REVIEW: A Path to the Atomic-Resolution Structures of Prokaryotic and Eukaryotic Ribosomes
Gulnara Yusupova1 and Marat Yusupov1,2,a*
1Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), INSERM U964, CNRS UMR7104, Université de Strasbourg, 67404 Illkirch, France2Institute of Fundamental Medicine and Biology, Kazan Federal University, 420008 Kazan, Russia
* To whom correspondence should be addressed.
Received May 6, 2021; Revised June 8, 2021; Accepted June 8, 2021
Resolving first crystal structures of prokaryotic and eukaryotic ribosomes by our group has been based on the knowledge accumulated over the decades of studies, starting with the first electron microscopy images of the ribosome obtained by J. Pallade in 1955. In 1983, A. Spirin, then a Director of the Protein Research Institute of the USSR Academy of Sciences, initiated the first study aimed at solving the structure of ribosomes using X-ray structural analysis. In 1999, our group in collaboration with H. Noller published the first crystal structure of entire bacterial ribosome in a complex with its major functional ligands, such as messenger RNA and three transport RNAs at the A, P, and E sites. In 2011, our laboratory published the first atomic-resolution structure of eukaryotic ribosome solved by the X-ray diffraction analysis that confirmed the conserved nature of the main ribosomal functional components, such as the decoding and peptidyl transferase centers, was confirmed, and eukaryote-specific elements of the ribosome were described. Using X-ray structural analysis, we investigated general principles of protein biosynthesis inhibition in eukaryotic ribosomes, along with the mechanisms of antibiotic resistance. Structural differences between bacterial and eukaryotic ribosomes that determine the differences in their inhibition were established. These and subsequent atomic-resolution structures of the functional ribosome demonstrated for the first time the details of binding of messenger and transport RNAs, which was the first step towards understanding how the ribosome structure ultimately determines its functions.
KEY WORDS: ribosome, structure, X-ray diffraction analysis, cryo-EMDOI: 10.1134/S0006297921080046
Abbreviations: cryo-EM, cryogenic electron microscopy; mRNA, messenger RNA; DC, decoding site; PTC, peptidyl transferase center; rRNA, ribosomal RNA; tRNA, transport RNA.
INTRODUCTION
Proteins are synthesized on ribosomes, universal ribonucleoprotein complexes that read genetic information encoded in the messenger RNA (mRNA) and catalyze formation of peptide bonds between amino acids in a programmed order, resulting in the formation of a new polypeptide chains [1]. These giant organelles with a molecular mass of ~2.5 MDa in bacteria and up to 4.5 MDa in higher eukaryotes consist of numerous proteins and long-chain ribosomal RNAs (rRNAs). Despite the fact that ribosomes and their main functions were described more than 60 years ago, large size and highly dynamic nature of these macromolecular ensembles have limited their structural studies. It has become obvious that at least theoretically, the only method that could produce structural data with the atomic resolution, which was ultimately sought for by the researchers, was X-ray diffraction analysis.
Our knowledge on the three-dimensional ribosomal structure in 1990s was based mainly on the fragmentary information obtained by nuclear magnetic resonance (NMR) and X-ray diffraction analysis of small rRNA domains and 10 individual ribosomal proteins [2-6]. The relations between the ribosomal structure and functions have been derived from the models generated by electron microscopy (EM), neutron diffraction, and molecular modeling based on the results of biophysical and biochemical experiments [7-9]. Later, crystals of ribosomal domains containing rRNA fragments and ribosomal proteins have been obtained and their structures have been resolved [3, 5].
The models for the asymmetric structures of the 30S subunit (V-shaped model) and the 50S subunit (crown-like model) were suggested in the laboratories of Viktor Vasiliev and James Lake [10-12]. Further progress in the investigation of the low-resolution structures of ribosome and ribosomal complexes has been achieved using the reconstruction of 3D images of ribosomes from the EM images of ribosomes in a thin layer of frozen liquid (cryo-EM) [13, 14]. In the end of 1990s, two research groups independently elucidated the structure of the Escherichia coli bacterial ribosome used the cryo-EM approach. The diameter of the bacterial ribosome (~220 Å) and all previously described classic features of the small subunit, including head, neck, gap, and platform, as well as three appendages in the upper part of the large subunit, were confirmed with a 25 Å resolution [11, 15]. The overall size and shape of the subunits and the general geometry of their association were described. Although some RNA helices were visible at this resolution, it was difficult to distinguish proteins and RNAs. Further development of this method and examination of more ribosomal complexes allowed to localize transport RNAs (tRNAs) on the inner surface of the ribosomal subunits, and the existence of the E site for the tRNA binding was clearly demonstrated [7].
Mutual arrangement of ribosomal proteins, identification of neighboring proteins and RNA sequences interacting with proteins, as well as rRNAs sequences and proteins participating in the formation of functional sites on the ribosome have been elucidated with the help of protein–protein and RNA–protein crosslinking, affinity linking, and modification [16-18]. Unfortunately, these approaches have provided controversial information, as they examine only a small fraction (sometimes <1%) of a ribosomal preparation, which might not be structurally homogenous. Nevertheless, several models of ribosomal subunits have been suggested based on the results of such experiments, which has stimulated further studies, including validation of the suggested models. In particular, the model of the 30S subunit proposed by Spirin, Vasiliev, and Serdyuk was based on the results of EM of ribosomal subunits, as well as on the data produced by immuno-electron microscopy and protein-protein crosslinking. This model was validated experimentally using the techniques of neutron and X-ray scattering in solution [19]. Another approach – molecular modeling of small ribosomal subunits – was based on the phylogenetically predicted secondary structure of 16S RNA [20, 21].
One of the most important achievements in the studies of ribosomal structure was a map describing location of centers of masses for 21 ribosomal proteins in the small subunit, which was obtained by the neutron scattering technique [22]. This approach allows to determine the distance between the centers of masses of two proteins in the ribosome. Although the model proposed based on these results was later found inadequate, it initiated the studies aimed at its validation and has prompted many fruitful discussions in the “ribosome community”.
The experiments on the step-wise extraction of proteins from the ribosomal subunits using gradually increasing LiCl concentration followed by examination of the obtained ribonucleoprotein particles by EM showed that the shape of the ribosome is defined by rRNA [23, 24]. These results were used for modeling both the small ribosomal subunit and the three-dimensional structure of subunits based on the predicted secondary structure of rRNA [25].
An integrated approach involving phylogenetically determined secondary structure of 16S RNA [25, 26], the map of the centers of masses of proteins in the 30S subunit generated by neutron scattering [27], chemical probing of rRNA at different stages of subunit assembly [20, 28, 29], and results of UV-crosslinking of rRNA [30] was used for modeling RNA in the structure of the 30S subunit.
The first structures of the entire bacterial ribosomes, bacterial 30S subunit, and archaeal 50S subunit were obtained in the end of 1990s and beginning of 2000s as a result of extensive X-ray crystallography studies. These discoveries have initiated an avalanche of new studies on the ribosomal protein synthesis, which have significantly expanded our understanding of the functioning of this molecular machine [31-33].
Substantial knowledge in experimental biochemistry and ribosome hydrodynamics acquired during our work in the Spirin laboratory at the Institute of Protein Research has led us to the development of new methods for the isolation and crystallization of bacterial and, later, eukaryotic ribosomes. Here, we describe the strategy and key steps in the research conducted primarily in our laboratory, which have resulted in the resolution of the first crystal structure of prokaryotic ribosome and its functional complexes, as well as to our recent progress in determining the first crystal structure of eukaryotic ribosome [33-38]. Resolving the crystal structure of eukaryotic 80S ribosome was a breakthrough in the structural and functional studies of eukaryotic ribosomes, which allowed to rationalize existing biochemical and genetic information, and, eventually, can stimulate the development of future experimental models for investigating different aspects of protein biosynthesis.
CRYSTALLIZATION OF BACTERIAL RIBOSOME AND RESOLUTION OF ITS
STRUCTURE
Bacterial ribosome with the sedimentation coefficient of 70S is formed by the small and large subunits (30S and 50S, respectively). Each ribosomal subunit is composed from rRNA, which comprise two thirds of its mass, and ribosomal proteins (one third of subunit mass). The large 50S subunit contains two rRNAs, 5S (120 nucleotides) and 23S (~ 2900 nucleotides), while the small 30S subunit contains only one 16S rRNA (~ 1500 nucleotides). The protein portion of the 30S and 50S subunits consists of 21 and 33 individual proteins, respectively [1].
The first attempts to produce the three-dimensional crystals of ribosomal subunits suitable for the X-ray diffraction examination were conducted in 1980s. The first crystals of the 50S subunits isolated from Bacillus stearothermophilus and Haloarcula marismortui were obtained in the Max Planck Institute in Berlin by the Ada Yonath group in collaboration with Heinz-Günter Wittman [39, 40]. These pioneer studies demonstrated a possibility of using X-ray crystallography for investigating ribosomes [41]. At this time, Alexander Sergeevich Spirin, then a Director of the Institute of Protein Research of the USSR Academy of Sciences, initiated creation of our research group tasked with the development of approaches for elucidating the structure of the ribosome. In 1983, the collaborative Ribosome Crystallography Project was started at the Institute of Protein Research and Institute of Crystallography of the USSR Academy of Sciences.
Now, 40 years after the start of this project, it can be definitely stated that the most challenging and lengthy part of our work was the search for the conditions ensuring generation of crystals from ribosomes and their subunits suitable for the X-ray diffraction examination (which is true for any crystallography project). We have developed techniques for purification and crystallization of ribosomes from the bacterium Thermus thermophilus (a novel model organism for laboratory studies), which is an extreme thermophile (the optimal growth temperature for this organism is 75°C) [42-44]. Later, this experimental model was used for resolving the structures of the small 30S subunit [32, 45] and of the entire 70S ribosome in a complex with its functional ligands, such as tRNA and mRNA [33, 46, 47]. This model is still used in modern crystallographic and cryo-EM studies of translation and its regulation [48-50].
