Catalytic Properties of Flavocytochrome c Sulfide Dehydrogenase from Haloalkaliphilic Bacterium Thioalkalivibrio paradoxus
Tamara V. Tikhonova1,a*, Anastasiya V. Lilina1, Evgenii M. Osipov1, Nikolay S. Shipkov1, Nataliya I. Dergousova1, Olga G. Kulikova1, and Vladimir O. Popov1
1Bach Institute of Biochemistry, Research Centre of Biotechnology, Russian Academy of Sciences, 119071 Moscow, Russia* To whom correspondence should be addressed.
Received September 25, 2020; Revised November 23, 2020; Accepted November 23, 2020
Flavocytochrome c sulfide dehydrogenase (FCC) is one of the central enzymes of the respiratory chain in sulfur-oxidizing bacteria. FCC catalyzes oxidation of sulfide and polysulfide ions to elemental sulfur accompanied by electron transfer to cytochrome c. The catalytically active form of the enzyme is a non-covalently linked heterodimer composed of flavin- and heme-binding subunits. The Thioalkalivibrio paradoxus ARh1 genome contains five copies of genes encoding homologous FCCs with an amino acid sequence identity from 36 to 54%. When growing on thiocyanate or thiosulfate as the main energy source, the bacterium synthesizes products of different copies of FCC genes. In this work, we isolated and characterized FCC synthesized during the growth of Tv. paradoxus on thiocyanate. FCC was shown to oxidize exclusively sulfide but not other reduced sulfur compounds, such as thiosulfate, sulfite, tetrathionate, and sulfur, and it also does not catalyze the reverse reaction of sulfur reduction to sulfide. Kinetic parameters of the sulfide oxidation reaction are characterized.
KEY WORDS: sulfur-oxidizing bacteria, flavocytochrome c sulfide dehydrogenase, thiocyanate dehydrogenase, enzyme kineticsDOI: 10.1134/S0006297921030111
Abbreviations: CytC, cytochrome c from horse heart; FCC, flavocytochrome c sulfide dehydrogenase; TcDH, thiocyanate dehydrogenase; TpFCC, flavocytochrome c sulfide dehydrogenase from the bacterium Thioalkalivibrio paradoxus ARh 1.
INTRODUCTION
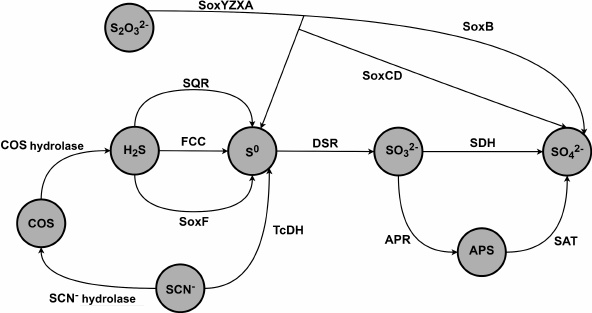
Gram-negative sulfur-oxidizing γ-proteobacteria of the genus Thioalkalivibrio are haloalkaliphilic obligate autotrophs that are found in soda lakes in Africa, Central Asia, and North America [1-4]. Microbial communities in these lakes have adapted to extreme conditions such as extreme alkaline pH levels (up to pH 11) and high salinity (up to 4 M Na+). Oxidation of the reduced sulfur compounds (thiosulfate, sulfide, polysulfide, and sulfur) serves as the main source of energy for the lithoautotrophic microbial communities in soda lakes [1]. Ten out of 85 Thioalkalivibrio strains isolated from soda lakes are able to grow on thiocyanate (SCN–) as a sole energy source [2, 5]. Thioalkalivibrio paradoxus ARh 1, Thioalkalivibrio nitratireducens ALEN 2, and Thioalkalivibrio thiocyanoxidans ARh 2T are the most well-characterized strains [6-11]. Figure 1 presents main metabolic pathways of sulfur compounds in the energy cycle of the bacteria of the Thioalkalivibrio genus based on the analysis of the genomes and transcriptomes of the Thioalkalivibrio bacteria [6].
Fig. 1. Schematic diagram of oxidation steps of the sulfur cycle in thiocyanate-oxidizing bacteria of the Thioalkalivibrio genus [6]. SQR is sulfide:quinone oxidoreductase; FCC is flavocytochrome c sulfide dehydrogenase; SoxYZXAB and SoxCD are components of the sulfur-oxidizing system Sox; TcDH is thiocyanate dehydrogenase; DSR is dissimilatory sulfite reductase; SDH is sulfite dehydrogenase; APR is adenosine 5′-phosphosulfate reductase; SAT is sulfate adenylyltransferase.
Flavocytochrome c sulfide dehydrogenase (FCC) is one of the central enzymes of sulfur metabolism in sulfur-oxidizing bacteria [12-16]. This enzyme catalyzes oxidation of sulfide and polysulfide ions to molecular sulfur accompanied by electron transfer to cytochrome c. The catalytically active form of FCC is a non-covalently linked heterodimer composed of a flavin-binding subunit (≈45 kDa) and a monoheme c (≈10 kDa) [13, 14] or diheme c (≈25 kDa) [12, 15, 16] binding subunit. The flavin-binding subunit of FCC contains a flavin adenine dinucleotide (FAD) molecule covalently bound through a thioether bond.
