REVIEW: rRNA Methylation and Antibiotic Resistance
I. A. Osterman1,2, O. A. Dontsova1,2,3, and P. V. Sergiev1,2,4,a*
1Center of Life Sciences, Skolkovo Institute of Science and Technology, 143028 Skolkovo, Russia2Faculty of Chemistry, Lomonosov Moscow State University, 119991 Moscow, Russia
3Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, 117997 Moscow, Russia
4Institute of Functional Genomics, Lomonosov Moscow State University, 119991 Moscow, Russia
* To whom correspondence should be addressed.
Received June 12, 2020; Revised July 16, 2020; Accepted July 16, 2020
Methylation of nucleotides in rRNA is one of the basic mechanisms of bacterial resistance to protein synthesis inhibitors. The genes for corresponding methyltransferases have been found in producer strains and clinical isolates of pathogenic bacteria. In some cases, rRNA methylation by housekeeping enzymes is, on the contrary, required for the action of antibiotics. The effects of rRNA modifications associated with antibiotic efficacy may be cooperative or mutually exclusive. Evolutionary relationships between the systems of rRNA modification by housekeeping enzymes and antibiotic resistance-related methyltransferases are of particular interest. In this review, we discuss the above topics in detail.
KEY WORDS: ribosome, methylation, antibiotics, macrolides, aminoglycosidesDOI: 10.1134/S000629792011005X
Abbreviations: DC, decoding center; PET, peptide exit tunnel; PTC, peptidyl transferase center; rRNA, ribosomal RNA.
INTRODUCTION
One of the major mechanisms of antibiotic action is inhibition of protein synthesis via binding to the ribosome functional sites: peptidyl transferase center (PTC), decoding center (DC), and peptide exit tunnel (PET) [1]. These centers also contain most of the modified ribosomal RNA (rRNA) nucleotides [2]. Methylation of the binding sites is one of the common mechanisms of antibiotic resistance that is ribosome-mediated. It has been detected in both antibiotic-producing strains and pathogenic bacteria causing various diseases in animals and humans.
At the same time, many modified rRNA nucleotides found in a broad range of bacteria or even ubiquitous are not associated with the action of antibiotics. However, some of these modified (mostly methylated) residues provide either moderate resistance to antibiotics or, on the contrary, determine susceptibility to antibiotics, thus affecting bacterial adaptation and resistance (the so-called fitness cost) and acting as evolutionary reservoirs for the emergence of resistance genes. In this article, we review various aspects of the effect of methylation on antibiotic resistance.
METHYLATION OF 23S rRNA NUCLEOTIDE RESIDUES IN THE PET
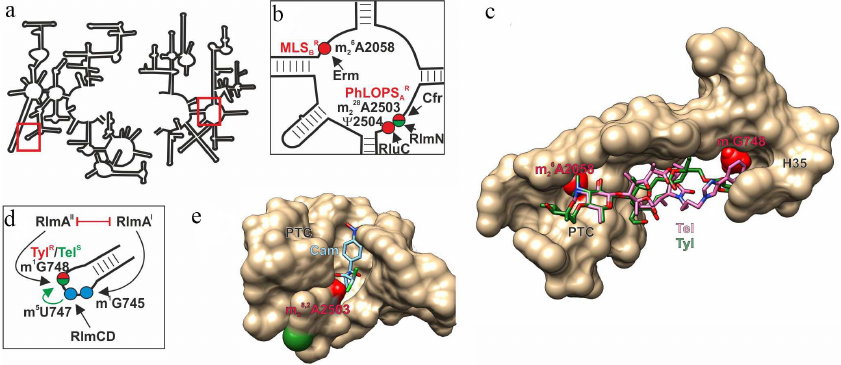
Nascent proteins leave ribosome through the PET that penetrates the large ribosomal subunit from the PTC to the opposite (cytoplasmic) side. PET interacts with various regulatory peptide sequences and some classes of antibiotics [3]. The most comprehensively studied (and most important from the medical point of view) modification is dimethylation of the PET-located 23S rRNA A2058 residue at the N6 atom (here and below, numeration is given according to Escherichia coli rRNA) by the Erm family methylases (Fig. 1, a-c; table) [4]. This modification confers resistance to a large group of antibiotics, including macrolides, lincosamides, and streptogramins B [4, 5]. 23S rRNA A2058 residue, which is located in the PET close to the PTC, interacts with antibiotics of all these three classes. Dimethylation (in the case of lincosamides monomethylation by TlrD) and some related enzymes [5] disrupts antibiotic binding due to inability of formation of hydrogen bond with A2058 and overlap with the methyl group (Fig. 1c). This resistance mechanism is common among macrolide producers and pathogenic strains, against which these antibiotics are traditionally used [6]. It is believed that the source of the erm gene family in bacteria, including pathogenic ones, has been horizontal gene transfer from the producer strains [7]. At the same time, the origin of ancestral erm genes in the producer strains remains unclear. The search for amino acid sequences homologous to the Erm protein sequences in bacteria [8] led to identification of the universally conserved 16S rRNA methyltransferase RsmA(KsgA) as the most evolutionarily close protein. Moreover, replacement of only two protein loops modifies substrate specificity of RsmA(KsgA), so that the enzyme acquires the ability to modify Erm substrates [9]. It is possible that the ancestor enzyme in the evolution of resistance mediated by Erm methyltransferases is 16S rRNA m26A1518/19 methyltransferase, which also demethylates adenosine residue at the exocyclic N6 atom.
Fig. 1. Effect of nucleotide modifications in PET and PTC on interaction with antibiotics. a) Secondary structure of 23S rRNA. Fragments in red boxes are show in other panels. b) Secondary structure of the 23S rRNA region V that forms a fragment of PET and PTC. Modified nucleotides discussed in the text are shown as circles: red circles, modifications of these nucleotides induce resistance to antibiotics (MLSBR, resistance to macrolides, lincosamides, and streptogramins B; PhLOPSAR, resistance to phenicols, lincosamides, oxazolidinones, pleuromycins, and streptogramins A); green circles, modification of these nucleotides facilitates antibiotic binding. Arrows indicate activity of the corresponding modifying enzymes. c) 3D structure of the PET-forming ribosome fragment (shown as molecular surface) with bound tylosin (green skeletal model) and telithromycin (pink skeletal model). Red van der Waals spheres correspond to the methyl groups of 23S rRNA m1G748 and m26A2058 residues overlapping with the tylosin-binding site and in the case of m26A2058, with the telithromycin-binding site. d) Secondary structure of the H35 helix of 23S rRNA that forms a fragment of the PET. Modified nucleotides discussed in the text are shown as circles: red/green circle, modification responsible for increased resistance to tylosin and susceptibility to telithromycin; blue circles, residues modified by housekeeping enzymes. Black arrows indicate activity of the corresponding modifying enzymes; green arrow indicates facilitation of one modification by another; red line shows mutually exclusive modifications. e) 3D structure of the ribosome fragment that forms PTC with bound chloramphenicol (blue skeletal model). Red and green van der Waals spheres correspond to the methyl groups of 23S rRNA m28,2A2503 residue that promote or prevent, respectively, binding of chloramphenicol and other antibiotics. The images, including overlapping 3D structures and models of modified nucleotide residues, were created with the UCSF Chimera software [10] using secondary rRNA structures [11] and 3D structures 1K9M [12], 4V7Z [13], and 6ND5 [14].
