Bacterial and Protozoan Lipoxygenases Could be Involved in Cell-to-Cell Signaling and Immune Response Suppression
G. F. Kurakin1,a*, A. M. Samoukina2, and N. A. Potapova3,4
1Department of Biochemistry and Laboratory Medicine, Tver State Medical University, Ministry of Health of the Russian Federation, 170100 Tver, Russia2Department of Microbiology, Virology, and Immunology, Tver State Medical University, Ministry of Health of the Russian Federation, 170100 Tver, Russia
3Kharkevich Institute for Information Transmission Problems, Russian Academy of Sciences, 127051 Moscow, Russia
4Faculty of Bioengineering and Bioinformatics, Lomonosov Moscow State University, 119234 Moscow, Russia
* To whom correspondence should be addressed.
Received June 19, 2020; Revised June 22, 2020; Accepted June 22, 2020
Lipoxygenases are found in animals, plants, and fungi, where they are involved in a wide range of cell-to-cell signaling processes. The presence of lipoxygenases in a number of bacteria and protozoa has been also established, but their biological significance remains poorly understood. Several hypothetical functions of lipoxygenases in bacteria and protozoa have been suggested without experimental validation. The objective of our study was evaluating the functions of bacterial and protozoan lipoxygenases by evolutionary and taxonomic analysis using bioinformatics tools. Lipoxygenase sequences were identified and examined using BLAST, followed by analysis of constructed phylogenetic trees and networks. Our results support the theory on the involvement of lipoxygenases in the formation of multicellular structures by microorganisms and their possible evolutionary significance in the emergence of multicellularity. Furthermore, we observed association of lipoxygenases with the suppression of host immune response by parasitic and symbiotic bacteria including dangerous opportunistic pathogens.
KEY WORDS: lipoxygenases, bacteria, protozoa, bioinformatics, phylogenetics, signaling, multicellularity, opportunistic pathogensDOI: 10.1134/S0006297920090059
INTRODUCTION
Lipoxygenases catalyze oxidation of polyunsaturated fatty acids (PUFAs) with the formation of hydroperoxides [1]. Eukaryotic lipoxygenases form a family of proteins with a conserved amino acid sequence. The active site of these enzymes contains non-heme iron or, rarely, manganese [2]. Lipoxygenases are abundant in animals, plants, and fungi [3, 4], where they perform the same function – synthesis of biologically active compounds oxylipins (oxidized PUFAs).
Arachidonic acid is the main substrate of vertebrate lipoxygenases. The resulting products – leukotrienes, hydroxy- and oxo-eicosatetraenoic acids, and lipoxins – control the inflammatory response, as well as apoptosis, thrombocyte function, and tumor development [4-8].
The products of lipoxygenase reaction in plants are involved in the organism response to damage and attack by pathogenic microorganisms [9-11]. The most known among them are jasmonates (cyclic oxylipins structurally similar to animal prostaglandins) that exhibit pronounced hormonal activity [10].
In addition to higher plants, lipoxygenases are also present in algae and mosses. It has been suggested that pheromones mediating chemoattraction between male and female gametes in brown algae are synthesized via the lipoxygenase pathway [12]. In mosses, the lipoxygenase pathway is likely required for normal development [13].
The most interesting feature of the lipoxygenase pathway in fungi is the presence of manganese lipoxygenases (MnLOXs) that contain manganese ion in the active site instead of iron ion. It is believed that these enzymes function as invasive factors and participate in quorum sensing and morphology switch in dimorphic fungi [12].
Hence, lipoxygenases in multicellular organisms are predominately involved in cell-to-cell communication, as well as the processes of antimicrobial defense and microbial aggression.
In view of this fact, the presence of lipoxygenases in some bacteria and protozoa seems important. Using molecular genetics and bioinformatics approaches, lipoxygenase genes have been identified in 0.5% of sequenced bacterial genomes (as of 2015) [14]. Only few of these enzymes have been investigated in vitro, including lipoxygenases from two cyanobacteria of the Nostoc genus [15, 16], cyanobacterium of the Anabaena genus [17], Myxococcus xanthus of myxobacteria [18], and Pseudomonas aeruginosa of the pseudomonad group [19, 20]. In Protozoa, the lipoxygenase sequence was identified only in the slime mold Dictyostelium discoideum [3]. At present, the functions of bacterial and protozoan lipoxygenases remain poorly understood [2, 14, 21].
Most of the data available on the lipoxygenase prevalence in bacteria are presented in the review by Horn et al. [14]; however, no association of their presence with the ecophysiology of respective organisms has been examined. A high prevalence of lipoxygenases in nitrifying bacteria was noted in this review and their association with nitrogen metabolism was suggested [14, 22]. A high prevalence of lipoxygenases in cyanobacteria was also mentioned, and an attempt was made to relate this fact to the Great Oxidation Event caused by their activity [14]. However, the biochemical mechanisms behind such association remain obscure.
The majority of data on the lipoxygenase functions have been accumulated for the enzyme from the rod-shaped pathogenic bacterium P. aeruginosa. This lipoxygenase is likely one of the bacterial pathogenic factors. It was found that it converts host fatty acids into lipoxins that exhibit the anti-inflammatory and local immunosuppressive activity. It was assumed based on this fact that P. aeruginosa uses lipoxygenase to suppress host immunity [19, 20]. The data became available recently that another lipoxygenase isozyme initiates ferroptosis of the bronchial epithelium by cleaving host cell phospholipids [23].
Horn et al. emphasized in their review that no lipoxygenases have been found in the majority of bacterial pathogens of humans. In particular, these enzymes are absent in staphylococci, streptococci, Mycobacterium tuberculosis and Mycobacterium leprae, Vibrio cholerae, Legionella pneumophila, Helicobacter pylori, and Klebsiella pneumoniae [14]. The authors concluded that the pathogenicity of these bacteria does not require the presence of lipoxygenase [14].
The existing hypotheses on the functions of bacterial lipoxygenases were comprehensively summarized by Goloshchapova et al. [24]:
a) cell-cell signaling – this function is postulated for P. aeruginosa and suggested for myxobacteria demonstrating complex patterns of social behavior;
b) invasive growth and disruption of cell membranes by the pathogen – this mechanism was suggested for P. aeruginosa;
c) evasion of immune response – this strategy was discussed above for P. aeruginosa;
d) lipoxygenases as oxygen sensors – this function is suggested for P. aeruginosa lipoxygenase based on its low oxygen affinity;
e) detoxification of PUFAs, which are detrimental for many bacteria.
However, no experimental data have been provided in this work to support one hypothesis or another.
Considering an insufficient amount of available experimental data, it seemed reasonable to conduct phylogenetic and taxonomic profiling of bacterial and protozoan lipoxygenases and compare obtained results with the literature data on the ecophysiology of respective organisms. However, only few of such studies have been reported so far.
The review by Horn et al. [14] only presents the taxonomic profile at the phylum level, which does not allow elucidating associations with the ecophysiology. Hansen et al. [25] conducted phylogenetic profiling of bacterial lipoxygenases, but compared the results with biochemical rather than ecophysiological data.
The objective of our study was to establish an association between the phylogenetic and taxonomic distribution of bacterial and protozoan lipoxygenases and ecophysiological features of the organisms.
MATERIALS AND METHODS
We searched for potential lipoxygenase sequences with BLAST [26] using UniProt (https://www.uniprot.org/blast/) [27] and NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) databases. The two databases were used to reduce the probability of false negative results. The main search among bacteria was carried out using the UniProt database, and the control search was done using the NCBI database. The main search among eukaryotes was performed using the NCBI database, and the control one – the UniProt database. The use of different databases for bacteria and protozoa was due to the fact that BLAST UniProt is optimized for visual control of a large number of alignments (more than 500 for bacteria), while NCBI is optimal for a targeted search among a relatively low number of sequences (which is the case for protozoa).
Lipoxygenase sequences from phylogenetically distant organisms in the Swiss-Prot database were used as queries: human arachidonate 12-lipoxygenase (UniProt ID: P18054) [28], manganese lipoxygenase from the fungus Gaeumannomyces graminis (UniProt ID: Q8X151) [29], and linoleate 9/13-lipoxygenase from the bacterium P. aeruginosa (UniProt ID: Q8RNT4) [30].
The functional activity of the identified proteins was determined indirectly based on the presence of conserved lipoxygenase domain, which was identified using the Batch-CD-Search online service [31] and CDD database [32] with the E-value threshold 0.01. In the case of inconclusive BLAST results (E-value close to one), the presence of the conserved metal-binding sites was confirmed manually.
Only sequences with the lipoxygenase domain and conserved metal-binding sites were used for further investigation. Since the presence of these sequences indicates a high probability of lipoxygenase activity of proteins, further in the paper, proteins that were found to contain them will be referred to as lipoxygenases and considered as such.
In order to evaluate the frequency of lipoxygenase occurrence from the taxonomical point of view, a relative occurrence frequency for each genus was calculated.