The genome of one of the thiocyanate-oxidizing strains, Thioalkalivibrio paradoxus ARh 1 [6], contains five copies of gene clusters encoding proteins homologous to FCC. The gene of one of the five potential FCCs (hereinafter TpFCC) is located adjacent to the operon responsible for biosynthesis of thiocyanate dehydrogenase (TcDH), which is a key enzyme of thiocyanate metabolism in bacteria of the Thioalkalivibrio genus [6, 7, 11]. Comparative analysis of the transcriptomes of the bacterium Thioalkalivibrio thiocyanoxidans ARh 2T grown on thiosulfate and thiocyanate as an energy source demonstrated that expression of the TcDH gene and neighboring genes significantly increased when the cells grew on thiocyanate. Maximum increase in the expression levels was achieved for the genes encoding TcDH (log2 of the gene expression level ratio during the growth on thiocyanate and thiosulfate was 7.5) and FCC (the corresponding logarithm of the expression level ratio was 6.94 for the flavin subunit of FCC and 5.82 for the heme-containing subunit of FCC). Other genes in the TcDH cluster show changes in the gene expression level in the range from 3.95 to 6.75 [6]. From this it follows that the expression of FCC gene in the bacterium Tv. thiocyanoxidans increases specifically and is strongly associated with the TcDH gene expression level. Presumably, a similar situation occurs with the closely related bacterium Tv. paradoxus ARh 1. Therefore, TpFCC is a required component of thiocyanate metabolism in the bacterium Tv. paradoxus. However, the role of TpFCC in the thiocyanate metabolism is unclear because thiocyanate decomposition (Fig. 1) does not involve formation of sulfide ions. It was suggested that TpFCC can be involved in the transfer of electrons generated in the process of thiocyanate oxidation by TcDH [6].
Previously, TpFCC was isolated as a major protein of the periplasmic fraction of the bacterium Tv. paradoxus grown on thiocyanate. The three-dimensional structure of TpFCC was determined by X-ray crystallography [17]. Comparative analysis of the three-dimensional structure of TpFCC and the structures of FCCs from other organisms confirmed that TpFCC exhibits all characteristic features of the active sites of sulfide dehydrogenases [FCC and SQR (sulfide:quinone oxidoreductase)], including redox-active cysteine residues involved in sulfide oxidation followed by electron transfer to FAD. Moreover, participation of TpFCC in the oxidation of sulfide ions to sulfur has been further confirmed by the presence of an additional sulfur atom as a product of the reaction at one of the catalytic cysteines [17]. Such structures containing chains of two or eight sulfur atoms in the active site were determined previously for FCC from Thermochromatium tepidum [16] and SQR from Aquifex aeolicus [18, 19]. The electron-transport subunit of TpFCC contains one domain that binds one heme c coordinated by His and Met residues. The polypeptide chain fold of the cytochrome subunit is similar to that of the N-terminal domains of the two-domain cytochrome subunits in other FCCs. In the three-dimensional structure determined in [17], TpFCC exists in complex with the copper-binding protein CopC, the gene of which is also characterized by the increased level of transcription in the cells growing on thiocyanate. It was suggested [17] that the TpFCC–CopC complex is a component of a larger periplasmic complex, which also includes TcDH. Role of the complex is to oxidize thiocyanate and utilize the resulting sulfur.
To test these assumptions, in the present study we performed functional characterization of TpFCC. It was shown that, like the FCCs described previously, TpFCC has a narrow substrate specificity and only catalyzes oxidation of sulfide ions. This enzyme does not catalyze oxidation of other anions containing sulfur in the reduced state (thiosulfate, tetrathionate, sulfite) as well as oxidation or reduction of molecular sulfur. Optimal conditions for the in vitro sulfide oxidation were determined. Kinetic parameters of the reaction were evaluated.
MATERIALS AND METHODS
Isolation and purification of TpFCC. Tv. paradoxus ARh 1 biomass was grown in flasks using a Na2CO3/NaHCO3 mineral medium supplemented with 0.6 M total Na+ at pH 9.75, as described previously [5] in the presence of sodium thiocyanate (10 mM) as an energy and nitrogen source. After all thiocyanate was consumed, another portion of thiocyanate was added to the growth medium up to concentration of 10 mM. Concentration of Cu2+ (as citrate) in the growth medium was 30 µg/liter.