rRNA methyltransferases affecting antibiotic resistance
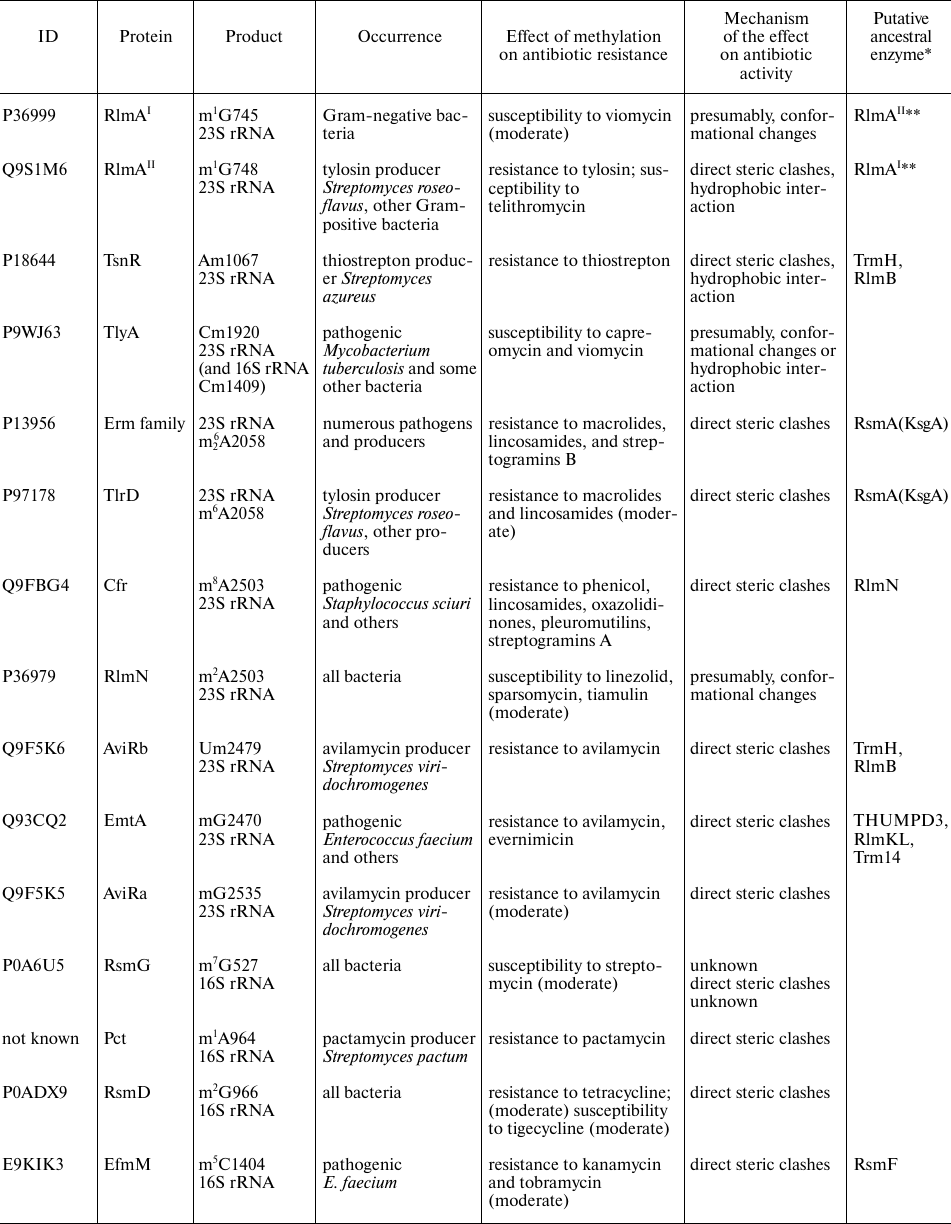
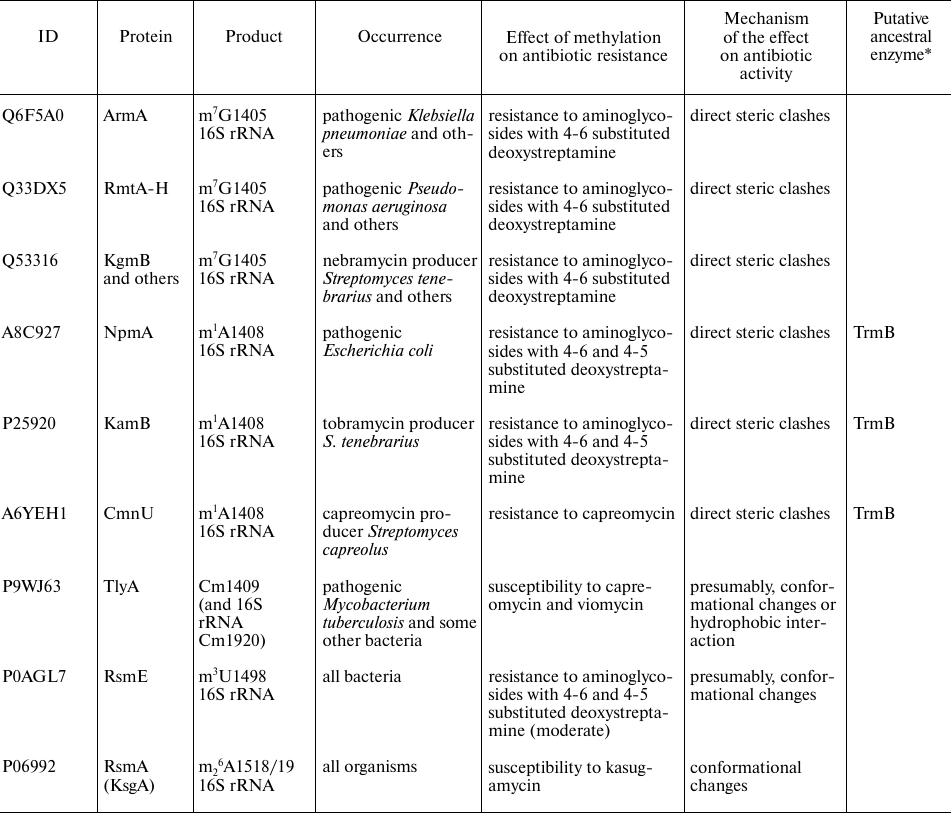
Notes. * Similarity of amino acid sequences was estimated with the BLAST
program [8]. ** RlmAI and
RlmAII are paralogs found in the Gram-negative and
Gram-positive bacteria, respectively.
The question arises on why the genes for Erm methyltransferases, unlike RsmA(KsgA), have not spread to other taxonomical groups. Apparently, the m26A2058 modification in 23S rRNA decreases adaptability of bacteria in the absence of antibiotics. Negative effect of methylation might be due to the fact that modified ribosomes synthesize protein in suboptimal proportions because of the interaction of the modified A2058 with amino acid residues of the nascent protein inside the ribosomal tunnel, rather than to the increased expenses for methyltransferase synthesis [15]. Cells with constitutively active ErmC methyltransferase and, therefore, carrying dimethylated A2058, are less competitive than the cells lacking this modification [15]. This might explain suppression of erm genes in the absence of macrolides not only in the majority of resistant strains, but also in the producer strains themselves [16]. The most common mechanism of the erm expression regulation involves halting of the ribosome on a short reading frame upstream of the methyltransferase gene [17-19], which makes translation initiation site of the resistance gene accessible for the binding with other ribosome. The alternative mechanisms for activation of erm genes in the presence of antibiotics are transcription attenuation [20], the use of specific transcription factor [22], and ketolide-induced frameshift [22].
Methylation of the 23S rRNA G748 residue, also located in the ribosome large subunit tunnel (Fig. 1, a, c, and d), at the N1 atom simultaneously with monomethylation of A2058 provides resistance of the producer bacteria to tylosin (16-membered macrolide with bulky sugar substituents) [23], while modification of either A2058 or G748 alone fails to confer the resistance. Formation of 23S rRNA m1G748 is catalyzed by methyltransferase RlmAII (TlrD), which is commonly found in Gram-positive bacteria. Gram-negative bacteria usually contain its homolog RlmAI that modifies the neighboring 23S rRNA G745 residue at the N1 position [24] (Fig. 1d; table). RlmAI is a housekeeping enzyme. It is possible that both these methyltransferases had originated from a common ancestor. Expression of the rlmAII gene in Bacillus subtilis is regulated by transcription and translation attenuation, probably, to minimize negative effect of methylation on the adaptability in the absence of antibiotic [25], which questions classification of such enzymes as housekeeping proteins.
Interestingly, the increasing resistance of the Streptococcus pneumoniae containing dimethylated A2058 to the erythromycin synthetic derivative telithromycin (2 to 32 µg/ml increase in the minimal inhibition concentration) requires inactivation of the rlmAII gene and, respectively, absence of the 23S rRNA G748 modification (unlike resistance to tylosin provided by rlmAII expression) [26]. This discrepancy could be explained by the fact that the binding site for telithromycin does not overlap with the methyl group; on the contrary, its location ensures antibiotic interaction with the modified nucleotide (Fig. 1c). The 23S rRNA m5U747 residue modified by the constitutive methyltransferase RlmCD [27] (Fig. 1d) and located next to G748, promotes recognition of the latter by methyltransferase RlmAII and, therefore, indirectly facilitates telithromycin binding [28].
As shown for macrolides, rRNA methylation could result in either antibiotic resistance because of the overlap of methyl groups with antibiotic-binding site or, on the contrary, induce antibiotic susceptibility due to the methyl group interaction with antibiotic. This example also illustrates mutually excluding phylogenetic distribution of rRNA methyltransferases (rlmAI and rlmAII) or their cooperative action (RlmCD and RlmAII).
METHYLATION OF 23S rRNA NUCLEOTIDES IN THE PTC
PTC is the major catalytic site of the ribosome, where formation of peptide bonds between amino acids attached to tRNAs takes place. Many PTC residues are modified by housekeeping enzymes [2]. Modification of nucleotide residues in the PTC often results in antibiotic resistance. Thus, modification of 23S rRNA A2503 (Fig. 1, b and e) blocks binding of several classes of antibiotics. Methylation of this residue at the C8 atom by methyltransferase Cfr induces cell resistance to phenicols, lincosamides, pleuromycins, oxazolidinones, and streptogramins A [29]. It is surprising that despite the wide occurrence of the cfr gene in Gram-positive and Gram-negative bacteria, it is absent in antibiotic producers. This modification has not been preserved in all bacteria. It would be logical to suggest that it exerts negative effect on the bacteria viability. However, it was found that expression of the cfr gene only slightly suppressed growth of bacterial cells [30]. Expression of the cfr gene has been studied in less detail than expression of the erm genes. The cfr reading frame in one of the pathogenic Staphylococcus aureus strains is preceded by a short, probably, regulatory, reading frame. In another case, the ermB gene was found upstream of the cfr gene, and the promoter of their common operon was induced by erythromycin, which resulted in the resistance to seven classes of antibiotics [31].
In addition to Cfr, the 23S rRNA A2503 is modified by the ubiquitous housekeeping methyltransferase RlmN that attaches methyl group at the C2 position [32]. Absence of this modification induces low-level resistance to linezolid [33]. Nevertheless, acquisition of the moderate antibiotic resistance in the absence of this modification most likely does not outweigh its benefit for the ribosome functioning. Cells deficient in RlmN do not express recombinant proteins at high levels and demonstrate moderate increase in the content of intermediates of ribosomal subunit assembly when grown at 20°C [34]. Interestingly, induction of the ermC methyltransferase gene in the cells with the A2503 residue unmodified at the C2 atom is less pronounced, which indicates possible interaction of the modified residue with the growing peptide and its involvement in regulation of the protein synthesis [35].
Evolutionary origin of the cfr gene is still a subject of debate. Identification of similar sequences revealed relation between Cfr and RlmN; it was suggested that Cfr had originated from RlmN through plastid RNA methyltransferases [36]. A common origin of these enzymes is supported by the fact that they modify the same residue, as well as function via the same rather rare radical mechanism [37, 38]. However, some authors [39, 40] suggested that Cfr originated from the methyltransferase of Clostridia related to RlmN but had the substrate different from the A2503 23S rRNA. Considering that RlmN exhibits dual specificity by methylating both rRNA and tRNA [41], it is possible that methyltransferases of Clostridia, which are related to both RlmN and Cfr, can act exclusively on tRNA, but not rRNA [39].
Out of multiple modifications of the PTC nucleotide residues, only one promotes antibiotic resistance, i.e., represents to some extent a constitutive resistance modification. It is pseudouridinylation of the U2504 residue by RluC [42], which increases resistance to tiamulin, clindamycin, and linezolid and, to a lesser extent, chloramphenicol, hygromycin A, and sparsomycin [43]. Despite the fact that RluC catalyzes conversion of three uridine residues (955, 2504, and 2580) into pseudouridine, modification of only ψ2504 (Fig. 1b) increases cell resistance to the abovementioned inhibitors of the peptidyl transferase reaction.