Two steps were found to be crucially important in the development of techniques for the isolation and purification of ribosomes from T. thermophilus. The first step is purification of ribosomes by sedimentation in a high-density solution of CsCl and sucrose, that was suggested by Zurab Gogia (Institute of Protein Research). The second step is hydrophobic chromatography on the TOYOPEARL Butyl column (Tosoh Bioscience, Germany) suggested by Sergei Trakhanov (Institute of Crystallography). This step allows to obtain ribosomes with tightly attached subunits suitable for crystallization.
This two-step protocol was used with minor modifications for the crystallization of three types of ribosome crystals. The first crystal form was produced at the Institute of Protein Research, thus confirming that X-ray analysis could be used for investigating the structure of the ribosome and its functional complexes [42-44]. The second crystal form was used for the elucidation of the structure of ribosome in a complex with mRNA and tRNA at a resolution of 7.8 Å [46] and then at 5.5 Å [33, 47]. These studies were conducted in collaboration with the group of Harry Noller from the Center for Molecular Biology of RNA, University of California, Santa Cruz. The third crystal form was produced in the laboratory of Venki Ramakrishnan and used for determining the ribosome structure with a resolution of 2.8 Å [51]. Earlier, the structure of E. coli ribosome was determined with a 3.5 Å resolution in the Jamie Cate laboratory [52].
Our group was the first to produce the crystals of T. thermophilus 30S subunit [53, 54]. After optimization of conditions for the crystal growth and acquisition of X-ray diffraction data, this crystal form was used for determining the structure of the 30S subunit in the laboratories of Venki Ramakrishnan and Ada Yonath [32, 45]. In the same manner, the crystals of the 50S subunit earlier obtained in the Ada Yonath laboratory [55] were used by Peter Moore and Thomas Steitz for elucidation of the structure of the 50S subunit of H. marismortui ribosome after optimization of crystal growth conditions and data acquisition [31, 56].
The crystals of individual ribosomal subunits (30S and 50S) were used for investigating their functional complexes with ligands mimicking mRNA and tRNA molecules, as well as for studying the decoding center (DC) and peptidyl transferase center (PTC) of the ribosome [57, 58]. The high-resolution structures of the 30S subunits from T. thermophilus [32] and 50S subunits from H. marismortui [31] and the electron density map of the entire 70S ribosome were used by our group for modeling the entire T. thermophilus ribosome [33, 46].
CRYSTAL STRUCTURE OF BACTERIAL RIBOSOME
The three-dimensional structures of the 70S ribosome and its subunits were characterized by different electron microscopy methods in 1980s [10, 11]. The general morphology of the small subunit includes the following main blocks: “head” and subunit “body”. The body includes the shoulder, the platform; and the structural element between the head and the body termed the neck (Fig. 1). The large subunit has a more compact structure consisting of a round base with three protuberances named the L1-protuberance, the central protuberance (CP), and the L7/L12 stalk. A significantly higher resolution was achieved in 1990s due to the introduction of the technique of the cryo-EM image reconstruction [59].
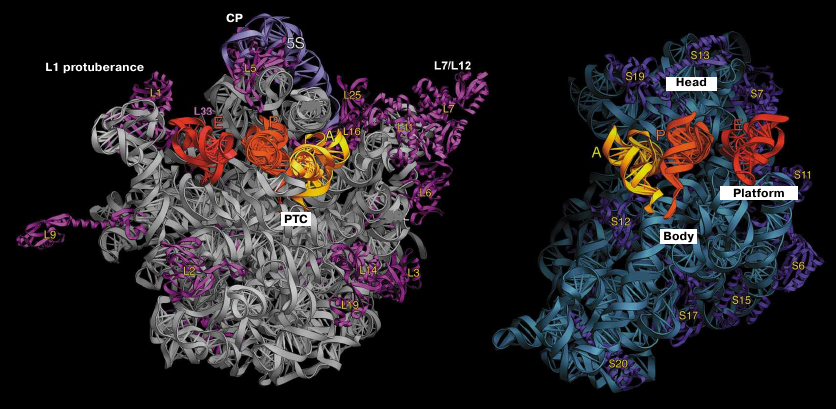
Fig. 1. Structure of T. thermophilus ribosomal subunits based on the X-ray diffraction data. Left panel, inner side of the subunit surface with tRNAs in the A (yellow), P (orange), and E sites (red); L1 protuberance, central protuberance (CP), L7/L12 stalk, and position of the PTC are shown in the large subunit. Right panel, head, body, and platform of the small subunit.
Crystallographic examination of bacterial ribosome demonstrated that the protuberance on the head of the small subunit consists exclusively of the h33 helix, while the CP of the large subunit is composed of the 5S rRNA, part of the 23S rRNA, and ribosomal proteins L5, L18, L25, and L33. More detailed analysis of the subunit structure allowed to identify the role of domains in the rRNA secondary structure. In particular, the 5′-domain of the 16S RNA forms a body of the small ribosomal subunit, the central part of the 16S RNA form the platform, and the 3′-domain forms the head of the subunit. Overall, the crystal structure of the ribosome corroborated the predicted secondary structure of the rRNAs [33].
Resolution of crystal structures of bacterial ribosomal subunits and the entire 70S ribosome allowed to describe more than 50 structures of individual ribosomal proteins and to localized them on the ribosome. The presence of the globular domain, which is usually located on the ribosomal subunit surface and of the long unstructured amino acid sequence that penetrates deep into the so-called subunit core consisting almost entirely of rRNAs, is a characteristic feature of most ribosomal proteins. Structural analysis revealed that binding of ribosomal proteins to various rRNA helices provides a connection between different domains of the ribosome.
The most surprising observation in the crystallographic analysis of the entire ribosome was most likely the fact that ribosomal proteins were absent on the contact surfaces of ribosomal subunit, which implied that the functional centers of the ribosomes consist entirely of the rRNA. Prior to that, it had been presumed by analogy with the cellular enzymes that the enzymatic activity of the ribosome is mediated exclusively by proteins. We were the first to demonstrate in 1986 the absence of ribosomal proteins on the contact surfaces of ribosome subunits using the hot tritium bombardment experiments [60].
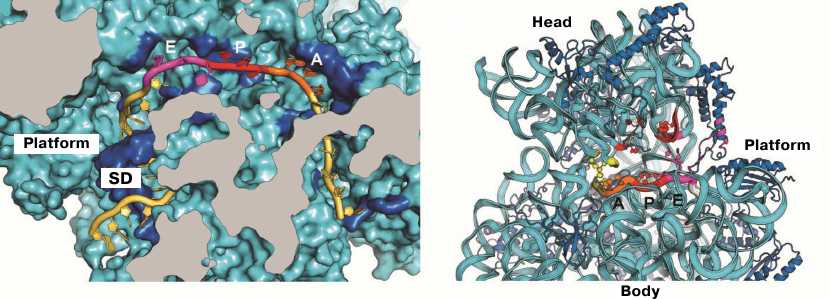
The first crystal structure of the entire 70S ribosome contained three transport RNAs in the A, P, and E sites and a 24-nucleotide mRNA fragment stabilized between the ribosomal subunits [33, 47]. These structures, as well as the data obtained in later crystallographic studies conducted in our laboratory, were used to elucidate the atomic structure of the ribosomal functional centers, such as tRNA-interacting centers [33, 61, 62]. The mRNA-binding sites and the three tRNA-binding sites (A, P, and E) are located at the interface between the subunits, where the DC of the small subunit is situated. The codon-anticodon coupling in the DC ensures the accuracy of mRNA decoding. Analysis of the structures obtained in our laboratory showed that mRNA reaches the ribosome interface through a tunnel located between the head and the shoulder and wraps around the neck of the small subunit [47]. The exit for the mRNA molecule (mRNA 5′-end) is located between the head and the platform. It was demonstrated in the same work that mRNA forms a bend between the A and P sites, which determines the possibility of simultaneous binding of two tRNA molecules. Moreover, it was also described how the Shine–Dalgarno sequence (SD) in the mRNA interacts with the anti-SD site in the rRNA, thus fixing exact position of the mRNA reading frame (Fig. 2), as well as how mRNA with this newly formed duplex moves during the first steps of protein synthesis [63-65].
Fig. 2. Location of mRNA on the small subunit. a) Cross section of the small subunit at the level of mRNA: Shine–Dalgarno sequence (SD) in mRNA interacts with the anti-SD site in rRNA; b) mRNA tunnel winding around the neck of the small subunit between the head and the body. Orientation of codons in the A, P, and E sites at the inner surface of the small subunit is shown.
In the studies that followed, we used X-ray diffraction analysis to reveal molecular events responsible for the decoding errors leading to the incorporation of erroneous amino acid into the nascent proteins, and proposed the geometric mimicry mechanism of erroneous decoding resulting from the combination of protonation and rare event of nucleotide tautomerization in the codon-anticodon duplexes [66-71]. By using the frontline technology of long-wavelength macromolecular crystallography, we were able for the first time to localize hundreds of potassium ions in the functional ribosome [49]. We also demonstrated that potassium ions not only participate in the formation of complexes between rRNA and ribosomal proteins, but also play an essential role in the ribosome functions, such as decoding and peptidyl transferase reaction.
The structural studies of new functional complexes have further extended our knowledge on the mechanism of tRNA binding in the presence of elongation factor (EF-Tu), processes of mRNA decoding and peptide bond formation, and mechanisms of GTP hydrolysis, translocation, termination, and ribosome recycling [37, 38, 72-75].