After full utilization of thiocyanate, the cells were precipitated from ten liters of the culture by centrifugation (Hitachi, USA) and washed with 20 mM MOPS buffer, pH 7.5, supplemented with 0.6 M NaCl. TpFCC was isolated from the periplasmic fraction. Isolation procedure included two consecutive anion-exchange chromatography steps and one size exclusion chromatography step. Periplasmic fraction was obtained by incubation of the cells in 42 ml of a lysis buffer (20 mM MOPS, pH 7.5, 1 mg/ml lysozyme, 500 mM sucrose) for 10 min at room temperature. Then Milli-Q water (63 ml) and 1 mM PMSF were added, and the mixture was incubated at 4°C for 15 min. The periplasmic fraction was separated from the spheroplasts by centrifugation at 8,000 rpm for 30 min and then loaded onto a MonoQ 10/100 anion-exchange column (GE Healthcare, USA) equilibrated with 25 mM MOPS, pH 7.5. Proteins were eluted with a linear gradient of 1 M NaCl in the same buffer from 0 to 70%. Chromatography was performed on an ACTA FPLC chromatograph (GE Healthcare). Fractions with sulfide dehydrogenase activity were collected at the 1 M NaCl content of 30-40%. The combined fractions were dialyzed against 25 mM MOPS, pH 7.5, and again applied to a MonoQ 10/100 column equilibrated with the same buffer. FCC was eluted with a stepwise gradient of 1 M NaCl in 25 mM MOPS, pH 7.5. The fractions with sulfide dehydrogenase activity were eluted in the presence of 42% NaCl. Final purification step involved size exclusion chromatography on a Superdex 75 10/300 column (GE Healthcare), which was equilibrated with 25 mM MOPS buffer, pH 7.5, supplemented with 150 mM NaCl.
Protein concentration was determined by the Bradford assay [20]. Purity of the protein was analyzed with SDS–PAGE [21]. The protein was identified by MALDI-TOF MS using an Ultraflex III MALDI-TOF/TOF mass spectrometer (Bruker, Germany).
Activity assay of TpFCC. TpFCC activity was measured at 30°C using a Cary 100 Bio spectrophotometer (Thermo Scientific, USA). The reaction mixture (1 ml) was composed of 50 mM glycine–NaOH buffer, pH 9.5, 10 µM sodium sulfide, and 14 nM enzyme; 20 µM horse heart cytochrome c (CytC, Sigma-Aldrich, USA) was used as an electron acceptor. The reaction was initiated with sodium sulfide. Initial enzyme reaction rates were calculated from the initial parts of kinetic curves of CytC reduction recorded at 550 nm (ε550 = 22500 M–1 cm–1). Correction for non-enzyme reaction was applied. For this purpose, the rate of CytC reduction with sodium sulfide was measured in a parallel experiment in the absence of FCC. All kinetic measurements were performed in triplicate. Sodium sulfide concentration in the solution was determined using Ellman’s reagent [DTNB, dithiobis(2-nitrobenzoic acid)]. DTNB reacts with free sulfhydryl groups to form a mixed disulfide and bright-yellow 2-nitro-5-thiobenzoic acid (ε412 = 14150 M–1 cm–1).
Kinetic parameters of the sulfide dehydrogenase reaction were calculated from the plots of the initial reaction rates as a function of the substrate concentration.
Activity of TpFCC in oxidation of other substrates (1 mM thiosulftate, sulfite, or tetrathionate) was evaluated under similar conditions as described in [13]. Activity of TpFCC in the oxidation of elemental sulfur was evaluated using sulfur from granules of Tv. paradoxus that were produced during isolation of TpFCC. Sulfur suspension was prepared according to the procedure described by Rühl et al. [22] with slight modifications. Sulfur (0.1 g) was suspended in 5 ml of 50 mM glycine–NaOH buffer, pH 9.5. Then the 2% suspension was ultrasonicated for 10 min. The reaction mixture containing 0.2% sulfur suspension, 20 µM CytC, and 52 nM TpFCC in 50 mM glycine–NaOH buffer, pH 9.5, was incubated in an Eppendorf ThermoMixer at 30°C. Aliquots were withdrawn from the reaction mixture at regular intervals and centrifuged. Absorption spectra of obtained supernatants were recorded to detect formation of the reduced CytC. A similar experiment was performed in the absence of TpFCC.
To evaluate ability of TpFCC to catalyze the reverse reaction of sulfur reduction to sulfide, a 0.2% sulfur suspension was added to TpFCC, which was pre-reduced with dithionite, and changes in absorption spectra of TpFCC were recorded. Oxidation of TpFCC in the presence of sulfur would be indicative of reduction activity [23]. This reaction was performed in an anaerobic glove box (Belle Technology, UK) under N2 atmosphere containing residual oxygen of at most 2 ppm. All solutions were pre-purged with high-purity argon and stored in the glove box for several hours.
Determination of the reaction product. To determine the product of sulfide oxidation, the enzyme reaction was carried out up to high degrees of conversions. Addition of sulfide was terminated once sulfur precipitation was visually observed. Sulfur granules of the bacterium Tv. paradoxus (standard) and the precipitate obtained in the reaction were dissolved in a chloroform:methanol mixture (1 : 9). The reaction product was identified by reversed-phase chromatography on a Nucleosil C18 column (Sigma-Aldrich) according to a procedure, which was proposed in [24] and described in detail in our previous study [11].
