REVIEW: Microbial Arsenal of Antiviral Defenses. Part II
Artem B. Isaev1,a*, Olga S. Musharova1,2,b, and Konstantin V. Severinov1,3,c*
1Skolkovo Institute of Science and Technology, 143028 Moscow, Skolkovo, Russia2Institute of Molecular Genetics, 119334 Moscow, Russia
3Waksman Institute of Microbiology, Piscataway, NJ 08854, USA
* To whom correspondence should be addressed.
Received August 30, 2020; Revised January 29, 2021; Accepted January 29, 2021
Bacteriophages or phages are viruses that infect bacterial cells (for the scope of this review we will also consider viruses that infect Archaea). The constant threat of phage infection is a major force that shapes evolution of microbial genomes. To withstand infection, bacteria had evolved numerous strategies to avoid recognition by phages or to directly interfere with phage propagation inside the cell. Classical molecular biology and genetic engineering had been deeply intertwined with the study of phages and host defenses. Nowadays, owing to the rise of phage therapy, broad application of CRISPR-Cas technologies, and development of bioinformatics approaches that facilitate discovery of new systems, phage biology experiences a revival. This review describes variety of strategies employed by microbes to counter phage infection. In the first part defense associated with cell surface, roles of small molecules, and innate immunity systems relying on DNA modification were discussed. The second part focuses on adaptive immunity systems, abortive infection mechanisms, defenses associated with mobile genetic elements, and novel systems discovered in recent years through metagenomic mining.
KEY WORDS: bacteriophages, phage–host interactions, antiviral defense, immunity systems, CRISPR-Cas, abortive infection, Toxin–Antitioxin, PICI, prokaryotic Argonaute, CBASS, retronsDOI: 10.1134/S0006297921040064
Abbreviations: Abi, abortive infection; Cap, CD-Ntase associated protein; CBASS, Cyclic-Oligonucleotide-Based Anti-Phage Signaling System; CD-NTase, oligonucleotide cyclase; CRISPR-Cas, Clustered Regularly Interspaced Palindromic Repeats and CRISPR-associated proteins; DRT, defense-associated RTase; MGE, mobile genetic element; DUF, Domain of Unknown Function; pAgo, prokaryotic Argonaute; PAM, protospacer adjacent motif; PICI, phage-inducible chromosomal islands; PLE, phage-inducible chromosomal island-like element; R-M, Restriction–Modification system; RTase, reverse transcriptase; SaPi, S. aureus pathogenicity island; TA, toxin–antitoxin.
INTRODUCTION
The first part of the review covered strategies that allow host cells to avoid recognition by phages, innate immunity mechanisms blocking early stages of infection, and systems that rely on DNA modification for self vs. non-self discrimination. Here, we will continue description of the variety of microbial antiviral systems.
CRISPR-Cas ADAPTIVE IMMUNITY SYSTEMS
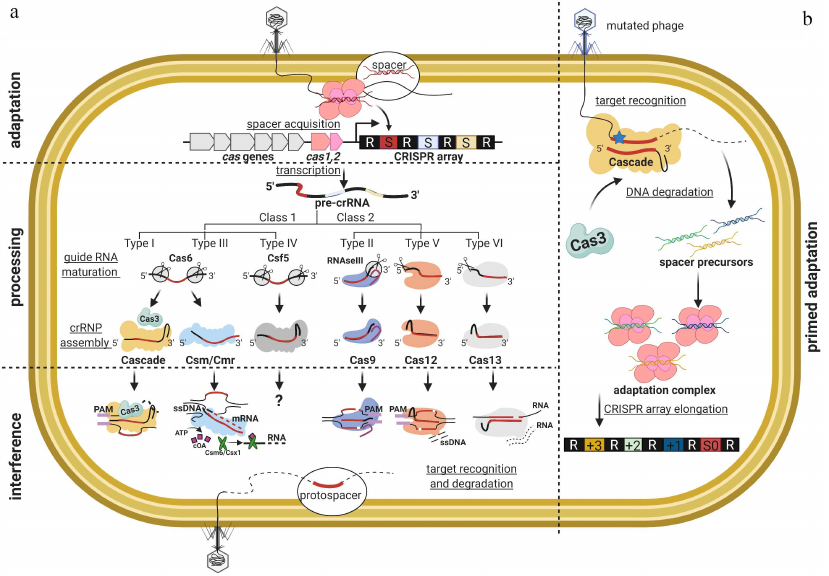
In contrast to the DNA modification-based innate immunity systems, where target recognition relies on interaction of defense proteins with a predetermined sequence within a phage genome, prokaryotes also possess adaptive immunity CRISPR-Cas (Clustered Regularly Interspaced Palindromic Repeats and CRISPR-associated proteins) systems. Here, the target nucleic acid recognition is driven by annealing of the complementary RNA molecule, and the system can generate and store guides for interference with novel sequences. An ability to preserve information about previous encounters with invaders is a feature that is shared between CRISPR-Cas systems and immunity of higher eukaryotes, such as humans. Unlike the case of mammals, the CRISPR-Cas immunity is inheritable. The CRISPR-Cas system consists of a CRISPR array (the number of arrays in prokaryotic genomes varies from one to several dozens) and associated cas genes [1-3]. A CRISPR array is a cluster of short repeated genomic DNA fragments separated by unique spacer sequences, at least some of which originate from the foreign DNA. An AT-rich leader region is located in front of the CRISPR array [1]. The cas genes encode protein components of the CRISPR-Cas mechanism. CRISPR-Cas systems are responsible for two different processes: adaptation and interference. CRISPR adaptation is the process of integration of new invader-derived spacers into the CRISPR array. In the course of elemental act of CRISPR adaptation, the array is expanded by one new spacer and one repeat. The proteins responsible for CRISPR adaptation generally are homologous in all CRISPR-Cas systems. Transcription of the array leads to the formation of a pre-crRNA that is processed into short crRNAs in such a way that each crRNA contains a spacer flanked by partial repeats. A crRNA bound by Cas proteins forms an effector complex capable of specific recognition of a protospacer – a DNA or RNA sequence complementary to the spacer part of the crRNA. Protospacer recognition is followed by the degradation of the target nucleic acid molecule that contains it. The process of target recognition and destruction is called CRISPR interference (Fig. 1).
Fig. 1. Mechanism of CRISPR-Cas adaptive immunity in prokaryotes. a) Fragments originating from foreign DNA could be integrated into a CRISPR array in the process of CRISPR adaptation. CRISPR array is elongated by one new spacer and one repeat. The CRISPR array is then transcribed with formation of the pre-crRNA that is processed into short crRNAs so that each crRNA contains a spacer flanked by partial repeats. The cas genes code for protein components of the CRISPR interference and CRISPR adaptation complexes. CRISPR interference complex consists of crRNA bound by Cas proteins and interacts with a protospacer, i.e., a DNA sequence complementary to the sequence of the crRNA spacer. Recognition of the protospacer by CRISPR effector complex leads to degradation of the target DNA molecule. Protein composition of the interference module is variable and used as a major criterion in CRISPR-Cas system classification. CRISPR-Cas systems are subdivided into two classes, six types, and several subtypes. The two classes are distinguished based on the composition of the interference complexes: CRISPR-Cas systems of Class 1 are multi-subunit, while systems of Class 2 contain only one protein. b) In the process of primed adaptation, new spacers are preferentially selected from the DNA targeted by the CRISPR effector complexes during interference.
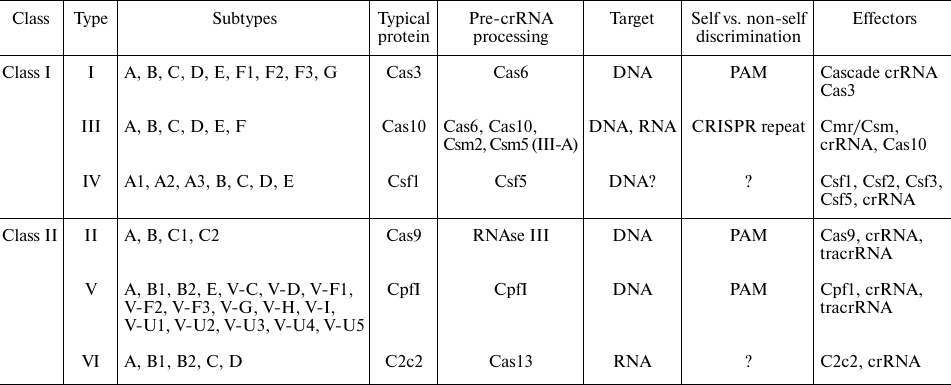
Diversity of CRISPR interference mechanisms. Classification of the CRISPR-Cas systems is based on the protein composition of effector complexes. According to the latest census, CRISPR-Cas systems can be subdivided into two classes, six types, and 33 subtypes [4]. Class 1 (Types I, III, and IV) systems utilize multisubunit effectors, while Class 2 (types II, V, and VI) effectors are single-subunit proteins (table). Different types of CRISPR-Cas systems are distinguished by the presence of specific “signature proteins” responsible for DNA degradation (Cas3, Csf1, Cas10, Cas9, Cpf1 and C2c2 for the Types I, IV, III, II, V, and VI, respectively [4]).
Diversity of interference mechanisms and classification of CRISPR-Cas
systems
CRISPR-Cas systems of Class I include three types: Type I, Type III, and Type IV. The effector complexes of Type I and Type III systems have been studied in detail. Architectural similarity of the effector complexes indicates common origin of these systems [5]. Type I effector comprises a large multisubunit protein complex called Cascade, which consists of the Repeat-Associated Mysterious Protein (RAMP) subunits in Cse11:Cse22:Cas76:Cas51:Cas61 stoichiometry. Cascade binds processed 61 nucleotide-long crRNA with 32-nt spacer [6-9]. Annealing of the Cascade-bound crRNA to the complementary protospacer leads to the localized melting of the target dsDNA and formation of an R-loop – a heteroduplex between the crRNA spacer and the “target” strand of the DNA protospacer, while the “non-target” DNA strand of the protospacer is displaced and remains in a single-stranded form. The obligatory condition for the target recognition is the presence of a short two-three nucleotide-long protospacer adjacent motif (PAM) sequence located at the 3′-end of the target strand, i.e., downstream from the protospacer. The requirement of PAM safeguards CRISPR-Cas systems from attack on the cell’s own genome, as PAM is never adjacent to the spacers in the CRISPR loci. Following the R-loop formation, the Cas3 nuclease/helicase is recruited to the complex [10]. Cas3 first introduces a single-stranded break in the “non-target” protospacer strand 11-15 nucleotides downstream from the PAM, then begins to unwind and cleave the DNA in 3′-5′ direction from the PAM.
In the Type III systems, the effector has a similar helicoid structure as that of Cascade [11, 12]. However, the Type III effector recognizes not dsDNA but RNA sequences complementary to the crRNA spacers [13, 14]. Recognition of the transcribing RNA target stimulates nonspecific DNA cleavage activity of the signature nuclease Cas10 HD (Histidine-Aspartate) domain, which results in the in situ degradation of DNA in the transcription bubble [15-20]. At the same time, the cyclic oligoadenylate (cOA) synthetase Palm domain of Cas10 is activated to produce cOA secondary messenger. cOA can be sensed by the auxiliary Cas ribonucleases (e.g., Csm6/Csx1) that degrade host and viral transcripts in a sequence non-specific manner [21-23]. The Type III systems do not rely on PAM for autoimmunity prevention, since the effector complex cannot target CRISPR array or crRNA, however, crRNA encodes 8-nt long sequence tag that inhibits Cas10 activity to avoid self-DNA cleavage in the case of CRISPR array transcription from the opposite strand [17]. If the target sequence is completely complementary to the spacer and the crRNA tag, the interference does not occur.
The exact immunity mechanism of CRISPR in the Type IV systems is not yet fully understood. The signature protein of such systems is Csf1. The Type IV CRISPR–Cas have been found localized on plasmids or in prophage genomes, implying the possibility for recurrent transfer of the CRISPR–Cas machinery to and from mobile genetic elements (MGEs) [24, 25]. The Type IV CRISPR-Cas signature genes are not accompanied by the adaptation module genes [26]. This leads to a suggestion that the Type IV proteins can be involved in cellular functions unrelated to the adaptive immunity [27, 28].
The CRISPR-Cas systems of Class II include three types: Type II, Type V and Type VI. In the Type II systems, the monomeric Cas9 protein in the complex with crRNA is responsible for the target dsDNA recognition and its degradation. Cas9 possesses two nuclease domains (RuvC and HNH), and is capable of double-stranded breaks generation [29, 30]. It represents a minimal interference system, and therefore became the preferred tool in the CRISPR–Cas-based genome engineering applications [31-33]. The Type V systems are characterized by the presence of Cpf1 effector protein. The Cpf1 contains the RuvC nuclease domain, similar to Cas9, while the HNH domain is absent [34]. The Type V effector is able to destroy the target double-stranded DNA in a PAM-specific manner [34, 35], while binding of the Cpf1 to the targets also unleashes its indiscriminate single-stranded DNase activity [36]. The majority of Type V systems contain Cpf1 effector, while in the V-F subtype it is replaced by Cas14. To date, Cas14 is the smallest of known CRISPR effectors. The Cas14a is an ssDNA-targeting CRISPR endonuclease that does not require PAM for activation [37]. Some V-U subtype effectors demonstrate phylogenetic similarity to the TnpB transposases [37, 38]. The Type VI CRISPR-Cas system was bioinformatically predicted in 2015 [39]. Soon, the effector protein C2c2 from the VI-A subtype was described. The VI-A locus in Leptotrichia shahii contains only three genes (cas1, cas2, c2c2) and a CRISPR array. The C2c2 nuclease with bound crRNA forms an effector complex, which is able to cleave the single-stranded RNA molecules. In contrast to all known CRISPR nucleases, C2c2 mediates RNA cleavage by the HEPN (higher eukaryotes and prokaryotes nucleotide) domain. Mutation in the catalytic centre of the HEPN domain leads to inactivation of the effector complex, but the RNA-binding activity of the resulting protein is retained [40]. Because of its ability to bind RNA molecules in a predetermined manner, C2c2 nuclease could be used as an effective tool for RNA editing and regulation of gene expression.
Origin of the CRISPR effectors diversity and their phylogenetic relations represent an interesting question. The effector complexes of Type I and Type III systems are quite similar in structure. It is assumed that the effector complex of the Type III system is more ancient. Here, cas genes are not always associated with the CRISPR arrays and cas1-cas2 adaptation module [41]. The standalone Cas1 homologs were detected in the mobile genetic elements named casposons. The Cas9 and the Cpf1 proteins, typical for the Type II and Type V, respectively, are similar to the TnpB transposon-encoded protein and contain an RuvC endonuclease domain [42]. The protein Cas13 (Type VI system) has RNAse HEPN domains. Thus, the CRISPR-Cas systems could have evolved by adopting interference and adaptation module genes from casposons, while effector nucleases may have originated from the cellular genomes or mobile genetic elements.
