gene). Several CYM sequences: yack (NW_005393277), wild yak (JH880704.1), camel (LOC105085668), sheep (NC_019458.2), and monkey (NC_013902) were selected. The sequences were aligned and the most conservative regions in the area that completely overlap the CYM gene exons were identified. For the maral CYM amplification, several pairs of primers for the conservative regions were designed, including degenerate primers. Amplification conditions were optimized by using temperature gradient at the annealing stage (Fig. 1) and by varying free magnesium ion concentration in the reagent mixture.
Fig. 1. Gel electrophoresis of PCR amplified products of the maral CYM gene with CYM_GL_9840F and CYM_GL_16024R pair of primers. Lines: 1) DNA size markers; 2-7) annealing temperatures of 54°C, 57°C, 60°C, 63°C, 66°C, and 69°C, respectively.
Finally, we selected optimal algorithm for the gene amplification. Two long overlapped fragments of the maral CYM were amplified, and their DNA after production and purification were sequenced from both ends by the Sanger method. The reads were assembled in the contig that included both CYM fragments. Central part of the CYM was directly amplified and sequenced to ensure the proper assembly.
Several polymorphisms were identified in the left fragment of the maral CYM gene: two regions of point deletions outside of the protein-coding regions and a pair of synonymous single nucleotide polymorphisms in exons. Gene alleles were separated by cloning of the CYM PCR product into the pJET vector and following sequencing of several DNA clones.
Finally, for the first time, we identified two allelic variants of the maral CYM gene, with their sizes of 11,161 nucleotides. The obtained nucleotide sequence was submitted to the GenBank database under the accession number MT225406. Final sequence of the gene with exons and introns annotation is presented in Fig. S1 in the Supplement.
Intron/exon boundaries were localized by comparing the sequence of interest with the mRNA and CYM gene sequences of mammals. Thus, the maral CYM mRNA sequence was derived.
Confirmation of DNA origin. As an additional control of affiliation of the isolated genomic DNA to the subspecies Cervus elaphus sibiricus (Severtzov, 1873), we amplified and sequenced a fragment of the mitochondrial genome, as described early [31]. It should be noted that there are several MT-CYB sequences of Cervus elaphus maral in the GenBank. A Latin word “maral” is also referred to the noble deer inhabiting the territory of Iran and Turkey, and this species is genetically different from the Altai maral. We compared the obtained sequence of C.e. sibiricus MT-CYB (GenBank ID MT331597) with the corresponding region of the mitochondrial DNA for 12 subspecies of Cervus elaphus available in the GenBank database. The resulting phylogenetic tree is shown in Fig. S2 (see Supplement). As expected, sequence of the MT-CYB belongs to Altai maral C.e. sibiricus since it is almost identical to the sequence of C.e. canadensis MT-CYB, which is a synonym of C.e. sibiricus, according to Pavlinov taxonomy [32]. These two sequences form a separate branch of the Cervis elaphus MT-CYB phylogenetic tree.
Producing of recombinant maral prochymosin. To produce recombinant analog of the maral prochymosin we used the expression system based on E. coli BL21(DE3) strain and pET plasmid expression vector. This system is one of the most effective for high yield production of heterologous proteins in prokaryotes. Target protein concentration can be as high as 40% of the total cell proteins. Previously, use of pET21 in E. coli BL21(DE3) allowed getting such expression levels for the bovine chymosin [33] and alpaca chymosin [21]. The synthesized nucleotide sequence of maral prochymosin with codon composition optimized for E. coli was integrated into the expression vector pET21a so that the N-terminal part of the gene contained the sequence of the T7 expression tag.