Bioinformatics methods. Search for FCC homologues was performed using the Protein BLAST algorithm with the NCBI website (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Alignment of the amino acid sequences of FCCs from different organisms was carried out with the BLAST COBALT program (http://www.st-va.ncbi.nlm.nih.gov/tools/cobalt/re_cobalt.cgi).
RESULTS AND DISCUSSION
FCCs homologs in the genome of Tv. paradoxus. The genome of the bacterium Tv. paradoxus, like the genome of the closely related bacterium Tv. nitratireducens, contains five copies of genes encoding homologous FCCs [6]. The percent of amino acid sequence identity between the flavin-binding subunits encoded by different copies of the FCC gene from Tv. paradoxus is 40-54% and between the heme-containing subunits – 35-40% (Table 1).
Table 1. Identity between the flavin- and
heme-binding subunits encoded by five FCC copies in the genome of
Tv. paradoxus ARh 1
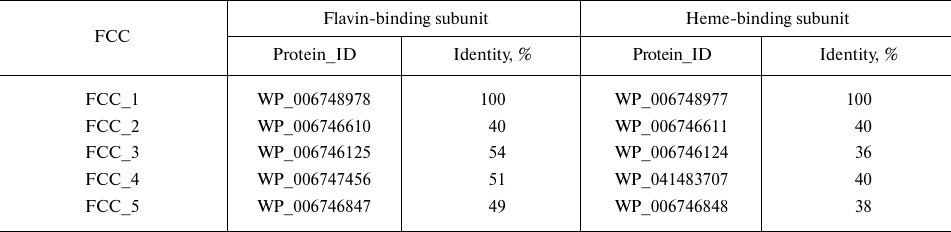
Only one protein (FCC_5 in Table 1) contains a diheme subunit; all other proteins are monoheme (the number of hemes was determined from the number of heme-binding motifs CXXCH in the amino acid sequence of the protein).
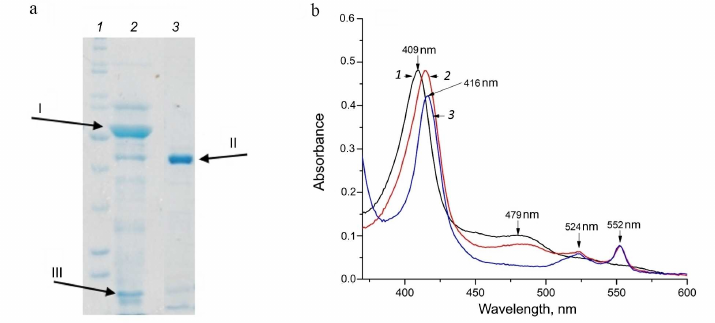
Different fcc genes were expressed during bacterial growth in the presence of different electron donors. According to our unpublished data, the proteins FCC_3 and FCC_5 are expressed in the presence of thiosulfate as an electron donor. During the growth on thiocyanate in the presence of Cu2+ ions, the bacterium Tv. paradoxus ARh 1 showed strong expression of the protein FCC_1 (hereinafter, TpFCC) (Table 1), which is one of the major proteins of the cell periplasmic fraction, along with the proteins TcDH and CopC under these conditions (Fig. 2a). These data are in agreement with the results of the transcriptome analysis performed in [7] and are indicative of the potential involvement of TpFCC in the thiocyanate decomposition. In order to characterize catalytic properties of the enzyme, TpFCC was isolated from the periplasmic fraction of Tv. paradoxus ARh 1.
Fig. 2. Characteristics of homogeneity and spectral properties of TpFCC. a) SDS–PAGE of the periplasmic fraction (lane 2) and purified TpFCC preparation (lane 3). In lanes 2 and 3, the arrows indicate TcDH (I), the FAD-containing subunit of TpFCC with a molecular weight of 42 kDa (II), and the copper-binding protein CopC (III); the heme-containing subunit of TpFCC migrated with the solvent front. Lane 1, molecular weight markers. b) Spectra of the oxidized (1) and reduced (2, 3) forms of TpFCC. Reduction was performed with (2) 1 mM Na2S and (3) 1 mM Na2S2O4. TpFCC concentration was 0.86 µM. Main absorption maxima of the oxidized and reduced forms are indicated in the spectra.
Physicochemical properties of TpFCC. According to the results of size exclusion chromatography on the Superdex 75 10/300 column, TpFCC has molecular weight of about 50 kDa. This enzyme consists of two subunits: the FAD-binding subunit with molecular weight of 42 kDa (Fig. 2a) and the heme c-binding subunit with molecular weight of 8 kDa.
The spectra of oxidized and reduced forms of TpFCC were recorded using sodium dithionite as a reducing agent (which can fully reduce FAD and heme c) and sodium sulfide (which is the main substrate for FCC).