CRISPR adaptation. The most conserved components among all CRISPR-Cas systems are Cas1 and Cas2, which are required at the stage of spacer acquisition [43]. As a rule, the cas1 and cas2 genes are located close to each other, and the encoded proteins form a stable complex [44, 45]. Deletion of cas1 and cas2 does not affect CRISPR interference and crRNA maturation in Type I [46-49], Type II [50, 51] and Type III [52] systems. Cas1 is an endonuclease [53, 54], which also has ability to resolve Holliday junctions. In vitro, Cas1 can promote DNA integration and recombination events [55]. Cas2 displays nuclease activity towards both, RNA and DNA, in vitro [56, 57]. However, CRISPR adaptation in vivo requires nuclease activity associated only with Cas1 [44]. The ability to assemble stable Cas1–Cas2 complex is also essential for in vivo adaptation. Mutations that disrupt complex formation in vitro interfere with the in vivo spacer acquisition [44]. During the process of new spacer incorporation, the Cas1-Cas2 complex introduces a single-strand break exactly at the leader–repeat junction in the CRISPR array by catalyzing nucleophilic attack of the 3′-OH end of the incoming spacer on the 5′-end of the first repeat. Similarly, the other strand is nicked at the first repeat–spacer junction, and the 5′-end of the repeat strand is joined to the 3′-end of the new spacer. As a result, the incorporated spacer is flanked by the single-stranded repeat sequences that are filled later due to the activity of the host repair machinery [58]. Similar intermediates are known for the transposase-mediated mobile element integration suggesting that the spacer acquisition and transposon integration reactions are mechanistically similar [59-62].
The ability of the Cas1–Cas2 adaptation complex to uptake new spacers independent from the activity of CRISPR effector complexes is known as the process of naïve adaptation. During naïve adaptation, new spacers may be acquired from the extrachromosomal DNA as well as from the host genome, and only 50% of the new spacers contain consensus PAM. Naïve adaptation is essential for subsequent targeting of unknown foreign DNA and seems to be a universal feature of all CRISPR-Cas systems. The process is known to be at least partially dependent on the activity of the host RecBCD complexes [63]. The RecBCD performs processing of the stalled replication forks and it is believed that the resulting DNA fragments may be used by the Cas1–Cas2 complex for insertion into the CRISPR array. The absence of RecBCD reduces naïve adaptation efficiency but does not stop it. Consequently, the Cas1–Cas2 complex can utilize other sources of spacers. The question of participation of other host proteins in the CRISPR adaptation and regulation of this process has only recently attracted attention of the researchers [64-67]. For example, it was shown that the DNA polymerase I is necessary for both naïve and primed adaptation (presumably to fill single-stranded repeats that arise during spacers embedding) [68]. Dorman and Bhriain hypothesized that the negative supercoiling could alter various stages of the CRISPR proteins interaction with DNA, including adaptation, expression of cas genes and CRISPR loci, and, in fact, interference [65].
The presence of PAM allows distinguishing the host genome containing a spacer in the CRISPR array and protospacer within the target molecule. However, mutations in the PAM (or seed) sequence may protect viruses from recognition and degradation by the effector complexes [30, 69-72]. Therefore, the CRISPR-Cas system should update its “memory” to avoid infection by the “escaper” phages. To achieve this goal, some types of the CRISPR-Cas systems rely on the primed adaptation – a highly efficient process of new spacers acquisition from the already “known”, previously encountered phages whose fragments have been stored in the CRISPR array as immunological memory. Primed adaptation was demonstrated in the I-E [48], I-F [49, 73], I-B [74, 75], I-C [76], I-U [77], and II-A [78] CRISPR-Cas systems. Primed adaptation leads to the highly efficient and targeted accumulation of new spacers located in cis-position to the “priming” protospacer recognized by the effector [79]. The observed efficiency of primed adaptation (measured as the number of extended CRISPR arrays in the population) is very low if the target protospacer fully matches the crRNA and contains a consensus interference-proficient PAM (AAG or ATG in the case of E. coli I-E system) [48, 80]. Efficiency of the primed adaptation is enhanced by the presence of PAM or protospacer mutations that decrease interference efficiency [30, 70, 73]. Yet, primed adaptation requires activity of Cas3 protein, suggesting functional link between the CRISPR interference and primed adaptation [71, 81]. Recent in vitro study suggests that Cascade, Cas1-Cas2 and Cas3 form a single priming complex with activity leading to the effective selection of new spacers [82]. Two alternative models have been suggested to explain such a link. One model postulates that the effectors bound to protospacers with certain PAMs assume a specific conformation that recruits adaptation machinery (Cas1 and Cas2) as well as Cas3 protein, followed by the directional scanning of the target and selection of the new spacers [83]. In contrast, the complexes formed on targets with the interference-proficient PAMs do not support Cas1-Cas2 recruitment, leading to interference only [84]. The second model postulates that the apparent difference in the efficiency of primed adaptation with different targets is a consequence of the dynamics of degradation of the less-than-optimal targets [81, 85]. Since most MGEs are able to replicate and have copy-number maintenance mechanisms of their own, competition between the attenuated CRISPR interference and copy number maintenance mechanisms could create a situation when degradation fragments of the MGE genomes are present in the cell for an extended time, allowing the presumably slower adaptation reaction to occur [86].
ARGONAUTE-MEDIATED INTERFERENCE
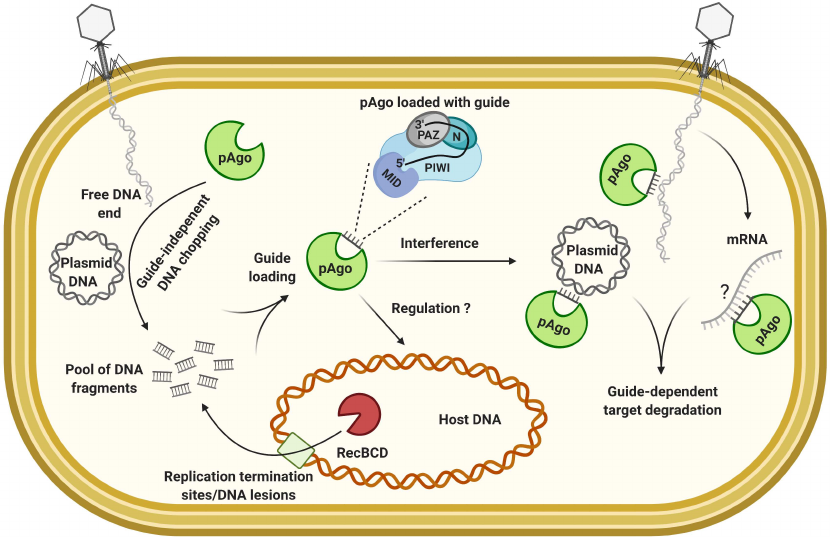
Argonaute proteins play a key role in the regulation of gene expression and anti-viral defense through RNA interference in eukaryotes. Members of this protein family are also widely encountered in Bacteria and Archaea [87, 88]. Functions of prokaryotic Argonautes (pAgo) are not yet fully understood, but these proteins are involved in the silencing of exogenous genetic material [89, 90]. pAgos use guide molecules to recognize nucleic acid targets, but in contrast to the CRISPR-Cas and eukaryotic Agos, they often exploit short single-stranded DNA [91], although RNA guides have also been described [92]. The 5′-end of the guide is loaded into the MID domain of pAgo, while the 3′-end interacts with the PAZ domain [93]. In vitro experiments with pAgos from different organisms demonstrated that recognition of the complementary target leads to its nucleolytic cleavage by the catalytic PIWI domain. pAgos mostly target DNA molecules [94, 95], while some may also target RNA in vitro [96-98]. However, it is not clear whether the RNA targeting might be important for the in vivo activities of pAgo. Nevertheless, all possible combinations of pAgos-mediated DNA/RNA guide-target interactions potentially exist [89, 93, 99]. In vivo, the presence of pAgo affects plasmids maintenance and inhibits transformation [92, 100]. While it is generally accepted that the pAgos are also involved in antiviral defense the only experimental evidence was recently obtained with the pAgo from Clostridium butyricum, whose heterologous expression in E. coli decreased the titers of the chronic phage M13 and lytic phage P1vir. However, the mechanisms were undefined [90]. Based on the architecture of the domains, pAgos are divided into classes and some proteins, surprisingly, contain the catalytically-inactive PIWI domain [89, 101]. Function of such variants, if any, remain to be determined.
One of the major questions associated with the pAgo interference is the mechanism of generation and the source of guide molecules, as well as the question on how self-targeting is avoided. Sequencing of the DNA guides bound to pAgo in vivo has shown that they are preferentially derived from the actively replicating or multicopy elements, including plasmids and transposons [90, 100]. The guide-independent nuclease activity, termed DNA chopping, was shown for different pAgo proteins [102, 103]. The sequence-independent plasmid chopping may generate a pool of different-size DNA fragments, and some of them may be further loaded as guides to activate more efficient sequence-dependent degradation of the complementary targets [103]. To generate guides, pAgos could target free DNA ends or replication intermediates more often present in exogenous DNA and, akin to CRISPR-Cas, cooperate with RecBCD [90]. The pAgo from Thermus thermophilus was shown to sequester guides from the replication termination regions similar to the pAgo from C. butyricum and, it is supposed to be involved in the host replication control together with the DNA gyrase by resolving catenated chromosomes [104]. It has been suggested that the DNA compaction characteristic for archaeal genomes can contribute to the self/non-self discrimination by the pAgos [102]. The RNA guides found associated with pAgo from Rhodobacter sphaeroides are presumed to be promiscuously incorporated from the degraded transcripts. Yet, the protein retains its specificity to the foreign DNA [92]. A current model of pAgo mechanism of action is presented in Fig. 2.
Fig. 2. Model of pAgo mechanism of action and schematic structure of the protein loaded with a guide.
The described mode of interference may not be very efficient against the quickly acting lytic phages and the pAgo defense may be specialized on controlling of the less harmful mobile elements or associated with other defense systems to enhance protection against viral infection. The fact that pAgo genes are often found coupled with the nuclease or cas genes within the defense islands supports the latter hypothesis [88, 101, 105].
INDUCED CELL DORMANCY OR SUICIDE – ABORTIVE INFECTION (Abi)
AND TOXIN–ANTITOXIN (TA) SYSTEMS
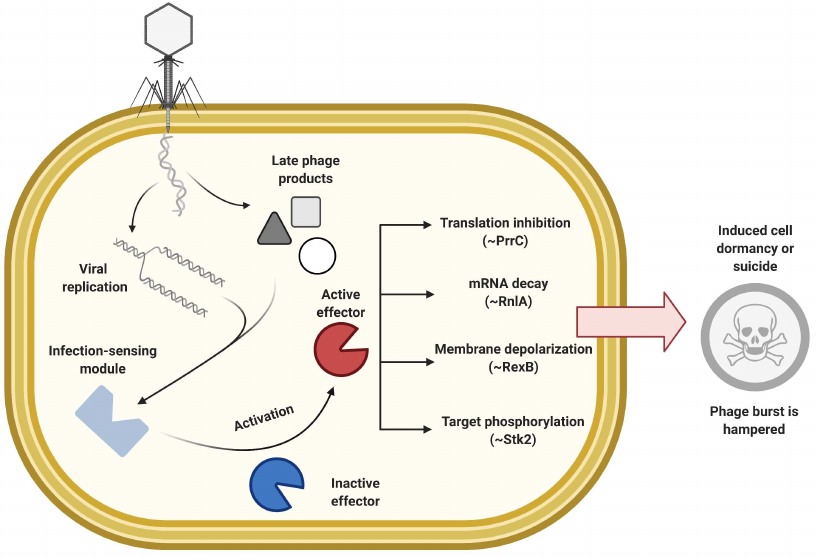
In this section we will consider abortive infection (Abi) in a broad sense – as cellular responses to infection that lead to cessation of the host metabolism (bacteriostatic effect) or cell death (bactericidal effect), prior to the completion of the viral life cycle, thus preventing production of active phage particles or decreasing phage burst size [106, 107]. The Abi systems are mechanistically very diverse. Generally, they are composed of two modules (Fig. 3): one module senses phage infection and transfers the signal, and another – effector module – shuts down host metabolism and/or causes cell suicide after receiving the signal [107, 108]. It is generally accepted that the induced dormancy provides more time for other defense mechanisms to deal with infection. It is also believed that some Abi systems may be the “last resort” of defense, i.e., they activate suicidal response at the later stages of viral infection when other immune mechanisms fail. The strategy of self-elimination by the infected cell stops spread of the infection at the community level and thus benefits a clonal population [109, 110]. Some systems whose action phenotypically resembles the Abi response may directly target phages and not per se cause active cell death. However, their action may be accompanied by the cell lysis caused by the vestigial viral toxic components.
Fig. 3. General principle of abortive infection response, examples of effectors with various mechanisms are shown.
Abi systems. Diversity of the plasmid-encoded systems with Abi mechanism had been historically investigated in the Gram-positive lactococci [106, 111]. Amongst 23 described systems, designated from AbiA to AbiZ, the mode of action was determined only for a few. For example, AbiZ protein cooperates with the phage φ31 holin and lysin causing premature cell lysis [112]; AbiK exhibits untemplated DNA polymerization activity [113]; AbiA, AbiK, and AbiF are thought to inhibit replication [106, 114]; AbiB and AbiQ activities are associated with mRNA decay [115, 116]; AbiD1 might interfere with the viral DNA packaging through inhibition of phage resolving nuclease [117]; and AbiT and AbiV are thought to target expression of the late phage proteins [118, 119]. The way phage infection is sensed is not clear for most of these systems. The Abi system with a sensing module that relies on protein phosphorylation was found in Staphylococci [120]. Phosphorylation, an efficient way to amplify a signal, is often exploited by eukaryotic antiviral systems. Staphylococcal serine/threonine kinase Stk2 is activated by phage φNM1 protein PacK and phosphorylates multiple target host proteins thus inhibiting core metabolism [120].
Plethora of Abi mechanisms have also been described in Gram-negative E. coli [121]. The Lit and PrrC proteins encoded by cryptic prophages are specific against the T4 phage infection. The Lit protease is activated upon interaction with the conserved Gol peptide of the T4 capsid protein and arrests translation through the specific proteolytic cleavage of the translation elongation factor EF-Tu [122]. The PrrC RNAse also inhibits translation by cleaving tRNALys. PrrC interacts with the Type I Restriction–Modification system (R-M) EcoprrI restriction complex and is activated only upon restriction complex inhibition caused by the T4 encoded peptide Stp [123]. Another interesting example includes F-plasmid encoded protein PifA, which provides protection against the phage T7 [124]. This membrane-associated protein is activated by T7 gp10 or gp1.2 and causes leakage of ATP and other small molecules from the infected cell by disrupting membrane integrity [124, 125]. The λ prophage-encoded system RexAB also acts by increasing membrane permeability [126, 127]. RexA is thought to recognize the DNA-protein intermediates of viral replication complexes and stimulate the membrane-associated RexB to form an ion channel, leading to the loss of membrane potential and inhibition of the energy-dependent processes [127].