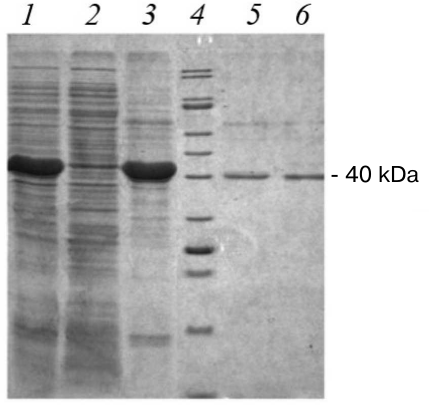
The recombinant strain producer was generated following transformation of competent cells by the pET21-CYM-Cer plasmid. Standard protocol was used for the target protein production, which included addition of IPTG inducer. For evaluating the efficiency of recombinant prochymosin (rProChn) synthesis and determining its localization, gel electrophoretic examination of different protein fractions derived from the recombinant strain was performed (Fig. 2)
Fig. 2. Results of SDS-PAGE of proteins from the recombinant strain. Lines: 1) biomass of E. coli strain containing pET21-CYM-Cer 24 h after inducer addition; 2) soluble fraction of E. coli biomass after treatment with STET buffer and centrifugation; 3) insoluble fraction of E. coli biomass after treatment with a lysis buffer and centrifugation (inclusion bodies); 4) molecular weight markers; 5) maral recombinant chymosin; 6) camel recombinant chymosin.
E. coli cells containing pET21-CYM-Cer plasmid and induced with IPTG had high content of a protein, which demonstrated the same electrophoretic mobility as that calculated for the maral prochymosin (41 kDa). The target protein content was ≥30% (Fig. 2, line 1) of the total protein mass. As can be seen the soluble fraction of E. coli biomass after treatment with STET buffer and centrifugation (Fig. 2, line 2) hardly contains any target protein, but the inclusion body fraction is almost completely represented by the rProChn (Fig. 2, line 3). Therefore, the conditions of gene expression in E. coli resulted in a highly effective synthesis of the target protein, with almost all protein accumulated in the inclusion bodies as an insoluble fraction. Hence, protein renaturation by stepwise dialysis was required.
Before the dialysis, activated maral rProChn demonstrated no coagulation activity. After the dialysis, activated rProChn demonstrated high milk-clotting activity indicating a successful refolding of the proper zymogen tertiary structure.
Recombinant prochymosin activation and obtaining of maral recombinant chymosin. For preventing autocatalytic activation [34] after isolation and purification, the rProChn preparations were stored in a weak base buffer (50 mM Tris-HCl, 150 mM NaCl, pH 8.0). For rProChn activation, titration procedure to pH 3.0 was selected [26]. Initial milk-clotting activity of the rProChn was less than 1.0 AU/ml, and after activation, it was 2329.8 AU/ml. Thereby, milk-clotting activity increased more than 2300-fold following activation indicating effectiveness of the conversion of rProChn to rChn. Concentration of the isolated maral rChn was about 44 mg/liter, which was close to the concentration of camel rChn produced in Pichia pastoris cells [35].
Comparison of milk-clotting activities of the maral rChn and commercial chymosin products is presented in Table 2.
Table 2. Total and relative milk-clotting
activities of recombinant chymosin of maral, cow, and camel
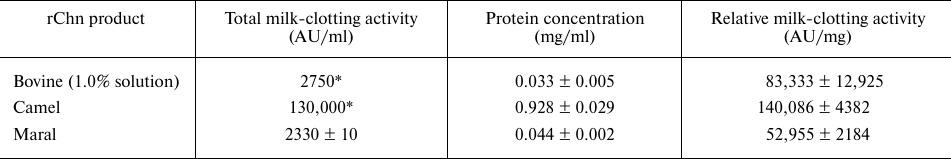
* MCA values declared by the manufacturers.
The relative milk-clotting activity of maral rChn appeared to be lower than the milk-clotting activities of bovine and camel rChn by 1.6- and 2.6-fold, respectively. It can be explained by the fact that the bovine and camel rChn are produced in Aspergillus niger var. Awamori (stain dgr246pyrG) expression system, providing more effective refolding and eukaryote-specific posttranslational modifications.