Frequency of occurrence = (Number of genera with lipoxygenases/Number of predicted proteomes) × 100
The number of predicted proteomes was evaluated with the UniProt database considering only non-redundant proteomes. The used equation mitigates the effect of differences in the number of available genomes/proteomes.
Multiple alignments were constructed with the online MAFFT v.7 software (https://mafft.cbrc.jp/alignment/server/) [33, 34] using the FFT-NS-i algorithm with iterative refinement [35]; the sequences with multiple insertions and deletions were removed using the MaxAlign server integrated with MAFFT v. 7 MaxAlign [36]. Phylogenetic trees were constructed with the MAFFT programs using the neighbor-joining method and MEGA [37] with the minimum evolution method. Conserved sites and Poisson substitution model were used in both methods. The bootstrap technique with 500 repetitions was used to test tree nodes. Visualization and analysis of phylogenetic trees were carried out using the iTOL online tools (https://itol.embl.de/) [38].
In addition to the phylogenetic trees, phylogenetic networks were constructed and examined using the SplitsTree program for their construction and visualization [39]. The trees and the networks were constructed for the entire set of discovered lipoxygenases and for certain groups for detailed analysis.
RESULTS AND DISCUSSION
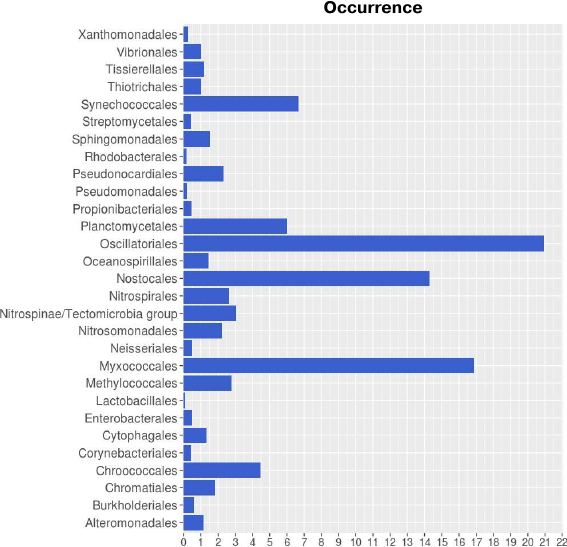
Taxonomic distribution of lipoxygenases corroborates the hypothesis on their association with cell-to-cell signaling. Taxonomic distribution of lipoxygenases at the phylum level was in general agreement with the study by Horn et al. [14]. However, we conducted a more detailed analysis at the order level. Lipoxygenases were found to be more prevalent among the representatives of the Oscillatoriales (20.93), Myxococcales (16.87), and Nostocales (14.29) orders (Fig. 1). The orders Oscillatoriales and Nostocales include cyanobacteria with filamentous morphology; the order Myxococcales includes bacteria that form fruiting bodies. The following taxonomic distribution of cyanobacteria lipoxygenases was determined: Oscillatoriales (20.93) > Nostocales (14.29) > Synechococcales (6.67) > Chroococcales (4.49), hence lipoxygenases are more prevalent in multicellular cyanobacteria than in unicellular ones. This suggests that bacterial lipoxygenases are associated with the formation of multicellular structures.
Fig. 1. Occurrence of lipoxygenases in different orders of bacteria. Image produced with the R Studio (v.1.2.1335).
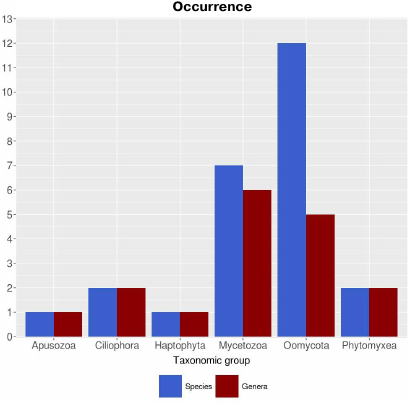
Calculating the prevalence of lipoxygenases in Protozoa with correction for proteomes is difficult due to the limited data available. According to the calculations with the uncorrected parameters, lipoxygenases are prevalent in the taxa Oomycota (12 species and 5 genera) and Mycetozoa (7 species and 6 genera) (Fig. 2). Formation of fungus-like mycelial forms is typical for oomycetes, while Mycetozoa representatives are alternatively called slime molds and form multicellular structures of true plasmodium or pseudoplasmodium and fruiting bodies. Only 1-2 species were identified as containing lipoxygenases in other groups of protozoa. This pattern is also in agreement with the suggestion on the association of lipoxygenases with multicellularity. Additional information was provided by phylogenetic analysis.
Fig. 2. Occurrence of lipoxygenases in Protozoa (parameters are nor corrected for the number of available proteomes). Image produced with the R Studio (v.1.2.1335).
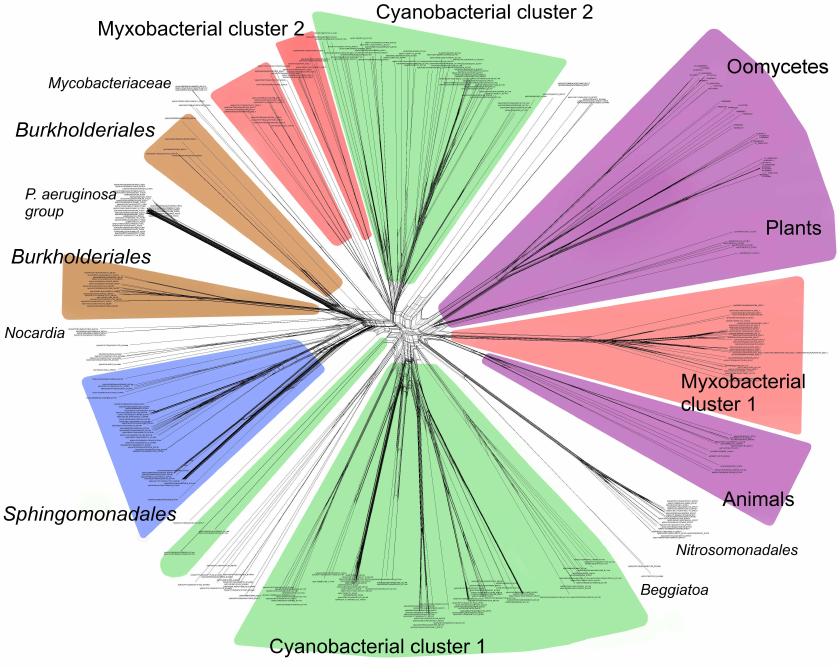
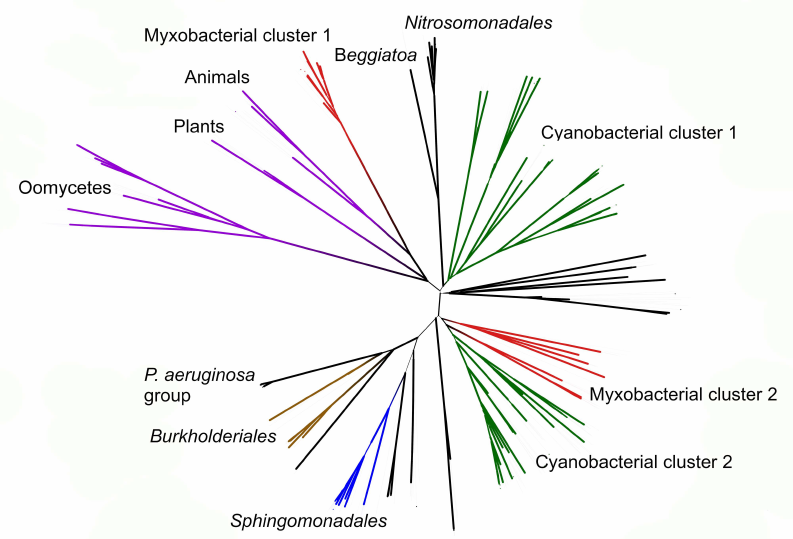
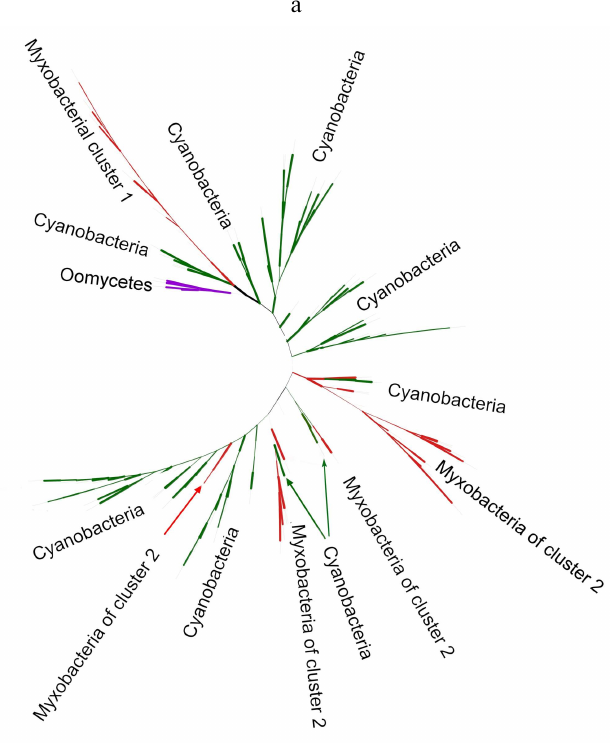
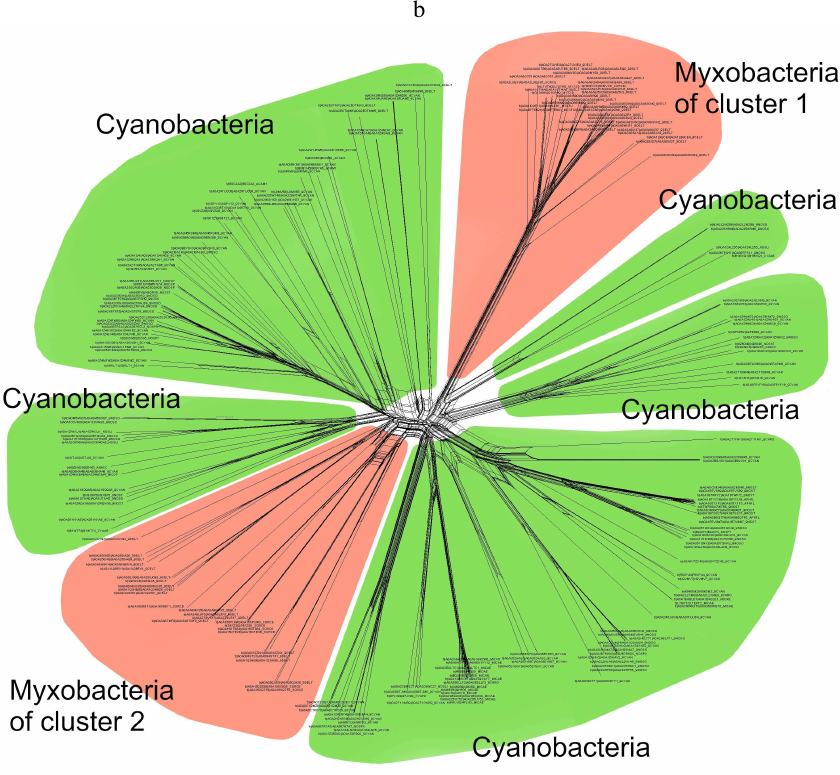
Phylogenetic analysis demonstrates close evolutionary association of lipoxygenases and multicellularity. Lipoxygenases from cyanobacteria form two large separate clusters in phylogenetic trees and networks, which are marked in the figures as cyanobacterial cluster I and cyanobacterial cluster II. They are most pronounced in the phylogenetic network (Fig. 3) and trees constructed using the minimum evolution method (Fig. 4). The fact that they are discovered using different methods makes their monophyly plausible despite a rather low bootstrap support (less than 59). Moreover, the cluster taxonomic structure overlaps to a large degree, and approximately 15-20% of species and strains are the same in both clusters, which could occur if the clusters have originated from a common ancestor that initially had two lipoxygenase isozymes.
Fig. 3. Phylogenetic network of all analyzed lipoxygenases. Image produced with the SplitsTree program [39].
Fig. 4. Initial phylogenetic tree for all analyzed lipoxygenases constructed with the minimum evolution method using the MEGA program [37]. Bootstrap support confidence levels are represented by the line thickness. Image produced using the iTOL server [38].
The remaining taxonomic groups – both bacterial and eukaryotic – are presented in the trees and phylogenetic networks by a single cluster or separate branches; moreover, all these branches gravitate towards one of the two cyanobacterial clusters. The tree segment between the clusters has very few branches, and there are no branches in the consensus tree. This suggests that lipoxygenases had first originated in cyanobacteria and next (after lipoxygenase gene duplication in their common ancestor) have been acquired by the rest of the groups via horizontal gene transfer. The fact that both cyanobacterial clusters branch out from the network center, while the rest of the groups are mainly combined into two net-like clusters at the periphery of the central node reflects this course of events in the phylogenetic network.