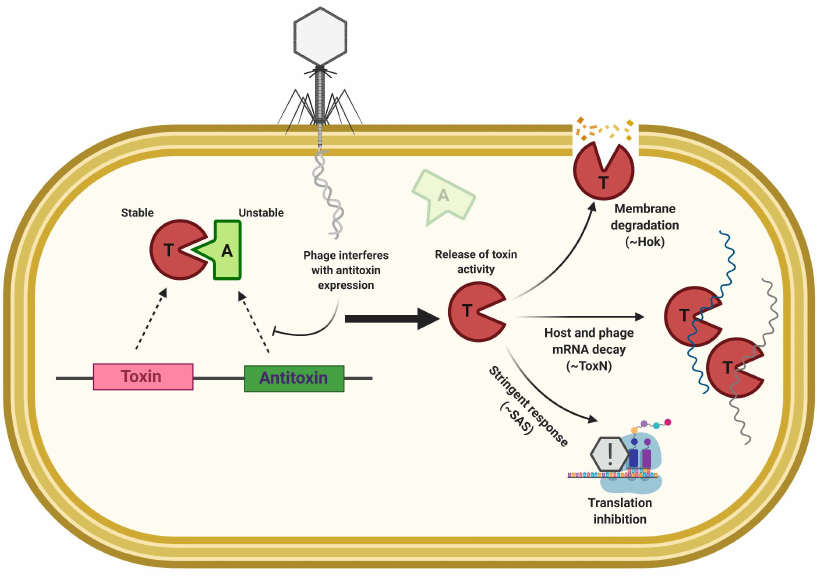
Toxin—Antitoxin (TA) based defense. Toxin–antitoxin systems are selfish elements comprising a stable toxin subunit and an unstable antitoxin. Under stress conditions, increased antitoxin degradation releases activity of the toxin, leading to the growth arrest [128, 129]. TA modules are involved in the stress response, biofilm formation and persistence (although the latter has been controversial [129]) but also may be implicated in the Abi anti-viral defense, since phage infection often interferes with the host metabolism in a way that may cause the loss of antitoxin (Fig. 4). TA modules are frequently found within the defense islands and there is an extensive domain exchange between the TA and Abi systems [130]. In fact, no clear boundary can be drawn between the Abi and TA systems, since Abi is a defense strategy, while TA is an organizational/mechanistic principle. Rather, certain Abi systems can be regarded as based on the TA mechanism, e.g., even some Abi systems discussed in the previous section can be considered as solitary toxins, while PrrC/EcoprrI can be considered as a bona fide toxin–antitoxin pair. Depending on the nature of toxin–antitoxin interaction, the TA systems are divided into 6 types, e.g., antitoxin might represent an RNA molecule that directly inhibits toxin protein (Type III) or regulates the level of toxin mRNA translation (Type I); in other types, antitoxin can be represented by a protein that inhibits toxin through the direct protein-protein interaction (Type II) or by counteracting toxin effect on the targets (Type IV) [129, 131].
Fig. 4. General principle of abortive infection based on the activity of TA modules. Several examples of toxin effectors are shown.
Examples of the Abi response based on TA modules include the ToxIN and RnlAB systems. ToxIN, originally identified as AbiQ in Lactococci, is frequently found in bacterial genomes, and functions as a Type III TA system, where activity of the RNAse toxin ToxN is blocked by interaction with ToxI RNA antitoxin [116, 132, 133]. RnlAB system from E. coli represents Type II TA module and protects against phage T4 infection [134]. The RnlA toxin is a stable RNase; the RnlB antitoxin is quickly degraded by host proteases. Thus, if phage infection interferes with the continuous gene expression, prevention of RnlB synthesis releases toxic activity of RnlA leading to the cellular mRNA decay [134]. RnlAB homologs are found in E. coli plasmids and the system is called LsoAB [135]. The T4 phage-encoded protein Dmd functions as an antitoxin for both of these systems [134, 135]. Many TA systems have a reversible effect and do not induce cell death. Still, temporal cessation of growth can provide phage resistance. AbiE, a Type IV TA system, is an example: AbiEii toxin transcribed from the abiE promoter does not directly interact with its antitoxin AbiEi. Instead, AbiEi binds to the promoter region and inhibits transcription of the entire TA operon [136]. AbiEii toxin belongs to the DNA polymerase β-like superfamily and exhibits nucleotidyltransferase activity [136]. Recently it was demonstrated that AbiEii homologue MenT3 from Mycobacterium tuberculosis can transfer pyrimidines to the acceptor stem of the specific tRNAs [137]. In agreement with this is the fact that overexpression of AbiE toxin in Serratia causes growth cessation and decrease in the tRNAs levels [138]. In E. coli, the Type II system MazEF interferes with phage P1 propagation [139], and the Type I TA system hok/sok reduces the burst size of T4 phage [140]. The latter system is based on the holin-like activity of the Hok toxin, while the antitoxin sok is an antisense RNA that inhibits Hok synthesis by binding to its mRNA [141]. The role of TA systems in phage defense remains controversial and poorly characterized [142, 143]. Based on the abundance of TA systems and their involvement in the Abi response in model bacteria it can be expected that TA-based phage defenses are widespread [130, 142, 144].
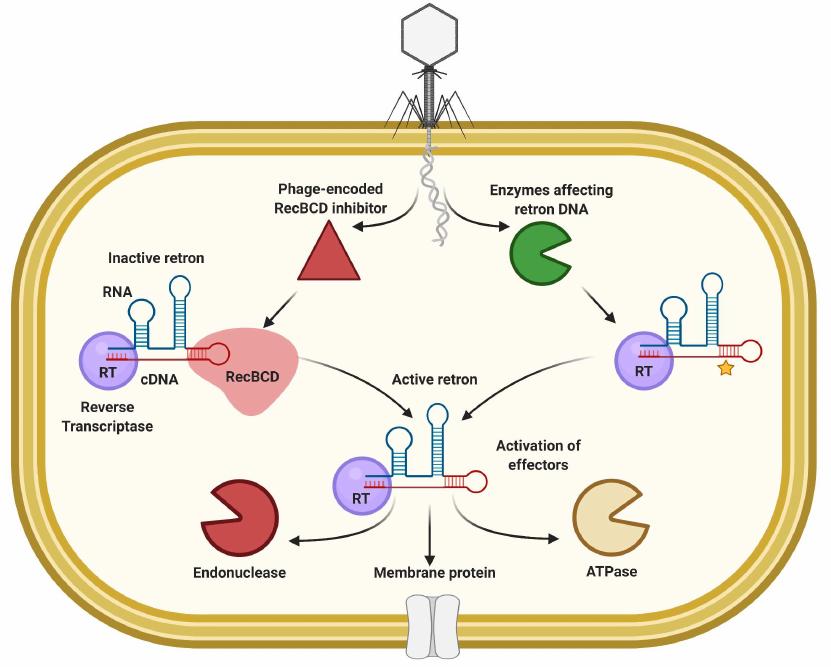
Retrons as immunity systems. Retrons are genetic elements that encode reverse transcriptase (RTase) and non-coding RNA (ncRNA) that is used by RTase to generate covalently linked RNA/DNA hybrids [145]. The functional role of retrons remained unknown until the recent works demonstrated that they are part of the tripartite TA systems that may be involved in phage defense through Abi [146-149]. The RTase complex with RNA/DNA hybrid is inactive under normal conditions, while phage infection causes its activation and signal transduction to the cognate toxin effectors (Fig. 5). Anti-phage activity was demonstrated for multiple retrons and mutations that affect the ncRNA secondary structure and branching site or RTase catalytic motif eliminated the defense [146, 149]. About 2000 retron systems were found within the defense islands with RTases that could be fused to or accompanied by ATPases, ribosyltransferases, and endonucleases as effector proteins [146, 149]. The Ec48 retron from E. coli “guards” the RecBCD enzyme, which is one of the main barriers for the foreign DNA uptake, and RecB inhibition by the phage-encoded proteins (e.g., Gam from phage λ or gp 5.9 of T7) activates the retron and releases activity of the cognate membrane-anchored effector that cause premature cell lysis [146]. It was shown for the Sen2 retron from S. enterica that perturbations of the DNA part of the complex activate the RcaT toxin [147, 148]. DNA degradation or methylation associated with the phage-encoded RecE endonuclease or Dam-methyltransferase activates the response, while some prophage-encoded proteins can serve as blockers of the retron activation [147].
Fig. 5. General principle of abortive infection based on retron elements.
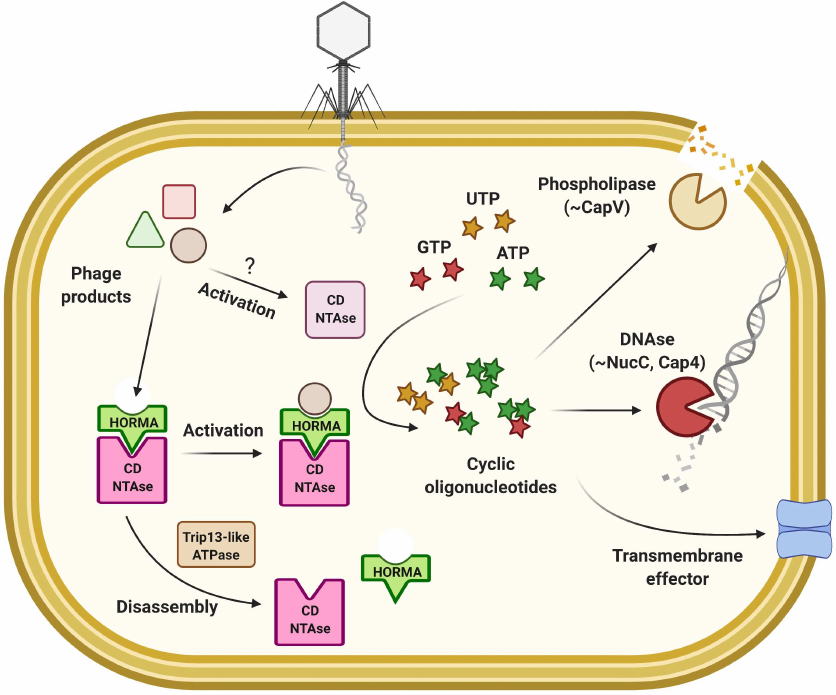
Cyclic-Oligonucleotide-Based Anti-Phage Signaling Systems (CBASS). A widely encountered family of systems that rely on the synthesis of cyclic oligonucleotides to activate the Abi response has been recently described [107, 150, 151]. In the presence of CBASS phage infection triggers synthesis of a secondary messenger (cyclic GMP-AMP, cyclic triadenylate, etc.) by the cGAS/DncV-like nucleotidyltransferases (CD-NTases), which activates various effectors that induce programmed cell death (Fig. 6) [150-153]. These systems provide another link between the immunity defense of eukaryotes and prokaryotes since cyclic GMP-AMP synthase (cGAS) in animals is involved in anti-viral and inflammatory response through the cGAS-STING pathway activated by sensing of cytosolic DNA [154]. Oligonucleotide cyclase genes are found in about 10% of prokaryotic genomes, and more than half reside within the defense islands. Diversity of CBASS systems can be classified based on the composition of operon, type of the effector or produced signalling molecule [153]. The Type I systems contain only CD-NTase (oligonucleotide cyclase) and effector, while other systems also carry auxiliary components: genes with ubiquitin-associated domains in Type II, HORMA and Trip13-like domain genes in Type III, or nucleotide modification domains in rarely encountered Type IV CBASS [153]. The Type II CBASS from V. cholerae was the first to be studied experimentally [151, 155, 156]. Core of the system is composed of two components: cGAS enzyme DncV and cyclic GMP-AMP (cGAMP) sensing phospholipase CapV. These two components are sufficient to provide defense against the P1 phage, yet, protection against other phages required two additional proteins carrying E1, E2, and JAB domains, typical of ubiquitinating enzymes [151]. The sensing mechanism is not yet determined, but the cells were shown to produce cGAMP upon phage infection, which triggered CapV phospholipase-mediated disruption of the cell membrane before completion of the viral life cycle. In addition to phospholipase, the known effectors of CBASS systems include endonucleases or transmembrane domain carrying proteins [153, 157]. An interesting group of CBASS effectors contain domain homologous to the eukaryotic STING (Stimulator of Interferon Genes). In Bacteria, sensing of cyclic oligonucleotides by STING activates the coupled Toll/interleukin-1 receptor (TIR) domain of the effector, which leads to NAD+ degradation [158]. Comparative analysis of the metazoan and bacterial STING domain structures reveals its transition from the direct effector role in CBASS to the regulatory functions in the immune response of higher animals [158].
Fig. 6. Model of the CBASS-mediated immune response.
Many bacterial CD-NTases were shown to be inactive in vitro [152] and recent work demonstrated importance of the auxiliary proteins for their in vivo activity [159]. The HORMA-domain proteins in eukaryotes bind to the specific closure motifs in the target proteins to assemble into the signaling complexes [160]. In the Type III CBASS of E. coli and P. aeruginosa, HORMA proteins activate CD-NTase leading to cyclic triadenylate (cAAA) second messenger production, which in turn activates promiscuous endonuclease activity of the NucC effector providing defense against phage infection via the Abi mechanism [159, 161]. Without infection, activity of the system is suppressed by the Trip13-like ATPase that is thought to disassemble the CD–NTase complex with HORMA. Recognition of the specific motifs in phage proteins is believed to change HORMA conformation and activate the CD–NTase activity of the complex. Intriguingly, the NucC effector can be found as an accessory endonuclease in the Type III CRISPR/Cas systems that also rely on the cyclic oligoadenylate signaling [21, 22, 161]. Another CBASS effector that can respond to different types of cyclic oligonucleotides is the Cap4 protein from Enterobacter cloacae [162], a founding member of an entire protein family. Cap4 proteins recognize secondary messengers through the divergent SAVED domains composed of 2 CRISPR-related CARF subunits, which induces oligomerization and release of the DUF4297 endonuclease activity [162].
Wide variety of bacterial CD-NTases and coupled effectors has been described in recent years, yet, many questions remain unanswered. How is phage infection sensed by these systems? What are the functions of auxiliary proteins? Are all these systems involved only in phage defense or can some of them perform other functions? What are the costs of the CBASS genes and are there additional mechanisms to restrain their possible self-toxicity, like in the case with the Trip13-like ATPase negative regulation?