Total milk-clotting activities of the liquid commercial chymosin products in the cheese-making industry (animal rennet products “Naturen® Premium 225”, “Chr. Hansen”, Denmark and “Clerici 96/04”, “Caglficio Clerici”, Italy; recombinant bovine and camel chymosins “Chy-Max® Extra” and “Chy-Max® M 1000”, “Chr. Hansen”, Denmark) are in the range 225-1000 IMCU/ml, which corresponds to (30-130) × 103 AU/ml. For production of liquid milk-clotting products with activity of more than 30 × 103 RU/ml, initial diluted enzyme solutions are concentrated by ultrafiltration. In some technologies, rProChn is secreted by strains-producers into the culture medium [19, 34, 36, 37]. If the components of the medium have no coagulation activity and do not contain any toxins released by the producing strain, isolation and purification are not necessary. Zymogen activation and transformation into the active chymosin occur in this case directly in the cell culture medium, which is later clarified and concentrated by ultrafiltration up to the necessary milk-clotting activity. The values of milk-clotting activity of the cell culture medium containing rChn lie are in the range 400-12,480 AU/ml [36, 37], so the medium have to be concentrated 10-75-fold. To increase milk-clotting activity of the maral rChn product, 13-56-fold concentration is necessary.
Therefore, the produced maral rChn displayed coagulation activity that was comparable to the activity of the average commercially available milk-clotting agents.
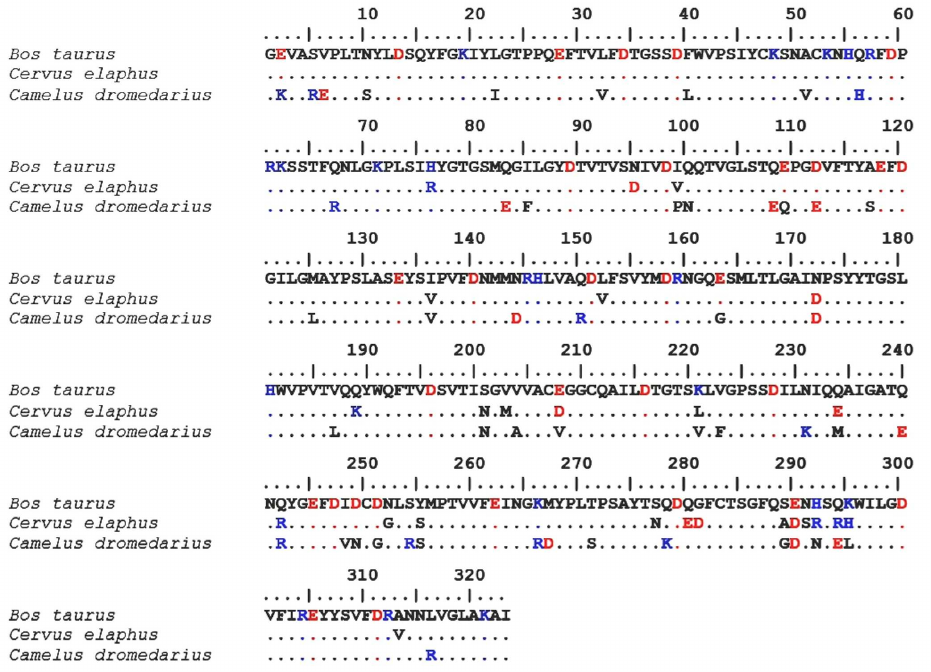
Comparison of the structures of maral, bovine and camel chymosins. Comparative analysis of the amino acid sequences of maral, bovine, and camel chymosins was conducted. The sequences of maral and bovine mature chymosins were found to have a 92.6% identity and were aligned without insertions or deletions (Fig. 3).
Fig. 3. Sequence alignment of the maral, bovine, and camel chymosins. Positively charged residues are shown in blue, negatively charged residues – in red. Identical residues are marked with dots.
Hence, it can be assumed that the tertiary structures of these proteins are also close, and homology modeling results are highly reliable. It is known that bovine and camel chymosins have similar tertiary structures, with the identity of their primary structures being just 84.8% [38].
Bovine chymosin contains 32 negatively charged residues and 19 positively charged residues, and maral chymosin has 36 and 22 of negatively and positively charged residues, respectively. Total charges of these chymosins are almost equal: –13 for the bovine chymosin and –14 for the maral chymosin. At the same time, total charge of the camel chymosin is –7: it has 33 negatively charged residues and 26 positively charged residues. Both the camel rChn and the maral rChn have a higher total number of charged amino acid residues than the bovine rChn.