The presence of two cyanobacterial clusters is the main difference of our results from the results reported by Hansen et al. [25], which show only one cluster of cyanobacteria with two large groups of proteobacteria located on both sides, while the trees and the networks constructed in our work contain two cyanobacterial clusters. This could be explained by a lower number of cyanobacterial sequences examined in the Hansen’s study (29 versus >500 in our work) [25], which either did not allow detecting the second cluster or resulted in combining all cyanobacterial lipoxygenases into a single cluster.
Nevertheless, two groups of proteobacterial lipoxygenases can be identified in our trees that are located on the opposite sides of the cyanobacterial clusters. Each of the proteobacterial clusters gravitates towards one of the cyanobacterial clusters.
Lipoxygenases from myxobacteria also form two clusters (one in each of the proteobacterial group). The first of them (myxobacterial cluster I) is monophyletic with a bootstrap support of 100 and includes the highest number of genera, which implies that it has most likely originated from a common ancestor of myxobacteria. It must be mentioned that it is represented in the tree by a long branch, while in the network, it is located at equal distances from both cyanobacterial clusters; hence, the distance between this cluster and cyanobacteria is large.
The myxobacterial cluster II does not have a common ancestor node and consists of at least two groups with a common ancestor branching from the cyanobacterial cluster II root. In some trees, it is divided into a larger number of branches. In the phylogenetic network, it is separated and is in close proximity to the cyanobacterial cluster II, which only indicates phylogenetic closeness of these two clusters.
Similarly to the cyanobacterial clusters, the taxonomic composition of the myxobacterial clusters overlaps, and some of the strains in these clusters coincide. This raises the question on which of these two taxonomic groups (cyanobacteria or myxobacteria) was the first to have lipoxygenases, and which acquired them via horizontal transfer. The third option also exists implying that both taxa have acquired lipoxygenases from a common ancestor; however, this seems very unlikely, as the topology of the trees and the network does not indicate the phylogeny of the main bacterial taxa (in particular, myxobacteria are not closely related to cyanobacteria). Furthermore, this hypothesis implies the presence of lipoxygenases in the latest common ancestor of bacteria and loss of these enzymes by the majority of present-day taxa, which seems unlikely.
To address this issue, we analyzed the trees that included myxobacteria and cyanobacteria only (Fig. 5a). It was found that the myxobacterial cluster I (the largest one) included a sister group of cyanobacterial lipoxygenases with a bootstrap support of 100. This group was absent in the trees constructed for the complete set of data because it was removed during alignment with the MaxAlign program. Similarly, this cluster was present in the phylogenetic network for the same subset of data (Fig. 5b). Based on these facts, horizontal transfer of lipoxygenases from a common myxobacterial ancestor to cyanobacteria seems very unlikely; hence, direct or indirect transfer from cyanobacteria to myxobacteria is a more plausible scenario.
Fig. 5. a) Phylogenetic tree targeting phylogenetic relations of cyanobacterial and myxobacterial lipoxygenases (nearest neighbor method, MAFFT program [33, 34]). Oomycetes were used as an outgroup. The levels of bootstrap support are represented by line thickness. Due to the presentation of additional groups, the cyanobacterial clusters are not well pronounced, while the myxobacterial clusters are still visible. Image produced with the iTOL server [38]. b) Phylogenetic network for myxobacterial and cyanobacterial lipoxygenases. Similarly to Fig. 5a, myxobacterial cluster I includes sister group of cyanobacteria. Myxobacterial cluster II is clearly visible. Image produced with the SplitsTree program [39].
The myxobacterial cluster II is represented in all trees by several trunks, which leads us to the most “economic” hypothesis. It assumes that lipoxygenases had originated in a common ancestor of cyanobacteria and later both its isozymes have been acquired by myxobacteria via horizontal transfer. It is likely that a single-time transfer of one of the isozymes to the common myxobacteria ancestor had occurred, which resulted in the emergence of the myxobacterial cluster I. Next, after splitting of myxobacteria into several branches (which in our models are presented as myxobacterial cluster II), a multiple transfer of the second isozyme to myxobacteria has occurred.
The branches formed by eukaryotes in our trees gravitate towards the cyanobacterial cluster I, but no additional data on their evolutionary relationships are available. Their topology varied in different trees, but in none of the trees, they formed a monophyletic cluster. In the phylogenetic network, they originate from a single net-like cluster; however, it could be a result of the long branch attraction. Hence, it is likely that eukaryotes have acquired their lipoxygenases as a result of several sequential horizontal gene transfer events.