PROPHAGE-MEDIATED DEFENSE
Temperate phages can integrate their genomes into the host chromosome to form prophages [163]. Most known bacteria carry prophages, and interaction of the prophage with the host can be considered as mutualistic: since prophage survival is dependent on the host, it is beneficial for the prophage to exclude secondary infection of lysogenized cell [164]. Indeed, prophages often can carry genes associated with antiviral defense (Fig. 7a) [165-168]. The simplest way by which infection with homoimmune phages can be inhibited is expression of a repressor protein – transcription factor that regulates the switch between lytic and lysogenic life strategies of the phage. Since the repressor protein is constantly present in the lysogenic cell to inhibit expression of lytic genes of the prophage, secondary lytic infection with a phage regulated by the same or similar repressor will be inhibited [169]. Although the repressor-associated defense has a narrow specificity range systematic studies of prophages in P. aeruginosa and M. smegmatis uncovered abundant defense genes that provide heterotypic protection [165, 166].
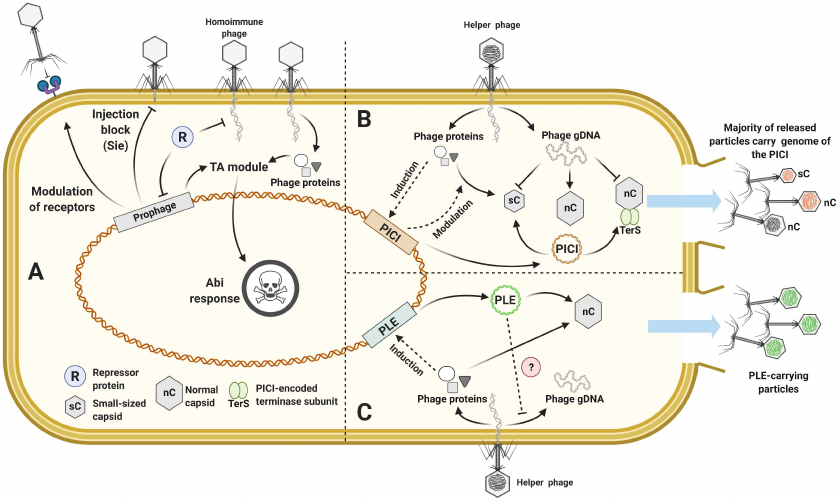
Fig. 7. Defense associated with prophages and mobile genetic elements. a) Prophage encoded defense systems; b) model of the PICI induction and interference; c) model of the PLE induction and interference.
Genes that are dispensable for the prophage survival but can increase fitness of the host were named “morons” (from adding “more on” to the phenotype). Morons can affect multiple host processes including motility, antibiotic resistance, metabolism, and phage defense [170, 171]. Some prophage-encoded TA systems and the Sie mechanism that can be considered as morons were already discussed. Morons can also affect cell surface to prevent receptors recognition. For example, P. aeruginosa phages D3 and φ297 can alter conformation of the O-antigen subunits in lipopolysaccharide (LPS) by encoding their own O-antigen polymerase [172-174], while some Shigella and E. coli prophages may block growth of the O-antigen chain by glucosylation or acetylation [175, 176].
Prophages of mycobacteria encode different superinfection exclusion systems. For example, phages Sbash and CarolAnn carry putative TA modules with membrane-associated effectors [177, 178]. Other mycobacteriophages were shown to encode restriction endonucleases, Sie membrane proteins, or Guanosine pentaphosphate [(p)ppGpp] synthases capable of defending the host [166]. Initially found as gp29 and gp30 proteins of the phage Phrann, the system encoding (p)ppGpp synthase and its cognate inhibitor is part of a novel family of TA modules relying on alarmone signaling found in multiple prophages [166, 179]. The toxic component is an enzyme (SAS, Small Alarmone Synthetase) that synthesize ppGpp or ppApp – signaling molecules typical for stress responses associated with starvation (stringent response) that cause growth cessation [180]. The antitoxins directly bind to the synthase or degrade the signaling alarmone [179]. Another Abi system widespread in the prophages of Gram-negative bacteria consists of the BstA effector that co-localizes with the replicating DNA of target phages and interferes with the replication process by an unknown mechanism [181]. This system provides an interesting example of self-immunity avoidance in prophages: BstA is inactivated by its binding to a specific anti-BstA locus (aba) within the prophage genome and thus does not prevent phage’ own lytic infection.
“Morons” can be very different even among the closely related strains and a recent study of P2- and P4-like prophages uncovered an unprecedented variety of the compact defense systems in the specific hotspot loci of their genomes [182]. In addition to the known antiviral systems, multiple gene clusters that carry predicted defense domains (~TIR, SIR2, Nuclease, ATPase) or domains of unknown function were reported in these hotspots. Defense activity has been validated for 14 novel systems. For example, PARIS system that consists of ATPase and DUF4435 protein is suggested to provide abortive infection response triggered by the phage T7 anti-restriction protein Ocr that inhibits host R-M and BREX systems [182-184]. Mining of the diversity hotspots in other prophages might represent a valuable and simple tool for detection of novel phage defense systems.
PHAGE’S PARASITES – MOLECULAR PIRACY OF THE PICIs AND
PLEs
MGE can be randomly transferred by phages during the process of generalized transduction. However, some MGEs evolved to hijack phage DNA packaging machinery and capsids to load their own genomes, a phenomenon sometimes called molecular piracy [185-188]. Phage-Inducible Chromosomal Islands (PICI) are widespread in the genomes of Gram-positive and Gram-negative bacteria. Their induction from the chromosome is dependent on the infection with the helper phages and from the host perspective they can be considered as a version of the Abi response [189, 190]. The infected cell is lysed eventually but the burst size of the superinfecting lytic phage progeny is suppressed and majority of the released particles carry the PICI genome instead of the phage genome [191]. Since the PICI elements do not undergo a lytic cycle on their own and limit the spread of helper phages, their presence can be beneficial for bacterial population.
PICI. The most studied group of PICIs was discovered in Staphylococci and is named SaPI (S. aureus pathogenicity islands). These chromosomal islands are less than 15 kbp in size; they encode an integrase, an excision protein, and replication machinery. Expression of these genes is strictly controlled by the master repressor Stl. SaPIs often carry accessory genes, including toxins and virulence factors, and their presence could contribute to the host pathogenicity [185, 192]. The life cycle of SaPI is coupled with the infection by a helper phage and sensing of the specific helper phage proteins relieves the Stl-mediated repression [193, 194]. Following activation, SaPIs could interfere with the helper phage reproduction via several different mechanisms (Fig. 7b). They facilitate assembly of the small capsids capable of accommodating the SaPI genome but excluding the larger genome of the helper phage [195]. SaPIs can also interfere with the viral genome packaging by inhibiting the small terminase subunit TerS, while loading of the SaPI own genome is supported by the SaPI-encoded terminase [195]. Finally, SaPI-encoded proteins can bind viral transcription factors impairing expression of the late genes [196]. While these mechanisms have been described for the staphylococcal SaPIs, PICIs are widely distributed in bacteria [190, 197] and may rely on the similar mechanisms for their parasitic lifestyles. For example, PmCI172 from Pasteurella multiocida directs production of the small-size capsids when the cell is infected by the Mu-like helper phage, while EcCICFT073 from E. coli encodes Rpp, a protein reprogramming phage λ TerS to package the genome of the PICI [190, 198].
PLE. Another type of the satellite MGEs specific to V. cholera and interfering with the phage ICP1 propagation is called PLE – Phage-inducible chromosomal Island-like Element [199]. Similar to PICIs, PLE senses helper phage ICP1 proteins to trigger their genome excision process and exploits molecular machinery and structural components of ICP1. Yet genome organization of the PLE is different and in contrast to PICIs, which only lower the phage burst size, no ICP1 infectious particles are produced when PLE is induced (Fig. 7c) [199-202]. ICP1 replication was shown to be severely inhibited in the PLE-carrying cells, while some PLEs can also modulate viral gene expression [202, 203]. An additional mechanism that may contribute to the ICP1 suppression is production of the PLE-encoded LidI, a protein that disrupts the lysis interference system of ICP1 and accelerates cell lysis [204]. It must be noted that ICP1 in its turn encodes a CRISPR-Cas system to target PLE [205].
SYSTEMS WITH PLAUSIBLE NOVEL MECHANISMS
Mining of the conserved gene clusters found within the defense islands of available prokaryotic genomes allowed predicting multiple novel types of the defense systems [149, 206-208]. Systematic verification of these predictions had been recently carried out. The pipeline included cloning of 28 candidate systems from several source organisms to Gram-negative E. coli or Gram-positive B. subtilis surrogate hosts followed by screening with a set of diverse phages. This work validated 10 novel systems that were named after mythological protective deities [207]. Druantia, Kiwa, and Zorya were validated in E. coli, while activities of Gabija, Hachiman, Lamassu, Thoeris, Septu, Shedu, and Wadjet were demonstrated in B. subtilis. Multiple protein domains distinct from that, known for already studied defense systems were shown to be involved in antiviral protection, which suggests novel mechanisms of action of the discovered systems.
The Zorya system is active against both ssDNA and dsDNA phages. This system encodes ZorA and ZorB proteins, which are homologous to the proton channel-forming unit MotAB of the bacterial flagellar motor [209]. In the type I Zorya systems, ZorAB could be accompanied by the predicted small nuclease ZorE, while in the Type II system – by the helicase/ATPase and Pfam00691-containing proteins ZorCD. Phage infection of the cells carrying Zorya initiates premature cell lysis likely due to the Abi response mediated by the ZorAB effector causing membrane depolarization. This point of view is further supported by the fact that mutations of amino acid residues predicted to be involved in the proton transport reduce the degree of defense provided by the Zorya system [207].
Another studied system – Thoeris – is composed of the ThsA protein with SIR2 and SLOG domains and Toll/interleukin-1 (TIR) domain protein ThsB [207, 210]. The structure of both proteins has been recently resolved [211]. Mutations in the NAD-binding pocket of the ThsA SIR2 domain results in the loss of in vitro activity of the protein and phage resistance in vivo, linking the Thoeris-mediated defense to NAD+ hydrolysis [207, 211]. The TIR domain could serve as a signal transducer in the eukaryotic immune pathways [212] and was also found in other prokaryotic systems (e.g., CBASS). In Thoeris system phage infection could trigger the TIR domain of ThsA to synthesize an isomer of cyclic ADP-ribose that transfers the signal to ThsB, causing abortive infection response associated with NAD+ depletion [210]. Multiple copies of thsB gene could be associated with the single thsA and it was shown that diversification of TIR domains provides defense against a broader range of infecting phages [210].
The Wadjet system was shown to provide defense not against phages but against plasmid transformation [207]. Wadjet is composed of four components: JetABC homologous to the SMC proteins (structural maintenance of the chromosome) involved in the plasmid and host genome segregation, and JetD protein with a putative topoisomerase VI domain [207, 213]. The phenomenon of plasmid maintenance suppression associated with non-canonical SMC proteins has been reported in M. smegmatis, where modulation of the plasmid supercoiling status impaired its segregation to the daughter cells [214]. It could be suggested that Wadjet system functions in a similar manner to exclude foreign extrachromosomal genetic elements.
Another recent study employed a similar strategy for experimental validation of the candidate systems but relied on the different prediction algorithm. Instead of assessing protein domains enrichment, all sequences in the 10-gene neighborhood of the known defense systems were analyzed for the probability to be localized within defense islands. More than 7000 candidate defense genes were predicted, many of which harbored unannotated domains or domains of unknown function. Following manual curation, 48 predicted systems were selected for further work. Heterologous expression in E. coli validated antiviral activities for 29 novel systems [149].
Association of retrons with phage defense systems was established independently. Moreover, reverse transcriptases (RTase) not associated with retron functions were also shown to be involved in the protection. Six groups of RTases were combined under the term DRT (defense-associated RTases). Some DRTs were shown to be associated with the auxiliary proteins, whereas DRTs from the Type I were fused to the nitrilase domain often involved in the small-molecule metabolism [215]. Mutations of the predicted catalytic residues in RTase or nitrilase inactivated the system. Analysis of the transcriptome identified that the DRTs of different types do not interfere with the early viral genes expression, while the Type I system affected accumulation of the late viral transcripts. The mechanism of DRTs protection remains undetermined.
The RADAR (restriction by adenosine deaminase acting on RNA) system was studied in more detail. The core of the system is composed of ATPse RdrA and adenosine deaminase RdrB, that could be accompanied with the ancillary proteins (SLATT, or Csx27 that is also involved in the Type VI CRISPR defense). Analysis of the transcriptome of the phage-infected cells identified A to G substitutions in the sequenced reads, consistent with the predicted RdrB-related formation of inosine in the host and viral RNAs. The RNA stem-loop secondary structures were shown to be preferential editing targets. Infection of the RADAR+ culture with phages at high multiplicity of infection (MOI) results in growth arrest, suggesting defense response through Abi mechanism. This behavior was attributed to the editing of the host transfer-messenger RNA (tmRNA) that rescues stalled ribosomes. It was further demonstrated that expression of the certain phage T2 DNA-binding proteins triggers RNA editing in the uninfected culture, which allowed suggesting a mechanism for the RADAR response activation [149].
NTPases from the STAND superfamily are involved in the programmed cell death signal transduction pathways in eukaryotes [216], but role of these proteins in prokaryotes have been long undetermined. Here, several STAND NTPases were shown to be active in protection against phage infection, and 5 types of such systems were combined under the name AVAST (AntiViral ATPases/NTPases from the STAND superfamily). The NTPase domains were found fused to the different putative effector domains (like nuclease previously known as DUF4297, protease, or SIR2) and were suggested to act through the Abi mechanism. Mutational analysis verified importance of ATPse and putative effectors for the AVAST-mediated defense [149].
Functional domains of other novel systems include nucleases (e.g., DUF4297 from the Lamassu system was shown to be a nuclease involved in CBASS), helicases, SIR2, DNA-binding proteins, phosphatases, and ATPases, as well as multiple unannotated domains [149, 207, 208, 217]. For example, the large Druantia system is composed primarily of the genes with unknown functions. These discoveries significantly expanded our understanding of the diversity of biochemical activities that may be involved in antiviral protection, and should pave the way for further experimental investigations of novel defense mechanisms.