To summarize, these data suggest evolution of the resistance-mediating rRNA methyltransferase (Cfr) from a constitutive rRNA/tRNA methyltransferase, as well as possible transformation of the resistance-mediating pseudouridine synthase into a constitutive modifying housekeeping enzyme (RluC).
METHYLATION OF 16S rRNA NUCLEOTIDE RESIDUES IN DC
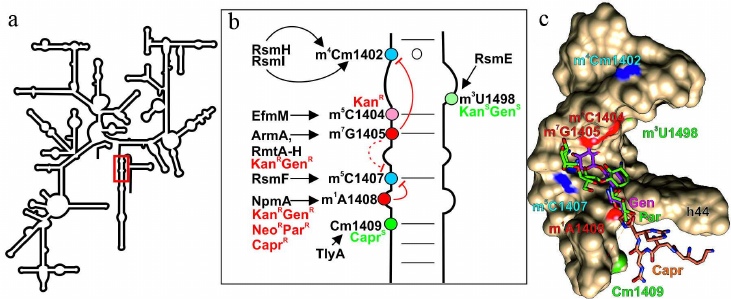
The second functionally important site of the ribosome is its decoding center (DC), which is located on the small subunit and formed mostly by the 16S rRNA helix 44. Many antibiotics, mostly aminoglycosides, interact with this ribosomal site and induce errors in decoding [44], as well as prevent translocation [45]. Resistance to aminoglycosides can be induced by methylation of the DC residues 16S rRNA G1405 at the N7 atom and 16S rRNA A1408 at the N1 atom [46] (Fig. 2; table). Enzymes catalyzing these modifications have been found in both aminoglycoside-producing strains and pathogenic bacteria that had most probably acquired them by the horizontal gene transfer [46]. Thus, the m7G1405 residue in the ribosomes of bacterial strains producing aminoglycosides with 4-6 substituted deoxystreptamine is formed in the reaction catalyzed by a large family of the structurally similar methyltransferases, including KgmB, Sgm, FmrO, GrmA, GrmB, GrmO, Kmr, NbrB, and Srm1 [47]. Commonly occurring N7 methyltransferases ArmA and RmtA-H catalyze formation of the 16S rRNA m7G1405 residue in pathogenic bacteria [48], inducing resistance to aminoglycosides containing 4-6 substituted deoxystreptamine, e.g., kanamycin and gentamycin [Fig. 2a, b and c; in panel (c), see positions of gentamycin (pink) and 16S rRNA m7G1405] [46]. The m1A1408 modification in 16S rRNA confers resistance to a broader spectrum of aminoglycosides containing either 4-6 or 4-5 substituted deoxystreptamine (e.g., neomycin and paromomycin) [Fig. 2a, b and c; in panel (c), compare location of gentamycin (pink) and paromomycin (green) and 16S rRNA m1A1408 residue]. These methyltransferases have been found in the strains producers of aminoglycosides (KamA, KamB, and KamC) and antituberculosis antibiotic capreomycin (CmnU) unrelated to aminoglycosides [47]. It was assumed for a long time that pathogenic bacteria do not have this type of resistance; however, in 2007, similar N1 A1408 methyltransferase NpmA was identified in Escherichia coli [49]. Based on the similarity of their amino acid sequences, methyltransferases modifying N1 atom of the A1408 residue are most likely related proteins, but evolutionary origin of this enzyme family remains obscure. It is possible that these proteins originate from the tRNA-specific guanine N7 methyltransferase TrmB [50].
Fig. 2. Effect of nucleotide modifications in the DC 16S rRNA on interactions with antibiotics. a) Secondary structure of 16S rRNA. Fragments in red box is shown in other panels. b) Secondary structure of the 16S rRNA helix H44 that forms a fragment of DC. Modified nucleotides discussed in the text are shown as circles: red circles, modifications of these nucleotides induce resistance to antibiotics (KanRGenR, resistance to kanamycin, gentamycin, and aminoglycosides containing 4-6 substituted deoxystreptamine; NeoRParR, resistance to neomycin, paromomycin, and aminoglycosides containing 4-5 substituted deoxystreptamine; CaprR, resistance to capreomycin); green circles, modification of these nucleotides facilitates antibiotic binding; blue circles, residues modified by the housekeeping enzymes. Arrows indicate activity of the corresponding modifying enzymes; red line shows mutually exclusive modifications; dotted red line, putative effect. c) 3D structure of the H44 helix of 16S rRNA that forms fragment of the DC (shown as molecular surface) and bound gentamycin (4-6 substituted aminoglycoside; pink skeletal model), paromomycin (4-5 substituted aminoglycoside; green skeletal model), capreomycin (orange skeletal model). Red, green, and blue van der Waals spheres correspond to the methyl groups of nucleotide residues preventing, promoting, or exhibiting no effect, respectively, on antibiotic binding. The images, including overlapping 3D structures and models of modified nucleotide residues, were created with the UCSF Chimera software [10] using secondary rRNA structures [11] and 3D structures 4V53 [51], 4V7M [52], and 4V51 [53].
Another mechanism of bacterial resistance to aminoglycosides (kanamycin and tobramycin, but not gentamycin) is associated with methylation of the 16S rRNA C1404 residue at the C5 atom [54] by methyltransferase EfmM found in the pathogenic strain of Enterococcus faecium. This protein resembles methyltransferase RsmF, from which it likely originates. Interestingly, methyltransferase RsmF in Thermus thermophilus modifies three nucleotide residues (C1400, C1404, and C1407) [55], indicating relatively easy evolution of the substrate specificity of these enzymes. The modified m5C1404 residue is located in close vicinity to the kanamycin-binding site [56], but does not overlap with it, which excludes steric clashes as a resistance mechanism. This is also corroborated by a small (12-fold in the case of efmM expression in E. coli) difference in the minimal inhibiting concentration between the presence and absence of the modification. Surprisingly, this modification does not disturb binding of gentamycin, which has bulky methyl group (vs. amino group in kanamycin) at the site of contact with the methyl group of m5C1404. It is believed that the resistance to kanamycin and tobramycin induced by the m5C1404 modification [54] is associated with the displacement of the antibiotic-bound water. In the case of gentamycin, on the contrary, formation of m5C1404 results in the emergence of additional hydrophobic contact [Fig. 2, b and c; in panel (c), see positions of gentamycin (pink) and 16S rRNA m5C1404 residue].
Formation of the hydrophobic contact could explain interaction of capreomycin, an antibiotic used for tuberculosis treatment, with the 2′-OMe group of Cm1409 [57]. Some pathogenic bacteria, including Mycobacterium sp., Campylobacter jejuni, and Brachyspira (Serpulina) hyodysenteriae, contain methyltransferase TlyA responsible for modification of the nucleotide residues 16S rRNA C1409 and 23S rRNA C1920 at the 2′-OH group [58, 59]. Deletion or mutation of the tlyA gene in Mycobacterium sp. induces resistance to capreomycin. Formation of the B2a bridge between the ribosomal subunits brings these two residues close to each other. Structural studies have shown that this is also the site of tuberactinomycins (viomycin and capreomycin) binding [52]. Methylation of both subunits makes ribosome more susceptible to these antibiotics [59]. In addition to the suggested hydrophobic contact between Cm1409 and capreomycin [Fig. 2, b and c; in panel (c), see positions of capreomycin (orange) and 16S rRNA Cm1409], the increased susceptibility of the TlyA-modified ribosomes to antibiotic could be explained by the changes in the position of ribosomal subunits relatively to each other during translocation, as tuberactinomycins affect this stage of ribosomal cycle [60]. The studies of the tlyA gene orthologs revealed existence of two protein variants – TlyAI, which modified only 23S rRNA C1920, and TlyAII, which modified residues in both subunits. Expression of the gene encoding TlyAII in E. coli makes the cells more susceptible to tuberactinomycins, but provides no advantages during the cell growth in the absence of the antibiotic [61]. Why do pathogenic bacteria carry the tlyA gene if it increases susceptibility to antibiotics? It is possible that TlyA has an additional function beside rRNA modification. The recombinant TlyA protein was found to exhibit hemolytic activity in vitro and was located in the cell membrane when expressed in E. coli [62]. Presence of the tlyA genes is one of the virulence factors in C. jejuni; mutations suppressing enzymatic activity of TlyA exert the same effect as deletions of the tlyA gene. Hence, methylation plays a principal role in pathogenicity [58]. Despite the close location of the TlyA target nucleotides to aminoglycoside-binding site, the presence or absence of this methyltransferase produces no effect on the efficacy of aminoglycoside amikacin. At the same time, the A1408G mutation of the residue located close to the methylation site provides resistance to both amikacin and capreomycin [63]. Interestingly, tlyA inactivation has virtually no effect on the growth rate of bacteria lacking the A1408G mutation in rRNA, while expression of tlyA increases adaptability of the bacteria with this mutation, which is a very unusual type of relationship between methylation and antibiotic resistance [63]. In this case, methylation reduces phenotypic cost of antibiotic resistance induced by mutation.
The capreomycin producer Saccharothrix mutabilis subsp. Capreolus was found to contains the cmnU gene providing resistance to capreomycin itself. This gene codes for a homolog of methyltransferases modifying A1408 residue [64]. The resistance to capreomycin in the case of identical methylation products can be explained by steric clashes preventing antibiotic binding [Fig. 2, b and c; in panel (c), see positions of capreomycin (orange) and 16S rRNA m1A1408].
The DC contains one more modification, the absence of which in Mycobacterium smegmatis results in resistance to aminoglycosides – methylation of the 16S rRNA U1498 at the N3 atom (Fig. 2) [65] by the housekeeping methyltransferase RsmE [66]. It should be noted that antibiotic resistance changes approximately 2-fold, which might be explained by some indirect effects. The absence of this modification produces very little effect on viability and functioning of the cells in the absence of antibiotic [66].