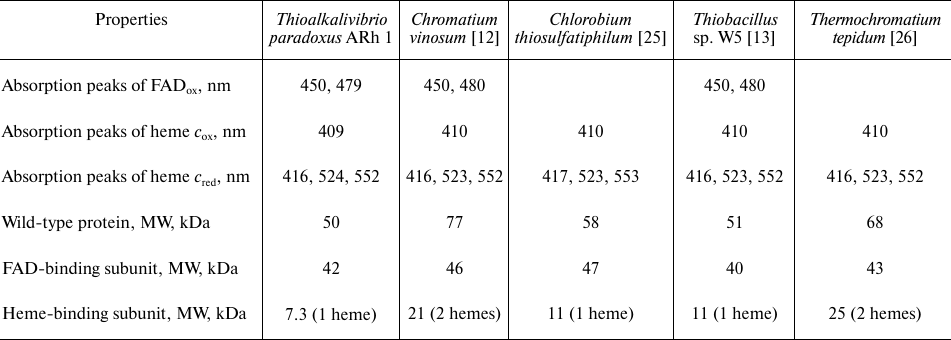
These spectra are very similar to the spectra of FCCs from other organisms published previously (Table 2). The spectrum of the oxidized form of TpFCC (Fig. 2b) shows characteristic absorption peak of the oxidized FAD at 480 nm. Another absorption band of FAD at 450 nm is less pronounced, like in the spectra of other FCCs [13, 26] and is, probably, partially overlapped with the more intense absorption peak of the heme-containing subunit with maximum at 409 nm. Reduction of TpFCC with sodium sulfide (1 mM) is accompanied by the partial reduction of FAD and the virtually complete reduction of heme c. Further addition of sodium sulfide does not lead to additional reduction of TpFCC. Apparently, under these conditions, FAD accepts two electrons from the sulfide, transfers one electron to the heme c, and remains in the semiquinone form, resulting in the partial retention of absorption at 480 nm. The addition of sodium dithionite (redox potential of –560 mV) to the reaction mixture does not change the degree of reduction of the heme c but leads to the complete reduction of FAD. The reduced form of TpFCC is characterized by absorption bands of the reduced heme c at 416, 524, and 552 nm.
Table 2. Spectral and molecular properties
of FCCs from different microorganisms
Characterization of the catalytic activity of TpFCC. The structural data indicate that TpFCC is a typical flavocytochrome c sulfide dehydrogenase. Hence, sulfide was considered as the main substrate for characterization of catalytic properties of TpFCC. Horse heart cytochrome c (CytC) was used as an electron acceptor, as in most studies on the catalytic properties of FCCs [12, 13, 25-27].
Optimization of the conditions for evaluation of catalytic activity. TpFCC is located in the periplasm of the haloalkaliphilic bacterium Tv. paradoxus, where the pH value is about 9.0 and salinity is up to 1 M [9]. To optimize conditions for evaluation of activity and compare them with the physiological conditions of TpFCC functioning, the effects of pH, nature of the buffer, and ionic strength were examined.
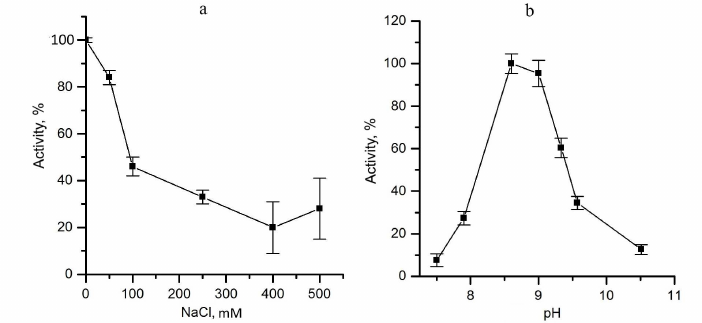
The effect of ionic strength was evaluated by addition of NaCl to the reaction mixture. Activity of the enzyme was about 50% of the initial value in 0.1 M NaCl (Fig. 3a). Apparently, increase of ionic strength leads to the weakening of ionic interactions between sulfide and the protein or hinders electrostatic interaction of CytC with the cytochrome subunit of FCC. Therefore, low ionic strength conditions are optimal for kinetic experiments.
Fig. 3. Factors affecting the catalytic activity of TpFCC. a) Effect of ionic strength (NaCl concentration) on the TpFCC activity. b) pH-Dependence of the TpFCC activity.
pH-Dependence of the TpFCC activity was determined in the pH range from 7 to 10.5 using the following two buffers: 50 mM Tris-HCl, pH from 7.0 to 8.6, and 50 mM glycine–NaOH, pH from 8.6 to 10.5 (Fig. 3b). TpFCC exhibits highest activity in the pH range from 8.5 to 9.0. These data are consistent with the fact that TpFCC is the periplasmic protein from the haloalkaliphilic bacterium with optimal growth conditions at pH 9-10 [9].
The enzyme exhibited the highest activity in the glycine buffer. Hence, all kinetic experiments were carried out in 25 mM glycine–NaOH, pH 8.6.