CONCLUSIONS
Interaction of viruses with their hosts is a highly dynamic process that drives generation of multiple offence and defense strategies. Prokaryotic genes associated with phage resistance are amongst the most quickly evolving, and high turnover rate of prokaryotic adaptations followed by viral counter-adaptations is often described in terms of the Red Queen hypothesis, e.g., both sides of this arms race need to constantly evolve just to keep the “status quo” [218, 219]. Long-term evolutionary studies show that mutation accumulation rate is higher when the phage co-evolve with the host, compared to the situation when phage evolves while the host maintains its genotype [220, 221]. One of the examples that allows estimating dynamics of these interactions is confrontation of the V. cholerae PLEs with the phage ICP1. Using V. cholerae stool samples collected since 1940’s the history of this competition can be tracked: five different types of PLE gradually replaced each other, likely by escaping interference from the ICP1-encoded CRISPR-Cas system [199, 205].
Only in recent years we began to appreciate the real abundance of the defense systems and diversity of their mechanisms. This immediately raises the question on how the presence of multiple defense systems in a single bacterium affects its survival and phage resistance? Some defense systems were shown to be highly specific, while other can target multiple phages, like some Abi systems that sense general perturbations in the host metabolism, or DNA-recognition systems that can adapt or evolve to interact with the new sequences [3, 222]. Defense systems target different stages of the viral life cycle, and, in general, three lines of defense can be indicated: systems that affect cell surface to prevent phage adsorption and genome entry; systems that degrade phage genetic material; systems that induce cell dormancy or suicide if the first two lines of defense were circumvented. It can be suggested that the simultaneous presence of different defense systems in one cell increases the chances of survival and broadens the range of the targeted parasites. Some defense systems are even known to act in co-operation, for example, DNA degradation by the Type I R-M systems generates the pool of DNA fragments that can be used as spacer precursors for the CRISPR-Cas system [64, 223, 224], or PrrC and retron Ec48 Abi systems that exploit “guarding” strategy and are activated only if the phage interferes with the function of other defense systems [146]. At the same time, presence of defense systems imposes fitness costs on the host: in the absence of phage infection, expression of the defense genes is energy-costly, while their unrestrained activity is often associated with self-toxicity [110, 219, 225]. Thus, the balance between the benefits of antiviral protection and accompanied fitness costs defines the amount of the active defense systems in the genome, and prevalence of the specific defense. Defense systems can be lost under conditions of low phage pressure, and their persistence in the population is facilitated by the extensive horizontal gene transfer (HGT), downregulation of gene expression, and phase variation [226, 227].
Novel discoveries allowed to highlight additional features of the defense systems that were not so evident before:
Functional modules can be exchanged between the different defense systems, and similar protein domains can be employed in different types of defense systems [208]. For example, Dnd system modification module could be associated with DndFGH or PblABCDE effectors, NucC nuclease could be associated with CBASS or Type III CRISPR-Cas, TIR domains are found in CBASS and Thoeris, etc. [161, 210, 228].
Defense systems are not unique for the prokaryotic genomes, as different types of MGEs often adopt defense systems for inter-MGE conflicts or for the host suppression [205, 226]. For example, recent metagenomic study of the giant phages uncovered occurrence of multiple CRISPR loci with spacers targeting other phages [229].
Proteins typical for the eukaryotic immunity systems were found to perform similar functions in prokaryotes, such as cGAS, pAgo, STING, TIR, STAND [105, 149, 151, 158, 210]. Recent work of Burroughs and Arravind adds more to these findings by tracing homologs of the eukaryotic Wnt, YEATS, TPR-S and other domains in prokaryotic immunity systems [208]. Phylogenetic history of these domains indicates that their common root resides within prokaryotes, which suggests that some immunity mechanisms originated before the eukaryote branching, and were inherited by the latter.
Acknowledgments. The authors would like to thank Andrey Kulbachinskiy for critical comments to the pAgo section. Illustrations were prepared using BioRender.com under the paid subscription.
Funding. The work was financially supported by the Russian Foundation for Basic Research (project no. 19-14-50560). AI was financially supported by the Russian Foundation for Basic Research (grant no. 19-34-90160), OM was financially supported by the Russian Science Foundation (grant no. 19-74-00118). Funding for open access charge: Skolkovo Institute of Science and Technology.
Authors’ contributions. OM&AI prepared CRISPR-Cas section, AI composed the rest of the manuscript, AI&OM prepared illustrations, KS revised the text.
Ethics declarations. The authors declare no conflict of interest in financial or any other sphere. This article does not contain any studies with human participants or animals performed by any of the authors.
Open access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
REFERENCES
1.Jansen, R., Embden, J. D. A., van Gaastra, W., and
Schouls, L. M. (2002) Identification of genes that are associated with
DNA repeats in prokaryotes, Mol. Microbiol., 43,
1565-1575.
2.Mojica, F. J. M., and Garrett, R. A. (2013)
Discovery and seminal developments in the CRISPR field, In
CRISPR-Cas Systems, Springer, pp. 1-31.
3.Nussenzweig, P. M., and Marraffini, L. A. (2020)
Molecular mechanisms of CRISPR-Cas immunity in bacteria, Annu. Rev.
Genet., 54, 93-120.
4.Makarova, K. S., Wolf, Y. I., Iranzo, J., Shmakov,
S. A., Alkhnbashi, O. S., et al. (2020) Evolutionary classification of
CRISPR-Cas systems: a burst of class 2 and derived variants, Nat.
Rev. Microbiol., 18, 67-83.
5.Jackson, R. N., and Wiedenheft, B. (2015) A
conserved structural chassis for mounting versatile CRISPR RNA-guided
immune responses, Mol. Cell, 58, 722-728.
6.Liu, T. Y., and Doudna, J. A. (2020) Chemistry of
Class 1 CRISPR-Cas effectors: binding, editing, and regulation, J.
Biol. Chem., 295, 14473-14487.
7.Reeks, J., Graham, S., Anderson, L., Liu, H.,
White, M. F., and Naismith, J. H. (2013) Structure of the archaeal
Cascade subunit Csa5: relating the small subunits of CRISPR effector
complexes, RNA Biol., 10, 762-769.
8.Jore, M. M., Lundgren, M., van Duijn, E., Bultema,
J. B., Westra, E. R., et al. (2011) Structural basis for CRISPR
RNA-guided DNA recognition by Cascade, Nat. Struct. Mol. Biol.,
18, 529-536.
9.Wiedenheft, B., Lander, G. C., Zhou, K., Jore, M.
M., Brouns, S.J. J., et al. (2011) Structures of the RNA-guided
surveillance complex from a bacterial immune system, Nature,
477, 486-489.
10.Sinkunas, T., Gasiunas, G., Waghmare, S. P.,
Dickman, M. J., Barrangou, R., et al. (2013) In vitro
reconstitution of Cascade‐mediated CRISPR immunity in
Streptococcus thermophilus, EMBO J., 32,
385-394.
11.Benda, C., Ebert, J., Scheltema, R. A., Schiller,
H. B., Baumgärtner, M., et al. (2014) Structural model of a CRISPR
RNA-silencing complex reveals the RNA-target cleavage activity in Cmr4,
Mol. Cell, 56, 43-54.
12.Rouillon, C., Zhou, M., Zhang, J., Politis, A.,
Beilsten-Edmands, V., et al. (2013) Structure of the CRISPR
interference complex CSM reveals key similarities with cascade, Mol.
Cell, 52, 124-134.
13.Hale, C. R., Zhao, P., Olson, S., Duff, M. O.,
Graveley, B. R., Wells, L., et al. (2009) RNA-guided RNA cleavage by a
CRISPR RNA–Cas protein complex, Cell, 139,
945-956.
14.Zhang, J., Kasciukovic, T., and White, M. F.
(2012) The CRISPR associated protein Cas4 Is a 5¢ to 3¢ DNA
exonuclease with an iron-sulfur cluster, PLoS One, 7,
e47232.
15.Elmore, J. R., Sheppard, N. F., Ramia, N.,
Deighan, T., Li, H., et al. (2016) Bipartite recognition of target RNAs
activates DNA cleavage by the Type III-B CRISPR-Cas system, Genes
Dev., 30, 447-459.
16.Estrella, M. A., Kuo, F.-T., and Bailey, S.
(2016) RNA-activated DNA cleavage by the type III-B CRISPR-Cas effector
complex, Genes Dev., 30, 460-470.
17.Kazlauskiene, M., Tamulaitis, G., Kostiuk, G.,
Venclovas, Č., and Siksnys, V. (2016) Spatiotemporal control of
type III-A CRISPR-Cas immunity: coupling DNA degradation with the
target RNA recognition, Mol. Cell, 62, 295-306.
18.Samai, P., Pyenson, N., Jiang, W., Goldberg, G.
W., Hatoum-Aslan, A., and Marraffini, L. A. (2015) Co-transcriptional
DNA and RNA cleavage during type III CRISPR-Cas immunity, Cell,
161, 1164-1174.
19.Liu, T., Pan, S., Li, Y., Peng, N., and She, Q.
(2017) Type III CRISPR-Cas system: introduction and its application for
genetic manipulations, Curr. Issues Mol. Biol., 26,
1-14.
20.Goldberg, G. W., Jiang, W., Bikard, D., and
Marraffini, L. A. (2014) Conditional tolerance of temperate phages via
transcription-dependent CRISPR-Cas targeting, Nature,
514, 633-637.
21.Kazlauskiene, M., Kostiuk, G., Venclovas,
Č., Tamulaitis, G., and Siksnys, V. (2017) A cyclic
oligonucleotide signaling pathway in type III CRISPR-Cas systems,
Science, 357, 605-609.
22.Niewoehner, O., Garcia-Doval, C., Rostøl,
J. T., Berk, C., Schwede, F., et al. (2017) Type III CRISPR-Cas systems
produce cyclic oligoadenylate second messengers, Nature,
548, 543-548.
23.You, L., Ma, J., Wang, J., Artamonova, D., Wang,
M., et al. (2019) Structure studies of the CRISPR-Csm complex reveal
mechanism of co-transcriptional interference, Cell, 176,
239-253.
24.Koonin, E. V, and Makarova, K. S. (2017) Mobile
genetic elements and evolution of CRISPR-Cas systems: all the way there
and back, Genome Biol. Evol., 9, 2812-2825.
25.Özcan, A., Pausch, P., Linden, A., Wulf, A.,
Schühle, K., et al. (2019) Type IV CRISPR RNA processing and
effector complex formation in Aromatoleum aromaticum, Nat.
Microbiol., 4, 89-96.
26.Makarova, K. S., Wolf, Y. I., Alkhnbashi, O. S.,
Costa, F., Shah, S. A., et al. (2015) An updated evolutionary
classification of CRISPR-Cas systems, Nat. Rev. Microbiol.,
13, 722-736.
27.Koonin, E. V, and Makarova, K. S. (2019) Origins
and evolution of CRISPR-Cas systems, Philos. Trans. R. Soc. B,
374, 20180087.
28.Pinilla-Redondo, R., Mayo-Muñoz, D.,
Russel, J., Garrett, R. A., Randau, L., et al. (2020) Type IV
CRISPR-Cas systems are highly diverse and involved in competition
between plasmids, Nucleic Acids Res., 48, 2000-2012.
29.Gasiunas, G., Barrangou, R., Horvath, P., and
Siksnys, V. (2012) Cas9-crRNA ribonucleoprotein complex mediates
specific DNA cleavage for adaptive immunity in bacteria, Proc. Natl.
Acad. Sci. USA, 109, E2579-E2586.
30.Jinek, M., Chylinski, K., Fonfara, I., Hauer, M.,
Doudna, J. A., and Charpentier, E. (2012) A programmable
dual-RNA-guided DNA endonuclease in adaptive bacterial immunity,
Science, 337, 816-821.
31.Hsu, P. D., Lander, E. S., and Zhang, F. (2014)
Development and applications of CRISPR-Cas9 for genome engineering,
Cell, 157, 1262-1278.
32.Mali, P., Esvelt, K. M., and Church, G. M. (2013)
Cas9 as a versatile tool for engineering biology, Nat. Methods,
10, 957-963.
33.Sander, J. D., and Joung, J. K. (2014) CRISPR-Cas
systems for editing, regulating and targeting genomes, Nat.
Biotechnol., 32, 347-355.
34.Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O.,
Slaymaker, I. M., Makarova, K. S., et al. (2015) Cpf1 is a single
RNA-guided endonuclease of a class 2 CRISPR-Cas system, Cell,
163, 759-771.
35.Yamano, T., Zetsche, B., Ishitani, R., Zhang, F.,
Nishimasu, H., and Nureki, O. (2017) Structural basis for the canonical
and non-canonical PAM recognition by CRISPR-Cpf1, Mol. Cell,
67, 633-645.
36.Chen, J. S., Ma, E., Harrington, L. B., Da Costa,
M., Tian, X., et al. (2018) CRISPR-Cas12a target binding unleashes
indiscriminate single-stranded DNase activity, Science,
360, 436-439.
37.Harrington, L. B., Burstein, D., Chen, J. S.,
Paez-Espino, D., Ma, E., et al. (2018) Programmed DNA destruction by
miniature CRISPR-Cas14 enzymes, Science, 362,
839-842.
38.Chylinski, K., Makarova, K. S., Charpentier, E.,
and Koonin, E. V (2014) Classification and evolution of type II
CRISPR-Cas systems, Nucleic Acids Res., 42,
6091-6105.
39.Shmakov, S., Abudayyeh, O. O., Makarova, K. S.,
Wolf, Y. I., Gootenberg, J. S., et al. (2015) Discovery and functional
characterization of diverse class 2 CRISPR-Cas systems, Mol.
Cell, 60, 385-397.
40.Abudayyeh, O. O., Gootenberg, J. S., Konermann,
S., Joung, J., Slaymaker, I. M., et al. (2016) C2c2 is a
single-component programmable RNA-guided RNA-targeting CRISPR effector,
Science, 353, aaf5573, doi: 10.1126/science.aaf5573.
41.Koonin, E. V, and Krupovic, M. (2015) Evolution
of adaptive immunity from transposable elements combined with innate
immune systems, Nat. Rev. Genet., 16, 184-192.
42.Koonin, E. V, Makarova, K. S., and Zhang, F.
(2017) Diversity, classification and evolution of CRISPR-Cas systems,
Curr. Opin. Microbiol., 37, 67-78.
43.Makarova, K. S., and Koonin, E. V (2013)
Evolution and classification of CRISPR-Cas systems and cas protein
families, in CRISPR-Cas System, Springer, pp. 61-91.
44.Nuñez, J. K., Kranzusch, P. J., Noeske,
J., Wright, A. V., Davies, C. W., and Doudna, J. A. (2014)
Cas1–Cas2 complex formation mediates spacer acquisition during
CRISPR-Cas adaptive immunity, Nat. Struct. Mol. Biol.,
21, 528.
45.Nuñez, J. K., Harrington, L. B.,
Kranzusch, P. J., Engelman, A. N., and Doudna, J. A. (2015) Foreign DNA
capture during CRISPR-Cas adaptive immunity, Nature, 527,
535-538.