A very interesting phenomenon is mutually exclusive formation of the constitutively methylated residues and methylation of the nucleotides responsible for antibiotic resistance (Fig. 2b). Methylation of G1405 at the N7 atom by methyltransferase RmtC, discovered in the pathogenic E. coli, decreases activity of the housekeeping methyltransferase RsmF that modifies C1407 at the C5 position [67]. There is virtually no phenotypic cost of G1405 methylation by RmtC [68]. Inactivation of the rsmF gene slows down slightly the growth of bacteria without inducing defects in the ribosome assembly [31]; however, absence of this methyltransferase significantly reduces ability of the bacteria to compete with the wild-type cells [67, 68] and increases resistance to aminoglycosides [68]. Later, this observation was questioned, as it was found that modification of the same G1405 residue at the N7 position by the related methyltransferase ArmA blocked the housekeeping methyltransferase RsmI [69], which modified the 2′-OH group of the C1402 residue rather than RsmF [70]. The absence of Cm1402 methylation can only partially explain significant decrease in the adaptability of bacteria expressing armA in the absence of antibiotics [69], because inactivation of RsmI results in a slight increase in the frequency of +1 and –1 reading frame shifts but does not noticeably affect growth rate of the bacteria [70].
Antagonism in the modification of 16S rRNA residues Cm1402 and m7G1405 was observed in Pseudomonas aeruginosa, which normally lacked methyltransferase RsmF. The methyltransferase RmtD catalyzing m7G1405 formation in this bacterium, also prevents modification of Cm1402 by RsmI. In turn, activity of RsmF responsible for 16S rRNA m5C1407 modification, is suppressed by the NpmA enzyme that methylates A1408 at the N1 position [69]. The effect of npmA expression on cell adaptability is less pronounced than the effect of armA expression [69]. The genes for enzymes modifying G1405 and A1408 residues have been found in both aminoglycoside-producing strains and pathogens, but their expression affects growth of bacteria differently. In the absence of antibiotic, pathogenic strains expressing resistance-providing methyltransferase genes completely disappear from the mixed culture during competitive cultivation, while expression of methyltransferase genes in the antibiotic producers has a considerably less pronounced negative effect on adaptability of bacteria [71].
Analysis of DC methylation leads to the interesting conclusions such as very unusual mutually exclusive interactions between the resistance-providing and housekeeping methyltransferases and on the possibility to suppress antibiotic binding in the absence of methyl group overlapping with the antibiotic by affecting the water-mediated interactions. Beneficial role of Cm1409 methylation in the binding of capreomycin, likely due to formation of the hydrophobic contact, is also worth mentioning.
RESISTANCE TO OTHER ANTIBIOTICS DUE TO METHYLATION-INDUCED STERIC
CLASHES PREVENTING ANTIBIOTIC BINDING
In addition to the abovementioned examples, when rRNA methylation induces resistance to antibiotic binding in the PET, PTC, and DC, rRNA methylation might confer resistance to antibiotics that bind outside of these functional centers. This type of methylation involves mostly ribosome modification in the producer species to protect them from the synthesized compounds.
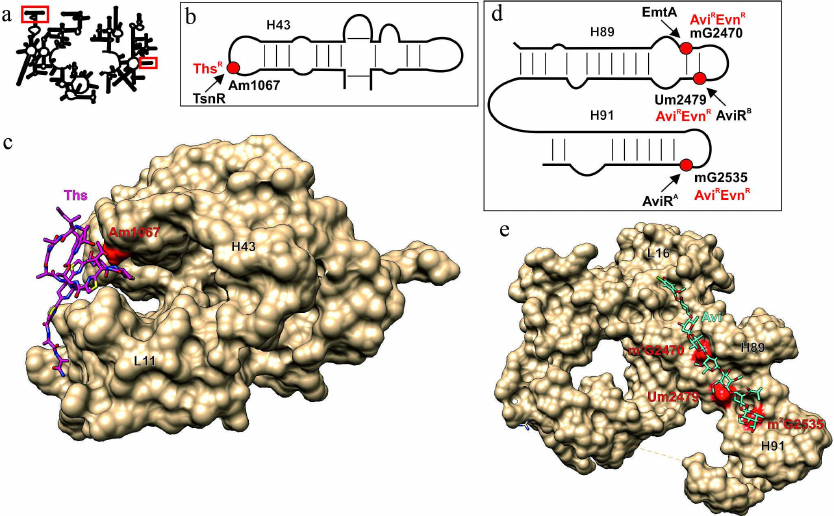
Methylation of 23S rRNA A1067 at the 2′-OH-group of ribose (Fig. 3, a and b; table) provides resistance to thiostrepton and micrococcin [72]. In nature, the tsnR resistance gene was found only in the thiostrepton producers, e.g., in Stretomyces aureus. The TsnR methyltransferase modifies only free 23S rRNA, while no modification takes place in the presence of thiostrepton or ribosomal protein L11 [73]. Structure of the complex of thiostrepton with the 50S ribosomal subunit of Deinococcus radiodurans (Fig. 3c) illustrates the resistance mechanism that involves methylation of ribose in A1067, which prevents close approaching of the loop 1 of the antibiotic and the 23S rRNA H43 helix [74]. The major advantage of this antibiotic is that it binds at a distance from the binding sites for most protein synthesis inhibitors and does not exhibit cross-resistance with other antibiotics. There are no data on the effect of this modification on adaptability; however, both modified and intact ribosomes ensure similar GTPase activity of the EF-G factor [75].
Fig. 3. Effect of modified 23S rRNA nucleotides on the interaction with thiostrepton and orthosomycins. a) Secondary structure of 23S rRNA. Fragments in red boxes are show in other panels. b) Secondary structure of region II of the 23S rRNA domain interacting with GTPase translation factors. Red circle, 23S rRNA Am1067 residue, methylation of which results in thiostrepton resistance (ThsR). Arrow indicates activity of the modifying enzyme TsnR. c) 3D structure of the ribosome fragment (shown as molecular surface) that forms thiostrepton binding site (pink skeletal model). Red van der Waals spheres correspond to the Am1067 methyl group overlapping with the thiostrepton-binding site. H43 helix of the 23S rRNA and L11 protein are indicated. d) Secondary structure of the 23S rRNA helices H89 and H91 that form the orthosomycin-binding site. Modified nucleotide residues determining resistance to evernimicin (EvnR) and avilamycin (AviR) are shown as red circles. Black arrows indicate activity of the corresponding modifying enzymes. e) 3D structure of the ribosome fragment (shown as molecular surface) that forms avilamycin binding site (blue skeletal model). Red van der Waals spheres correspond to the methyl groups of nucleotide residues overlapping with the avilamycin-binding site. H89 and H91 helices and L16 protein are indicated. The images, including overlapping 3D structures and models of the modified nucleotide residues, were created with the UCSF Chimera software [10] using secondary rRNA structures [11] and 3D structures 3CF5 [74] and 5KCR [76].
The avilamycin biosynthesis cluster discovered in the genome of Streptomyces viridochromogenes strain Tü57 contains genes for two methyltransferases (aviRa and aviRb) whose expression provides resistance to this antibiotic. Expression of aviRa in Streptomyces lividans TK66 confers very low antibiotic resistance (10 µg/ml), while expression of aviRb ensures a much high resistance to avilamycin (250 µg/ml) [77]. Both methyltransferases modify rRNA residues: AviRa methylates G2535 at positions N1 or N2 (as suggested based on the data of reverse transcriptase halting), while AviRb modifies U2479 at the 2′-OH position of ribose [78] (Fig. 3d; table). The resistance against avilamycin and related evernimicin is also induced by modification of G2470 by methyltransferase EmtA (minimal inhibiting concentration over 256 µg/ml) [79] encoded in the pathogenic bacterium plasmid. The structure of E. coli ribosome complexes with avilamycin and evernimicin obtained by cryoelectron microscopy explains the effect of methylation (Fig. 3e). When attached to the 2′-OH group of U2479, the methyl group blocks the antibiotic-binding site. Exact positions of the methyl groups attached to G2535 and G2470 remain unknown, but based on the emergence of antibiotic resistance and inhibition of reverse transcription, these residues are likely modified at the N1 or N2 atoms. Based the structure of avilamycin complex with the ribosome [76], overlapping of the methyl groups attached by either AviRa or EmtA with binding site of this group of antibiotics most likely requires methylation at the N2 amino group. This suggestion is also supported by the fact that the BLAST search for the enzymes with the sequences similar to that of EmtA reveals possible relationship of this protein with RNA guanine-N2-methyltransferase. No data are available so far on the effect of these modifications on the growth rate of bacteria in the absence of antibiotics.
Methylation of 16S RNA A964 at the N1 position in the pactamycin producer Streptomyces pactum (table) induces antibiotic resistance by inhibiting binding of this antibiotic [80]. Antibiotic-resistance mutations A694G, C795U, and C796U [81] are located in close vicinity to the binding site identified by X-ray analysis [82]. The effect of A964 methylation on bacterial growth remains unknown, although A964 is located close to the residues G966 and C967 involved in the regulation of the ratio between transcription and translation rates and methylated by the housekeeping methyltransferases RsmD [83] and RsmB [84, 85], respectively [86]. No functional relation between modifications of A964 and neighboring residues have been revealed yet.
Based on the above data, we can make the following trivial conclusion: rRNA methylation could facilitate antibiotic resistance even if antibiotic binds far from the regions often containing methylated nucleotides modified by the housekeeping enzymes.