The TpFCC activity with other substrates was evaluated under these conditions. TpFCC did not catalyze oxidation of thiosulfate, sulfite, tetrathionate, and sulfur. Previously, similar results were obtained for FCCs from other sources [13]. Since the first step of the respiratory chain in Tv. paradoxus growing on thiocyanate involves oxidative degradation of thiocyanate producing elemental sulfur, it was suggested that FCC can catalyze the reverse reaction of sulfur reduction to form sulfide followed by the oxidation of the latter, e.g., by the reverse dissimilatory sulfite reductase (DSR in Fig. 1), which catalyzes interconversion of sulfite to sulfide (or sulfur) and vice versa. However, our experiment demonstrated that sulfur did not oxidize the reduced form of TpFCC, i.e., TpFCC did not transfer electrons to sulfur. It is worth noting that the surface of the FAD-containing subunit bears a positive charge at the entrance to the sulfide-binding site [17], which is unfavorable for binding a hydrophobic uncharged substrate, such as sulfur. Therefore, TpFCC does not catalyze the reverse reaction of sulfur reduction to sulfide and, consequently, it cannot be involved in the utilization of sulfur that is generated via oxidation of thiocyanate. Since we found sulfide as the only substrate for TpFCC, which is consistent with the data for other FCCs [12, 13], we characterized kinetic parameters of this reaction.
Determination of kinetic parameters. The kinetics of oxidation of sodium sulfide and reduction of CytC in the absence of the reaction products is described by the following equation for a two-substrate reaction with formation of a ternary complex [28]:
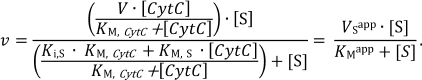 (1)
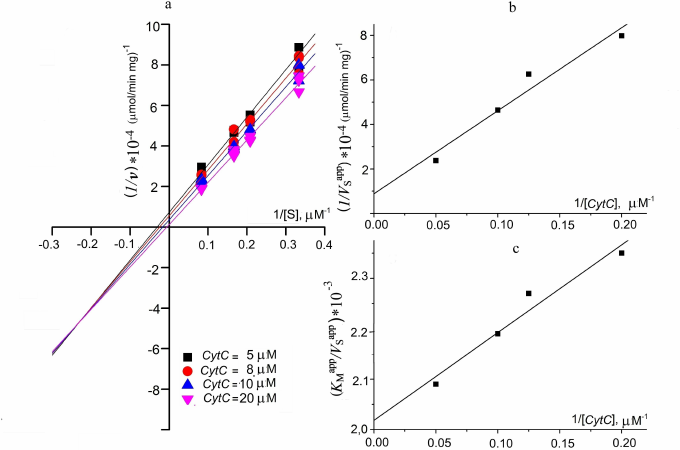
(1)The apparent Michaelis constants (KMapp) are 1.9 ± 0.4 and 8 ± 3 µM for sulfide and CytC, respectively, at a fixed concentration of the second substrate. These data are in good agreement with the Michaelis constants measured for FCC from Thiobacillus (KM is 1.7 ± 0.4 µM for sulfide and 3.8 ± 0.8 µM for cytochrome) [13]. The maximum specific activity at these concentrations was 54 ± 9 µmol (CytC)/min per milligram of the protein. However, these constants are apparent because they are functions of the concentration of the second substrate. Experimental determination of the true kinetic constants included measurement of the kinetic constants with respect to sulfide (Vsapp, KMapp) at different fixed concentrations of CytC (5, 8, 10, 20 µM):
where Ki,S is the dissociation constant of the enzyme–substrate (ES) complex.
The results are presented as the primary Lineweaver–Burk plots (Fig. 4a). The straight lines corresponding to the fixed CytC concentrations intersect at a point with coordinates ((1–KM,S/Ki,S)/V; –1/Ki,S). Intersection with the X axis corresponds to –1/KMapp; with the Y axis – to 1/VSapp. This plot is typical for the enzyme reactions involving formation of a ternary complex.
The values of 1/V were determined from the intersection of the Y axis of the secondary plot in the coordinates (1/VSapp; 1/[CytC]) and KM,CytC/V based on the slope (Fig. 4b):
The true Michaelis constant with respect to sulfide was determined from the plot in the coordinates (KMapp/VSapp; 1/[CytC]). The value of KM,S/V corresponds to the intersection of the straight line with the Y axis. The constant can be evaluated from KM,S/V and the known maximum reaction rate (Fig. 4c).
The following kinetic parameters were obtained based on the experimental data:
V = 80 ± 40 µmol CytC/min per mg of the protein, kcat = 0.67 s–1;
KM,CytC = 26 ± 1 µM;
KM,S = 157 ± 9 µM.
Specific properties of the substrates did not allow us to perform experiments at saturation concentrations because at high CytC concentrations, the absorbance is too high, while at high sulfide concentrations, contribution of the non-enzyme reduction of CytC becomes significant. In other studies of FCCs, the kinetic parameters were also measured at non-saturating concentrations of the substrates, which makes it impossible to directly compare the measured values. This also accounts for the difference in the observed and true values of KM and V.