46.Brouns, S. J. J., Jore, M. M., Lundgren, M.,
Westra, E. R., Slijkhuis, R. J. H., et al. (2008) Small CRISPR RNAs
guide antiviral defense in prokaryotes, Science, 321,
960-964.
47.Yosef, I., Goren, M. G., and Qimron, U. (2012)
Proteins and DNA elements essential for the CRISPR adaptation process
in Escherichia coli, Nucleic Acids Res.,
40, 5569-5576.
48.Datsenko, K. A., Pougach, K., Tikhonov, A.,
Wanner, B. L., Severinov, K., and Semenova, E. (2012) Molecular memory
of prior infections activates the CRISPR/Cas adaptive bacterial
immunity system, Nat. Commun., 3, 1-7.
49.Vorontsova, D., Datsenko, K. A., Medvedeva, S.,
Bondy-Denomy, J., Savitskaya, E. E., et al. (2015) Foreign DNA
acquisition by the IF CRISPR-Cas system requires all components of the
interference machinery, Nucleic Acids Res., 43,
10848-10860.
50.Deltcheva, E., Chylinski, K., Sharma, C. M.,
Gonzales, K., Chao, Y., et al. (2011) CRISPR RNA maturation by
trans-encoded small RNA and host factor RNase III, Nature,
471, 602-607.
51.Sapranauskas, R., Gasiunas, G., Fremaux, C.,
Barrangou, R., Horvath, P., and Siksnys, V. (2011) The
Streptococcus thermophilus CRISPR/Cas system provides
immunity in Escherichia coli, Nucleic Acids Res.,
39, 9275-9282.
52.Hatoum-Aslan, A., Maniv, I., Samai, P., and
Marraffini, L. A. (2014) Genetic characterization of antiplasmid
immunity through a type III-A CRISPR-Cas system, J. Bacteriol.,
196, 310-317.
53.Babu, M., Beloglazova, N., Flick, R., Graham, C.,
Skarina, T., et al. (2011) A dual function of the CRISPR-Cas system in
bacterial antivirus immunity and DNA repair, Mol. Microbiol.,
79, 484-502.
54.Wiedenheft, B., Zhou, K., Jinek, M., Coyle, S.
M., Ma, W., and Doudna, J. A. (2009) Structural basis for DNase
activity of a conserved protein implicated in CRISPR-mediated genome
defense, Structure, 17, 904-912.
55.Beloglazova, N., Lemak, S., Flick, R., and
Yakunin, A. F. (2015) Analysis of nuclease activity of Cas1 proteins
against complex DNA substrates, in CRISPR, Springer, pp.
251-264.
56.Nam, K. H., Ding, F., Haitjema, C., Huang, Q.,
DeLisa, M. P., and Ke, A. (2012) Double-stranded endonuclease activity
in Bacillus halodurans clustered regularly interspaced short
palindromic repeats (CRISPR)-associated Cas2 protein, J. Biol.
Chem., 287, 35943-35952.
57.Dixit, B., Ghosh, K. K., Fernandes, G., Kumar,
P., Gogoi, P., and Kumar, M. (2016) Dual nuclease activity of a Cas2
protein in CRISPR-Cas subtype I‐B of Leptospira
interrogans, FEBS Lett., 590, 1002-1016.
58.Savitskaya, E. E., Musharova, O. S., and
Severinov, K. V (2016) Diversity of CRISPR-Cas-mediated mechanisms of
adaptive immunity in prokaryotes and their application in
biotechnology, Biochemistry (Moscow), 81, 653-661.
59.Mizuuchi, K., and Adzuma, K. (1991) Inversion of
the phosphate chirality at the target site of Mu DNA strand transfer:
evidence for a one-step transesterification mechanism, Cell,
66, 129-140.
60.Krupovic, M., Makarova, K. S., Forterre, P.,
Prangishvili, D., and Koonin, E. V (2014) Casposons: a new superfamily
of self-synthesizing DNA transposons at the origin of prokaryotic
CRISPR-Cas immunity, BMC Biol., 12, 36.
61.Béguin, P., Charpin, N., Koonin, E. V.,
Forterre, P., and Krupovic, M. (2016) Casposon integration shows strong
target site preference and recapitulates protospacer integration by
CRISPR-Cas systems, Nucleic Acids Res., 44,
10367-10376.
62.Rollie, C., Graham, S., Rouillon, C., and White,
M. F. (2018) Prespacer processing and specific integration in a type IA
CRISPR system, Nucleic Acids Res., 46, 1007-1020.
63.Levy, A., Goren, M. G., Yosef, I., Auster, O.,
Manor, M., et al. (2015) CRISPR adaptation biases explain preference
for acquisition of foreign DNA, Nature, 520, 505-510.
64.Weissman, J. L., Stoltzfus, A., Westra, E. R.,
and Johnson, P. L. F. (2020) Avoidance of Self during CRISPR
Immunization, Trends Microbiol., 28, 543-553, doi:
10.1016/j.tim.2020.02.005.
65.Dorman, C. J., and Bhriain, N. N. (2020)
CRISPR-Cas, DNA supercoiling, and nucleoid-associated proteins,
Trends Microbiol., 28, 19-27.
66.Kurilovich, E., Shiriaeva, A., Metlitskaya, A.,
Morozova, N., Ivancic-Bace, I., et al. (2019) Genome maintenance
proteins modulate autoimmunity mediated primed adaptation by the
Escherichia coli type IE CRISPR-cas system, Genes
(Basel), 10, 872.
67.Radovčić, M., Killelea, T.,
Savitskaya, E., Wettstein, L., Bolt, E. L., and
Ivančić-Baće, I. (2018) CRISPR-Cas adaptation in
Escherichia coli requires RecBCD helicase but not
nuclease activity, is independent of homologous recombination, and is
antagonized by 5′ ssDNA exonucleases, Nucleic Acids Res.,
46, 10173-10183.
68.Ivančić-Baće, I., Cass, S.
D., Wearne, S. J., and Bolt, E. L. (2015) Different genome stability
proteins underpin primed and naive adaptation in E. coli
CRISPR-Cas immunity, Nucleic Acids Res., 43,
10821-10830.
69.Deveau, H., Barrangou, R., Garneau, J. E.,
Labonté, J., Fremaux, C., et al. (2008) Phage response to
CRISPR-encoded resistance in Streptococcus thermophilus, J.
Bacteriol., 190, 1390-1400.
70.Semenova, E., Jore, M. M., Datsenko, K. A.,
Semenova, A., Westra, E. R., et al. (2011) Interference by clustered
regularly interspaced short palindromic repeat (CRISPR) RNA is governed
by a seed sequence, Proc. Natl. Acad. Sci. USA, 108,
10098-10103.
71.Fineran, P. C., Gerritzen, M. J. H.,
Suárez-Diez, M., Künne, T., Boekhorst, J., et al. (2014)
Degenerate target sites mediate rapid primed CRISPR adaptation,
Proc. Natl. Acad. Sci. USA, 111, E1629-E1638.
72.Westra, E. R., Semenova, E., Datsenko, K. A.,
Jackson, R. N., Wiedenheft, B., et al. (2013) Type IE CRISPR-cas
systems discriminate target from non-target DNA through base
pairing-independent PAM recognition, PLoS Genet., 9,
e1003742.
73.Richter, C., Dy, R. L., McKenzie, R. E., Watson,
B. N. J., Taylor, C., et al. (2014) Priming in the Type IF CRISPR-Cas
system triggers strand-independent spacer acquisition, bi-directionally
from the primed protospacer, Nucleic Acids Res., 42,
8516-8526.
74.Garrett, S., Shiimori, M., Watts, E. A., Clark,
L., Graveley, B. R., and Terns, M. P. (2020) Primed CRISPR DNA uptake
in Pyrococcus furiosus, Nucleic Acids Res.,
48, 6120-6135.
75.Li, M., Wang, R., and Xiang, H. (2014)
Haloarcula hispanica CRISPR authenticates PAM of a target
sequence to prime discriminative adaptation, Nucleic Acids Res.,
42, 7226-7235.
76.Rao, C., Chin, D., and Ensminger, A. W. (2017)
Priming in a permissive type IC CRISPR-Cas system reveals distinct
dynamics of spacer acquisition and loss, RNA, 23,
1525-1538.
77.Almendros, C., Nobrega, F. L., McKenzie, R. E.,
and Brouns, S. J. J. (2019) Cas4–Cas1 fusions drive efficient PAM
selection and control CRISPR adaptation, Nucleic Acids Res.,
47, 5223-5230.
78.Nussenzweig, P. M., McGinn, J., and Marraffini,
L. A. (2019) Cas9 cleavage of viral genomes primes the acquisition of
new immunological memories, Cell Host Microbe, 26,
515-526.
79.Savitskaya, E., Semenova, E., Dedkov, V.,
Metlitskaya, A., and Severinov, K. (2013) High-throughput analysis of
type IE CRISPR/Cas spacer acquisition in E. coli, RNA
Biol., 10, 716-725.
80.Xue, C., Seetharam, A. S., Musharova, O.,
Severinov, K., Brouns, S. J., et al. (2015) CRISPR interference and
priming varies with individual spacer sequences, Nucleic Acids
Res., 43, 10831-10847.
81.Künne, T., Kieper, S. N., Bannenberg, J. W.,
Vogel, A. I. M., Miellet, W. R., et al. (2016) Cas3-derived target DNA
degradation fragments fuel primed CRISPR adaptation, Mol. Cell,
63, 852-864.
82.Dillard, K. E., Brown, M. W., Johnson, N. V.,
Xiao, Y., Dolan, A., et al. (2018) Assembly and translocation of a
CRISPR-Cas primed acquisition complex, Cell, 175,
934-946.
83.Redding, S., Sternberg, S. H., Marshall, M.,
Gibb, B., Bhat, P., et al. (2015) Surveillance and processing of
foreign DNA by the Escherichia coli CRISPR-Cas system,
Cell, 163, 854-865.
84.Sashital, D. G., Wiedenheft, B., and Doudna, J.
A. (2012) Mechanism of foreign DNA selection in a bacterial adaptive
immune system, Mol. Cell, 46, 606-615.
85.Semenova, E., Savitskaya, E., Musharova, O.,
Strotskaya, A., Vorontsova, D., et al. (2016) Highly efficient primed
spacer acquisition from targets destroyed by the Escherichia
coli type IE CRISPR-Cas interfering complex, Proc. Natl.
Acad. Sci. USA, 113, 7626-7631.
86.Severinov, K., Ispolatov, I., and Semenova, E.
(2016) The influence of copy-number of targeted extrachromosomal
genetic elements on the outcome of CRISPR-Cas defense, Front. Mol.
Biosci., 3, 45.
87.Shabalina, S., and Koonin, E. (2008) Origins and
evolution of eukaryotic RNA interference, Trends Ecol. Evol.,
23, 578-587.
88.Makarova, K. S., Wolf, Y. I., van der Oost, J.,
and Koonin, E. V (2009) Prokaryotic homologs of Argonaute proteins are
predicted to function as key components of a novel system of defense
against mobile genetic elements., Biol. Direct, 4,
29.
89.Willkomm, S., Makarova, K. S., and Grohmann, D.
(2018) DNA silencing by prokaryotic Argonaute proteins adds a new layer
of defense against invading nucleic acids, FEMS Microbiol. Rev.,
42, 376-387.
90.Kuzmenko, A., Oguienko, A., Esyunina, D., Yudin,
D., Petrova, M., et al. (2020) DNA targeting and interference by a
bacterial Argonaute nuclease, Nature, 587, 632-637, doi:
10.1038/s41586-020-2605-1.
91.Wang, Y., Juranek, S., Li, H., Sheng, G., Tuschl,
T., and Patel, D. J. (2008) Structure of an argonaute silencing complex
with a seed-containing guide DNA and target RNA duplex, Nature,
456, 921-926.
92.Olovnikov, I., Chan, K., Sachidanandam, R.,
Newman, D. K., and Aravin, A. A. (2013) Bacterial argonaute samples the
transcriptome to identify foreign DNA, Mol. Cell, 51,
594-605.
93.Lisitskaya, L., Aravin, A. A., and Kulbachinskiy,
A. (2018) DNA interference and beyond: structure and functions of
prokaryotic Argonaute proteins, Nat. Commun., 9,
1-12.
94.Swarts, D. C., Hegge, J. W., Hinojo, I.,
Shiimori, M., Ellis, M. A., et al. (2015) Argonaute of the archaeon
Pyrococcus furiosus is a DNA-guided nuclease that targets
cognate DNA, Nucleic Acids Res., 43, 5120-5129.
95.Kuzmenko, A., Yudin, D., Ryazansky, S.,
Kulbachinskiy, A., and Aravin, A. A. (2019) Programmable DNA cleavage
by Ago nucleases from mesophilic bacteria Clostridium
butyricum and Limnothrix rosea, Nucleic Acids
Res., 47, 5822-5836.
96.Yuan, Y.-R., Pei, Y., Ma, J.-B., Kuryavyi, V.,
Zhadina, M., et al. (2005) Crystal structure of A. aeolicus
argonaute, a site-specific DNA-guided endoribonuclease, provides
insights into RISC-mediated mRNA cleavage, Mol. Cell, 19,
405-419.
97.Wang, Y., Juranek, S., Li, H., Sheng, G., Wardle,
G. S., et al. (2009) Nucleation, propagation and cleavage of target
RNAs in Ago silencing complexes, Nature, 461,
754-761.
98.Kaya, E., Doxzen, K. W., Knoll, K. R., Wilson, R.
C., Strutt, S. C., et al. (2016) A bacterial Argonaute with
noncanonical guide RNA specificity, Proc. Natl. Acad. Sci. USA,
113, 4057-4062.
99.Willkomm, S., Zander, A., Gust, A., and Grohmann,
D. (2015) A prokaryotic twist on argonaute function, Life,
5, 538-553.
100.Swarts, D. C., Jore, M. M., Westra, E. R., Zhu,
Y., Janssen, J. H., et al. (2014) DNA-guided DNA interference by a
prokaryotic Argonaute, Nature, 507, 258-261.
101.Ryazansky, S., Kulbachinskiy, A., and Aravin,
A. A. (2018) The expanded universe of prokaryotic Argonaute proteins,
MBio, 9, e01935-18, doi: 10.1128/mBio.01935-18.
102.Zander, A., Willkomm, S., Ofer, S., Van
Wolferen, M., Egert, L., et al. (2017) Guide-independent DNA cleavage
by archaeal Argonaute from Methanocaldococcus jannaschii,
Nat. Microbiol., 2, 1-10.
103.Swarts, D. C., Szczepaniak, M., Sheng, G.,
Chandradoss, S. D., Zhu, Y., et al. (2017) Autonomous generation and
loading of DNA guides by bacterial Argonaute, Mol. Cell,
65, 985-998.