THE STRUCTURE OF EUKARYOTIC RIBOSOME
The information obtained from the high-resolution crystal structures of prokaryotic ribosomes has expanded our knowledge on protein synthesis in bacteria; however, our understanding of molecular basis of the eukaryotic ribosome functioning remained very limited. As a result of evolution, eukaryotic ribosomes have a larger mass (by 40% in lower eukaryotes) than prokaryotic ribosomes [35, 76]. Despite the presence of conserved regions in rRNA and a number of common proteins, prokaryotic and eukaryotic ribosomes differ significantly. These differences are due to the occurrence of approximately 30 additional eukaryote-specific proteins and over 50 nucleotide sequences named RNA expansion segments, that are present in the evolutionary conserved rRNA core. Until 2010, our knowledge on the structural arrangement of eukaryotic ribosomes had been based only on the low-resolution EM images. It was commonly was believed that the X-ray diffraction could not be used to produce structural information at the atomic resolution, as obtaining the crystals of such gigantic molecule (molecular weight, ~3.3 MDa in lower eukaryotes) for the generation of high-resolution diffraction patterns was impossible.
The first reports that the eukaryotic ribosome can form crystals were published already in 1966, when the two-dimensional ribosome crystals were observed in chicken embryo tissues under hypothermia [77]. However, all attempts to obtain crystals of eukaryotic ribosomes had been unsuccessful until recently, despite the competition between groups working in this area of structural biology.
In 2010, our group succeeded in obtaining the crystals of the entire 80S eukaryotic ribosome from Saccharomyces cerevisiae and elucidating the first X-ray diffraction-derived structure of eukaryotic ribosome with a resolution of 4.15 Å and then – 3.0 Å [34, 78]. Successful crystallization of the yeast ribosome became possible due to the newly developed methods for the purification of intact eukaryotic ribosomes. First of all, we used a known experimental fact that glucose starvation of growing yeast cells inhibits translation initiation and results in the accumulation of uniform ligand-lacking ribosomes. Hence, yeast cells were subjected to glucose starvation in order to increase the initial amount of ribosome monomers. Next, a very mild protocol for ribosome purification that guaranteed retention of all components in the ribosome in their undamaged state was developed. Adam Ben-Shem introduced fractionation of the 30S cell extract with 20,000 polyethylene glycol followed by purification of 80S ribosomes in the sucrose gradient under non-dissociating conditions [78]. Using these approaches, highly uniform ribosome preparations were isolated that produced crystals with strong X-ray diffraction.
The crystal structures of eukaryotic ribosomes from S. cerevisiae have significantly improved the understanding of the mechanisms of protein synthesis and translation regulation in the cell. In particular, analysis of yeast ribosomes isolated from the cells subjected to glucose starvation showed that the non-ribosomal protein Stm1 bound to the 80S ribosome, thus creating an additional bridge between the two ribosomal subunits that stabilized ribosome under crystallization conditions. This stress protein completely blocked the binding of main functional ligands, such as mRNA and tRNA, by covering their binding sites on the ribosome. Crystallography-derived structures of eukaryotic ribosome complexes with eukaryote-specific and broad-spectrum inhibitors not only allowed researchers to determine the general principles of protein synthesis inhibition and resistance to inhibitors, but also made it possible to improve the quality of crystals and X-ray diffraction data and to increase resolution to 2.8 Å [79-83].
Ribosomal 40S and 60S subunits from the eukaryotic organism Tetrahymena thermophila were crystallized with protein factors in the Nenad Ban laboratory and their structures were determined with a resolution of 3.8 and 3.6 Å, respectively [84, 85].
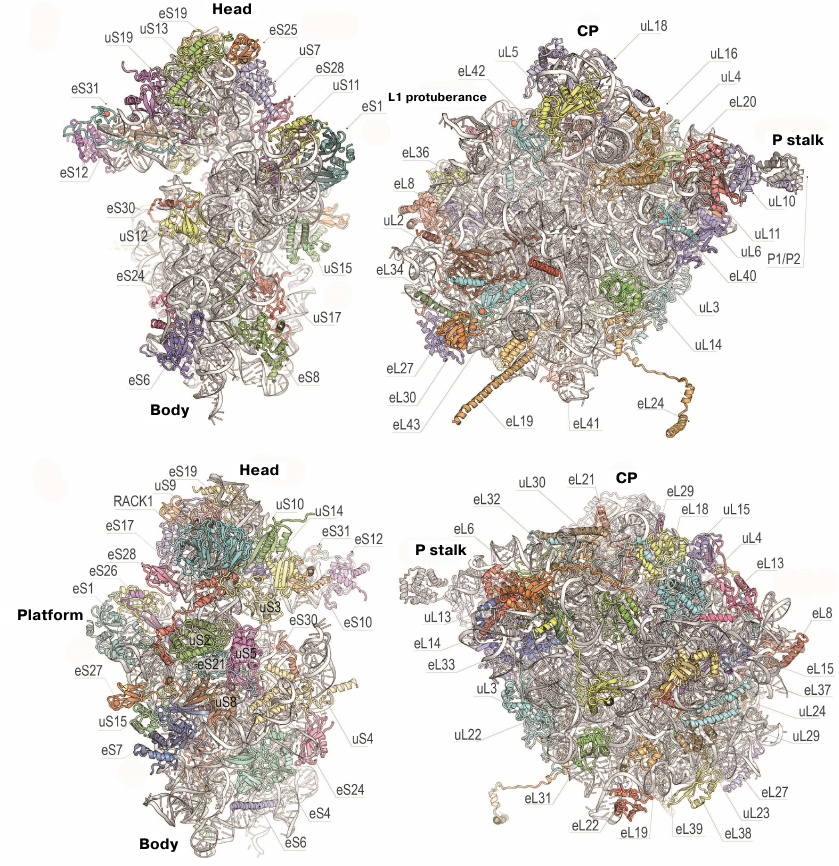
The 80S ribosome is an asymmetric complex that includes 80 different proteins and RNA molecules (Fig. 3). Each ribosomal component is present as a single copy, except proteins of the P stalk, which are present in four copies. It was also shown that bacterial and eukaryotic ribosomes have the same structural core consisting of 34 conserved proteins (15 – in the small subunit and 19 – in the large subunit) and a ~4400-nt RNA that together form the main functional centers of the ribosome, such as the DC, PTC, and tRNA-binding sites [76].
Fig. 3. Structure of yeast ribosomal 40S (left) and 60S (right) subunits. Upper and bottom panels, inner and outer surfaces of the subunits (using new protein nomenclature). Head, body, and platform of the 40S subunit and CP, L1 protuberance, and P stalk of the 60S subunit are indicated.
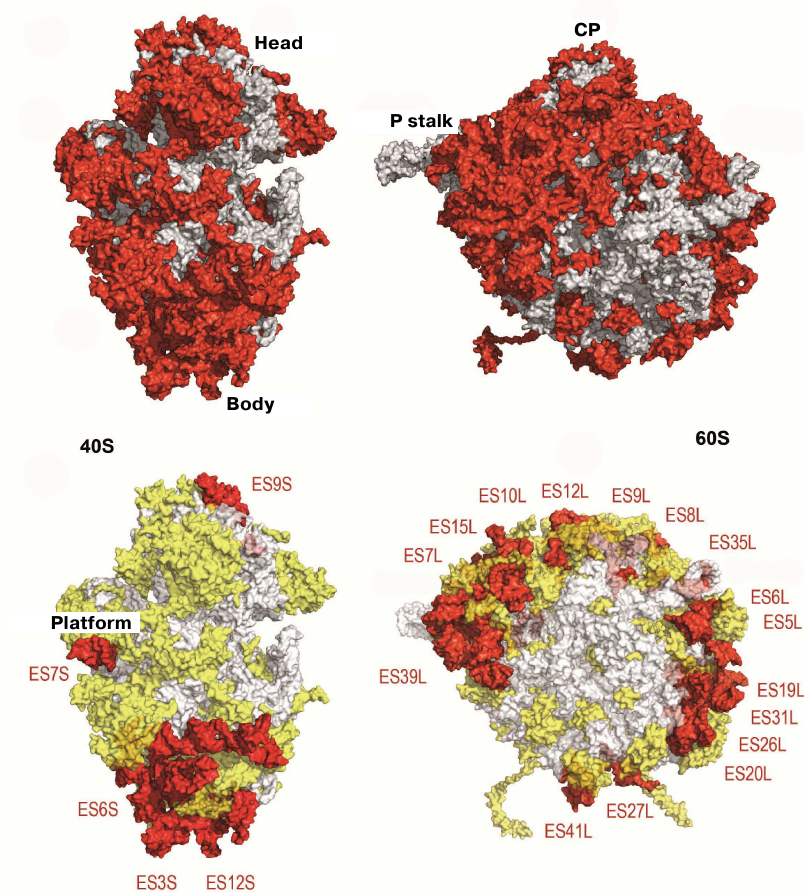
The structure of the evolutionary conserved ribosomal core could be determined by comparing the model of the yeast 80S ribosome with its bacterial analogue (Fig. 4) [34, 76]. The vast majority of the ~1.35-MDa eukaryote-specific components are located on the ribosomal surface and surround the evolutionary conserved core: In the eukaryotic ribosome, rRNA expansion segments comprise 350 kDa, eukaryote-specific domains present in the conserved proteins comprise 200 kDa, and proteins that are totally absent in bacteria comprise 800 kDa (Fig. 4)
Fig. 4. Characteristics of the yeast ribosome subunit outer surface. Upper panel, conserved subunit core (grey) and specific elements of eukaryotic ribosome (red). Bottom, eukaryote-specific protein fragments (yellow) and rRNA expansion segment (ES, red).