Fig. 4. Determination of kinetic constants of the sulfide oxidation by TpFCC. a) Double reciprocal plot of the initial reaction rate as function of sulfide concentration at different fixed CytC concentrations. b) Double reciprocal plot of the maximum reaction rate as function of the CytC concentration. c) Dependence of KMapp/VSapp on the reciprocal CytC concentration
In summary, it can be stated that TpFCC is a typical sulfide dehydrogenase that catalyzes sulfide oxidation producing elemental sulfur accompanied by electron transfer to cytochrome c. The role of this enzyme in thiocyanate metabolism remains unknown. Nevertheless, our preliminary experiments (the results not shown) demonstrated that the presence of catalytic amounts of TpFCC in the TcDH-catalyzed thiocyanate oxidation results in the change of the kinetic curve shape and increase in the reaction rate. The mechanism of the observed phenomenon remains unclear and needs further research.
Funding. This study was financially supported by the Russian Science Foundation (project no. 20-14-00314; characterization of the enzyme, enzyme kinetics) and by the Ministry of Science and Higher Education of the Russian Federation (isolation of the enzyme).
Ethics declarations. The authors declare no conflict of interest in financial or any other sphere. This article does not contain any studies with human participants or animals performed by any of the authors.
REFERENCES
1.Sorokin, D. Y., Berben, T., Melton, E. D.,
Overmars, L., Vavourakis, C. D., and Muyzer, G. (2014) Microbial
diversity and biogeochemical cycling in soda lakes,
Extremophiles, 18, 791-809, doi:
10.1007/s00792-014-0670-9.
2.Sorokin, D. Y., Banciu, H., Robertson, L. A.,
Kuenen, J. G., Muntyan, M. S., and Muyzer, G. (2013) in The
Prokaryotes: Prokaryotic Physiology and Biochemistry (Rosenberg,
E., DeLong, E. F., Lory, S., Stackebrandt, E., and Thompson, F., eds)
Springer, Berlin, pp. 529-554, doi: 10.1007/978-3-642-30141-4_77.
3.Sorokin, D. Y., Kuenen, J. G., and Muyzer, G.
(2011) The microbial sulfur cycle at extremely haloalkaline conditions
of soda lakes, Front. Microbiol., 2, 44, doi:
10.3389/fmicb.2011.00044.
4.Sorokin, D. Y., and Kuenen, J. G. (2005)
Haloalkaliphilic sulfur-oxidizing bacteria in soda lakes, FEMS
Microbiol. Rev., 29, 685-702, doi:
10.1016/j.femsre.2004.10.005.
5.Sorokin, D. Y., Tourova, T. P., Lysenko, A. M., and
Kuenen, J. G. (2001) Microbial thiocyanate utilization under highly
alkaline conditions, Appl. Environ. Microbiol., 67,
528-538, doi: 10.1128/AEM.67.2.528-538.2001.
6.Berben, T., Overmars, L., Sorokin, D. Y., and
Muyzer, G. (2017) Comparative genome analysis of three thiocyanate
oxidizing Thioalkalivibrio species isolated from soda lakes,
Front. Microbiol., 8, 254, doi:
10.3389/fmicb.2017.00254.
7.Berben, T., Balkema, C., Sorokin, D. Y., and
Muyzer, G. (2017) Analysis of the genes involved in thiocyanate
oxidation during growth in continuous culture of the haloalkaliphilic
sulfur-oxidizing bacterium Thioalkalivibrio thiocyanoxidans ARh
2T using transcriptomics, mSystems, 2, e00102-17, doi:
10.1128/mSystems.00102-17.
8.Berben, T., Overmars, L., Sorokin, D. Y., and
Muyzer, G. (2019) Diversity and distribution of sulfur
oxidation-related genes in Thioalkalivibrio, a genus of
chemolithoautotrophic and haloalkaliphilic sulfur-oxidizing bacteria,
Front. Microbiol., 10, 160, doi:
10.3389/fmicb.2019.00160.
9.Sorokin, D. Y., Tourova, T. P., Lysenko, A. M.,
Mityushina, L. L., and Kuenen, J. G. (2002) Thioalkalivibrio
thiocyanoxidans sp. nov. and Thioalkalivibrio paradoxus sp.
nov., novel alkaliphilic, obligately autotrophic, sulfur-oxidizing
bacteria capable of growth on thiocyanate, from soda lakes, Int. J.
Syst. Evol. Microbiol., 52, 657-664, doi:
10.1099/00207713-52-2-657.
10.Sorokin, D. Y., Tourova, T. P., Sjollema, K. A.,
and Kuenen, J. G. (2003) Thialkalivibrio nitratireducens
sp. nov., a nitrate-reducing member of an autotrophic denitrifying
consortium from a soda lake, Int. J. Syst. Evol. Microbiol.,
53, 1779-1783, doi: 10.1099/ijs.0.02615-0.