104.Jolly, S. M., Gainetdinov, I., Jouravleva, K.,
Zhang, H., Strittmatter, L., et al. (2020) Thermus
thermophilus Argonaute functions in the completion of DNA
replication, Cell, 182, 1545-1559.
105.Koonin, E. V. (2017) Evolution of RNA- and
DNA-guided antivirus defense systems in prokaryotes and eukaryotes:
common ancestry vs convergence, Biol. Direct, 12,
1-14.
106.Chopin, M.-C., Chopin, A., and Bidnenko, E.
(2005) Phage abortive infection in lactococci: variations on a theme,
Curr. Opin. Microbiol., 8, 473-9.
107.Lopatina, A., Tal, N., and Sorek, R. (2020)
Abortive infection: bacterial suicide as an antiviral immune strategy,
Annu. Rev. Virol., 7, 371-384.
108.Labrie, S. J., Samson, J. E., and Moineau, S.
(2010) Bacteriophage resistance mechanisms, Nat. Rev.
Microbiol., 8, 317-327.
109.Fukuyo, M., Sasaki, A., and Kobayashi, I.
(2012) Success of a suicidal defense strategy against infection in a
structured habitat, Sci. Rep., 2, 238.
110.Van Houte, S., Buckling, A., and Westra, E. R.
(2016) Evolutionary ecology of prokaryotic immune mechanisms,
Microbiol. Mol. Biol. Rev., 80, 745-763.
111.Barrangou, R., and Horvath, P. (2011) Lactic
acid bacteria defenses against phages, in Stress Responses of Lactic
Acid Bacteria, Springer, pp. 459-478.
112.Durmaz, E., and Klaenhammer, T. R. (2007)
Abortive phage resistance mechanism AbiZ speeds the lysis clock to
cause premature lysis of phage-infected Lactococcus
lactis, J. Bacteriol., 189, 1417-1425.
113.Wang, C., Villion, M., Semper, C., Coros, C.,
Moineau, S., and Zimmerly, S. (2011) A reverse transcriptase-related
protein mediates phage resistance and polymerizes untemplated DNA in
vitro, Nucleic Acids Res., 39, 7620-7629.
114.Tangney, M., and Fitzgerald, G. F. (2002)
Effectiveness of the lactococcal abortive infection systems AbiA, AbiE,
AbiF and AbiG against P335 type phages, FEMS Microbiol. Lett.,
210, 67-72.
115.Parreira, R., Ehrlich, S. D., and Chopin, M.
(1996) Dramatic decay of phage transcripts in lactococcal cells
carrying the abortive infection determinant AbiB, Mol.
Microbiol., 19, 221-230.
116.Samson, J. E., Spinelli, S., Cambillau, C., and
Moineau, S. (2013) Structure and activity of AbiQ, a lactococcal
endoribonuclease belonging to the type III toxin–antitoxin
system, Mol. Microbiol., 87, 756-768.
117.Bidnenko, E., Ehrlich, D., and Chopin, M.-C.
(1995) Phage operon involved in sensitivity to the Lactococcus
lactis abortive infection mechanism AbiD1, J. Bacteriol.,
177, 3824-3829.
118.Bouchard, J. D., Dion, E., Bissonnette, F., and
Moineau, S. (2002) Characterization of the two-component abortive phage
infection mechanism AbiT from Lactococcus lactis, J.
Bacteriol., 184, 6325-6332.
119.Haaber, J., Samson, J. E., Labrie, S. J.,
Campanacci, V., Cambillau, C., et al. (2010) Lactococcal abortive
infection protein AbiV interacts directly with the phage protein SaV
and prevents translation of phage proteins, Appl. Environ.
Microbiol., 76, 7085-7092.
120.Depardieu, F., Didier, J.-P., Bernheim, A.,
Sherlock, A., Molina, H., et al. (2016) A eukaryotic-like
serine/threonine kinase protects staphylococci against phages, Cell
Host Microbe, 20, 471-481.
121.Snyder, L. (1995) Phage-exclusion enzymes: a
bonanza of biochemical and cell biology reagents? Mol.
Microbiol., 15, 415-20.
122.Yu, Y. T., and Snyder, L. (1994) Translation
elongation factor Tu cleaved by a phage-exclusion system, Proc.
Natl. Acad. Sci. USA, 91, 802-806.
123.Levitz, R., Chapman, D., Amitsur, M., Green,
R., Snyder, L., and Kaufmann, G. (1990) The optional E. coli prr
locus encodes a latent form of phage T4‐induced anticodon
nuclease, EMBO J., 9, 1383-1389.
124.Cheng, X., Wang, W., and Molineux, I. J. (2004)
F exclusion of bacteriophage T7 occurs at the cell membrane,
Virology, 326, 340-352.
125.Schmitt, C. K., Kemp, P., and Molineux, I. J.
(1991) Genes 1.2 and 10 of bacteriophages T3 and T7 determine the
permeability lesions observed in infected cells of Escherichia
coli expressing the F plasmid gene pifA, J. Bacteriol.,
173, 6507-6514.
126.Toothman, P., and Herskowitz, I. (1980)
Rex-dependent exclusion of lambdoid phages II. Determinants of
sensitivity to exclusion, Virology, 102, 147-160.
127.Parma, D. H., Snyder, M., Sobolevski, S.,
Nawroz, M., Brody, E., and Gold, L. (1992) The Rex system of
bacteriophage lambda: tolerance and altruistic cell death, Genes
Dev., 6, 497-510.
128.Yamaguchi, Y., Park, J.-H., and Inouye, M.
(2011) Toxin–antitoxin systems in bacteria and archaea, Annu.
Rev. Genet., 45, 61-79.
129.Page, R., and Peti, W. (2016)
Toxin–antitoxin systems in bacterial growth arrest and
persistence, Nat. Chem. Biol., 12, 208-214.
130.Makarova, K. S., Wolf, Y. I., and Koonin, E. V.
(2013) Comparative genomics of defense systems in archaea and bacteria,
Nucleic Acids Res., 41, 4360-4377.
131.Harms, A., Brodersen, D. E., Mitarai, N., and
Gerdes, K. (2018) Toxins, targets, and triggers: an overview of
toxin–antitoxin biology, Mol. Cell, 70,
768-784.
132.Fineran, P. C., Blower, T. R., Foulds, I. J.,
Humphreys, D. P., Lilley, K. S., and Salmond, G. P. C. (2009) The phage
abortive infection system, ToxIN, functions as a protein–RNA
toxin–antitoxin pair, Proc. Natl. Acad. Sci. USA,
106, 894-899.
133.Short, F. L., Akusobi, C., Broadhurst, W. R.,
and Salmond, G. P. C. (2018) The bacterial Type III
toxin–antitoxin system, ToxIN, is a dynamic protein–RNA
complex with stability-dependent antiviral abortive infection activity,
Sci. Rep., 8, 1-10.
134.Koga, M., Otsuka, Y., Lemire, S., and Yonesaki,
T. (2011) Escherichia coli rnlA and rnlB compose a novel
toxin–antitoxin system, Genetics, 187, 123-130.
135.Otsuka, Y., and Yonesaki, T. (2012) Dmd of
bacteriophage T4 functions as an antitoxin against Escherichia
coli LsoA and RnlA toxins, Mol. Microbiol., 83,
669-681.
136.Dy, R. L., Przybilski, R., Semeijn, K.,
Salmond, G. P. C., and Fineran, P. C. (2014) A widespread bacteriophage
abortive infection system functions through a type IV
toxin–antitoxin mechanism, Nucleic Acids Res., 42,
4590-4605.
137.Cai, Y., Usher, B., Gutierrez, C., Tolcan, A.,
Mansour, M., Fineran, P. C., et al. (2020) A nucleotidyltransferase
toxin inhibits growth of Mycobacterium tuberculosis
through inactivation of tRNA acceptor stems, Sci. Adv.,
6, eabb6651.
138.Hampton, H. G., Smith, L. M., Ferguson, S.,
Meaden, S., Jackson, S. A., and Fineran, P. C. (2020) Functional
genomics reveals the toxin–antitoxin repertoire and AbiE activity
in Serratia, Microb. Genom., 6, mgen000458, doi:
10.1099/mgen.0.000458.
139.Hazan, R., and Engelberg-Kulka, H. (2004)
Escherichia coli mazEF-mediated cell death as a defense
mechanism that inhibits the spread of phage P1, Mol. Genet.
Genomics, 272, 227-234.
140.Pecota, D. C., and Wood, T. K. (1996) Exclusion
of T4 phage by the hok/sok killer locus from plasmid R1, J.
Bacteriol., 178, 2044-2050.
141.Hayes, F. (2003) Toxins-antitoxins: plasmid
maintenance, programmed cell death, and cell cycle arrest,
Science, 301, 1496-1499.
142.Koonin, E. V., Makarova, K. S., and Wolf, Y. I.
(2017) Evolutionary genomics of defense systems in archaea and
bacteria, Annu. Rev. Microbiol., 71, 233-261.
143.Song, S., and Wood, T. K. (2018)
Post-segregational killing and phage inhibition are not mediated by
cell death through toxin/antitoxin systems, Front. Microbiol.,
9, 814.
144.Sberro, H., Fremin, B. J., Zlitni, S., Edfors,
F., Greenfield, N., et al. (2019) Large-scale analyses of human
microbiomes reveal thousands of small, novel genes, Cell,
178, 1245-1259.e14.
145.Simon, A. J., Ellington, A. D., and
Finkelstein, I. J. (2019) Retrons and their applications in genome
engineering, Nucleic Acids Res., 47, 11007-11019.
146.Millman, A., Bernheim, A., Stokar-Avihail, A.,
Fedorenko, T., Voichek, M., et al. (2020) Bacterial retrons function in
anti-phage defense, Cell, 183, 1551-1561.e12, doi:
10.1016/j.cell.2020.09.065.
147.Bobonis, J., Mitosch, K., Mateus, A., Kritikos,
G., Elfenbein, J. R., et al. (2020) Phage proteins block and trigger
retron toxin/antitoxin systems, BioRxiv, doi:
10.1101/2020.06.22.160242.
148.Bobonis, J., Mateus, A., Pfalz, B.,
Garcia-Santamarina, S., Galardini, M., et al. (2020) Bacterial retrons
encode tripartite toxin/antitoxin systems, BioRxiv, doi:
10.1101/2020.06.22.160168.
149.Gao, L., Altae-Tran, H., Böhning, F.,
Makarova, K. S., Segel, M., et al. (2020) Diverse enzymatic activities
mediate antiviral immunity in prokaryotes, Science, 369,
1077-1084.
150.Burroughs, A. M., Zhang, D., Schäffer, D.
E., Iyer, L. M., and Aravind, L. (2015) Comparative genomic analyses
reveal a vast, novel network of nucleotide-centric systems in
biological conflicts, immunity and signaling, Nucleic Acids
Res., 43, 10633-10654.
151.Cohen, D., Melamed, S., Millman, A., Shulman,
G., Oppenheimer-Shaanan, Y., et al. (2019) Cyclic GMP-AMP signalling
protects bacteria against viral infection, Nature, 574,
691-695.
152.Whiteley, A. T., Eaglesham, J. B., de Oliveira
Mann, C. C., Morehouse, B. R., Lowey, B., et al. (2019) Bacterial
cGAS-like enzymes synthesize diverse nucleotide signals, Nature,
567, 194-199.
153.Millman, A., Melamed, S., Amitai, G., and
Sorek, R. (2020) Diversity and classification of
cyclic-oligonucleotide-based anti-phage signalling systems, Nat.
Microbiol., 5, 1608-1615, doi:
10.1038/s41564-020-0777-y.
154.Ablasser, A., and Chen, Z. J. (2019) cGAS in
action: expanding roles in immunity and inflammation, Science,
363, eaat8657, doi: 10.1126/science.aat8657.
155.Davies, B. W., Bogard, R. W., Young, T. S., and
Mekalanos, J. J. (2012) Coordinated regulation of accessory genetic
elements produces cyclic di-nucleotides for V. cholerae
virulence, Cell, 149, 358-370.
156.Severin, G. B., Ramliden, M. S., Hawver, L. A.,
Wang, K., Pell, M. E., et al. (2018) Direct activation of a
phospholipase by cyclic GMP-AMP in El Tor Vibrio
cholerae, Proc. Natl. Acad. Sci. USA, 115,
E6048-E6055.
157.Yoon, S. H., and Waters, C. M. (2021) The
ever-expanding world of bacterial cyclic oligonucleotide second
messengers, Curr. Opin. Microbiol., 60, 96-103.
158.Morehouse, B. R., Govande, A. A., Millman, A.,
Keszei, A. F. A., Lowey, B., et al. (2020) STING cyclic dinucleotide
sensing originated in bacteria, Nature, 586, 429-433.
159.Ye, Q., Lau, R. K., Mathews, I. T., Birkholz,
E. A., Watrous, J. D., et al. (2020) HORMA domain proteins and a
Trip13-like ATPase regulate bacterial cGAS-like enzymes to mediate
bacteriophage immunity, Mol. Cell, 77, 709-722.e7.
160.Rosenberg, S. C., and Corbett, K. D. (2015) The
multifaceted roles of the HORMA domain in cellular signaling, J.
Cell Biol., 211, 745-755.
161.Lau, R. K., Ye, Q., Birkholz, E. A., Berg, K.
R., Patel, L., et al. (2020) Structure and mechanism of a cyclic
trinucleotide-activated bacterial endonuclease mediating bacteriophage
immunity, Mol. Cell, 77, 723-733.e6.
162.Lowey, B., Whiteley, A. T., Keszei, A. F. A.,
Morehouse, B. R., Mathews, I. T., et al. (2020) CBASS immunity uses
CARF-related effectors to sense 3′-5′-and
2′-5′-linked cyclic oligonucleotide signals and protect
bacteria from phage infection, Cell, 182, 38-49.
163.Casjens, S. (2003) Prophages and bacterial
genomics: what have we learned so far? Mol. Microbiol.,
49, 277-300.
164.Canchaya, C., Proux, C., Fournous, G., Bruttin,
A., and Brüssow, H. (2003) Prophage genomics, Microbiol. Mol.
Biol. Rev., 67, 238-276.
165.Bondy-Denomy, J., Qian, J., Westra, E. R.,
Buckling, A., Guttman, D. S., et al. (2016) Prophages mediate defense
against phage infection through diverse mechanisms, ISME J.,
10, 2854-2866.
166.Dedrick, R. M., Jacobs-Sera, D., Bustamante, C.
A. G., Garlena, R. A., Mavrich, T. N., et al. (2017) Prophage-mediated
defence against viral attack and viral counter-defence, Nat.
Microbiol., 2, 1-13.
167.Roux, S., Krupovic, M., Daly, R. A., Borges, A.
L., Nayfach, S., et al. (2019) Cryptic inoviruses revealed as pervasive
in bacteria and archaea across Earth’s biomes, Nat.