Yeast ribosome contains 46 eukaryote-specific proteins (18 in the 40S subunit and 28 in the 60S subunit), as well as eukaryote-specific fragments present in the majority of main proteins. rRNA contains several expansion segments in its conserved strands with a total length of over 900 nucleotides [34]. The majority of these rRNA expansion segments and protein inserts surround the core from the solvent side and are accessible for potential interactions with molecular partners, such as translation factors and chaperone proteins. The size of the 80S ribosome in different eukaryotic organisms varies in the range of 0.5-1 MDa, mostly due to the size of inserts in the expansion segments ES7L, ES15L, ES27L, and ES39L in 25S–28S rRNA [76].
The small 40S subunit has the same structural elements as the 30S prokaryotic ribosome subunit (head, body, and platform, Fig. 3). The information on the functional sites of eukaryotic ribosome was obtained by comparing its structure with the crystal structures of functional complexes of bacterial ribosomes [33, 62].
The large ribosomal subunit has a crown-like shape and includes the CP, L1 protuberance, and P stalk (Fig. 3). The tRNA-binding sites (A, P, and E) and the PTC, where formation of peptide bonds is catalyzed, are located on the inner side of the large ribosomal subunit. The PTC is adjacent to the tunnel entrance, through which the nascent polypeptide chain moves before exiting the ribosome at the solvent side.
The absence of the class-specific fragments on the subunit inner surfaces has become obvious as a result of comparison of ribosomal structures, which was in agreement with the conservatism of universal functions of ribosomes (Fig. 4). This structural conservatism is observed in the DC, PTC, and around the peptide tunnel at the solvent side, which is involved on the ribosome association with the membranes during protein synthesis. Significant differences in the structural organization of the small and large ribosomal subunits at the solvent-facing side are likely related to a significantly more complex process of translation initiation in eukaryotic cells in comparison with prokaryotes.
In conclusion, it should be mentioned that our model of the first and, at the moment only, high-resolution crystal structure of eukaryotic ribosome includes ~90.5% (~ 13,000) amino acid residues of the ribosomal proteins and ~95.5% of rRNA nucleotides (out of ~5500). This atomic model demonstrates a unique topography of the eukaryote-specific elements, as well as the pattern of their interactions with the universally conserved core, which includes all eukaryote-specific bridges between the ribosomal subunits.
NEW NOMENCLATURE OF RIBOSOMAL PROTEINS
To simplify comparison of different types of ribosomes, a new nomenclature based on the protein names was introduced (Fig. 3). Since the ribosomal proteins from E. coli were isolated and sequenced first, their archaeal and eukaryotic homologues were named accordingly. Proteins found in ribosomes from all three domains of life (Bacteria, Archaea, and Eukaryotes) have the prefix “u” (universal) followed by the name of the E. coli protein. Bacterial proteins that do not have eukaryotic homologues have the prefix “b” (bacterial) and eukaryote-specific proteins have prefix “e” (eukaryotic) [86].
EXPANSION SEGMENTS OF rRNA
rRNA expansion segments are located predominantly on the outer surface of both subunits of the eukaryotic ribosome (Figs. 4 and 5a). The interface between the ribosome subunits is highly conserved, as well as the regions of mRNA entrance and polypeptide exit tunnel; therefore, they contain almost no expansion segments and eukaryote-specific proteins. The subunits differ significantly in the spatial arrangement of the rRNA expansion segments. In particular, the major portion of the eukaryote-specific rRNAs in the small subunit are located in the bottom part, where the two large expansion segments ES3S and ES6S interact. On the contrary, numerous expansion segments in the large subunit form almost continuous ring that circles the polypeptide exit tunnel and extends further from the P stalk in the direction of the L1 protein.
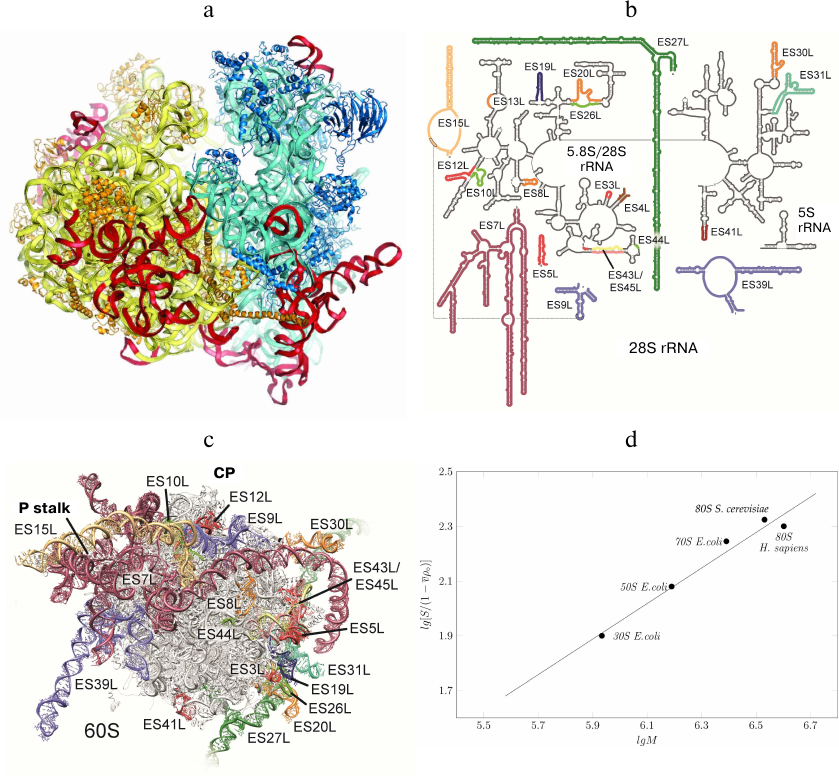
Fig. 5. rRNA expansion segments. a) Yeast ribosome with the expansion segments shown in red; the model is based on the electron density data of the X-ray diffraction analysis. b) Secondary structure of human 28S RNA; expansion fragments ES3S, ES6S, ES7L, ES15L, ES27L, ES30L, and ES39L are shown in color. Due to their unstructured nature, these fragments cannot be interpreted in the cryo-EM reconstructions. c) Molecular model of human ribosomal 60S subunit with theoretically predicted rRNA expansion segments (not verified experimentally) [87]. CP, L1 protuberance, and P stalk are indicated. The image of the human 60S subunit was kindly provided by D. Wilson. d) Dependence of the ribosomal subunit sedimentation coefficient on molecular mass (kindly provided by S. Agalarov).
Information on the expansion segments was obtained in the cryo-EM experiments with Drosophila and human 80S ribosomes [87, 88]. Suggested theoretical models of all expansion segments included 9 segments in the 40S subunit and 21 segments in the 60S subunit (Fig. 5, b and c). Expansion segments of the human 28S rRNA are similar to the yeast expansion segments, but have a significantly greater length. In particular, yeast ES3S, ES7L, ES9L, ES15L, ES27L, and ES39L segments contain 110, 200, 70, 20, 160, and 140 nucleotides, respectively, while the same segments in the human ribosome are longer by 50, 670, 40, 170, 550, and 100 nucleotides, respectively. Moreover, human rRNA expansion segments ES3S, ES6S, ES7L, ES15L, ES27L, ES30L, and ES39L cannot be interpreted in the cryo-EM reconstructions due to their unstructured nature. Hence, the secondary and tertiary rRNA structures presented in the model were calculated based on the primary structures (Fig. 5, b and c).
Comparison of the sedimentation coefficients of bacterial 50S and 30S ribosome subunits, entire 70S bacterial ribosome, and yeast and human 80S ribosomes demonstrated that human ribosome is partly unfolded (Fig. 5d), as the sedimentation coefficients for the human and yeast ribosomes were 78S and 80S, respectively. However, the molecular mass of the human ribosome is by 500,000 Da larger than that of the yeast ribosome due to the long expansion segments in the 28S RNA. Apparently, partly unfolded human ribosome expansion segments hinder crystallization of this ribosome and other structural and functional studies.
BRIDGES BETWEEN THE SUBUNITS
The bridges between the subunits are undoubtedly important, as they maintain structural integrity of the entire ribosome and facilitate communication between the small and large subunits during protein synthesis. Translocation of mRNA and tRNA, protein synthesis termination, and other processes are accompanied by the global conformational rearrangements of the ribosome in the course of translation. These conformational changes include rotation of subunits relative to each other, including the turn of the head domain in the small subunit. Thus, such rotated state was observed by us in the crystal structure of the 80S ribosome [78].
The first several eukaryote-specific bridges were revealed in the low-resolution cryo-EM structures of the yeast ribosome [89, 90]. Our model with the atomic resolution of 3.0 Å has provided more accurate and detailed information on the molecular components mediating these contacts between the subunits. The high degree of evolutionary conservatism of bridges between the subunits in the ribosomal core is especially remarkable. Seven “conserved” bridges and several bacteria- and eukaryote-specific bridges have been identified [33, 34, 51, 52], virtually all components of the additional bridges of both subunits being eukaryote-specific. It should be mentioned that unlike bacterial ribosomes, the dominating role in the formation of contacts between the subunits in eukaryotes belong to proteins [33]. Eukaryote-specific bridges are located at the periphery of the subunit interface, which significantly increases the surface area of the interactions [34, 90, 91].