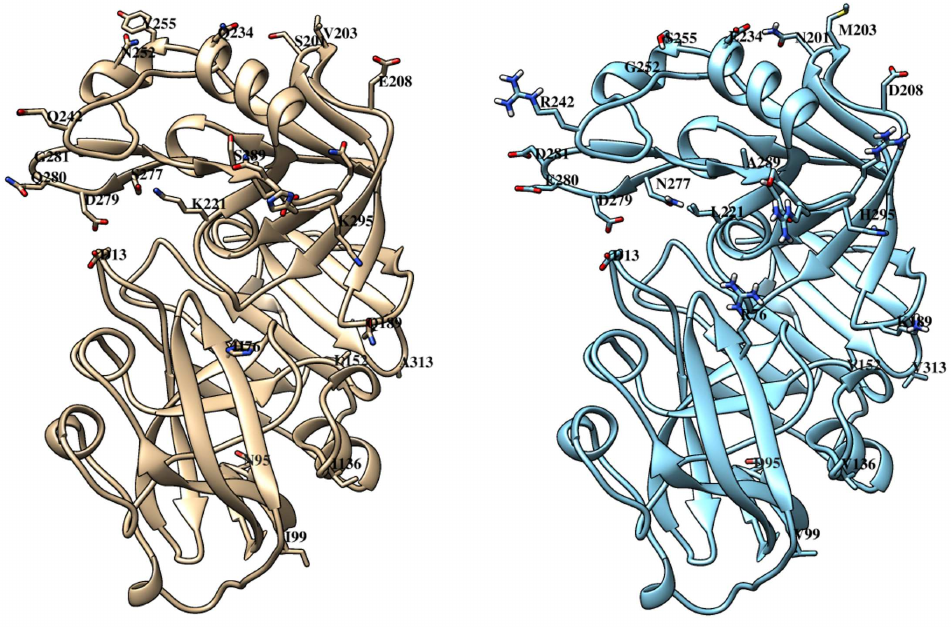
Like other similar peptidases, chymosin consists of two domains with predominantly beta-stranded structure and one catalytic residue in each (Fig. 4).
Fig. 4. Tertiary structures of bovine (left) and maral (right) chymosins. Catalytic residues D13 and D279 are marked. Positions with amino acid substitutions and boundaries of the region 289-295 with five substitutions are marked. C-terminal domain is on top, N-terminal domain is below.
Comparison of the structures of bovine and maral chymosins reveals that most of the substitutions are in the C-terminal domain. All substitutions in the N-terminal domain are quite far from the catalytic residue D13. However, in the C-terminal domain, as much as three substitutions are located in the same loop with the catalytic residue D279, with two of them (Q280E and G281D) generating charge changes. It can be assumed that these substitutions affect significantly catalytic properties of the chymosin. Interestingly, camel chymosin has only one substitution (Q278K) in this region that also changes the charge, but in the opposite direction.
Some substitutions (Q189K, K221L, Q242R) are located in the substrate-binding cleft of the maral chymosin and six substitutions in the region of amino acid residues 289–295. Such substitutions can affect the enzyme binding to κ-CS and, subsequently, the observed catalytic activity. Camel chymosin also has several substitutions in the substrate-binding cleft, compared with the bovine chymosin [38].
Michaelis–Menten kinetic parameters. Chymosin-mediated hydrolysis of the synthetic fluorogenic substrate containing the Chn-sensitive region of the bovine κ-CS results in the nonlinear increase of the fluorescence intensity depending on the substrate concentration that reflects the rate of enzymatic reaction. The data presented in Table 3 show that the Michaelis–Menten kinetic parameters of maral rChn and the commercial liquid product of the camel rChn are close. However, these parameters are significantly different from the kinetics constants reported for the bovine rChn.