ANTIBIOTIC RESISTANCE INDUCED BY THE ABSENCE OF
METHYLATION
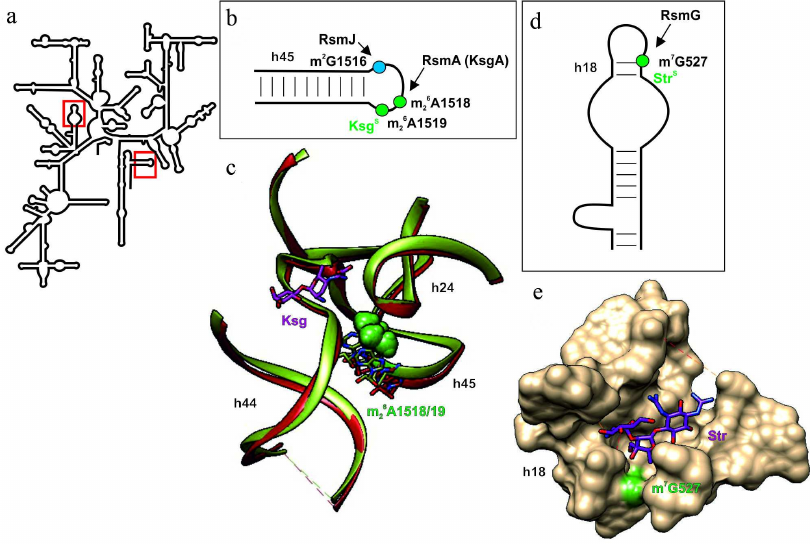
In addition to antibiotic resistance caused by steric clashes resulting from introduction of methyl group into the rRNA region responsible for antibiotic binding by methyltransferases, there are also the opposite examples (Fig. 4; table). The most well studied of them is resistance to kasugamycin in the absence of dimethylation of nucleotide residues 16S rRNA A1518 and A1519 at the N6 position (Fig. 4, a and b; table) [87]. The structure of kasugamycin complex with the ribosome (Fig. 4c) revealed that A1518/19 residues do not form contacts with antibiotic; therefore, it is highly unlikely that the presence or absence of methyl groups directly affects antibiotic binding via formation of hydrophobic contacts [88]. Later experiments confirmed the possibility of kasugamycin interaction with ribosomes lacking methylated 16S rRNA A1518/19 residues [89]. Despite the fact that the cells acquire resistance to kasugamycin in the absence of A1518/19 methylation, and, therefore, exhibit certain advantages (at least in the presence of this antibiotic in the medium), almost all pro- and eukaryotes contain methyltransferase orthologs. The structure of the ribosome with nonmodified A1518/19 demonstrated that methylation is essential for DC formation in the small subunit [Fig. 4c; compare the structure of 16S rRNA helices in the ribosomes containing (green) or lacking (red) methyl groups introduced by RsmA(KsgA)] [90]. Apparently, changes in the structure of the 30S ribosomal subunit are the reason behind kasugamycin resistance observed upon inactivation of the rsmA(ksgA) gene. Besides, methyltransferase RsmA(KsgA) participates in the 30S subunit synthesis as a molecular switch protein [91]. Bacteria deficient in this methyltransferase display decreased growth rate; Yersinia pestis cells lacking this enzyme have reduced virulence [92].
Fig. 4. Effect of nucleotide modifications in the 16S rRNA on the interaction with kasugamycin and streptomycin. a) Secondary structure of 16S rRNA. Fragments in red boxes are show in other panels. b) Secondary structure of the 16S rRNA helix H45. Modified nucleotides discussed in the text are shown as circles: green circles, modifications of these nucleotides facilitate kasugamycin binding; blue circles, modification of these nucleotides by the housekeeping enzymes does not affect antibiotic activity. c) 3D structure of the ribosome fragment (shown as a tube model, 16S rRNA helices are indicated), conformation of which depends on methylation of the m26A1518/19 residues. The structure of ribosome containing methylated or nonmethylated A1518/19 residues (skeletal models of corresponding colors) is shown in green and red, respectively. Methyl groups are shown as green van der Waals spheres. Kasugamycin interacting with the ribosome containing methylated nucleotide residues is shown as pink skeletal model. d) Secondary structure of the H18 helix of 16S rRNA that forms the streptomycin-binding site. The nucleotide residue m7G527, modification of which promotes action of streptomycin, is shown as green circle. Black arrow indicates activity of the modifying RsmG enzyme. e) 3D structure of the ribosome fragment (shown as molecular surface) that forms the streptomycin binding site (purple skeletal model). Green van der Waals spheres correspond to the methyl groups of 16S rRNA m7G527. The images, including overlapping 3D structures and models of modified nucleotide residues, were created with the UCSF Chimera software [10] using secondary rRNA structures [11] and 3D structures 1J5E [93], 3OTO [90], 4V4H [94], 4NXM [95]
Unlike for the most aminoglycosides, resistance to streptomycin could not be induced by methylation of residues in the 16S rRNA helix H44 because of the different binding site (Fig. 4e). However, inactivation of the housekeeping methyltransferase RsmG gene, which modifies 16S rRNA G527 residue at the N7 position (Fig. 4, a and d; table) ensures moderate resistance to this antibiotic in Streptomyces coelicolor [96]. It was shown that deletion of the gidB gene (rsmG homolog that also modifies G527) in the pathogenic M. tuberculosis and Salmonella enterica also provides slight resistance to streptomycin [97, 98]. The structure of the ribosome complex with streptomycin (Fig. 4e) revealed that G527 is located in close vicinity to the antibiotic-binding site, but position of the methyl group does not overlap with the position of antibiotic. The hypothesis on induction of conformational changes by methylation of 16S rRNA G527 has not been confirmed experimentally [95]. Despite the lack of significant effect of the rsmG deletion on bacterial growth [34], it was shown that rsmG-deficient Salmonella cells were outgrown by the wild-type bacteria in the competitive growth assay [98]. Although inactivation of the rsmG gene provides only slight resistance to streptomycin compared to mutation in the ribosomal S12 protein gene, inactivation of the former is an important prerequisite for emergence of the latter. The rsmG deletion in Bacillus subtilis increased accuracy of translation; at the same time, frequency of the mutations ensuring streptomycin resistance in the S12-encoding rpsL gene increased significantly (200-fold) in these strains [96]. Similar results were obtained in E. coli. Deletion of the rsmG gene that provided moderate resistance to streptomycin was accompanied by the increase in the frequency of mutations in the rpsL (S12) gene, which further promoted antibiotic resistance [100].
As mentioned earlier, activity of RlmAII induces tylosin resistance in Bacillus subtilis. In E. coli, deletion of the rlmAI gene paralog responsible for formation of the 23S rRNA m1G745 results in the resistance to viomycin [101] despite the absence of contacts between viomycin and this modified nucleotide. According to the results of footprinting experiments, deletion of the rlmAI gene most probably, prevents antibiotic binding. Similar to the deletion of housekeeping methyltransferase genes, rlmAI inactivation negatively affects bacterial adaptability, i.e., growth rate decreases and ability of the ribosomal subunits for association is reduced, as well as translation efficiency [101]. It is possible that structural changes that decrease the efficiency of subunit interaction are also responsible for the resistance to viomycin, as its binding site is located in close vicinity to the intersubunit B2a bridge.
The tetracycline binding site contains m2G966 nucleotide residue [102] modified by methyltransferase RsmD [83]. Inactivation of this housekeeping methyltransferase in S. pneumoniae slightly increases resistance to tigecycline [103] (table), while deletion of rsmD in E. coli increases susceptibility to tetracycline [104]. Despite the discrepancy of these observations, this is one of the rare examples, when a housekeeping methyltransferase provides certain resistance to antibiotics. Also, susceptibility to antibiotics often requires modification of the nucleotide residues by housekeeping methyltransferases.
CONCLUSION
Antibiotics are an example of “tools” that living organisms use in the competition for resources and habitats. The mechanisms of antibiotic resistance allow the species to either produce antibiotics or to survive in their presence. Most known inhibitors of protein synthesis bind to the functional sites of the ribosome, which usually contain numerous modified (mostly methylated) nucleotide residues [2]. rRNA methylation is one of the major mechanisms of antibiotic resistance, when introduced methyl groups spatially overlap with the antibiotic-binding site. However, generation of steric claches is not the only mechanism of methylation-induced antibiotic resistance. Thus, methylation by EfmM supposedly results in the displacement of water molecule mediating formation of additional contact between antibiotic and rRNA.
At first glance, facilitation of antibiotic binding by rRNA methylation might seem unusual. However, it happens quite often. It is possible that in some cases, antibiotic directly interacts with the methyl group by forming hydrophobic or van der Waals contacts, e.g., in the interaction of capreomycin with 16S rRNA Cm1409 or telithromycin with m1G748. In the absence of direct contact between the methyl group in rRNA and antibiotic, activity of the latter might be affected by the methylation-induced conformational changes in rRNA. This phenomenon has been well documented for the effect of 16S rRNA m26A1518/19 on the activity of kasugamycin. It is possible that the same mechanism takes place during the interactions between linezolid, sparsomycin, and tiamulin and 23S rRNA m2A2503, aminoglycosides and 16S m3U1498, and viomycin and 23S rRNA m1G745. It is more difficult to explain how methyl group in rRNA could facilitate antibiotic binding in the absence of contacts with antibiotic or without generating conformational changes (as in the case of 16S rRNA m7G527 16S and streptomycin). Such interactions or conformational changes might take place in some other, probably, transient state of the translating ribosome; methylation might also affect antibiotic binding indirectly, e.g., via displacement of water molecules.
Evolutionary relation of housekeeping methyltransferases and methyltransferases involved in antibiotic resistance are of particular interest. According to the commonly accepted hypothesis, the latter spread “vertically”, i.e., evolve together with the corresponding taxa. Antibiotic resistance methyltransferases either are typical for a narrow group of the corresponding producer strains or spread by the horizontal gene transfer, given that they originate from the corresponding producer bacteria. While considering emergence of methyltransferases in antibiotic-producing and, presumably, some other bacteria, it is possible to imagine scenario in which housekeeping enzymes serve as a reservoir for evolution of antibiotic resistance. Such scenario is most probable for evolution of the Erm family methyltransferases from RsmA(KsgA). Is the opposite process (i.e., transformation of antibiotic resistance methyltransferase into a housekeeping methyltransferase) possible? Both antibiotics and mechanisms of antibiotic resistance have been evolving for a long time; theoretically, biosynthesis of antibiotics could evolve to avoid antibiotic resistance. It is possible that ubiquitous presence of the resistance genes can lead to their transformation into the variants of housekeeping methyltransferases. This might happen if methylation causes no negative consequences or might be even beneficial for the bacterial adaptation in the absence of antibiotics. For bacteria the “cost” of methylation providing antibiotic resistance could vary from a significant loss of adaptability (e.g., upon methylation of 23S rRNA A2058 by the Erm family enzymes) to almost no effect (e.g., upon methylation of A1408 by NpmA). An interesting phenomenon is the reduction of the cost of amikacin resistance caused by the A1408G mutation in 16S rRNA facilitated by methylation of the neighboring residue by methyltransferase TlyA.