Only one eukaryote-specific bridge between the subunits was found in the ribosome center – bridge eB14. It is formed by eL41, which is the smallest protein in yeast cells (25 aa) (Fig. 3) consisting of a single alpha-helix. eL41 protrudes from the 60S subunit and connects with the 40S subunit in a close vicinity of the DC. It is almost entirely submerged into the binding pocket formed by the helices h27, h45, and h44. The structure of this bridge has two notable features. First, the eL41 binding pocket in the small subunit is highly conserved across eukaryotes and bacteria. Second, eL41 has stronger connection with the 40S subunit than with the 60S subunit in the ribosome. However, despite weaker contacts with the 60S subunit, eL41 remains associated with the large subunit during ribosome dissociation. The only example of such unusual bridge has been identified in bacteria, where it is formed by the ribosomal protein of the large subunit and attaches to the small subunit via functionally significant fragments of its structure [62]. This unusual bridge is formed by the bL31 protein, which is conserved among bacteria and connects the CP of the large subunit with the labile head domain of the small subunit. The eukaryote-specific features of the ribosome include two bridges of the large subunit, eB12 and eB13, formed by the long alpha-helices of the eL19 and eL24 proteins (Fig. 3).
INHIBITION OF EUKARYOTIC RIBOSOME
Decades of studies on antibacterial agents (antibiotics) have demonstrated the variety of mechanisms of protein synthesis inhibition [92]. The atomic structures of bacterial ribosomes have become a basis for the development of novel ribosome inhibitors, which not only can be used for pharmacological purposes, but might also serve as important tools for investigating the mechanisms of protein synthesis in bacteria. Similarly, eukaryotic ribosome is the main target of eukaryote-specific inhibitors of natural origin. Despite yet poor understanding of molecular mechanisms of their action, eukaryote-specific ribosome inhibitors have found an increasing use in research studies and potentially represent novel therapeutics against a broad spectrum of infectious, oncological diseases, and genetic disorders [93–96]. Some eukaryote-specific ribosome inhibitors have been investigated using the crystals of the 50S subunit from the archaeon H. marismortui due to its close (although not total) resemblance to some fragments of the eukaryotic ribosome [97, 98].
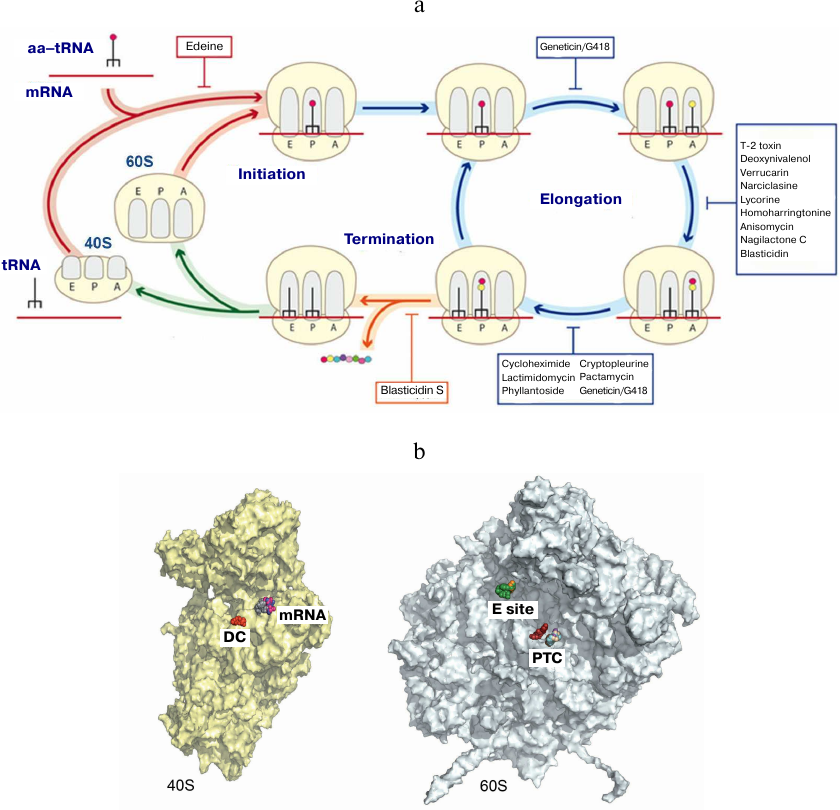
We also investigated the mechanisms of eukaryotic ribosome inhibition using X-ray diffraction analysis of 16 complexes of S. cerevisiae ribosome with eukaryote-specific inhibitors (Fig. 6) [79]. Broad-spectrum inhibitors target the PTC on the large subunit (blasticidin S), the DC (geneticin/G418), and the mRNA-tRNA binding site at the small subunit (pactamycin, edeine). It was shown that the mechanisms of protein synthesis inhibition in eukaryotes are fundamentally different from those in prokaryotes, as they involve the tRNA-binding E site that is absent in bacteria. Various inhibitors specific to eukaryotes have been described that include (i) cycloheximide, lactimidomycin, and phyllantoside interacting with the tRNA-binding E site and (ii) T-2 toxin, deoxynivalenol, verrucarin, narciclasine, lycorine, nagilactone C, anisomycin, homoharringtonine, and blasticidin S interacting with the PTC (Fig. 6). Glutarimide inhibitors cycloheximide and lactimidomycin were localized to the E site formed by the highly conserved nucleotides of the 25S rRNA and the site of the eukaryote-specific protein eL42. Among those two, lactimidomycin carries an additional lactone ring located above eL42 and directed toward the subunit interface. Despite differences in the chemical properties with glutarimides, phyllantoside forms contacts with the same rRNA nucleotides and interacts with eL42 in a manner resembling interaction with the CCA-end of tRNA. Strict selectivity of the E site inhibitors toward eukaryotes can be explained by the presence of two specific rRNA residues that form the binding pocket. Although cycloheximide and lactimidomycin bind to the same site and likely, compete with tRNA, they affect translation differently. Lactimidomycin predominantly inhibits ribosome operation during formation of the first peptide bond, while cycloheximide arrests ribosome in the process of continuing translation [99, 100].
Fig. 6. Inhibition of eukaryotic ribosome. a) Schematic representation of protein synthesis in eukaryotes with indicated inhibitor-binding sites; b) inhibitor binding sites in the functional centers at the inner surface of the small (left) and large (right) ribosomal subunits. DC, decoding center (geneticin/G418); mRNA, mRNA channel (pactamycin, cryptopleurine, edeine); E site, tRNA-binding site (cycloheximide, lactimidomycin, phyllantoside), PTC, peptidyl transferase center (blasticidin S, T-2 toxin, deoxynivalenol, verrucarin, narciclasine, lycorine, nagilactone C, anisomycin, homoharringtonine).
The PTC is formed exclusively by the rRNA nucleotides and located on the large subunit. Formation of peptide bonds requires correct alignment of the two substrates, peptidyl-tRNA and aminoacyl-tRNA, in the A and P sites, respectively. While blasticidin S binds to the P site of the large subunit in a similar way in eukaryotes, bacteria, and archaea, numerous eukaryote-specific inhibitors bind to the A site in the PTC (Fig. 4). Furthermore, the chemical nature of binding of these inhibitors inside the pocket is similar. All A site inhibitors induced similar structural rearrangements upon binding with the ribosome either in the close vicinity or at a distance (up to 15 Å) from the PTC. It was shown that the U2873 (U2504 in E. coli) and C2824 (C2452 in E. coli) nucleotides participate in the interaction with inhibitors in yeast. Different orientation of these nucleotides in bacteria occurring due to the different organization of the “secondary” and, likely, following layers of the binding pocket prevents the binding of these inhibitors to prokaryotic ribosomes [79, 101].
The DC is located on the small subunit and represents a rigid fixed pocket, where precise selection of aminoacyl-tRNA takes place according to complementary of the anticodon to the mRNA codon located at the A site. In bacteria, aminoglycoside antibiotics reduce the accuracy of translation and inhibit tRNA translocation, resulting in the disruption of nucleotide conformation in the DC. In addition to their use against Gram-negative bacteria, aminoglycosides can find application in the therapy of hereditary diseases caused by nonsense mutations due to their ability to suppress premature translation termination resulting in the peptide abortion [96, 102]. The canonic binding site for aminoglycosides is located in the upper part of the h44 helix in the 18S rRNA and contains conserved nucleotides A1755 and A1756 (A1492 and A1493 in E. coli, respectively) of the DC. The two nucleotides G1645 (A1408 in E. coli) and A1754 (G1491 in E. coli) located in a close vicinity are different in bacteria and eukaryotes, but identical in yeasts and humans. We demonstrated [79] how this difference in the DC nucleotides can explain the selectivity of some aminoglycoside antibiotics toward bacterial, but not human ribosomes [79]. We also showed that aminoglycoside antibiotics, such as paromomycin, G418, TC007, and gentamycin, can bind to the sites on the eukaryotic ribosome other than the DC [103].
Our X-ray crystallography studies have demonstrated that inhibitors such as pactamycin, cryptopleurine, and edeine bind in the mRNA channel at the E site of the small subunit and interact only with the 18S rRNA [79]. Moreover, pactamycin and edeine are broad-spectrum inhibitors. Thus, the conserved pactamycin-binding site was reported in bacteria and eukaryotes. The binding of edeine also occurs at the conserved site; however, the conformation acquired by the inhibitor in the small subunits of yeast and bacterial ribosomes is different. On the contrary, cryptopleurine was described as a eukaryote-specific inhibitor [104], although the structure of the cryptopleurine complex with the yeast ribosome provides no explanation for its specificity. It can be suggested based on the position of edeine, pactamycin, and cryptopleurine in their complexes with the ribosome that these antibiotics affect translocation of deacetylated tRNA from the P site to the E site. It is likely that they also affect translation initiation in eukaryotes. Using the previously developed model of the eukaryotic ribosome, we continue our crystallographic studies of novel inhibitors with the anticancer activity [80-83].
CONCLUSIONS
Advances in the development of new technologies in the X-ray crystallography and cryo-EM provide unique opportunities for detailed investigation of the mechanisms of ribosome functioning in various organisms, including pathogenic ones, and open new possibilities for developing novel strategies to fight pathogens such as Candida albicans, Staphylococcus aureus, and others.