Table 3. Michaelis–Menten kinetic
parameters for recombinant chymosins

The maral rChn demonstrated the lowest Km value, indicating the highest affinity to the used fluorogenic substrate. The maral rChn substrate saturation rate was estimated to be 3.4-fold higher than the one of the bovine rChn. The camel rChn substrate affinity was estimated to be 3.1-fold higher than that of the bovine rChn. Similar results were obtained when Km values for the chromogenic undecapeptide substrate containing the bovine Chn-sensitive κ-CS region were compared for the bovine and the camel rCYMs [16]. The camel rChn Km values obtained in the pH range 5.50-6.65 with the same fluorogenic substrate as in the current research were also 2.1-3.1-fold lower than the Km of for the commercial bovine rChn [39].
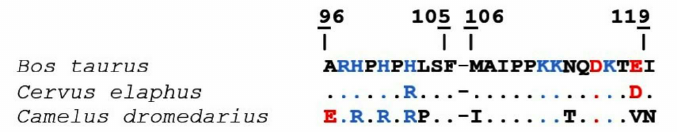
The analysis of Chn-sensitive camel and bovine κ-CS regions (96-119) demonstrated that they differed in nine positions, including the M106I substitution in the hydrolyzed peptide bond 105-106. Sequence identity of the bovine and the camel 96-119 κ-CS regions is 62.5%. On the contrary, amino acid sequences of the bovine and maral Chn-sensitive κ-CS regions differ only in two positions indicating sequence identity of 91.7%. It can be suggested that the higher substrate affinity of the maral and camel rChn in comparison with the bovine rChn is likely due to the differences in their substrate-binding cleft structures and their Chn-sensitive κ-CS regions (Fig. 5).
Fig. 5. Alignment of Chn-sensitive regions of bovine, maral, and camel κ-CS. Positively charged residues are shown in blue, negatively charged – in red. Identical residues are marked by dots. The 105-106 sequences are marked with whitespaces.
The mechanisms facilitating increased affinity of the maral and camel chymosins to the used fluorogenic substrate could involve slowing down the release of reaction products from the enzymes, which is manifested by the observed decrease of the catalytic rate constant kcat and catalytic effectiveness kcat/Km.
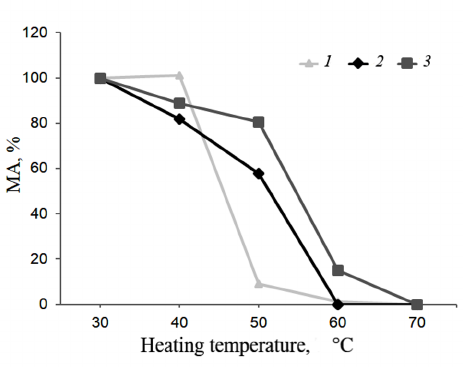
Thermal stability. Shown in Figure 6 are the results of the TS investigations of bovine, camel, and maral rChn.
Fig. 6. Dependence of the enzyme residual activity on heating temperature (TS). Curves: 1) commercial bovine rChn, 2) commercial camel rChn, 3) maral rChn.
At 40°C, milk-clotting activities of the bovine and camel rChn were 101 and 82%, respectively. The bovine rChn was almost completely inactivated at 50°C with residual activity being only 9%, but the camel rChn retained 58% of milk-clotting activity at this temperature. Complete thermal inactivation of the camel rChn was observed at 60°C. According to the chosen criteria (see “Materials and Methods” section), the range of TS for the bovine and camel chymosins was 30-40°C. However, at temperatures above 40°C, the camel enzyme was inactivated slower. The obtained results are in agreement with the results reported by Kappeler et al. [16], who also noted higher TS of the camel rChn as compared to the bovine rChn. The maral rChn proved to be thermally stable in a wider temperature range (30-50°C). After heating at 40°C and 50°C, the enzyme retained 89 and 91% of the milk-clotting activity, respectively. At 60°C, the maral rChn demonstrated only 15% of the initial coagulation activity, and at 70°C, the enzyme was completely inactivated. Therefore, threshold of the maral rChn thermal inactivation was 50°C, which was 10°C higher than that of the control rChn proteins.