Microbiol., 4, 1895-1906.
168.Benler, S., and Koonin, E. V (2020) Phage
lysis‐lysogeny switches and programmed cell death: danse
macabre, BioEssays, 42, e2000114, doi:
10.1002/bies.202000114.
169.Waldor, M. K., and Friedman, D. I. (2005) Phage
regulatory circuits and virulence gene expression, Curr. Opin.
Microbiol., 8, 459-465.
170.Juhala, R. J., Ford, M. E., Duda, R. L.,
Youlton, A., Hatfull, G. F., and Hendrix, R. W. (2000) Genomic
sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism
in the lambdoid bacteriophages, J. Mol. Biol., 299,
27-51.
171.Taylor, V. L., Fitzpatrick, A. D., Islam, Z.,
and Maxwell, K. L. (2019) The diverse impacts of phage morons on
bacterial fitness and virulence, in Advances in Virus Research,
Elsevier, Vol. 103, pp. 1-31.
172.Newton, G. J., Daniels, C., Burrows, L. L.,
Kropinski, A. M., Clarke, A. J., and Lam, J. S. (2001)
Three‐component‐mediated serotype conversion in
Pseudomonas aeruginosa by bacteriophage D3, Mol. Microbiol.,
39, 1237-1247.
173.Krylov, S. V, Kropinski, A. M., Shaburova, O.
V, Miroshnikov, K. A., Chesnokova, E. N., and Krylov, V. N. (2013) New
temperate Pseudomonas aeruginosa phage, phi297: specific
features of genome structure, Russ. J. Genet., 49,
806-818.
174.Taylor, V. L., Hoage, J. F. J., Thrane, S. W.,
Huszczynski, S. M., Jelsbak, L., and Lam, J. S. (2016) A
bacteriophage-acquired O-antigen polymerase (Wzyβ) from P.
aeruginosa serotype O16 performs a varied mechanism compared to its
cognate Wzyα, Front. Microbiol., 7, 393.
175.Lehane, A. M., Korres, H., and Verma, N. K.
(2005) Bacteriophage-encoded glucosyltransferase GtrII of
Shigella flexneri: membrane topology and identification
of critical residues, Biochem. J., 389, 137-143.
176.Perry, L. L., SanMiguel, P., Minocha, U.,
Terekhov, A. I., Shroyer, M. L., et al. (2009) Sequence analysis of
Escherichia coli O157: H7 bacteriophage ΦV10 and
identification of a phage-encoded immunity protein that modifies the
O157 antigen, FEMS Microbiol. Lett., 292, 182-186.
177.Gentile, G. M., Wetzel, K. S., Dedrick, R. M.,
Montgomery, M. T., Garlena, R. A., et al. (2019) More evidence of
collusion: a new prophage-mediated viral defense system encoded by
mycobacteriophage sbash, MBio, 10, e00196-19, doi:
10.1128/mBio.00196-19.
178.Montgomery, M. T., Guerrero Bustamante, C. A.,
Dedrick, R. M., Jacobs-Sera, D., and Hatfull, G. F. (2019) Yet more
evidence of collusion: a new viral defense system encoded by gordonia
phage carolann, MBio, 10, 1-18.
179.Jimmy, S., Saha, C. K., Kurata, T.,
Stavropoulos, C., Oliveira, S. R. A., et al. (2020) A widespread
toxin–antitoxin system exploiting growth control via alarmone
signaling, Proc. Natl. Acad. Sci. USA, 117,
10500-10510.
180.Ferullo, D. J., and Lovett, S. T. (2008) The
stringent response and cell cycle arrest in Escherichia
coli, PLoS Genet., 4, e1000300.
181.Owen, S. V, Wenner, N., Dulberger, C. L.,
Rodwell, E. V., Bowers-Barnard, A., et al. (2020) Prophage-encoded
phage defence proteins with cognate self-immunity, bioRxiv, doi:
10.1101/2020.07.13.199331.
182.Rousset, F., Dowding, J., Bernheim, A., Rocha,
E., and Bikard, D. (2021) Prophage-encoded hotspots of bacterial immune
systems, bioRxiv, doi: 10.1101/2021.01.21.427644.
183.Atanasiu, C., Su, T. J., Sturrock, S. S., and
Dryden, D. T. F. (2002) Interaction of the ocr gene 0.3 protein of
bacteriophage T7 with EcoKl restriction/modification enzyme, Nucleic
Acids Res., 30, 3936-3944.
184.Isaev, A., Drobiazko, A., Sierro, N., Gordeeva,
J., Yosef, I., et al. (2020) Phage T7 DNA mimic protein Ocr is a potent
inhibitor of BREX defence, Nucleic Acids Res., 48,
5397-5406.
185.Penadés, J. R., and Christie, G. E.
(2015) The phage-inducible chromosomal islands: a family of highly
evolved molecular parasites, Annu. Rev. Virol., 2,
181-201.
186.Christie, G. E., and Dokland, T. (2012) Pirates
of the Caudovirales, Virology, 434, 210-221.
187.Dokland, T. (2019) Molecular piracy:
redirection of bacteriophage capsid assembly by mobile genetic
elements, Viruses, 11, 1003.
188.Fillol-Salom, A., Miguel-Romero, L., Marina,
A., Chen, J., and Penadés, J. R. (2020) Beyond the CRISPR-Cas
safeguard: PICI-encoded innate immune systems protect bacteria from
bacteriophage predation, Curr. Opin. Microbiol., 56,
52-58.
189.Novick, R. P., Christie, G. E., and
Penadés, J. R. (2010) The phage-related chromosomal islands of
Gram-positive bacteria, Nat. Rev. Microbiol., 8,
541-551.
190.Fillol-Salom, A., Martínez-Rubio, R.,
Abdulrahman, R. F., Chen, J., Davies, R., and Penadés, J. R.
(2018) Phage-inducible chromosomal islands are ubiquitous within the
bacterial universe, ISME J., 12, 2114-2128.
191.Mitarai, N. (2020) How pirate phage interferes
with helper phage: comparison of the two distinct strategies, J.
Theor. Biol., 486, 110096.
192.Novick, R. P. (2019) Pathogenicity islands and
their role in staphylococcal biology, Microbiol. Spectr.,
7, doi: 10.1128/microbiolspec.GPP3-0062-2019.
193.Tormo-Más, M. Á., Mir, I.,
Shrestha, A., Tallent, S. M., Campoy, S., Lasa, Í., et al.
(2010) Moonlighting bacteriophage proteins derepress staphylococcal
pathogenicity islands, Nature, 465, 779-782.
194.Mir‐Sanchis, I.,
Martínez‐Rubio, R., Martí, M., Chen, J., Lasa,
Í., et al. (2012) Control of Staphylococcus aureus
pathogenicity island excision, Mol. Microbiol., 85,
833-845.
195.Ram, G., Chen, J., Kumar, K., Ross, H. F.,
Ubeda, C., et al. (2012) Staphylococcal pathogenicity island
interference with helper phage reproduction is a paradigm of molecular
parasitism, Proc. Natl. Acad. Sci. USA, 109, 16300-5.
196.Ram, G., Chen, J., Ross, H. F., and Novick, R.
P. (2014) Precisely modulated pathogenicity island interference with
late phage gene transcription, Proc. Natl. Acad. Sci. USA,
111, 14536-14541.
197.Martínez-Rubio, R., Quiles-Puchalt, N.,
Martí, M., Humphrey, S., Ram, G., et al. (2017) Phage-inducible
islands in the Gram-positive cocci, ISME J., 11,
1029-1042.
198.Fillol-Salom, A., Bacarizo, J., Alqasmi, M.,
Ciges-Tomas, J. R., Martínez-Rubio, R., et al. (2019) Hijacking
the hijackers: Escherichia coli pathogenicity islands
redirect helper phage packaging for their own benefit, Mol.
Cell, 75, 1020-1030.
199.O’Hara, B. J., Barth, Z. K., McKitterick,
A. C., and Seed, K. D. (2017) A highly specific phage defense system is
a conserved feature of the Vibrio cholerae mobilome,
PLoS Genet., 13, 1-17.
200.McKitterick, A. C., and Seed, K. D. (2018)
Anti-phage islands force their target phage to directly mediate island
excision and spread, Nat. Commun., 9, 1-8.
201.McKitterick, A. C., Hays, S. G., Johura, F. T.,
Alam, M., and Seed, K. D. (2019) Viral satellites exploit phage
proteins to escape degradation of the bacterial host chromosome,
Cell Host Microbe, 26, 504-514.e4.
202.Barth, Z. K., Silvas, T. V, Angermeyer, A., and
Seed, K. D. (2020) Genome replication dynamics of a bacteriophage and
its satellite reveal strategies for parasitism and viral restriction,
Nucleic Acids Res., 48, 249-263.
203.Barth, Z. K., Netter, Z., Angermeyer, A.,
Bhardwaj, P., and Seed, K. D. (2020) A family of viral satellites
manipulates invading virus gene expression and can affect cholera toxin
mobilization, mSystems, 5, e00358-20, doi:
10.1128/mSystems.00358-20.
204.Hays, S. G., and Seed, K. D. (2020) Dominant
Vibrio cholerae phage exhibits lysis inhibition sensitive
to disruption by a defensive phage satellite, Elife, 9,
e53200.
205.McKitterick, A. C., LeGault, K. N., Angermeyer,
A., Alam, M., and Seed, K. D. (2019) Competition between mobile genetic
elements drives optimization of a phage-encoded CRISPR-Cas system:
insights from a natural arms race, Philos. Trans. R. Soc. B,
374, 20180089.
206.Makarova, K. S., Wolf, Y. I., Snir, S., and
Koonin, E. V. (2011) Defense islands in bacterial and archaeal genomes
and prediction of novel defense systems, J. Bacteriol.,
193, 6039-6056.
207.Doron, S., Melamed, S., Ofir, G., Leavitt, A.,
Lopatina, A., et al. (2018) Systematic discovery of antiphage defense
systems in the microbial pangenome, Science, 359,
eaar4120, doi: 10.1126/science.aar4120.
208.Burroughs, A. M., and Aravind, L. (2020)
Identification of uncharacterized components of prokaryotic immune
systems and their diverse eukaryotic reformulations, J.
Bacteriol., 202, e00365-20, doi: 10.1128/JB.00365-20.
209.Santiveri, M., Roa-Eguiara, A., Kuhne, C.,
Wadhwa, N., Berg, H. C., et al. (2020) Structure and function of stator
units of the bacterial flagellar motor, Cell, 183,
244-257.
210.Ofir, G., Herbst, E., Baroz, M., Cohen, D.,
Millman, A., et al. (2021) Antiviral activity of bacterial TIR domains
via signaling molecules that trigger cell death, bioRxiv, doi:
10.1101/2021.01.06.425286.
211.Ka, D., Oh, H., Park, E., Kim, J., and Bae, E.
(2020) Structural and functional evidence of bacterial antiphage
protection by Thoeris defense system via NAD+ degradation,
Nat. Commun., 11, 2816, doi:
10.1038/s41467-020-16703-w.
212.Bayless, A. M., and Nishimura, M. T. (2020)
Enzymatic functions for Toll/Interleukin-1 receptor domain proteins in
the plant immune system, Front. Genet., 11, 539.
213.Badrinarayanan, A., Le, T. B. K., and Laub, M.
T. (2015) Bacterial chromosome organization and segregation, Annu.
Rev. Cell Dev. Biol., 31, 171-199.
214.Panas, M. W., Jain, P., Yang, H., Mitra, S.,
Biswas, D., et al. (2014) Noncanonical SMC protein in
Mycobacterium smegmatis restricts maintenance of
Mycobacterium fortuitum plasmids, Proc. Natl. Acad.
Sci. USA, 111, 13264-13271.
215.Pace, H. C., and Brenner, C. (2001) The
nitrilase superfamily: classification, structure and function,
Genome Biol., 2, 1-9.
216.Mekhedov, S. L., Makarova, K. S., and Koonin,
E. V. (2017) The complex domain architecture of SAMD9 family proteins,
predicted STAND-like NTPases, suggests new links to inflammation and
apoptosis, Biol. Direct, 12, 13.
217.Forsberg, K. J., and Malik, H. S. (2018)
Microbial genomics: the expanding universe of bacterial defense
systems, Curr. Biol., 28, R361-R364.
218.Van, V., and Van Valen, L. (1973) A New
Evolutionary Law, The University of Chicago, Illinois.
219.Stern, A., and Sorek, R. (2011) The phage-host
arms race: shaping the evolution of microbes, Bioessays,
33, 43-51.
220.Paterson, S., Vogwill, T., Buckling, A.,
Benmayor, R., Spiers, A. J., et al. (2010) Antagonistic coevolution
accelerates molecular evolution, Nature, 464,
275-278.
221.Ignacio-Espinoza, J. C., Ahlgren, N. A., and
Fuhrman, J. A. (2020) Long-term stability and Red Queen-like strain
dynamics in marine viruses, Nat. Microbiol., 5,
265-271.
222.Furuta, Y., Kawai, M., Uchiyama, I., and
Kobayashi, I. (2011) Domain movement within a gene: a novel
evolutionary mechanism for protein diversification, PLoS One,
6, e18819.
223.Dupuis, M.-È., Villion, M.,
Magadán, A. H., and Moineau, S. (2013) CRISPR-Cas and
restriction-modification systems are compatible and increase phage
resistance, Nat. Commun., 4, 1-7.
224.Hynes, A. P., Villion, M., and Moineau, S.
(2014) Adaptation in bacterial CRISPR-Cas immunity can be driven by
defective phages, Nat. Commun., 5, 1-6.
225.Bernheim, A., and Sorek, R. (2020) The
pan-immune system of bacteria: antiviral defence as a community
resource, Nat. Rev. Microbiol., 18, 113-119.
226.Koonin, E. V., Makarova, K. S., Wolf, Y. I.,
and Krupovic, M. (2020) Evolutionary entanglement of mobile genetic
elements and host defence systems: guns for hire, Nat. Rev.
Genet., 21, 119-131.
227.Høyland-Kroghsbo, N. M., Paczkowski, J.,
Mukherjee, S., Broniewski, J., Westra, E., et al. (2017) Quorum sensing
controls the Pseudomonas aeruginosa CRISPR-Cas adaptive immune system,
Proc. Natl. Acad. Sci. USA, 114, 131-135.
228.Xiong, L., Liu, S., Chen, S., Xiao, Y., Zhu,
B., et al. (2019) A new type of DNA phosphorothioation-based antiviral
system in archaea, Nat. Commun., 10, 1-11.
229.Al-Shayeb, B., Sachdeva, R., Chen, L.-X., Ward,
F., Munk, P., et al. (2020) Clades of huge phages from across
Earth’s ecosystems, Nature, 578, 425-431.