Unfortunately, ribosome functioning in higher eukaryotes has been investigated insufficiently and requires further biochemical and hydrodynamic studies, including generation of crystals suitable for the X-ray diffraction analysis. The main feature of human ribosomes (molecular weight, 4.5 MDa) that distinguishes them from the ribosomes of lower eukaryotes (yeast ribosome, molecular weight 3.3 MDa) is the emergence of the rRNA extension segments located predominantly on the surface of the 60S subunit, which increases its mobility. Recent cryo-EM studies of human ribosomes have shown that most rRNA extension segments (molecular weight, ~1 MDa) are unstructured and, hence, could not be visualized by X-ray diffraction [87, 88, 105]. Therefore, we still lack the comprehensive data on the atomic structure of human ribosome.
The development of stabilization techniques that can be used for the ribosomes of higher organisms remains an open issue.
Acknowledgments. The authors are grateful to Sultan Agalarov (Institute of Protein Research, Russian Academy of Sciences) and Daniel Wilson (Hamburg University, Germany) for providing Fig. 3.
Funding. This work was supported by the Russian Science Foundation (project no. 20-65-47031).
Ethics declarations. The authors declare no conflict of interest in financial or any other sphere. This article does not contain description of studies with human participants or animals performed by any of the authors.
REFERENCES
1.Spirin, A. S. (2002) Ribosome as a molecular
machine, FEBS Lett., 514, 2-10.
2.Fourmy, D., Recht, M. I., Blanchard, S. C., and
Puglisi, J. D. (1996) Structure of the A site of Escherichia
coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic,
Science, 274, 1367-1371, doi:
10.1126/science.274.5291.1367.
3.Agalarov, S. C., Sridhar Prasad, G., Funke, P. M.,
Stout, C. D., and Williamson, J. R. (2000) Structure of the
S15,S6,S18-rRNA complex: assembly of the 30S ribosome central domain,
Science, 288, 107-113, doi:
10.1126/science.288.5463.107.
4.Ramakrishnan, V., and White, S. W. (1998) Ribosomal
protein structures: insights into the architecture, machinery and
evolution of the ribosome, Trends Biochem. Sci., 23,
208-212, doi: 10.1016/s0968-0004(98)01214-6.
5.Wimberly, B. T., White, S. W., and Ramakrishnan, V.
(1997) The structure of ribosomal protein S7 at 1.9 Å resolution
reveals a beta-hairpin motif that binds double-stranded nucleic acids,
Structure, 5, 1187-1198, doi:
10.1016/s0969-2126(97)00269-4.
6.Bocharov, E. V., Gudkov, A. T., Budovskaya, E. V.,
and Arseniev, A. S. (1998) Conformational independence of N- and
C-domains in ribosomal protein L7/L12 and in the complex with protein
L10, FEBS Lett., 423, 347-350, doi:
10.1016/s0014-5793(98)00121-5.
7.Agrawal, R. K., Penczek, P., Grassucci, R. A., Li,
Y., Leith, A., et al. (1996) Direct visualization of A-, P-, and E-site
transfer RNAs in the Escherichia coli ribosome,
Science, 271, 1000-1002, doi:
10.1126/science.271.5251.1000.
8.Svergun, D. I., Burkhardt, N., Pedersen, J. S.,
Koch, M. H., Volkov, V. V., et al. (1997) Solution scattering
structural analysis of the 70 S Escherichia coli ribosome
by contrast variation. I. Invariants and validation of electron
microscopy models, J. Mol. Biol., 271, 588-601, doi:
10.1006/jmbi.1997.1190.
9.Green, R., and Noller, H. F. (1997) Ribosomes and
translation, Annu. Rev. Biochem., 66, 679-716, doi:
10.1146/annurev.biochem.66.1.679.
10.Vasiliev, V. D. (1974) Morphology of the
ribosomal 30S subparticle according to electron microscopic data,
Acta Biol. Med. Ger., 33, 779-793.
11.Lake, J. A. (1976) Ribosome structure determined
by electron microscopy of Escherichia coli small
subunits, large subunits and monomeric ribosomes, J. Mol. Biol.,
105, 131-139.
12.Vasiliev, V. D., Selivanova, O. M., Baranov, V.
I., and Spirin, A. S. (1983) Structural study of translating 70 S
ribosomes from Escherichia coli. I. Electron microscopy,
FEBS Lett., 155, 167-172, doi:
10.1016/0014-5793(83)80232-4.
13.Frank, J. (1989) Image analysis of single
macromolecules, Electron. Microsc. Rev., 2, 53-74, doi:
10.1016/0892-0354(89)90010-5.
14.Stark, H., Mueller, F., Orlova, E. V., Schatz,
M., Dube, P., et al. (1995) The 70S Escherichia coli
ribosome at 23 Å resolution: fitting the ribosomal RNA,
Structure, 3, 815-821, doi:
10.1016/s0969-2126(01)00216-7.
15.Vasiliev, V. D., and Kotliansky, V. E. (1977) The
30 S ribosomal subparticle retains its main morphological features
after removal of half the proteins, FEBS Lett., 76,
125-128.
16.Sergiev, P., Leonov, A., Dokudovskaya, S.,
Shpanchenko, O., Dontsova, O., et al. (2001) Correlating the X-ray
structures for halo- and thermophilic ribosomal subunits with
biochemical data for the Escherichia coli ribosome,
Cold Spring Harb. Symp. Quant. Biol., 66, 87-100, doi:
10.1101/sqb.2001.66.87.
17.Graifer, D., and Karpova, G. (2015) Interaction
of tRNA with eukaryotic ribosome, Int. J. Mol. Sci., 16,
7173-7194, doi: 10.3390/ijms16047173.
18.Graifer, D., and Karpova, G. (2015) Roles of
ribosomal proteins in the functioning of translational machinery of
eukaryotes, Biochimie, 109, 1-17, doi:
10.1016/j.biochi.2014.11.016.
19.Spirin, A. S., Serdyuk, I. N., Shpungin, J. L.,
and Vasiliev, V. D. (1979) Quaternary structure of the ribosomal 30S
subunit: model and its experimental testing, Proc. Natl. Acad. Sci.
USA, 76, 4867-4871, doi: 10.1073/pnas.76.10.4867.
20.Stern, S., Powers, T., Changchien, L. M., and
Noller, H. F. (1989) RNA-protein interactions in 30S ribosomal
subunits: folding and function of 16S rRNA, Science, 244,
783-790, doi: 10.1126/science.2658053.
21.Brimacombe, R. (1991) RNA–protein
interactions in the Escherichia coli ribosome,
Biochimie, 73, 927-936, doi:
10.1016/0300-9084(91)90134-m.
22.Moore, P. B., Engelman, D. M., Langer, J. A.,
Ramakrishnan, V. R., Schindler, D. G., et al. (1984) Neutron scattering
and the 30 S ribosomal subunit of E. coli, Basic Life Sci,
27, 73-91, doi: 10.1007/978-1-4899-0375-4_4.
23.Vasiliev, V. D., Selivanova, O. M., and
Koteliansky, V. E. (1978) Specific selfpacking of the ribosomal 16 S
RNA, FEBS Lett., 95, 273-276, doi:
10.1016/0014-5793(78)81009-6.
24.Vasiliev, V. D., and Zalite, O. M. (1980)
Specific compact selfpacking of the ribosomal 23 S RNA, FEBS
Lett., 121, 101-104, doi: 10.1016/0014-5793(80)81275-0.
25.Noller, H. F. (1984) Structure of ribosomal RNA,
Annu. Rev. Biochem., 53, 119-162, doi:
10.1146/annurev.bi.53.070184.001003.
26.Woese, C. R., Gutell, R., Gupta, R., and Noller,
H. F. (1983) Detailed analysis of the higher-order structure of
16S-like ribosomal ribonucleic acids, Microbiol. Rev.,
47, 621-669.
27.Capel, M. S., Engelman, D. M., Freeborn, B. R.,
Kjeldgaard, M., Langer, J. A., et al. (1987) A complete mapping of the
proteins in the small ribosomal subunit of Escherichia
coli, Science, 238, 1403-1406, doi:
10.1126/science.3317832.
28.Stern, S., Moazed, D., and Noller, H. F. (1988)
Structural analysis of RNA using chemical and enzymatic probing
monitored by primer extension, Methods Enzymol., 164,
481-489, doi: 10.1016/s0076-6879(88)64064-x.
29.Powers, T., and Noller, H. F. (1995) A
temperature-dependent conformational rearrangement in the ribosomal
protein S4.16 S rRNA complex, J. Biol. Chem., 270,
1238-1242, doi: 10.1074/jbc.270.3.1238.
30.Brimacombe, R., Gornicki, P., Greuer, B.,
Mitchell, P., Osswald, M., et al. (1990) The three-dimensional
structure and function of Escherichia coli ribosomal RNA,
as studied by cross-linking techniques, Biochim. Biophys. Acta,
1050, 8-13, doi: 10.1016/0167-4781(90)90133-m.
31.Ban, N., Nissen, P., Hansen, J., Moore, P. B.,
and Steitz, T. A. (2000) The complete atomic structure of the large
ribosomal subunit at 2.4 Å resolution, Science,
289, 905-920, doi: 10.1126/science.289.5481.905.
32.Wimberly, B. T., Brodersen, D. E., Clemons, W.
M., Jr., Morgan-Warren, R. J., Carter, A. P., et al. (2000) Structure
of the 30S ribosomal subunit, Nature, 407, 327-339, doi:
10.1038/35030006.