Higher TS of the maral rChn could be due to the peculiarities of its amino acid composition. Proteins of thermophilic microorganisms are known to have a higher number of charged residues in comparison with their orthologues from mesophilic microorganisms [40]. For example, phenylalanyl-tRNA synthetase from thermophilic microorganism Thermus thermophilus has a higher content of arginine and asparagine residues in its comparison as compared to its orthologue from Escherichia coli [41]. The fact that the maral and camel chymosins have more charged residues than the bovine chymosin is likely the reason for their higher TS.
The chymosin gene of the Altai maral [Cervus elaphus sibiricus (Severtzov, 1873)] was characterized in this study, and its recombinant analog was produced in the prokaryotic expression system. Some structural features and biochemical properties of the new recombinant MCE were investigated and analyzed. Our research team is planning further examination of the properties of the recombinant chymosins produced in both prokaryotic and eukaryotic expression systems.
Acknowledgements. The authors are grateful to the Genomics Core Facility (ICBFM SB) of the Siberian Branch of the Russian Academy of Sciences for DNA sequencing.
Funding. The work was financially supported by the Ministry of Science and Higher Education of the Russian Federation (project No. FZMW-2020-0002 “Development of recombinant enzyme producers for cheese making”).
Ethics declarations. The authors declare no conflict of interests in financial or any other sphere. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Electronic supplementary material. Supplementary materials are available in the electronic version on the journal website (http://protein.bio.msu.ru/biokhimiya) and Springer site (Link.springer.com).
REFERENCES
1.Harboe, M., Broe, M. L., and Qvist, K. B. (2010)
The production, action and application of rennet and coagulants, in
Technology of Cheesemaking (Law, B. A., and Tamime, A. Y., eds.)
2nd Edn., pp. 98-129.
2.Belov, A. N., Koval, A. D., Avdonina, E. A., and
Yelchaninov, V. V. (2009) Influence of milk-converting preparation on
organoleptic indicators of cheese, Cheese-Making and
Butter-Making, 1, 22-24.
3.Jacob, M., Jaros, D., and Rohm, H. (2011) Recent
advances in milk clotting enzymes, Int. J. Dairy Technol.,
64, 14-33.
4.Feijoo-Siota, L., Blasco, L., Rodriguez-Rama, J.
L., Barros-Velázquez, J., de Miguel, T.,
Sánchez-Pérez, A., and Villa, T. G. (2014) Recent patents
on microbial proteases for the dairy industry, Recent Adv. DNA Gene
Seq., 8, 44-55.
5.Kimberlin, R. H. (1996). Bovine spongiform
encephalopathy and public health: some problems and solutions in
assessing the risk, in 3rd International Symposium on Transmissible
Subacute Spongiform Encephalopathies: Prion Diseases, Paris (Court,
L., and Dodet, B., eds.) Elsevier, Amsterdam, pp. 487-502.
6.Lebedev, L. R., Kosogova, T. A., Teplyakova, T. V.,
Kriger, A. V., Elchaninov, V. V., Belov, A. N., and Koval, A. D. (2016)
Study of technological properties of milk-clotting enzyme from Irpex
lacteus (Irpex lacteus (fr.) fr.), Foods Raw
Mater., 4, 58-65.
7.Yelchaninov, V. V. (2006) Milk-clotting enzyme from
reindeer rennet, Cheese-Making and Butter-Making, 4,
42-44.
8.Yelchaninov, V. V. (2006) Investigation of a
milk-clotting enzyme from reindeer rennet, Author’s abstract
of Candidate’s dissertation, Kemerovo technological Institute of
food industry, Kemerovo, p. 172.
9.Uchiyama, H., Uozumi, T., Beppu, T., and Arima, K.
(1980) Purification of prorennin mRNA and its translation in
vitro, Agricult. Biol. Chem., 44, 1373-1381.
10.Flamm, E. L. (1991) How FDA approved chymosin: a
case history, Biotechnology, 9, 349.
11.Pungercar, J., Strukelj, B., Gubensek, F., Turk,
V., and Kregar, I. (1990) Complete primary structure of lamb
preprochymosin deduced from cDNA, Nucleic Acids Res., 18,
4602.