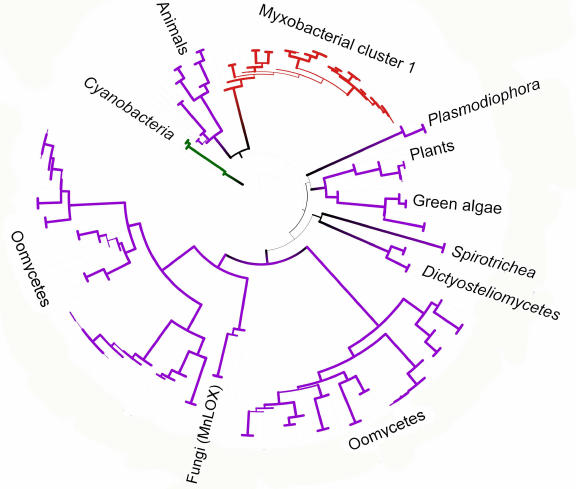
We constructed a tree for the data subset including eukaryotes and myxobacteria from cluster I. It can be seen that oomycete lipoxygenase and fungal manganese lipoxygenases have a common origin (bootstrap support 93) (Fig. 6), which is especially interesting considering similar life forms and ecology of these organisms. It also can be stated with confidence that lipoxygenases from plants and green algae have originated from a common ancestor (bootstrap support 95), which can be expected from the general biology point of view. Close relation of lipoxygenases from the Oxytrichia trifallax and Stylonichia lemnae ciliates and the cellular slime molds (Dictyosteliomycetes) can be also suggested (bootstrap support, 63.6). The latter also form a monophyletic cluster with a bootstrap support 100, although the lipoxygenase sequence from the mycophagous slime mold Planoprotostelium fungivorum has been found outside of this cluster, and its origin is unknown.
Fig. 6. Phylogenetic tree targeting phylogenetic relations of lipoxygenases from eukaryotes and cluster I myxobacteria (minimum evolution method, MEGA program [37], visualized with iTOL [38]). Cyanobacteria are used as an outgroup; bootstrap values are presented by line thickness.
Close phylogenetic relation between animal lipoxygenases and enzymes from the myxobacterial cluster I is probably our most interesting finding (Fig. 6). These two groups in the constructed trees have a common ancestor with a bootstrap support up to 76; moreover, animal lipoxygenases do not cluster with other eukaryotes. The myxobacterial cluster I in the phylogenetic network is located inside the net-like cluster of eukaryotes. Such close relations suggest either horizontal transfer of lipoxygenase genes to myxobacteria from animals, or the reverse process.
Large lipoxygenase clusters in the reconstructed evolutionary history were found closely related to the multicellular life forms. This trend can be observed starting from the enzyme origin in the common ancestor of cyanobacteria presented in the analysis; most likely it was a multicellular organism [40, 41]. The distribution pattern of lipoxygenases in their unicellular and multicellular descendants is similar to the distribution of regulators required for differentiation [41]: high occurrence frequency in multicellular life-forms and low occurrence in unicellular organisms.
The suggested evolutionary path for lipoxygenases follows the emergence of multicellularity in different taxa. Lipoxygenases are associated both taxonomically and phylogenetically with the formation of multicellular structures. Considering that in “true” multicellular organisms, lipoxygenases are involved in cell signaling, it can be suggested that they have the same function in bacteria and protists forming multicellular structures. Hence, our data support the hypothesis on the association of bacterial and protozoan lipoxygenases with cell-cell signaling [hypothesis (a) in the Introduction].
Few experimental data on the functions of lipoxygenases in microorganisms could provide additional corroboration of this hypothesis. Even P. aeruginosa that does not form multicellular structures, uses oxylipins as quorum sensing mediators [42]; lipoxygenase products in M. xanthus that forms multicellular structures, enhance twitching motility [43]. The culmination phase of the fruiting body formation is disrupted in the lipoxygenase mutant of D. discoideum belonging (Mycetozoa) [44].
Taxonomic distribution of lipoxygenases supports the hypothesis on their participation in the bacteria–host signaling. In addition to the enzymes from cyanobacteria, myxobacteria, and some eukaryotes, large clusters are formed only by lipoxygenases from sphingobacteria (Sphingomonadales) and representatives of the Burkholderiales order. The rest of lipoxygenases are found sporadically in individual representatives of various genera, families, or orders. In phylogenetic trees, they form individual branches or small taxonomically heterogenous clusters. Further, we will refer to this phenomenon as sporadic occurrence.
Below, we will show that the representatives of the Burkholderiales order exhibit the same ecophysiological properties as organisms with sporadic presence of lipoxygenases and, hence, they will be considered as one group.
Many bacteria from this group are pathogenic (table). Multiple lipoxygenase sequences were found in P. aeruginosa, which was in agreement with the available experimental data. We found lipoxygenases in many other bacteria with similar pathogenicity profiles. Almost all of them are opportunistic or nosocomial pathogens, and only rarely they cause community-acquired infections in immunocompetent patients. The identified lipoxygenase carriers are capable of infecting many organs, but the majority of them are associated with surgical infections of skin and soft tissues, lung infections (in patients with accompanying lung pathology), sepsis, and catheter-associated infections [45-50]. Many of them demonstrate antibiotic resistance.
Lipoxygenase occurrence in parasitic and symbiotic bacteria

Note. Opportunistic human bacterial pathogens are shown in red, plant
pathogens and symbionts – in green, animal parasites and
symbionts – in blue, bacteria carrying molecular indicators
of opportunistic pathogenicity are shown in orange. Species shown in
two colors play two physiological roles. Bacteria with unknown
ecophysiology are shown on grey background. Genus name in square
brackets indicate recent re-classification.
The group includes both widely known opportunistic pathogens (P. aeruginosa, Acinetobacter baumannii [51], Enterobacter cloacae [52]) and emerging causative agents of infection with similar properties. Nocardia pseudobrasiliensis [53], Rhodococcus erythropolis [54], and such outstanding representative as recently described Cedecea lapagei [55, 56], belong to the latter. Some representatives of the Burkholderia genus also belong to this group, such as B. stagnalis and B. singularis initially isolated form respiratory samples [57, 58], Burkholderia gladioli previously known as plant pathogen and recently described as an agent causing infections in humans [59-61], and Burkholderia thailandensis [62].
As in the review by Horn et al. [14], we did not identify lipoxygenases in specific obligate pathogens [14]. The only difference was that lipoxygenase was found in K. pneumoniae. However, this bacterium belongs to the Klebsiella-Enterobacter-Serratia group of predominantly opportunistic pathogens. Its high implication in community-acquired infections is considered an exception due to the presence of massive polysaccharide capsule [45].
Hence, sporadic occurrence of lipoxygenases was found to be associated with pathogenicity but not with specific highly pathogenic species (which is in general agreement with the conclusions of Horn et al. [14]). Lipoxygenases are more typical for bacteria with diminished pathogenicity that affect only immunocompromised hosts. This can be demonstrated more clearly for lipoxygenase-containing B. thailandensis, which is close to melioidosis causative agent Burkholderia pseudomallei (lacks lipoxygenase) but has significantly lower virulence [62]. The same trend is observed for the lipoxygenase-carrying opportunistic pathogen Burkholderia, while Burkholderia mallei that causes glanders and in B. pseudomallei that causes melioidosis lack the enzyme.
Not only opportunistic pathogens, but also bacteria associated with animals or plants have been identified among the lipoxygenase carriers. They can behave as pathogens or as symbionts. Lipoxygenase carriers with symbiotic functions usually have a specific host range. Species associated with plants often are growth-promoting. Among them are Rhodococcus 66b [63], Variovorax paradoxus [64, 65], Gynuella sunshinyii [66], and Pseudomonas fluorescens (can infect humans but it is not common and occurs very rarely) [46, 67, 68].
On the contrary, pathogenic representatives infect both plants and humans. B. gladioli, Burkholderia cepacia [50], and P. aeruginosa itself, which is phytopathogen infecting a wide range of hosts, are examples of such bacteria [69, 70]. Hence, pathogenic carriers of lipoxygenases are the so-called cross-kingdom pathogens. A detailed review devoted to these pathogens was published [71], which discussed molecular prerequisites of such properties. The cross-kingdom pathogens usually cause opportunistic or nosocomial infections in humans because their pathogenicity factors that function well in plants, cannot completely overcome human defense systems [71, 72]. It is likely that lipoxygenase is another typical virulence factor making cross-kingdom host jump possible, leading to opportunistic pathogenicity.
The discovery of lipoxygenases in bacteria with similar pathogenic properties corroborates the hypothesis on their association with virulence. The majority of the above-described bacteria are considered as analogues of P. aeruginosa, in which lipoxygenase helps to evade host immune response. The data obtained in this work allow expanding this suggestion onto many other species. Hence, the data on the prevalence of lipoxygenases are in agreement with the hypothesis on the participation of these enzymes in the microbial invasion [hypotheses (b) and (c) in Introduction).
This suggestion could be also extended to some lipoxygenase-containing bacteria with unknown ecophysiology. Lipoxygenase gene or protein sequences identified in these species indicate the possibility of transfer of genes providing opportunistic pathogenicity or virulence from the causative agents of opportunistic infections. These bacteria include:
1) Grimontia indica that has a set of virulence factors, which is slightly smaller in comparison with the typical pathogen Grimontia hollisae allowing considering it as a molecular indicator of opportunistic pathogenicity [73];
2) Oligoflexus tunisiensis that carries a gene for RND (resistance-nodulation-division) efflux pump that displays 67% identity with the multidrug resistance genes in Achromobacter xylosooxidans and P. aeruginosa [74];
3) Pseudomonads in the P. aeruginosa group that have unclear species assignment: Pseudomonas sp. HMSC059F05, Pseudomonas sp. HMSC065H01, and Pseudomonas sp. RW410. According to our data produced during the BLAST search, exotoxin A and elastase from these strains have a high degree of identity with the corresponding proteins from P. aeruginosa (99.5% and above).