Very few examples can be found in the literature that do not contradict suggested evolutionary origin of the housekeeping enzymes from methyltransferases responsible for antibiotic resistance of cells. Activity of the modifying RsmD and RluC proteins, which belong to the housekeeping enzymes, provides a certain level of resistance to tetracycline and tiamulin/clindamycin/linezolid, respectively. Methyltransferase RlmAII, which is required for formation of m1G748 23S rRNA and is abundant in Gram-positive bacteria, provides some resistance to tylosin. It is still unclear whether these proteins originated from the enzymes ensuring resistance to ancestral antibiotics that bound approximately at the same sites as their modern versions.
Although previously investigation of rRNA methyltransferases has been limited mostly to the model organisms and pathogens, while the search for antibiotics has been dominated by purely utilitarian approach dictated by the needs of medical practice, we believe that the future belongs to the studies on coevolution of ribosome modification systems and biosynthesis of antibacterial compounds.
Funding. This work was financially supported by the Russian Foundation for Basic Research (project nos. 20-04-00736 and 20-54-53014).
Ethics declarations. The authors declare no conflict of interest in financial or any other sphere. This article does not contain any studies with human participants or animals performed by any of the authors.
REFERENCES
1.Arenz, S., and Wilson, D. N. (2016) Bacterial
protein synthesis as a target for antibiotic inhibition, Cold Spring
Harb. Perspect. Med., 6, a025361, doi:
10.1101/cshperspect.a025361.
2.Sergiev, P. V., Aleksashin, N. A., Chugunova, A.
A., Polikanov, Y. S., and Dontsova, O. A. (2018) Structural and
evolutionary insights into ribosomal RNA methylation, Nat. Chem.
Biol., 14, 226-235.
3.Bogdanov, A. A., Sumbatyan, N. V., Shishkina, A.
V., Karpenko, V. V., and Korshunova, G. A. (2010) Ribosomal tunnel and
translation regulation, Biochemistry (Moscow), 75,
1501-1516.
4.Skinner, R., Cundliffe, E., and Schmidt, F. J.
(1983) Site of action of a ribosomal RNA methylase responsible for
resistance to erythromycin and other antibiotics, J. Biol.
Chem., 258, 12702-12706.
5.Pernodet, J. L., Fish, S., Blondelet-Rouault, M.
H., and Cundliffe, E. (1996) The macrolide-lincosamide-streptogramin B
resistance phenotypes characterized by using a specifically deleted,
antibiotic-sensitive strain of Streptomyces lividans,
Antimicrob. Agents Chemother., 40, 581-585.
6.Roberts, M. C., Sutcliffe, J., Courvalin, P.,
Jensen, L. B., Rood, J., and Seppala, H. (1999) Nomenclature for
macrolide and macrolide-lincosamide-streptogramin B resistance
determinants, Antimicrob. Agents Chemother., 43,
2823-2830.
7.Arthur, M., Brisson-Noël, A., and Courvalin,
P. (1987) Origin and evolution of genes specifying resistance to
macrolide, lincosamide and streptogramin antibiotics: data and
hypotheses, J. Antimicrob. Chemother., 20, 783-802.
8.Altschul, S. F., Gish, W., Miller, W., Myers, E.
W., and Lipman, D. J. (1990) Basic local alignment search tool, J.
Mol. Biol., 215, 403-410.
9.Bhujbalrao, R., and Anand, R. (2019) Deciphering
determinants in ribosomal methyltransferases that confer antimicrobial
resistance, J. Am. Chem. Soc., 141, 1425-1429.
10.Pettersen, E. F., Goddard, T. D., Huang, C. C.,
Couch, G. S., Greenblatt, D. M., Meng, E. C., and Ferrin, T. E. (2004)
UCSF Chimera – a visualization system for exploratory research
and analysis, J. Comput. Chem., 25, 1605-1612.
11.The RNAcentral Consortium (2019) RNAcentral: a
hub of information for non-coding RNA sequences, Nucleic Acids
Res., 47, D221-D229.
12.Hansen, J. L., Ippolito, J. A., Ban, N., Nissen,
P., Moore, P. B., and Steitz, T. A. (2002) The structures of four
macrolide antibiotics bound to the large ribosomal subunit, Mol.
Cell, 10, 117-128.
13.Bulkley, D., Innis, C. A., Blaha, G., and Steitz,
T. A. (2010) Revisiting the structures of several antibiotics bound to
the bacterial ribosome, Proc. Natl. Acad. Sci. USA, 107,
17158-17163.
14.Svetlov, M. S., Plessa, E., Chen, C.-W., Bougas,
A., Krokidis, M. G., Dinos, G. P., and Polikanov, Y. S. (2019)
High-resolution crystal structures of ribosome-bound chloramphenicol
and erythromycin provide the ultimate basis for their competition,
RNA, 25, 600-606.
15.Gupta, P., Sothiselvam, S.,
Vázquez-Laslop, N., and Mankin, A. S. (2013) Deregulation of
translation due to post-transcriptional modification of rRNA explains
why erm genes are inducible, Nat. Commun., 4, 1984.
16.Subramanian, S. L., Ramu, H., and Mankin, A. S.
(2012) Inducible resistance to macrolide antibiotics, in Antibiotic
Discovery and Development (Dougherty, T. J., and Pucci, M. J.,
eds.) Springer US, Boston, MA, pp. 455-484.
17.Vazquez-Laslop, N., Thum, C., and Mankin, A. S.
(2008) Molecular mechanism of drug-dependent ribosome stalling, Mol.
Cell, 30, 190-202.
18.Arenz, S., Ramu, H., Gupta, P., Berninghausen,
O., Beckmann, R., Vázquez-Laslop, N., Mankin, A. S., and Wilson,
D. N. (2014) Molecular basis for erythromycin-dependent ribosome
stalling during translation of the ErmBL leader peptide, Nat.
Commun., 5, 3501.
19.Weisblum, B. (1995) Insights into erythromycin
action from studies of its activity as inducer of resistance,
Antimicrob. Agents Chemother., 39, 797-805.
20.Kwak, J. H., Choi, E. C., and Weisblum, B. (1991)
Transcriptional attenuation control of ermK, a
macrolide-lincosamide-streptogramin B resistance determinant from
Bacillus licheniformis, J. Bacteriol., 173,
4725-4735.
21.Morris, R. P., Nguyen, L., Gatfield, J.,
Visconti, K., Nguyen, K., et al. (2005) Ancestral antibiotic resistance
in Mycobacterium tuberculosis, Proc. Natl. Acad. Sci.
USA, 102, 12200-12205.
22.Gupta, P., Kannan, K., Mankin, A. S., and
Vázquez-Laslop, N. (2013) Regulation of gene expression by
macrolide-induced ribosomal frameshifting, Mol. Cell, 52,
629-642.
23.Liu, M., and Douthwaite, S. (2002) Resistance to
the macrolide antibiotic tylosin is conferred by single methylations at
23S rRNA nucleotides G748 and A2058 acting in synergy, Proc. Natl.
Acad. Sci. USA, 99, 14658-14663.
24.Liu, M., and Douthwaite, S. (2002) Methylation at
nucleotide G745 or G748 in 23S rRNA distinguishes Gram-negative from
Gram-positive bacteria, Mol. Microbiol., 44, 195-204.
25.Yakhnin, H., Yakhnin, A. V., Mouery, B. L.,
Mandell, Z. F., Karbasiafshar, C., Kashlev, M., and Babitzke, P. (2019)
NusG-dependent RNA polymerase pausing and tylosin-Dependent ribosome
stalling are required for tylosin resistance by inducing 23S rRNA
methylation in Bacillus subtilis, mBio, 10,
e02665-19.
26.Takaya, A., Sato, Y., Shoji, T., and Yamamoto, T.
(2013) Methylation of 23S rRNA nucleotide G748 by RlmAII
methyltransferase renders Streptococcus pneumoniae telithromycin
susceptible, Antimicrob. Agents Chemother., 57,
3789-3796.
27.Desmolaize, B., Fabret, C., Brégeon, D.,
Rose, S., Grosjean, H., and Douthwaite, S. (2011) A single
methyltransferase YefA (RlmCD) catalyses both m5U747 and m5U1939
modifications in Bacillus subtilis 23S rRNA, Nucleic Acids Res.,
39, 9368-9375.
28.Shoji, T., Takaya, A., Sato, Y., Kimura, S.,
Suzuki, T., and Yamamoto, T. (2015) RlmCD-mediated U747 methylation
promotes efficient G748 methylation by methyltransferase RlmAII in 23S
rRNA in Streptococcus pneumoniae; interplay between two rRNA
methylations responsible for telithromycin susceptibility, Nucleic
Acids Res., 43, 8964-8972.
29.Long, K. S., Poehlsgaard, J., Kehrenberg, C.,
Schwarz, S., and Vester, B. (2006) The Cfr rRNA methyltransferase
confers resistance to Phenicols, Lincosamides, Oxazolidinones,
Pleuromutilins, and Streptogramin A antibiotics, Antimicrob. Agents
Chemother., 50, 2500-2505.
30.LaMarre, J. M., Locke, J. B., Shaw, K. J., and
Mankin, A. S. (2011) Low fitness cost of the multidrug resistance gene
cfr, Antimicrob. Agents Chemother., 55,
3714-3719.
31.Smith, L. K., and Mankin, A. S. (2008)
Transcriptional and translational control of the mlr operon, which
confers resistance to seven classes of protein synthesis inhibitors,
Antimicrob. Agents Chemother., 52, 1703-1712.
32.Toh, S.-M., Xiong, L., Bae, T., and Mankin, A. S.
(2008) The methyltransferase YfgB/RlmN is responsible for modification
of adenosine 2503 in 23S rRNA, RNA, 14, 98-106.
33.LaMarre, J. M., Howden, B. P., and Mankin, A. S.
(2011) Inactivation of the indigenous methyltransferase RlmN in
Staphylococcus aureus increases linezolid resistance,
Antimicrob. Agents Chemother., 55, 2989-2991.