12.Rogelj, I., Perko, B., Francky, A., Penca, V.,
and Pungerčar, J. (2001) Recombinant lamb chymosin as an
alternative coagulating enzyme in cheese production, J. Dairy
Sci., 84, 1020-1026.
13.Vega-Hernández, M. C.,
Gómez-Coello, A., Villar, J., and Claverie-Martı́n,
F. (2004) Molecular cloning and expression in yeast of caprine
prochymosin, J. Biotechnol., 114, 69-79.
14.Eskandari, M. H., Hosseini, A., and Aminlari, M.
(2008) Nucleotide sequence of cDNA encoding for preprochymosin in
native goat (Capra hircus) from Iran, Iranian J.
Veterinary Res., 9, 262-265.
15.Kappeler, S., Farah, Z., van den Brink, J. M.,
Rahbek-Nielsen, H., and Budtz, P. (2016) U.S. Patent No. 9,307,775,
Washington, DC, U.S. Patent and Trademark Office.
16.Kappeler, S. R., Rahbek-Nielsen, H., Farah, Z.,
Puhan, Z., Hansen, E. B., and Johansen, E. (2006) Characterization of
recombinant camel chymosin reveals superior properties for the
coagulation of bovine and camel milk, Biochem. Biophys. Res.
Commun., 342, 647-654.
17.Ersöz, F., and İnan, M. (2019)
Large-scale production of yak (Bos grunniens) chymosin A in Pichia
pastoris, Protein Expr. Purif., 154, 126-133.
18.Luo, F., Jiang, W. H., Yang, Y. X., Li, J., and
Jiang, M. F. (2016) Cloning and expression of yak active chymosin in
Pichia pastoris, Asian-Australas J. Anim. Sci.,
29, 1363.
19.Vallejo, J. A., Ageitos, J. M., Poza, M., and
Villa, T. G. (2008). Cloning and expression of buffalo active chymosin
in Pichia pastoris, J. Agric. Food Chem., 56,
10606-10610.
20.Belenkaya, S. V., Rudometov, A. P., Shcherbakov,
D. N., Balabova, D. V., Krieger, A. V., Belov, A. N., Koval, A. D., and
Yelchaninov, V. V. (2018) Some biochemical properties of recombinant
Alpaca chymosin (Vicugna pacos L.), Appl. Biochem. Microbiol.,
54, 585-593.
21.Vallejo, J. A., Ageitos, J. M., Poza, M., and
Villa, T. G. (2012) A comparative analysis of recombinant chymosins,
J. Dairy Sci., 95, 609-613.
22.Bansal, N., Drake, M. A., Piraino, P., Broe, M.
L., Harboe, M., Fox, P. F., and McSweeney, P. L. H. (2009) Suitability
of recombinant camel (Camelus dromedarius) chymosin as a
coagulant for Cheddar cheese, Int. Dairy J., 19,
510-517.
23.Wei, C., Tang, B., Zhang, Y., and Yang, K. (1999)
Oxidative refolding of recombinant prochymosin, Biochem. J.,
340, 345.
24.Laemmli, U. K. (1970) Cleavage of structural
proteins during the assembly of the head of bacteriophage T4,
Nature, 227, 680.
25.Bradford, M. M. (1976) A rapid and sensitive
method for the quantitation of microgram quantities of protein
utilizing the principle of protein-dye binding, Anal. Biochem.,
72, 248-254.
26.Rudometov, A. P., Belenkaya, S. V., Shcherbakov,
D. N., Balabova, D. V., Krieger, A. V., and Yelchaninov, V. V. (2017)
Study of enzymatic stability of the liquid preparations of the
recombinant bovine (Bos taurus taurus L.) chymosin, Cheese-Making
and Butter-Making, 6, 40-43.
27.Hall, T. A. (1999) BioEdit: a user-friendly
biological sequence alignment editor and analysis program for Windows
95/98/NT, in Nucleic Acids Symposium Series, Information Retrieval
Ltd., London, UK, 41, 95-98.