11.Tikhonova, T. V., Sorokin, D. Y., Hagen, W. R.,
Khrenova, M. G., Muyzer, G., et al. (2020) Trinuclear copper
biocatalytic center forms an active site of thiocyanate dehydrogenase,
Proc. Natl. Acad. Sci. USA, 117, 5280-5290, doi:
10.1073/pnas.1922133117.
12.Fukumori, Y., and Yamanaka, T. (1979)
Flavocytochrome c of Chromatium vinosum. Some enzymatic
properties and subunit structure, J. Biochem., 85,
1405-1414, doi: 10.1093/oxfordjournals.jbchem.a132467.
13.Visser, J. M., de Jong, G., Robertson, L. A., and
Kuenen, J. G. (1997) A novel membrane-bound flavocytochrome c
sulfide dehydrogenase from the colourless sulfur bacterium
Thiobacillus sp. W5, Arch. Microbiol., 167,
295-301, doi: 10.1007/s002030050447.
14.Kostanjevecki, V., Brigé, A., Meyer, T.
E., Cusanovich, M. A., Guisez, Y., and van Beeumen, J. (2000) A
membrane-bound flavocytochrome c-sulfide dehydrogenase from the
purple phototrophic sulfur bacterium Ectothiorhodospira
vacuolata, J. Bacteriol., 182, 3097-3103, doi:
10.1128/jb.182.11.3097-3103.2000.
15.Chen, Z., Koh, M., Van Driessche, G., Van
Beeumen, J., Bartsch, R., et al. (1994) The structure of
flavocytochrome c sulfide dehydrogenase from a purple
phototrophic bacterium, Science, 266, 430-432, doi:
10.1126/science.7939681.
16.Hirano, Y., Kimura, Y., Suzuki, H., Miki, K., and
Wang, Z.-Y. (2012) Structure analysis and comparative characterization
of the cytochrome c and flavocytochrome c from
thermophilic purple photosynthetic bacterium Thermochromatium
tepidum, Biochemistry, 51', 6556-6567, doi:
10.1021/bi3005522.
17.Osipov, E. M., Lilina, A. V., Tsallagov, S. I.,
Safonova, T. N., Sorokin, D. Y., et al. (2018) Structure of the
flavocytochrome c sulfide dehydrogenase associated with the
copper-binding protein CopC from the haloalkaliphilic sulfur-oxidizing
bacterium Thioalkalivibrio paradoxus ARh 1, Acta Cryst.,
D74, doi: 10.1107/S2059798318005648.
18.Marcia, M., Ermler, U., Peng, G., and Michel, H.
(2009) The structure of Aquifex aeolicus sulfide:quinone
oxidoreductase, a basis to understand sulfide detoxification and
respiration, Proc. Natl. Acad. Sci. USA, 106,
9625-9630.
19.Brito, J. A., Sousa, F. L., Stelter, M.,
Bandeiras, T. M., Vonrhein, C., et al. (2009) Structural and functional
insights into sulfide:quinone oxidoreductase, Biochemistry,
48, 5613-5622.
20.Bradford, M. M. (1976) A rapid and sensitive
method for the quantitation of microgram quantities of protein
utilizing the principle of protein-dye binding, Anal. Biochem.,
72, 248-254.
21.He, F. (2011) Laemmli-SDS-PAGE, Bio-101,
e80, doi: 10.21769/BioProtoc.80.
22.Rühl, P., Pöll, U., Braun, J., Klingl,
A., and Kletzin, A. (2017) A sulfur oxygenase from the haloalkaliphilic
bacterium Thioalkalivibrio paradoxus with atypically low
reductase activity, J. Bacteriol., 199, e00675-16, doi:
10.1128/JB.00675-16.
23.Ma, K., and Adams, M. W. (1994) Sulfide
dehydrogenase from the hyperthermophilic archaeon Pyrococcus
furiosus: a new multifunctional enzyme involved in the reduction of
elemental sulfur, J. Bacteriol., 176, 6509-6517, doi:
10.1128/jb.176.21.6509-6517.1994.
24.Alberta Environment and Parks (2015)
Development and Validation of Analytical Methods for Elemental
Sulfur in Alberta Soils, Prepared by Maxxam Analytics International
Corporation for Alberta Environment and Parks, Edmonton, Alberta.
25.Yamanaka, T. (1976) The subunits of
Chlorobium flavocytochrome c, J. Biochem.,
79, 655-660, doi: 10.1093/oxfordjournals.jbchem.a131110.
26.Castillo, M., Bih-Show, L., and Ondrias, M. R.
(1994) Characterization of flavocytochrome c 552 from the
thermophilic photosynthetic bacterium Chromatium tepidum,
Arch. Biochem. Biophys., 315, 262-266.
27.Sorokin, D. Yu., de Jong, G. A. H., Robertson, L.
A., and Kuenen, G. J. (1998) Purification and characterization of
sulfide dehydrogenase from alkaliphilic chemolithoautotrophic
sulfur-oxidizing bacteria, FEBS Lett., 427, 11-14.
28.Cornish-Bowden, A. (2012) Fundamentals of
enzyme kinetics, Wiley-Blackwel, Fourth Edition, p. 422.