Lipoxygenase carriers associated with animals have been mostly found in marine species. In particular, Endozoicomonas numazuensis [75] and Candidatus Entotheonella palauensis [76] are symbionts of sponges; Pseudobacteriovorax antillogorgiicola [77] and Enterovibrio coralii [78] were isolated from corals; Enterovibrio nigricans [79] and Enterovibrio norvegicus [80] were isolated from fish gut. Nocardia seriolae was characterized as a causative agent of nocardiosis in fish [81].
One of the animal-associated species – Photorhabdus temperata – is simultaneously a symbiont of entomopathogenic nematode and a pathogen for its host [82]. In general, lipoxygenase carriers demonstrate both symbiotic and parasitic strategies. This makes the hypothesis (c) from Introduction (immune response evasion) more plausible than the hypothesis (b) associated with invasive growth. Suppression of the host immunity is required for both symbiont and pathogen existence, while symbiont does not need invasive growth factors.
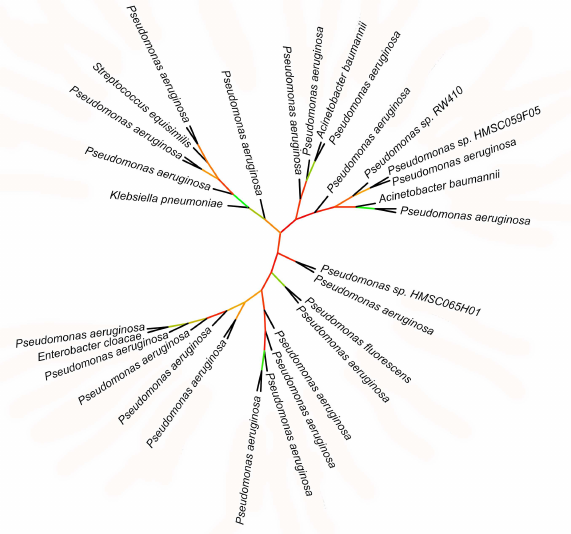
Phylogenetic data support association of lipoxygenases with symbiosis and pathogenicity. Lipoxygenases from some opportunistic pathogens form a cluster in phylogenetic trees and networks that contain closely related sequences differing by a few amino acid substitutions only. The bootstrap support level of this cluster is 100. The majority of sequences in this cluster belonged to P. aeruginosa, hence, it was termed “P. aeruginosa group”. In addition to this species, the group includes lipoxygenases of such dangerous nosocomial pathogens as Streptococcus dysgalactiae, K. pneumoniae, E. cloacae, A. baumannii, as well as lipoxygenase from P. fluorescens.
Because of slight differences between the sequences in the P. aeruginosa group, the phylogenetic relations within this group are unclear (Fig. 7): bootstrap support of the majority of nodes in the phylogenetic tree is extremely low (30 and below). On this background, the leaves formed by lipoxygenases from S. dysgalactiae, K. pneumoniae, E. cloacae, P. fluorescens, and one of the A. baumannii lipoxygenases stand out: together with the P. aeruginosa sister sequences, they split from the nodes with a relatively high bootstrap support level (>65%). This indicates rather recent (in evolutionary terms) horizontal transfers of lipoxygenase gene from P. aeruginosa to other nosocomial pathogens. Hence, lipoxygenase genes can be spread between pathogens similarly to the antibiotic resistance genes (it must be mentioned that antibiotic resistance and presence of lipoxygenase most often co-exist in the pathogens from our list).
Fig. 7. Initial phylogenetic tree constructed for P. aeruginosa group using the minimum evolution method with the MEGA program [37]. Unlike in previous figures, the bootstrap support level is presented by color (red – low support, green – high support). Visualization with iTOL [38].
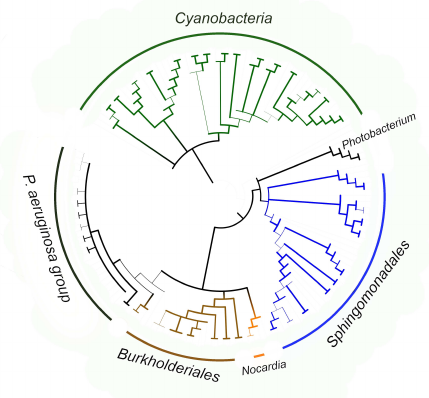
The fact that the P. aeruginosa group itself is located within the clade formed by pathogenic and symbiotic representatives of the Burkholderiales order (Fig. 8) is an additional confirmation of lipoxygenase association with pathogenesis/symbiosis. Pathogenic representatives of the Nocardia genus are also in this clade. The bootstrap support level of the node at the basis of the clade is 90 for the tree constructed using MAFFT, and above 99 in the tree constructed with the MEGA program. It is very likely that the existence of this clade reflects lipoxygenase gene transfer from one pathogenic/symbiotic bacterium to another.
Fig. 8. Consensus tree constructed with the MEGA program [37] using the minimum evolution method for representatives of P. aeruginosa group, Burkholderiales and Sphingomonadales orders, and Nocardia and Photobacterium genera. Cyanobacteria were used as an outgroup to calculate the bootstrap support. The length of branches in this tree does not correspond to the evolutionary distances (they are extremely low in P. aeruginosa group). The levels of bootstrap support are represented by the line thickness. Visualization with iTOL [38].
We also conducted an additional phylogenetic investigation of lipoxygenase from the emerging nosocomial pathogen C. lapagei. It was possible to examine only one isozyme due to a large number of insertions and deletions. It was demonstrated using two methods (construction of phylogenetic tree with the minimum evolution method and construction of phylogenetic network) that this lipoxygenase is closely related to lipoxygenase from another nosocomial pathogen, Pluralibacter gergoviae. This is in agreement with the suggestion considering lipoxygenase as a horizontally transferred virulence factor, and demonstrates evolutionary connection between lipoxygenases and pathogenicity.
Concluding remarks and unsolved issues. No lipoxygenases were found in archaea by using the BLAST search with the same queries as for bacteria and protists. This is in agreement with the data reported by Horn et al. [14] and can be explained from the biochemical point of view: the lipoxygenase pathway starts with phospholipase A2 that cleaves phospholipids in the cytoplasmic membrane, which is impossible in archaea because of different membrane structure. This fact is also in agreement with lipoxygenase origin in bacteria (cyanobacteria in particular) and its further acquisition by eukaryotes.
Unfortunately, many bacteria carrying lipoxygenase have been discovered only recently, and their ecophysiology is still poorly understood. Due to this, no conclusions could be made on the bacteria from the Alteromonadales, Bdellovibrionales, Cytophagales, Chromatiales, Sphingomonadales orders and from representatives of the Photobacterium and Enterovibrio genera. Further investigations could possibly provide answers to this question. It was not possible to draw any conclusions regarding the functional role of lipoxygenases in the protist group that includes Oxytrichia trifalax, S. lemnae, Emiliania huxleyi, and Thecamonas trahens species. Future studies in this direction are especially promising as they could provide additional confirmation or disprove our conclusions.
The discovered lipoxygenases from the Plasmiodiophorida slime molds and representatives of the Phytophtora genus are of interest as they fit both hypotheses: association of lipoxygenases with multicellularity and pathogenicity. On one hand, Phytophthora has a fungus-like structure, while slime molds form syncytial plasmodium, which can be considered as a tendency to multicellularity. On the other hand, these representatives of the two taxa are plant pathogens. The question on the role of lipoxygenase in these organisms still remains open.
In the beginning of this manuscript, we summarized existing hypotheses on the functional significance of bacterial and protozoan lipoxygenases. Bioinformatics analysis was performed to validate them.
The results of taxonomic data processing and construction of evolutionary trees and networks are in agreement with two of the suggested hypotheses – association with multicellularity and cell-to-cell signaling [hypothesis (a) in Introduction] and suppression of the host immune response by a parasite or a symbiont [hypothesis (c) in Introduction].
The first hypothesis has evolutionary significance. The emergence of lipoxygenases in organisms with primitive cell cooperation and differentiation could indicate their involvement in the origin of multicellularity. Elucidation of particular signaling mechanisms mediated by the lipoxygenase products requires further investigation.
Inter-kingdom signaling mechanisms that suppress host immune response have been on the other hand investigated in detail using P. aeruginosa as a model organism. Our data allow expanding this hypothesis to a larger number of bacteria and to define their features. It is very likely that similar strategy is used by many nosocomial and opportunistic infection agents that have acquired lipoxygenase from each other as a result of horizontal gene transfer. This virulence factor is as dangerous as antibiotic resistance genes, which dictates that close attention should be paid to lipoxygenases in pathogenic organisms.