33.Yusupov, M. M., Yusupova, G. Z., Baucom, A.,
Lieberman, K., Earnest, T. N., et al. (2001) Crystal structure of the
ribosome at 5.5 Å resolution, Science, 292,
883-896, doi: 10.1126/science.1060089.
34.Ben-Shem, A., Garreau de Loubresse, N., Melnikov,
S., Jenner, L., Yusupova, G., and Yusupov, M. (2011) The structure of
the eukaryotic ribosome at 3.0 Å resolution, Science,
334, 1524-1529, doi: 10.1126/science.1212642.
35.Yusupova, G., and Yusupov, M. (2014)
High-resolution structure of the eukaryotic 80S ribosome, Annu. Rev.
Biochem., 83, 467-486, doi:
10.1146/annurev-biochem-060713-035445.
36.Yusupova, G., and Yusupov, M. (2015) Ribosome
biochemistry in crystal structure determination, RNA, 21,
771-773, doi: 10.1261/rna.050039.115.
37.Jenner, L., Melnikov, S., Garreau de Loubresse,
N., Ben-Shem, A., Iskakova, M., et al. (2012) Crystal structure of the
80S yeast ribosome, Curr. Opin. Struct. Biol., 22,
759-767, doi: 10.1016/j.sbi.2012.07.013.
38.Demeshkina, N., Jenner, L., Yusupova, G., and
Yusupov, M. (2010) Interactions of the ribosome with mRNA and tRNA,
Curr. Opin. Struct. Biol., 20, 325-332, doi:
10.1016/j.sbi.2010.03.002.
39.Yonath, A., Mussig, J., and Wittmann, H. G.
(1982) Parameters for crystal growth of ribosomal subunits, J. Cell
Biochem., 19, 145-155, doi: 10.1002/jcb.240190205.
40.Makowski, I., Frolow, F., Saper, M. A., Shoham,
M., Wittmann, H. G., and Yonath, A. (1987) Single crystals of large
ribosomal particles from Halobacterium marismortui diffract to 6
Å, J. Mol. Biol., 193, 819-822, doi:
10.1016/0022-2836(87)90362-7.
41.Von Bohlen, K., Makowski, I., Hansen, H. A.,
Bartels, H., Berkovitch-Yellin, Z., et al. (1991) Characterization and
preliminary attempts for derivatization of crystals of large ribosomal
subunits from Haloarcula marismortui diffracting to 3 Å
resolution, J. Mol. Biol., 222, 11-15, doi:
10.1016/0022-2836(91)90730-t.
42.Trakhanov, S., Yusupov, M., Shirokov, V., Garber,
M., Mitschler, A., et al. (1989) Preliminary X-ray investigation of 70
S ribosome crystals from Thermus thermophilus, J. Mol.
Biol., 209, 327-328, doi: 10.1016/0022-2836(89)90282-9.
43.Yusupova, G., Yusupov, M., Spirin, A., Ebel, J.
P., Moras, D., Ehresmann, C., and Ehresmann, B. (1991) Formation and
crystallization of Thermus thermophilus 70S ribosome/tRNA
complexes, FEBS Lett., 290, 69-72, doi:
10.1016/0014-5793(91)81228-z.
44.Yusupov, M. M., Garber, M. B., Vasiliev, V. D.,
and Spirin, A. S. (1991) Thermus thermophilus ribosomes for
crystallographic studies, Biochimie, 73, 887-897, doi:
10.1016/0300-9084(91)90130-s.
45.Schluenzen, F., Tocilj, A., Zarivach, R., Harms,
J., Gluehmann, M., et al. (2000) Structure of functionally activated
small ribosomal subunit at 3.3 angstroms resolution, Cell,
102, 615-623, doi: 10.1016/s0092-8674(00)00084-2.
46.Cate, J. H., Yusupov, M. M., Yusupova, G. Z.,
Earnest, T. N., and Noller, H. F. (1999) X-ray crystal structures of
70S ribosome functional complexes, Science, 285,
2095-2104, doi: 10.1126/science.285.5436.2095.
47.Yusupova, G. Z., Yusupov, M. M., Cate, J. H., and
Noller, H. F. (2001) The path of messenger RNA through the ribosome,
Cell, 106, 233-241, doi:
10.1016/s0092-8674(01)00435-4.
48.Flygaard, R. K., Boegholm, N., Yusupov, M., and
Jenner, L. B. (2018) Cryo-EM structure of the hibernating
Thermus thermophilus 100S ribosome reveals a
protein-mediated dimerization mechanism, Nat. Commun., 9,
4179, doi: 10.1038/s41467-018-06724-x.
49.Rozov, A., Khusainov, I., El Omari, K., Duman,
R., Mykhaylyk, V., et al. (2019) Importance of potassium ions for
ribosome structure and function revealed by long-wavelength X-ray
diffraction, Nat. Commun., 10, 2519, doi:
10.1038/s41467-019-10409-4.
50.Hussain, T., Llacer, J. L., Wimberly, B. T.,
Kieft, J. S., and Ramakrishnan, V. (2016) Large-scale movements of IF3
and tRNA during bacterial translation initiation, Cell,
167, 133-144.e113, doi: 10.1016/j.cell.2016.08.074.
51.Selmer, M., Dunham, C. M., Murphy, F. V.,
Weixlbaumer, A., Petry, S., et al. (2006) Structure of the 70S ribosome
complexed with mRNA and tRNA, Science, 313, 1935-1942,
doi: 10.1126/science.1131127.
52.Schuwirth, B. S., Borovinskaya, M. A., Hau, C.
W., Zhang, W., Vila-Sanjurjo, A., et al. (2005) Structures of the
bacterial ribosome at 3.5 Å resolution, Science,
310, 827-834, doi: 10.1126/science.1117230.
53.Yusupov, M. M., Trakhanov, S. D., Barinin, V. V.,
Boroviagin, B. D., Garber, M. B., et al. (1987) Crystallization of the
30S subunits of Thermus thermophilus ribosomes, Dokl.
Akad. Nauk SSSR, 292, 1271-1274.
54.Trakhanov, S. D., Yusupov, M., Agalarov, S.,
Garber, M., Ryazantzev, S., et al. (1987) Crystallization of 70S
ribosomes and 30S ribosomal subunits from Thermus
thermophilus, FEBS Lett., 220, 319-322.
55.Harel, M., Shoham, M., Frolow, F., Eisenberg, H.,
Mevarech, M., et al. (1988) Crystallization of halophilic malate
dehydrogenase from Halobacterium marismortui, J. Mol.
Biol., 200, 609-610, doi: 10.1016/0022-2836(88)90547-5.
56.Ban, N., Freeborn, B., Nissen, P., Penczek, P.,
Grassucci, R. A., et al. (1998) A 9 Å resolution X-ray
crystallographic map of the large ribosomal subunit, Cell,
93, 1105-1115, doi: 10.1016/s0092-8674(00)81455-5.
57.Nissen, P., Hansen, J., Ban, N., Moore, P. B.,
and Steitz, T. A. (2000) The structural basis of ribosome activity in
peptide bond synthesis, Science, 289, 920-930, doi:
10.1126/science.289.5481.920.
58.Ogle, J. M., Brodersen, D. E., Clemons, W. M.,
Jr., Tarry, M. J., Carter, A. P., and Ramakrishnan, V. (2001)
Recognition of cognate transfer RNA by the 30S ribosomal subunit,
Science, 292, 897-902, doi: 10.1126/science. 1060612.
59.Frank, J., Penczek, P., Grassucci, R., and
Srivastava, S. (1991) Three-dimensional reconstruction of the 70S
Escherichia coli ribosome in ice: the distribution of
ribosomal RNA, J. Cell Biol., 115, 597-605.
60.Yusupov, M. M., and Spirin, A. S. (1986) Are
there proteins between the ribosomal subunits? Hot tritium bombardment
experiments, FEBS Lett., 197, 229-233, doi:
10.1016/0014-5793(86)80332-5.
61.Jenner, L., Demeshkina, N., Yusupova, G., and
Yusupov, M. (2010) Structural rearrangements of the ribosome at the
tRNA proofreading step, Nat. Struct. Mol. Biol., 17,
1072-1078, doi: 10.1038/nsmb.1880.
62.Jenner, L. B., Demeshkina, N., Yusupova, G., and
Yusupov, M. (2010) Structural aspects of messenger RNA reading frame
maintenance by the ribosome, Nat. Struct. Mol. Biol., 17,
555-560, doi: 10.1038/nsmb.1790.
63.Yusupova, G., Jenner, L., Rees, B., Moras, D.,
and Yusupov, M. (2006) Structural basis for messenger RNA movement on
the ribosome, Nature, 444, 391-394, doi:
10.1038/nature05281.
64.Jenner, L., Romby, P., Rees, B., Schulze-Briese,
C., Springer, M., et al. (2005) Translational operator of mRNA on the
ribosome: how repressor proteins exclude ribosome binding,
Science, 308, 120-123, doi: 10.1126/science.1105639.
65.Jenner, L., Rees, B., Yusupov, M., and Yusupova,
G. (2007) Messenger RNA conformations in the ribosomal E site revealed
by X-ray crystallography, EMBO Rep., 8, 846-850, doi:
10.1038/sj.embor.7401044.
66.Demeshkina, N., Jenner, L., Westhof, E., Yusupov,
M., and Yusupova, G. (2012) A new understanding of the decoding
principle on the ribosome, Nature, 484, 256-259, doi:
10.1038/nature10913.
67.Demeshkina, N., Jenner, L., Westhof, E., Yusupov,
M., and Yusupova, G. (2013) New structural insights into the decoding
mechanism: translation infidelity via a G.U pair with
Watson–Crick geometry, FEBS Lett., 587, 1848-1857,
doi: 10.1016/j.febslet.2013.05.009.