34.Pletnev, P., Guseva, E., Zanina, A., Evfratov,
S., Dzama, M., et al. (2020) Comprehensive functional analysis of
Escherichia coli ribosomal RNA methyltransferases, Front.
Genet., 11, 97.
35.Vázquez-Laslop, N., Ramu, H., Klepacki,
D., Kannan, K., and Mankin, A. S. (2010) The key function of a
conserved and modified rRNA residue in the ribosomal response to the
nascent peptide, EMBO J., 29, 3108-3117.
36.Kaminska, K. H., Purta, E., Hansen, L. H.,
Bujnicki, J. M., Vester, B., and Long, K. S. (2010) Insights into the
structure, function and evolution of the radical-SAM 23S rRNA
methyltransferase Cfr that confers antibiotic resistance in bacteria,
Nucleic Acids Res., 38, 1652-1663.
37.Giessing, A. M. B., Jensen, S. S., Rasmussen, A.,
Hansen, L. H., Gondela, A., Long, K., Vester, B., and Kirpekar, F.
(2009) Identification of 8-methyladenosine as the modification
catalyzed by the radical SAM methyltransferase Cfr that confers
antibiotic resistance in bacteria, RNA, 15, 327-336.
38.Yan, F., LaMarre, J. M., Röhrich, R.,
Wiesner, J., Jomaa, H., Mankin, A. S., and Fujimori, D. G. (2010) RlmN
and Cfr are radical SAM enzymes involved in methylation of ribosomal
RNA, J. Am. Chem. Soc., 132, 3953-3964.
39.Atkinson, G. C., Hansen, L. H., Tenson, T.,
Rasmussen, A., Kirpekar, F., and Vester, B. (2013) Distinction between
the Cfr methyltransferase conferring antibiotic resistance and the
housekeeping RlmN methyltransferase, Antimicrob. Agents
Chemother., 57, 4019-4026.
40.Stojković, V., Noda-Garcia, L., Tawfik, D.
S., and Fujimori, D. G. (2016) Antibiotic resistance evolved via
inactivation of a ribosomal RNA methylating enzyme, Nucleic Acids
Res., 44, 8897-8907.
41.Benítez-Páez, A., Villarroya, M.,
and Armengod, M.-E. (2012) The Escherichia coli RlmN
methyltransferase is a dual-specificity enzyme that modifies both rRNA
and tRNA and controls translational accuracy, RNA, 18,
1783-1795.
42.Conrad, J., Sun, D., Englund, N., and Ofengand,
J. (1998) The rluC gene of Escherichia coli codes for a
pseudouridine synthase that is solely responsible for synthesis of
pseudouridine at positions 955, 2504, and 2580 in 23 S ribosomal RNA,
J. Biol. Chem., 273, 18562-18566.
43.Toh, S.-M., and Mankin, A. S. (2008) An
indigenous posttranscriptional modification in the ribosomal peptidyl
transferase center confers resistance to an array of protein synthesis
inhibitors, J. Mol. Biol., 380, 593-597.
44.Davies, J., Gorini, L., and Davis, B. D. (1965)
Misreading of RNA codewords induced by aminoglycoside antibiotics,
Mol. Pharmacol., 1, 93-106.
45.Hausner, T. P., Geigenmüller, U., and
Nierhaus, K. H. (1988) The allosteric three-site model for the
ribosomal elongation cycle. New insights into the inhibition mechanisms
of aminoglycosides, thiostrepton, and viomycin, J. Biol. Chem.,
263, 13103-13111.
46.Doi, Y., and Arakawa, Y. (2007) 16S ribosomal RNA
methylation: emerging resistance mechanism against aminoglycosides,
Clin. Infect. Dis., 45, 88-94.
47.Grosjean, H. (2009) DNA and RNA Modification
Enzymes: Structure, Mechanism, Function and Evolution, Landes
Bioscience, Austin, Tex.
48.Doi, Y., Wachino, J., and Arakawa, Y. (2016)
Aminoglycoside resistance, Infect. Dis. Clin. North Am.,
30, 523-537.
49.Wachino, J., Shibayama, K., Kurokawa, H., Kimura,
K., Yamane, K., Suzuki, S., Shibata, N., Ike, Y., and Arakawa, Y.
(2007) Novel plasmid-mediated 16S rRNA m1A1408 methyltransferase, NpmA,
found in a clinically isolated Escherichia coli strain resistant
to structurally diverse aminoglycosides, Antimicrob. Agents
Chemother., 51, 4401-4409.
50.Macmaster, R., Zelinskaya, N., Savic, M., Rankin,
C. R., and Conn, G. L. (2010) Structural insights into the function of
aminoglycoside-resistance A1408 16S rRNA methyltransferases from
antibiotic-producing and human pathogenic bacteria, Nucleic Acids
Res., 38, 7791-7799.
51.Borovinskaya, M. A., Pai, R. D., Zhang, W.,
Schuwirth, B. S., Holton, J. M., et al. (2007) Structural basis for
aminoglycoside inhibition of bacterial ribosome recycling, Nat.
Struct. Mol. Biol., 14, 727-732.
52.Stanley, R. E., Blaha, G., Grodzicki, R. L.,
Strickler, M. D., and Steitz, T. A. (2010) The structures of the
anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S
ribosome, Nat. Struct. Mol. Biol., 17, 289-293.
53.Selmer, M., Dunham, C. M., Murphy, F. V.,
Weixlbaumer, A., Petry, S., Kelley, A. C., Weir, J. R., and
Ramakrishnan, V. (2006) Structure of the 70S ribosome complexed with
mRNA and tRNA, Science, 313, 19351942.
54.Galimand, M., Schmitt, E., Panvert, M.,
Desmolaize, B., Douthwaite, S., Mechulam, Y., and Courvalin, P. (2011)
Intrinsic resistance to aminoglycosides in Enterococcus faecium
is conferred by the 16S rRNA m5C1404-specific methyltransferase EfmM,
RNA, 17, 251-262.
55.Demirci, H., Larsen, L. H. G., Hansen, T.,
Rasmussen, A., Cadambi, A., Gregory, S. T., Kirpekar, F., and Jogl, G.
(2010) Multi-site-specific 16S rRNA methyltransferase RsmF from
Thermus thermophilus, RNA, 16, 1584-1596.
56.François, B., Russell, R. J. M., Murray,
J. B., Aboul-ela, F., Masquida, B., Vicens, Q., and Westhof, E. (2005)
Crystal structures of complexes between aminoglycosides and decoding A
site oligonucleotides: role of the number of rings and positive charges
in the specific binding leading to miscoding, Nucleic Acids
Res., 33, 5677-5690.
57.Maus, C. E., Plikaytis, B. B., and Shinnick, T.
M. (2005) Mutation of tlyA confers capreomycin resistance in
Mycobacterium tuberculosis, Antimicrob. Agents
Chemother., 49, 571-577.
58.Sałamaszyńska-Guz, A., Rose, S.,
Lykkebo, C. A., Taciak, B., Bącal, P., Uśpieński,
T., and Douthwaite, S. (2017) Biofilm formation and motility are
promoted by Cj0588-directed methylation of rRNA in Campylobacter
jejuni, Front. Cell. Infect. Microbiol., 7, 533.
59.Johansen, S. K., Maus, C. E., Plikaytis, B. B.,
and Douthwaite, S. (2006) Capreomycin binds across the ribosomal
subunit interface using tlyA-encoded 2′-O-methylations in 16S and
23S rRNAs, Mol. Cell, 23, 173-182.
60.Ermolenko, D. N., Spiegel, P. C., Majumdar, Z.
K., Hickerson, R. P., Clegg, R. M., and Noller, H. F. (2007) The
antibiotic viomycin traps the ribosome in an intermediate state of
translocation, Nat. Struct. Mol. Biol., 14, 493-497.
61.Monshupanee, T., Johansen, S. K., Dahlberg, A.
E., and Douthwaite, S. (2012) Capreomycin susceptibility is increased
by TlyA-directed 2′-O-methylation on both ribosomal subunits,
Mol. Microbiol., 85, 1194-1203.
62.Rahman, A., Srivastava, S. S., Sneh, A., Ahmed,
N., and Krishnasastry, M. V. (2010) Molecular characterization of
tlyA gene product, Rv1694 of Mycobacterium tuberculosis:
a non-conventional hemolysin and a ribosomal RNA methyl transferase,
BMC Biochem., 11, 35.
63.Freihofer, P., Akbergenov, R., Teo, Y.,
Juskeviciene, R., Andersson, D. I., and Böttger, E. C. (2016)
Nonmutational compensation of the fitness cost of antibiotic resistance
in mycobacteria by overexpression of tlyA rRNA methylase,
RNA, 22, 1836-1843.
64.Felnagle, E. A., Rondon, M. R., Berti, A. D.,
Crosby, H. A., and Thomas, M. G. (2007) Identification of the
biosynthetic gene cluster and an additional gene for resistance to the
antituberculosis drug capreomycin, Appl. Environ. Microbiol.,
73, 4162-4170.
65.Bijpuria, S., Sharma, R., and Taneja, B. (2020)
Deletion of RsmE 16S rRNA methyltransferase leads to low level increase
in aminoglycoside resistance in Mycobacterium smegmatis,
bioRxiv, doi: 10.1101/2020.01.15.907279.
66.Basturea, G. N., Rudd, K. E., and Deutscher, M.
P. (2006) Identification and characterization of RsmE, the founding
member of a new RNA base methyltransferase family, RNA,
12, 426-434.
67.Andersen, N. M., and Douthwaite, S. (2006) YebU
is a m5C methyltransferase specific for 16 S rRNA nucleotide 1407,
J. Mol. Biol., 359, 777-786.
68.Gutierrez, B., Escudero, J. A., San Millan, A.,
Hidalgo, L., Carrilero, L., et al. (2012) Fitness cost and interference
of Arm/Rmt aminoglycoside resistance with the RsmF housekeeping
methyltransferases, Antimicrob. Agents Chemother., 56,
2335-2341.