Ethics declarations. The authors declare no conflict of interest in financial or any other sphere. This article does not contain any studies with animals or human participants performed by any of the authors.
REFERENCES
1.Boldyrev, A. A., Kyaivyaryainen, E. I., and
Ilyukha, V. A. (2017) Biomembranologiya: Uchebnoe Posobie
(Biological Membranes: Manual) INFRA-M, Moscow.
2.Garreta, A., Val-Moraes, S. P.,
García-Fernández, Q., Busquets, M., Juan, C., Oliver A.,
Ortiz A., Gaffney, B. J., Fita, I., Manresa, A., and Carpena, X. (2013)
Structure and interaction with phospholipids of a prokaryotic
lipoxygenase from Pseudomonas aeruginosa, FASEB J.,
27, 4811-4821, doi: 10.1096/fj.13-235952.
3.Brash, A. R. (1999) Lipoxygenases: occurrence,
functions, catalysis, and acquisition of substrate, J. Biol.
Chem., 274, 23679-23682,
doi: 10.1074/jbc.274.34.23679.
4.Mashima, R., and Okuyama, T. (2015) The role of
lipoxygenases in pathophysiology; new insights and future perspectives,
Redox Biol., 6, 297-310,
doi: 10.1016/j.redox.2015.08.006.
5.Yokomizo, T. (2014) Two distinct leukotriene B4
receptors, BLT1 and BLT2, J. Biochem., 157, 65-71,
doi: 10.1093/jb/mvu078.
6.Kanaoka, Y., and Boyce, J. A. (2004) Cysteinyl
leukotrienes and their receptors: cellular distribution and function in
immune and inflammatory responses, J. Immunol., 173,
1503-1510, doi: 10.4049/jimmunol.173.3.1503.
7.Powell, W. S., and Rokach, J. (2015) Biosynthesis,
biological effects, and receptors of hydroxyeicosatetraenoic acids
(HETEs) and oxoeicosatetraenoic acids (oxo-ETEs) derived from
arachidonic acid, Biochim. Biophys. Acta, 1851, 340-355,
doi: 10.1016/j.bbalip.2014.10.008.
8.Kieran, N. E., Maderna, P., and Godson, C. (2004)
Lipoxins: potential anti-inflammatory, proresolution, and antifibrotic
mediators in renal disease, Kidney Int., 65, 1145-1154,
doi: 10.1111/j.1523-1755.2004.00487.x.
9.Brodhun, F., and Feussner, I. (2011) Oxylipins in
fungi, FEBS J., 278, 1047-1063,
doi: 10.1111/j.1742-4658.2011.08027.x.
10.Heldt, H.-W. (2011) Plant Biochemistry,
Academic Press, London.
11.Wasternack, C. (2007) Jasmonates: an update on
biosynthesis, signal transduction and action in plant stress response,
growth and development, Ann. Bot., 100, 681-697,
doi: 10.1093/aob/mcm079.
12.Andreou, A., Brodhun, F., and Feussner, I. (2009)
Biosynthesis of oxylipins in non-mammals, Prog. Lipid Res.,
48, 148-170, doi: 10.1016/j.plipres.2009.02.002.
13.De León, I. P., Hamberg, M., and
Castresana, C. (2015) Oxylipins in moss development and defense,
Front. Plant. Sci., 6, 483,
doi: 10.3389/fpls.2015.00483.
14.Horn, T., Adel, S., Schumann, R., Sur, S.,
Kakularam, K. R., Polamarasetty, A., Redanna, P., Kuhn, H., and
Heydeck, D. (2015) Evolutionary aspects of lipoxygenases and genetic
diversity of human leukotriene signaling, Prog. Lipid Res.,
57, 13-39, doi: 10.1016/j.plipres.2014.11.001.
15.Andreou, A. Z., Vanko, M., Bezakova, L., and
Feussner, I. (2008) Properties of a mini 9R-lipoxygenase from
Nostoc sp. PCC 7120 and its mutant forms, Phytochemistry,
69, 1832-1837, doi: 10.1016/j.phytochem.2008.03.002.
16.Lang, I., Göbel, C., Porzel, A., Heilmann,
I., and Feussner, I. (2008) A lipoxygenase with linoleate diol synthase
activity from Nostoc sp. PCC 7120, Biochem. J.,
410, 347-357, doi: 10.1042/BJ20071277.
17.Wang, X., Lu, F., Zhang, C., Lu, Y., Bie, X.,
Ren, D., and Lu, Z. (2014) Peroxidation radical formation and
regiospecificity of recombinated Anabaena sp. lipoxygenase and
its effect on modifying wheat proteins, J. Agric. Food. Chem.,
62, 1713-1719, doi: 10.1021/jf405425c.
18.An, J. U., Hong, S. H., and Oh, D. K. (2018)
Regiospecificity of a novel bacterial lipoxygenase from Myxococcus
xanthus for polyunsaturated fatty acids, Biochim. Biophys. Acta
Mol. Cell Biol. Lipids, 1863, 823-833,
doi: 10.1016/j.bbalip.2018.04.014.
19.Vance, R. E., Hong, S., Gronert, K., Serhan, C.
N., and Mekalanos, J. J. (2004) The opportunistic pathogen
Pseudomonas aeruginosa carries a secretable arachidonate
15-lipoxygenase, Proc. Natl. Acad. Sci. USA, 101,
2135-2139, doi: 10.1073/pnas.0307308101.
20.Banthiya, S., Kalms, J., Yoga, E. G., Ivanov, I.,
Carpena, X., Hamberg, M., Kuhn, H., and Scheerer, P. (2016) Structural
and functional basis of phospholipid oxygenase activity of bacterial
lipoxygenase from Pseudomonas aeruginosa, Biochim. Biophys.
Acta, 1861, 1681-1692,
doi: 10.1016/j.bbalip.2016.08.002.
21.Porta, H., and Rocha-Sosa, M. (2001) Lipoxygenase
in bacteria: a horizontal transfer event? Microbiology,
147, 3199-3200, doi: 10.1099/00221287-147-12-3199.
22.Koeduka, T., Kajiwara, T., and Matsui, K. (2007)
Cloning of lipoxygenase genes from a cyanobacterium, Nostoc
punctiforme, and its expression in Eschelichia coli,
Curr. Microbiol., 54, 315-319,
doi: 10.1007/s00284-006-0512-9.
23.Dar, H. H., Tyurina, Y. Y., Mikulska-Ruminska,
K., Shrivastava, I., Ting, H. C., et al. (2019) Pseudomonas
aeruginosa utilizes host polyunsaturated phosphatidylethanolamines
to trigger theft-ferroptosis in bronchial epithelium, J. Clin.
Invest., 128, 4639-4653, doi: 10.1172/JCI99490.
24.Goloshchapova, K., Stehling, S., Heydeck, D.,
Blum, M., and Kuhn, H. (2019) Functional characterization of a novel
arachidonic acid 12S-lipoxygenase in the halotolerant bacterium
Myxococcus fulvus exhibiting complex social living patterns,
MicrobiologyOpen, 8, e00775,
doi: 10.1002/mbo3.775.
25.Hansen, J., Garreta, A., Benincasa, M.,
Fusté, M. C., Busquets, M., and Manresa, A. (2013) Bacterial
lipoxygenases, a new subfamily of enzymes? A phylogenetic approach,
Appl. Microbiol. Biotechnol., 97, 4737-4747,
doi: 10.1007/s00253-013-4887-9.
26.Altschul, S. F., Madden, T. L., Schäffer, A.
A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997) Gapped
BLAST and PSI-BLAST: a new generation of protein database search
programs, Nucleic Acids Res., 25, 3389-3402,
doi: 10.1093/nar/25.17.3389.
27.UniProt Consortium (2018) UniProt: the universal
protein knowledgebase, Nucleic Acids Res., 46, 2699,
doi: 10.1093/nar/gky092.
28.Yoshimoto, T., Yamamoto, Y., Arakawa, T., Suzuki,
H., Yamamoto, S., Yokoyama, C., Tanabe, T., and Toh, H. (1990)
Molecular cloning and expression of human arachidonate 12-lipoxygenase,
Biochem. Biophys. Res. Commun., 172, 1230-1235,
doi: 10.1016/0006-291x(90)91580-l.
29.Hörnsten, L., Su, C., Osbourn, A. E.,
Hellman, U., and Oliw, E. H. (2002) Cloning of the manganese
lipoxygenase gene reveals homology with the lipoxygenase gene family,
Eur. J. Biochem., 269, 2690-2697,
doi: 10.1046/j.1432-1033.2002.02936.x.
30.Vidal-Mas, J., Busquets, M., and Manresa, A.
(2005) Cloning and expression of a lipoxygenase from Pseudomonas
aeruginosa 42A2, Antonie van Leeuwenhoek, 87,
245-251, doi: 10.1007/s10482-004-4021-1.