28.Waterhouse, A., Bertoni, M., Bienert, S., Studer,
G., Tauriello, G., Gumienny, R., and Lepore, R. (2018) SWISS-MODEL:
homology modelling of protein structures and complexes, Nucleic
Acids Res., 46, W296-W303.
29.Pettersen, E. F., Goddard, T. D., Huang, C. C.,
Couch, G. S., Greenblatt, D. M., Meng, E. C., and Ferrin, T. E. (2004)
UCSF Chimera – a visualization system for exploratory research
and analysis, J. Comput. Chem., 25, 1605-1612.
30.Johnson, K. A., and Goody, R. S. (2011) The
original Michaelis constant: translation of the 1913
Michaelis–Menten paper, Biochemistry, 50,
8264-8269.
31.Naidu, A., Fitak, R. R., Munguia-Vega, A., and
Culver, M. (2011) Novel primers for complete mitochondrial cytochrome
b gene sequencing in mammals, Mol. Ecol. Res., 12,
191-196.
32.Pavlinov, I. Y. (2006) Systematics of Recent
Mammals, Moscow Univ. Publ., Moscow.
33.Shcherbakov, D. N., Rudometov, A. P.,
Yelchaninov, V. V., Belenkaya, S. V., Krieger, A. V., and Ilichev, A.
A. (2006) Recombinant plasmid ret21a-ProChym providing the synthesis of
the chimeric protein prochymosin in Bos Taurus, and
Escherichia coli strain BL21(DE3)pLysE
ret21a-ProChym – producer of the chimeric protein
Prochymosin in Bos Taurus, Russian Patent No. 2670071, SPK S12N 9/6483
(2006.01); S12N 15/64 (2006.01); S12N 15/70 (2006.01); S12Y 304/23004
(2006.01).
34.Pedersen, V. B., Christensen, K. A., and
Foltmann, B. (1979) Investigations on the activation of bovine
prochymosin, Eur. J. Biochem., 94, 573-580.
35.Wang, N., Wang, K. Y., Li, G., Guo, W., and Liu,
D. (2015) Expression and characterization of camel chymosin in
Pichia pastoris, Protein Expr. Purif., 111,
75-81.
36.Jiang, X. P., Yin, M. L., Chen, P., and Yang, Q.
(2012) Constitutive expression, purification and characterization of
bovine prochymosin in Pichia pastoris GS115, World J. Microbiol.
Biotechnol., 28, 2087-2093.
37.Noseda, D. G., Recúpero, M. N., Blasco,
M., Ortiz, G. E., and Galvagno, M. A. (2013) Cloning, expression and
optimized production in a bioreactor of bovine chymosin B in Pichia
(Komagataella) pastoris under AOX1 promoter, Protein Expr.
Purif., 9, 235-244.
38.Jensen, J. L., Mølgaard, A., Navarro
Poulsen, J. C., Harboe, M. K., Simonsen, J. B., Lorentzen, A. M., and
Larsen, S. (2013) Camel and bovine chymosin: the relationship between
their structures and cheese-making properties, Acta Crystallogr. D
Biol. Crystallogr., 69, 901-913.
39.Jensen, J. L., Jacobsen, J., Moss, M. L.,
Rasmussen, F., Qvist, K. B., Larsen, S., and van den Brink, J. M.
(2015) The function of the milk-clotting enzymes bovine and camel
chymosin studied by a fluorescence resonance energy transfer assay,
J. Dairy Sci., 98, 2853-2860.
40.Zhou, X. X., Wang, Y. B., Pan, Y. J., and Li, W.
F. (2008) Differences in amino acids composition and coupling patterns
between mesophilic and thermophilic proteins, Amino Acids,
34, 25-33.
41.Bobkova, E. V., Gedrovich, A. V., Ankilova, V.
N., Lavrik, O. I., Baratova, L. A., and Shishkov, A. V. (1990)
Comparative study of phenylalanyl-tRNA synthetases from
Escherichia coli and Thermus thermophilus by
tritium topography, Biochemistry, 55, 1570-1577.
Supplementary Figures S1, S2 (PDF)