69.Lioy, V. S., Goussard, S., Guerineau, V., Yoon,
E.-J., Courvalin, P., Galimand, M., and Grillot-Courvalin, C. (2014)
Aminoglycoside resistance 16S rRNA methyltransferases block endogenous
methylation, affect translation efficiency and fitness of the host,
RNA, 20, 382-391.
70.Kimura, S., and Suzuki, T. (2010) Fine-tuning of
the ribosomal decoding center by conserved methyl-modifications in the
Escherichia coli 16S rRNA, Nucleic Acids Res., 38,
1341-1352.
71.Vidučić, D., Obranić, S.,
Matovina, M., Babić, F., and Vlahoviček, G. M. (2014)
Host fitness effects of aminoglycoside resistance 16S rRNA G1405 and
A1408 methyltransferases from clinical pathogens and natural antibiotic
producers, FEBS J., 281, Suppl. s1, 285.
72.Thompson, J., Schmidt, F., and Cundliffe, E.
(1982) Site of action of a ribosomal RNA methylase conferring
resistance to thiostrepton, J. Biol. Chem., 257,
7915-7917.
73.Lentzen, G., Klinck, R., Matassova, N.,
Aboul-ela, F., and Murchie, A. I. H. (2003) Structural basis for
contrasting activities of ribosome binding thiazole antibiotics,
Chem. Biol., 10, 769-778.
74.Harms, J. M., Wilson, D. N., Schluenzen, F.,
Connell, S. R., Stachelhaus, T., et al. (2008) Translational regulation
via L11: molecular switches on the ribosome turned on and off by
thiostrepton and micrococcin, Mol. Cell, 30, 26-38.
75.Cundliffe, E., and Thompson, J. (1981) The mode
of action of nosiheptide (multhiomycin) and the mechanism of resistance
in the producing organism, J. Gen. Microbiol., 126,
185-192.
76.Arenz, S., Juette, M. F., Graf, M., Nguyen, F.,
Huter, P., Polikanov, Y. S., Blanchard, S. C., and Wilson, D. N. (2016)
Structures of the orthosomycin antibiotics avilamycin and evernimicin
in complex with the bacterial 70S ribosome, Proc. Natl. Acad. Sci.
USA, 113, 7527-7532.
77.Weitnauer, G., Gaisser, S., Trefzer, A.,
Stockert, S., Westrich, L., Quiros, L. M., Mendez, C., Salas, J. A.,
and Bechthold, A. (2001) An ATP-binding cassette transporter and two
rRNA methyltransferases are involved in resistance to avilamycin in the
producer organism Streptomyces viridochromogenes Tü57,
Antimicrob. Agents Chemother., 45, 690-695.
78.Treede, I., Jakobsen, L., Kirpekar, F., Vester,
B., Weitnauer, G., Bechthold, A., and Douthwaite, S. (2003) The
avilamycin resistance determinants AviRa and AviRb methylate 23S rRNA
at the guanosine 2535 base and the uridine 2479 ribose, Mol.
Microbiol., 49, 309-318.
79.Mann, P. A., Xiong, L., Mankin, A. S., Chau, A.
S., Mendrick, C. A., et al. (2001) EmtA, a rRNA methyltransferase
conferring high-level evernimicin resistance, Mol. Microbiol.,
41, 1349-1356.
80.Ballesta, J. P., and Cundliffe, E. (1991)
Site-specific methylation of 16S rRNA caused by pct, a pactamycin
resistance determinant from the producing organism, Streptomyces
pactum, J. Bacteriol., 173, 7213-7218.
81.Mankin, A. S. (1997) Pactamycin resistance
mutations in functional sites of 16 S rRNA, J. Mol. Biol.,
274, 8-15.
82.Polikanov, Y. S., Osterman, I. A., Szal, T.,
Tashlitsky, V. N., Serebryakova, M. V., et al. (2014) Amicoumacin a
inhibits translation by stabilizing mRNA interaction with the ribosome,
Mol. Cell, 56, 531-540.
83.Lesnyak, D. V., Osipiuk, J., Skarina, T.,
Sergiev, P. V., Bogdanov, A. A., et al. (2007) Methyltransferase that
modifies guanine 966 of the 16 S rRNA: functional identification and
tertiary structure, J. Biol. Chem., 282, 5880-5887.
84.Gu, X. R., Gustafsson, C., Ku, J., Yu, M., and
Santi, D. V. (1999) Identification of the 16S rRNA m5C967
methyltransferase from Escherichia coli, Biochemistry,
38, 4053-4057.
85.Tscherne, J. S., Nurse, K., Popienick, P.,
Michel, H., Sochacki, M., and Ofengand, J. (1999) Purification,
cloning, and characterization of the 16S RNA m5C967 methyltransferase
from Escherichia coli, Biochemistry, 38,
1884-1892.
86.Prokhorova, I. V., Osterman, I. A., Burakovsky,
D. E., Serebryakova, M. V., Galyamina, M. A., et al. (2013) Modified
nucleotides m(2)G966/m(5)C967 of Escherichia coli 16S rRNA are
required for attenuation of tryptophan operon, Sci. Rep.,
3, 3236.
87.Helser, T. L., Davies, J. E., and Dahlberg, J. E.
(1972) Mechanism of kasugamycin resistance in Escherichia coli,
Nature New Biol., 235, 6-9.
88.Schluenzen, F., Takemoto, C., Wilson, D. N.,
Kaminishi, T., Harms, J. M., et al. (2006) The antibiotic kasugamycin
mimics mRNA nucleotides to destabilize tRNA binding and inhibit
canonical translation initiation, Nat. Struct. Mol. Biol.,
13, 871-878.
89.Vila-Sanjurjo, A., Squires, C. L., and Dahlberg,
A. E. (1999) Isolation of kasugamycin resistant mutants in the 16 S
ribosomal RNA of Escherichia coli, J. Mol. Biol.,
293, 1-8.
90.Demirci, H., Murphy, F., Belardinelli, R.,
Kelley, A. C., Ramakrishnan, V., et al. (2010) Modification of 16S
ribosomal RNA by the KsgA methyltransferase restructures the 30S
subunit to optimize ribosome function, RNA, 16,
2319-2324.
91.Connolly, K., Rife, J. P., and Culver, G. (2008)
Mechanistic insight into the ribosome biogenesis functions of the
ancient protein KsgA, Mol. Microbiol., 70, 1062-1075.
92.Mecsas, J., Bilis, I., and Falkow, S. (2001)
Identification of attenuated Yersinia pseudotuberculosis strains
and characterization of an orogastric infection in BALB/c mice on day 5
postinfection by signature-tagged mutagenesis, Infect. Immun.,
69, 2779-2787.
93.Wimberly, B. T., Brodersen, D. E., Clemons, W.
M., Morgan-Warren, R. J., Carter, A. P., Vonrhein, C., Hartsch, T., and
Ramakrishnan, V. (2000) Structure of the 30S ribosomal subunit,
Nature, 407, 327-339.
94.Schuwirth, B. S., Day, J. M., Hau, C. W.,
Janssen, G. R., Dahlberg, A. E., Cate, J. H. D., and Vila-Sanjurjo, A.
(2006) Structural analysis of kasugamycin inhibition of translation,
Nat. Struct. Mol. Biol, 13, 879-886.
95.Demirci, H., Murphy, F. V., Murphy, E. L.,
Connetti, J. L., Dahlberg, A. E., Jogl, G., and Gregory, S. T. (2014)
Structural analysis of base substitutions in Thermus
thermophilus 16S rRNA conferring streptomycin resistance,
Antimicrob. Agents Chemother., 58, 4308-4317.
96.Nishimura, K., Hosaka, T., Tokuyama, S., Okamoto,
S., and Ochi, K. (2007) Mutations in rsmG, encoding a 16S rRNA
methyltransferase, result in low-level streptomycin resistance and
antibiotic overproduction in Streptomyces coelicolor A3(2),
J. Bacteriol., 189, 3876-3883.
97.Okamoto, S., Tamaru, A., Nakajima, C., Nishimura,
K., Tanaka, Y., Tokuyama, S., Suzuki, Y., and Ochi, K. (2007) Loss of a
conserved 7-methylguanosine modification in 16S rRNA confers low-level
streptomycin resistance in bacteria, Mol. Microbiol., 63,
1096-1106.
98.Mikheil, D. M., Shippy, D. C., Eakley, N. M.,
Okwumabua, O. E., and Fadl, A. A. (2012) Deletion of gene encoding
methyltransferase (gidB) confers high-level antimicrobial resistance in
Salmonella, J. Antibiot., 65, 185-192.
99.Carter, A. P., Clemons, W. M., Brodersen, D. E.,
Morgan-Warren, R. J., Wimberly, B. T., and Ramakrishnan, V. (2000)
Functional insights from the structure of the 30S ribosomal subunit and
its interactions with antibiotics, Nature, 407,
340-348.
100.Benítez-Páez, A.,
Cárdenas-Brito, S., Corredor, M., Villarroya, M., and Armengod,
M. E. (2013) Mutaciones en genes modificadores de ARN ribosómico
y la resistencia a aminoglucósidos: el caso del gen rsmG,
Biomédica, 34, 41, doi:
10.7705/biomedica.v34i0.1702.
101.Gustafsson, C., and Persson, B. C. (1998)
Identification of the rrmA gene encoding the 23S rRNA m1G745
methyltransferase in Escherichia coli and characterization of an
m1G745-deficient mutant, J. Bacteriol., 180,
359-365.
102.Jenner, L., Starosta, A. L., Terry, D. S.,
Mikolajka, A., Filonava, L., et al. (2013) Structural basis for potent
inhibitory activity of the antibiotic tigecycline during protein
synthesis, Proc. Natl. Acad. Sci. USA, 110,
3812-3816.
103.Lupien, A., Gingras, H., Leprohon, P., and
Ouellette, M. (2015) Induced tigecycline resistance in Streptococcus
pneumoniae mutants reveals mutations in ribosomal proteins and
rRNA, J. Antimicrob. Chemother., 70, 2973-2980.
104.Nichols, R. J., Sen, S., Choo, Y. J., Beltrao,
P., Zietek, M., et al. (2011) Phenotypic landscape of a bacterial cell,
Cell, 144, 143-156.