31.Marchler-Bauer, A., and Bryant, S. H. (2004)
CD-Search: protein domain annotations on the fly, Nucleic Acids
Res., 32 (suppl. 2), W327-W331,
doi: 10.1093/nar/gkh454.
32.Marchler-Bauer, A., Bo, Y., Han, L., He, J.,
Lanczycki, C. J., et al. (2017) CDD/SPARCLE: functional classification
of proteins via subfamily domain architectures, Nucleic Acids
Res., 45, D200-D203, doi: 10.1093/nar/gkw1129.
33.Katoh, K., Rozewicki, J., and Yamada, K. D.
(2019) MAFFT online service: multiple sequence alignment, interactive
sequence choice and visualization, Brief. Bioinform., 20,
1160-1166, doi: 10.1093/bib/bbx108.
34.Kuraku, S., Zmasek, C. M., Nishimura, O., and
Katoh, K. (2013) aLeaves facilitates on-demand exploration of metazoan
gene family trees on MAFFT sequence alignment server with enhanced
interactivity, Nucleic Acids Res., 41, W22-W28,
doi: 10.1093/nar/gkt389.
35.Katoh, K., Misawa, K., Kuma, K. I., and Miyata,
T. (2002) MAFFT: a novel method for rapid multiple sequence alignment
based on fast Fourier transform, Nucleic Acids Res., 30,
3059-3066, doi: 10.1093/nar/gkf436.
36.Gouveia-Oliveira, R., Sackett, P. W., and
Pedersen, A. G. (2007) MaxAlign: maximizing usable data in an
alignment, BMC Bioinformatics, 8, 312,
doi: 10.1186/1471-2105-8-312.
37.Kumar, S., Stecher, G., Li, M., Knyaz, C., and
Tamura, K. (2018) MEGA X: molecular evolutionary genetics analysis
across computing platforms, Mol. Biol. Evol., 35,
1547-1549, doi: 10.1093/molbev/msy096.
38.Letunic, I., and Bork, P. (2016) Interactive tree
of life (iTOL) v3: an online tool for the display and annotation of
phylogenetic and other trees, Nucleic Acids Res., 44,
W242-W245, doi: 10.1093/nar/gkw290.
39.Huson, D. H., and Bryant, D. (2006) Application
of phylogenetic networks in evolutionary studies, Mol. Biol.
Evol., 23, 254-267, doi: 10.1093/molbev/msj030.
40.Schirrmeister, B. E., Antonelli, A., and Bagheri,
H. C. (2011) The origin of multicellularity in cyanobacteria, BMC
Evol. Biol., 11, 45, doi: 10.1186/1471-2148-11-45.
41.Elhai, J., and Khudyakov, I. (2018) Ancient
association of cyanobacterial multicellularity with the regulator HetR
and an RGSGR pentapeptide‐containing protein (PatX), Mol.
Microbiol., 110, 931-954, doi: 10.1111/mmi.14003.
42.Martínez, E., Cosnahan, R. K., Wu, M.,
Gadila, S. K., Quick, E. B., Mobley, J. A., and Campos-Gómez, J.
(2019) Oxylipins mediate cell-to-cell communication in Pseudomonas
aeruginosa, Commun. Biol., 2, 1-10,
doi: 10.1038/s42003-019-0310-0.
43.An, J. U., and Oh, D. K. (2018) Stabilization and
improved activity of arachidonate 11S-lipoxygenase from proteobacterium
Myxococcus xanthus, J. Lipid Res., 59, 2153-2163,
doi: 10.1194/jlr.M088823.
44.Basu, S., Fey, P., Pandit, Y., Dodson, R., Kibbe,
W. A., and Chisholm, R. L. (2012) DictyBase 2013: integrating multiple
Dictyostelid species, Nucleic Acids Res., 41, D676-D683,
doi: 10.1093/nar/gks1064.
45.Levinson, W. (2008) Medical Microbiology and
Immunology, 8th Edn., McGraw-Hill/Appleton & Lange, New
York.
46.Boitsov, A. G., and Vasil’ev, O. D. (2013)
Klinicheskaya Laboratornaya Diagnostika: Natsional’noe
Rukovodstvo (Dolgov, V. V., and Menshikov, V. V., eds.)
GEOTAR-Media, Moscow, pp. 380-388.
47.Kaftyreva, L. A., Boitsov, A. G., and Makarova M.
A. (2013) Klinicheskaya Laboratornaya Diagnostika:
Natsional’noe Rukovodstvo (Dolgov, V. V., and Menshikov, V.
V., eds.) GEOTAR-Media, Moscow, pp. 342-365.
48.Totolyan, A. A., Burova, L. A., Dmitriev, A. V.,
and Suvorov, A. N. (2013) Klinicheskaya Laboratornaya Diagnostika:
Natsional’noe Rukovodstvo (Dolgov, V. V., and Menshikov, V.
V., eds.) GEOTAR-Media, Moscow, pp. 417-435.
49.Van’t Wout, E. F., van Schadewijk, A., van
Boxtel, R., Dalton, L. E., Clarke, H. J., Tommassen, J., Marciniak, S.
J., and Hiemstra, P. S. (2015) Virulence factors of Pseudomonas
aeruginosa induce both the unfolded protein and integrated stress
responses in airway epithelial cells, PLoS Pathog., 11,
doi: 10.1371/journal.ppat.1004946.
50.Stoyanova, M., Pavlina, I., Moncheva, P., and
Bogatzevska, N. (2007) Biodiversity and incidence of
Burkholderia species, Biotechnology and Biotechnological
Equipment, 21, 306-310,
doi: 10.1080/13102818.2007.10817465.
51.Antunes, L., Visca, P., and Towner, K. J. (2014)
Acinetobacter baumannii: evolution of a global pathogen,
Pathog. Dis., 71, 292-301,
doi: 10.1111/2049-632X.12125.
52.Davin-Regli, A. (2015) Enterobacter
aerogenes and Enterobacter cloacae; versatile bacterial
pathogens confronting antibiotic treatment, Front. Microbiol.,
6, 392, doi: 10.3389/fmicb.2015.00392.
53.Lebeaux, D., Lanternier, F., Degand, N.,
Catherinot, E., Podglajen, I., Rubio, M. T., Suarez, F., Lecuit, M.,
Mainardi, J.-L., and Lortholary, O. (2010) Nocardia
pseudobrasiliensis as an emerging cause of opportunistic infection
after allogeneic hematopoietic stem cell transplantation, J. Clin.
Microbiol., 48, 656-659, doi: 10.1128/JCM.01244-09.
54.Baba, H., Nada, T., Ohkusu, K., Ezaki, T.,
Hasegawa, Y., and Paterson, D. L. (2009) First case of bloodstream
infection caused by Rhodococcus erythropolis, J. Clin.
Microbiol., 47, 2667-2669,
doi: 10.1128/JCM.00294-09.
55.Hong, S. K., Lee, J. S., and Kim, E. C. (2015)
First Korean case of Cedecea lapagei pneumonia in a patient with
chronic obstructive pulmonary disease, Ann. Lab. Med.,
35, 266-268, doi: 10.3343/alm.2015.35.2.266.
56.Herrera, V. R. C., De Silva, M. F. R., Alcaraz,
H. O., Espiritu, G. C., Peña, K. C., and Melnikov, V. (2018)
Death related to Cedecea lapagei in a soft tissue bullae
infection: a case report, J. Med. Case Rep., 12, 328,
doi: 10.1186/s13256-018-1866-x.
57.Vandamme, P., Peeters, C., De Smet, B., Price, E.
P., Sarovich, D. S., Henry, D. A., Hird, T. J., Zlosnik, J. E. A.,
Mayo, M., Warner, J., Baker, A., Currie, B. J., and Carlier, A. (2017)
Comparative genomics of Burkholderia singularis sp. nov., a low
G+C content, free-living bacterium that defies taxonomic dissection of
the genus Burkholderia, Front. Microbiol., 8,
1679, doi: 10.3389/fmicb.2017.01679.
58.De Smet, B., Mayo, M., Peeters, C., Zlosnik, J.
E., Spilker, T., Hird, T. J., LiPuma, J. J., Kidd, T. J., Kaestli, M.,
Ginther, J. L., Wagner, D. M., Keim, P., Bell, S. C., Jacobs, J. A.,
Currie, B. J., and Vandamme, P. (2015) Burkholderia stagnalis
sp. nov. and Burkholderia territorii sp. nov., two novel
Burkholderia cepacia complex species from environmental and
human sources, Int. J. Syst. Evol. Microbiol., 65,
2265-2271, doi: 10.1099/ijs.0.000251.
59.Segonds, C., Clavel-Batut, P., Thouverez, M.,
Grenet, D., Le Coustumier, A., Plésiat, P., and Chabanon, G.
(2009) Microbiological and epidemiological features of clinical
respiratory isolates of Burkholderia gladioli, J. Clin.
Microbiol., 47, 1510-1516,
doi: 10.1128/JCM.02489-08.
60.Quon, B. S., Reid, J. D., Wong, P., Wilcox, P.
G., Javer, A., Wilson, J. M., and Levy, R. D. (2011) Burkholderia
gladioli – a sessorictor of poor outcome in cystic
fibrosis patients who receive lung transplants? A case of locally
invasive rhinosinusitis and persistent bacteremia in a 36-year-old lung
transplant recipient with cystic fibrosis, Can. Respir. J.,
18, e64-e65, doi: 10.1155/2011/304179.
61.Imataki, O., Kita, N., Nakayama-Imaohji, H.,
Kida, J. I., Kuwahara, T., and Uemura, M. (2014) Bronchiolitis and
bacteraemia caused by Burkholderia gladioli in a non-lung
transplantation patient, New Microbes New Infect., 2,
175, doi: 10.1002/nmi2.64.
62.Haraga, A., West, T. E., Brittnacher, M. J.,
Skerrett, S. J., and Miller, S. I. (2008) Burkholderia
thailandensis as a model system for the study of the
virulence-associated type III secretion system of Burkholderia
pseudomallei, Infect. Immun., 76, 5402-5411,
doi: 10.1128/IAI.00626-08.
63.Thatcher, L. F., Myers, C. A., O’Sullivan,
C. A., and Roper, M. M. (2017) Draft genome sequence of
Rhodococcus sp. strain 66b, Genome Announc.,
5, e00229-17, doi: 10.1128/genomeA.00229-17.
64.Han, J. I., Spain, J. C., Leadbetter, J. R.,
Ovchinnikova, G., Goodwin, L. A., Han, C. S., Woyke, T., Davenport, K.
W., and Orwin, P. M. (2013) Genome of the root-associated plant
growth-promoting bacterium Variovorax paradoxus strain
EPS, Genome Announc., 1, e00843-13,
doi: 10.1128/genomeA.00843-13.
65.Han, J. I., Choi, H. K., Lee, S. W., Orwin, P.
M., Kim, J., LaRoe, S. L., Kim, T.-G., O’Neil, J., Leadbetter, J.
R., Lee, S. Y., Hur, C.-G., Spain, J. C., Ovchinnikova, G., Goodwin,
L., and Han, C. (2011) Complete genome sequence of the metabolically
versatile plant growth-promoting endophyte Variovorax paradoxus
S110, J. Bacteriol., 193, 1183-1190,
doi: 10.1128/JB.00925-10.
66.Chung, E. J., Park, J. A., Jeon, C. O., and
Chung, Y. R. (2015) Gynuella sunshinyii gen. nov., sp. nov., an
antifungal rhizobacterium isolated from a halophyte, Carex
scabrifolia Steud, Int. J. Syst. Evol. Microbiol.,
65, 1038-1043, doi: 10.1099/ijs.0.000060.
67.Gibb, A. P., Martin, K. M., Davidson, G. A.,
Walker, B., and Murphy, W. G. (1995) Rate of growth of Pseudomonas
fluorescens in donated blood, J. Clin. Pathol., 48,
717-718, doi: 10.1136/jcp.48.8.717.
68.Gershman, M. D., Kennedy, D. J., Noble-Wang, J.,
Kim, C., Gullion, J., Kacica, M., Jensen, B., Pascoe, N., Saiman, L.,
McHale, J., Wilkins, M., Schoonmaker-Bopp, D., Clayton, J., Arduino,
M., Srinivasan, A., and Pseudomonas fluorescens Investigation Team
(2008) Multistate outbreak of Pseudomonas fluorescens
bloodstream infection after exposure to contaminated heparinized saline
flush prepared by a compounding pharmacy, Clin. Infect. Dis.,
47, 1372-1379, doi: 10.1086/592968.
69.Walker, T. S., Bais, H. P., Déziel, E.,
Schweizer, H. P., Rahme, L. G., Fall, R., and Vivanco, J. M. (2004)
Pseudomonas aeruginosa-plant root interactions. Pathogenicity,
biofilm formation, and root exudation, Plant Physiol.,
134, 320-331, doi: 10.1104/pp.103.027888.
70.Rahme, L. G., Ausubel, F. M., Cao, H., Drenkard,
E., Goumnerov, B. C., Lau, G. W., Mahajan-Miklos, S., Plotnikova, J.,
Tan, M. W., Tsongalis, J., Walendziewicz, C. L., and Tompkins, R. G.
(2000) Plants and animals share functionally common bacterial virulence
factors, Proc. Natl. Acad. Sci. USA, 97, 8815-8821,
doi: 10.1073/pnas.97.16.8815.
71.Van Baarlen, P., Van Belkum, A., Summerbell, R.
C., Crous, P. W., and Thomma, B. P. (2007) Molecular mechanisms of
pathogenicity: how do pathogenic microorganisms develop cross-kingdom
host jumps? FEMS Microbiol. Rev., 31, 239-277,
doi: 10.1111/j.1574-6976.2007.00065.x.
72.Relman, D. A., and Falkow, S. (2015) A molecular
perspective of microbial pathogenicity, in Principles and Practice
of Infectious Diseases, Eighth Edition (Bennett, J. E.,
Dolin, R., and Blaser, M. J., eds.) Elsevier Saunders, Philadelphia,
pp. 1-10.
73.Singh, A., Vaidya, B., Khatri, I., Srinivas, T.
N. R., Subramanian, S., Korpole, S., and Pinnaka, A. K. (2014)
Grimontia indica AK16T, sp. nov., isolated
from a seawater sample reports the presence of pathogenic genes similar
to Vibrio genus, PLoS One, 9, e85590,
doi: 10.1371/journal.pone.0085590.
74.Nakai, R., Fujisawa, T., Nakamura, Y., Baba, T.,
Nishijima, M., Karray, F., Sami Sayadi, S., Isoda, N.,
Naganuma, T., and Niki, H. (2016) Genome sequence and overview
of Oligoflexus tunisiensis Shr3 T in the eighth class
Oligoflexia of the phylum Proteobacteria, Stand. Genomic. Sci.,
11, 90, doi: 10.1186/s40793-016-0210-6.
75.Nishijima, M., Adachi, K., Katsuta, A., Shizuri,
Y., and Yamasato, K. (2013) Endozoicomonas numazuensis sp. nov.,
a gammaproteobacterium isolated from marine sponges, and emended
description of the genus Endozoicomonas Kurahashi and Yokota
2007, Int. J. Syst. Evol. Microbiol., 63, 709-714,
doi: 10.1099/ijs.0.042077-0.
76.Schmidt, E. W., Obraztsova, A. Y., Davidson, S.
K., Faulkner, D. J., and Haygood, M. G. (2000) Identification of the
antifungal peptide-containing symbiont of the marine sponge
Theonella swinhoei as a novel
δ-proteobacterium,“Candidatus Entotheonella
palauensis”, Marine Biol., 136, 969-977,
doi: 10.1007/s002270000273.
77.McCauley, E. P., Haltli, B., and Kerr, R. G.
(2015) Description of Pseudobacteriovorax antillogorgiicola gen.
nov., sp. nov., a bacterium isolated from the gorgonian octocoral
Antillogorgia elisabethae, belonging to the family
Pseudobacteriovoracaceae fam. nov., within the order Bdellovibrionales,
Int. J. Syst. Evol. Microbiol., 65, 522-530,
doi: 10.1099/ijs.0.066266-0.
78.Thompson, F. L., Thompson, C. C., Naser, S.,
Hoste, B., Vandemeulebroecke, K., Munn, C., Bourne, D., and Swings, J.
(2005) Photobacterium rosenbergii sp. nov. and Enterovibrio
coralii sp. nov., vibrios associated with coral bleaching, Int.
J. Syst. Evol. Microbiol., 55, 913-917,
doi: 10.1099/ijs.0.63370-0.
79.Pascual, J., Macian, M. C., Arahal, D. R., Garay,
E., and Pujalte, M. J. (2009) Description of Enterovibrio
nigricans sp. nov., reclassification of Vibrio
calviensis as Enterovibrio calviensis comb. nov. and emended
description of the genus Enterovibrio Thompson et al. 2002,
Int. J. Syst. Evol. Microbiol., 59, 698-704,
doi: 10.1099/ijs.0.001990-0.
80.Thompson, F. L., Hoste, B., Thompson, C. C.,
Goris, J., Gomez-Gil, B., Huys, L., De Los, P., and Swings, J. (2002)
Enterovibrio norvegicus gen. nov., sp. nov., isolated from the
gut of turbot (Scophthalmus maximus) larvae: a new member of the
family Vibrionaceae, Int. J. Syst. Evol. Microbiol., 52,
2015-2022, doi: 10.1099/00207713-52-6-2015.
81.URL: https://thefishsite.com/articles/nocardia-seriolae-a-chronic-problem.
82.Hurst, S., Rowedder, H., Michaels, B., Bullock,
H., Jackobeck, R., Abebe-Akele, F., Durakovic, U., Gately, J., Janicki,
E., and Tisa, L. S. (2015) Elucidation of the Photorhabdus
temperata genome and generation of a transposon mutant library to
identify motility mutants altered in pathogenesis, J.
Bacteriol., 197, 2201-2216,
doi: 10.1128/JB.00197-15.