68.Rozov, A., Demeshkina, N., Westhof, E., Yusupov,
M., and Yusupova, G. (2015) Structural insights into the translational
infidelity mechanism, Nat. Commun., 6, 7251, doi:
10.1038/ncomms8251.
69.Rozov, A., Westhof, E., Yusupov, M., and
Yusupova, G. (2016) The ribosome prohibits the G*U wobble geometry at
the first position of the codon-anticodon helix, Nucleic Acids
Res., 44, 6434-6441, doi: 10.1093/nar/gkw431.
70.Rozov, A., Demeshkina, N., Khusainov, I.,
Westhof, E., Yusupov, M., and Yusupova, G. (2016) Novel base-pairing
interactions at the tRNA wobble position crucial for accurate reading
of the genetic code, Nat. Commun., 7, 10457, doi:
10.1038/ncomms10457.
71.Rozov, A., Wolff, P., Grosjean, H., Yusupov, M.,
Yusupova, G., and Westhof, E. (2018) Tautomeric G*U pairs within the
molecular ribosomal grip and fidelity of decoding in bacteria,
Nucleic Acids Res., 46, 7425-7435, doi:
10.1093/nar/gky547.
72.Voorhees, R. M., and Ramakrishnan, V. (2013)
Structural basis of the translational elongation cycle, Annu. Rev.
Biochem., 82, 203-236, doi:
10.1146/annurev-biochem-113009-092313.
73.Schmeing, T. M., and Ramakrishnan, V. (2009) What
recent ribosome structures have revealed about the mechanism of
translation, Nature, 461, 1234-1242, doi:
10.1038/nature08403.
74.Rozov, A., Demeshkina, N., Westhof, E., Yusupov,
M., and Yusupova, G. (2016) New structural insights into translational
miscoding, Trends Biochem. Sci., 41, 798-814, doi:
10.1016/j.tibs.2016.06.001.
75.Noller, H. F., Yusupov, M. M., Yusupova, G. Z.,
Baucom, A., and Cate, J. H. (2002) Translocation of tRNA during protein
synthesis, FEBS Lett., 514, 11-16.
76.Melnikov, S., Ben-Shem, A., Garreau de Loubresse,
N., Jenner, L., Yusupova, G., and Yusupov, M. (2012) One core, two
shells: bacterial and eukaryotic ribosomes, Nat. Struct. Mol.
Biol., 19, 560-567, doi: 10.1038/nsmb.2313.
77.Milligan, R. A., and Unwin, P. N. (1982) In
vitro crystallization of ribosomes from chick embryos, J. Cell
Biol., 95, 648-653, doi: 10.1083/jcb.95.2.648.
78.Ben-Shem, A., Jenner, L., Yusupova, G., and
Yusupov, M. (2010) Crystal structure of the eukaryotic ribosome,
Science, 330, 1203-1209, doi:
10.1126/science.1194294.
79.Garreau de Loubresse, N., Prokhorova, I.,
Holtkamp, W., Rodnina, M. V., Yusupova, G., and Yusupov, M. (2014)
Structural basis for the inhibition of the eukaryotic ribosome,
Nature, 513, 517-522, doi: 10.1038/nature13737.
80.Pellegrino, S., Meyer, M., Konst, Z. A., Holm,
M., Voora, V. K., et al. (2019) Understanding the role of
intermolecular interactions between lissoclimides and the eukaryotic
ribosome, Nucleic Acids Res., 47, 3223-3232, doi:
10.1093/nar/gkz053.
81.Pellegrino, S., Meyer, M., Zorbas, C., Bouchta,
S. A., Saraf, K., et al. (2018) The Amaryllidaceae alkaloid
haemanthamine binds the eukaryotic ribosome to repress cancer cell
growth, Structure, 26, 416-425.e414, doi:
10.1016/j.str.2018.01.009.
82.Konst, Z. A., Szklarski, A. R., Pellegrino, S.,
Michalak, S. E., Meyer, M., et al. (2017) Synthesis facilitates an
understanding of the structural basis for translation inhibition by the
lissoclimides, Nat. Chem., 9, 1140-1149, doi:
10.1038/nchem.2800.
83.McClary, B., Zinshteyn, B., Meyer, M., Jouanneau,
M., Pellegrino, S., et al. (2017) Inhibition of eukaryotic translation
by the antitumor natural product agelastatin A, Cell Chem.
Biol., 24, 605-613 e605, doi:
10.1016/j.chembiol.2017.04.006.
84.Klinge, S., Voigts-Hoffmann, F., Leibundgut, M.,
Arpagaus, S., and Ban, N. (2011) Crystal structure of the eukaryotic
60S ribosomal subunit in complex with initiation factor 6,
Science, 334, 941-948, doi: 10.1126/science.1211204.
85.Rabl, J., Leibundgut, M., Ataide, S. F., Haag,
A., and Ban, N. (2011) Crystal structure of the eukaryotic 40S
ribosomal subunit in complex with initiation factor 1, Science,
331, 730-736, doi: 10.1126/science.1198308.
86.Ban, N., Beckmann, R., Cate, J. H., Dinman, J.
D., Dragon, F., et al. (2014) A new system for naming ribosomal
proteins, Curr. Opin. Struct. Biol., 24, 165-169, doi:
10.1016/j.sbi.2014.01.002.
87.Armache, J. P., Jarasch, A., Anger, A. M., Villa,
E., Becker, T., et al. (2010) Cryo-EM structure and rRNA model of a
translating eukaryotic 80S ribosome at 5.5 Å resolution, Proc.
Natl. Acad. Sci. USA, 107, 19748-19753, doi:
10.1073/pnas.1009999107.
88.Armache, J. P., Jarasch, A., Anger, A. M., Villa,
E., Becker, T., et al. (2010) Localization of eukaryote-specific
ribosomal proteins in a 5.5 Å cryo-EM map of the 80S eukaryotic
ribosome, Proc. Natl. Acad. Sci. USA, 107, 19754-19759,
doi: 10.1073/pnas.1010005107.
89.Spahn, C. M., Beckmann, R., Eswar, N., Penczek,
P. A., Sali, A., et al. (2001) Structure of the 80S ribosome from
Saccharomyces cerevisiae – tRNA-ribosome and
subunit–subunit interactions, Cell, 107, 373-386,
doi: 10.1016/s0092-8674(01)00539-6.
90.Spahn, C. M., Gomez-Lorenzo, M. G., Grassucci, R.
A., Jorgensen, R., Andersen, G. R., et al. (2004) Domain movements of
elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA
translocation, EMBO J., 23, 1008-1019, doi:
10.1038/sj.emboj.7600102.
91.Budkevich, T., Giesebrecht, J., Altman, R. B.,
Munro, J. B., Mielke, T., et al. (2011) Structure and dynamics of the
mammalian ribosomal pretranslocation complex, Mol. Cell,
44, 214-224, doi: 10.1016/j.molcel.2011.07.040.
92.Wilson, D. N. (2014) Ribosome-targeting
antibiotics and mechanisms of bacterial resistance, Nat. Rev.
Microbiol., 12, 35-48, doi: 10.1038/nrmicro3155.
93.Hobbie, S. N., Kaiser, M., Schmidt, S.,
Shcherbakov, D., Janusic, T., et al. (2011) Genetic reconstruction of
protozoan rRNA decoding sites provides a rationale for paromomycin
activity against Leishmania and Trypanosoma, PLoS Negl. Trop.
Dis., 5, e1161, doi: 10.1371/journal.pntd.0001161.
94.Lu, W., Roongsawang, N., and Mahmud, T. (2011)
Biosynthetic studies and genetic engineering of pactamycin analogs with
improved selectivity toward malarial parasites, Chem. Biol.,
18, 425-431, doi: 10.1016/j.chembiol.2011.01.016.
95.Santagata, S., Mendillo, M. L., Tang, Y. C.,
Subramanian, A., Perley, C. C., et al. (2013) Tight coordination of
protein translation and HSF1 activation supports the anabolic malignant
state, Science, 341, 1238303, doi:
10.1126/science.1238303.
96.Bidou, L., Allamand, V., Rousset, J. P., and
Namy, O. (2012) Sense from nonsense: therapies for premature stop codon
diseases, Trends Mol. Med., 18, 679-688, doi:
10.1016/j.molmed.2012.09.008.
97.Gurel, G., Blaha, G., Steitz, T. A., and Moore,
P. B. (2009) Structures of triacetyloleandomycin and mycalamide A bind
to the large ribosomal subunit of Haloarcula marismortui,
Antimicrob. Agents Chemother., 53, 5010-5014, doi:
10.1128/AAC.00817-09.
98.Gurel, G., Blaha, G., Moore, P. B., and Steitz,
T. A. (2009) U2504 determines the species specificity of the A-site
cleft antibiotics: the structures of tiamulin, homoharringtonine, and
bruceantin bound to the ribosome, J. Mol. Biol., 389,
146-156, doi: 10.1016/j.jmb.2009.04.005.
99.Schneider-Poetsch, T., Ju, J., Eyler, D. E.,
Dang, Y., Bhat, S., et al. (2010) Inhibition of eukaryotic translation
elongation by cycloheximide and lactimidomycin, Nat. Chem.
Biol., 6, 209-217, doi: 10.1038/nchembio.304.
100.Ingolia, N. T., Ghaemmaghami, S., Newman, J.
R., and Weissman, J. S. (2009) Genome-wide analysis in vivo of
translation with nucleotide resolution using ribosome profiling,
Science, 324, 218-223, doi: 10.1126/science.1168978.
101.Mailliot, J., Garreau de Loubresse, N.,
Yusupova, G., Meskauskas, A., Dinman, J. D., and Yusupov, M. (2016)
Crystal structures of the uL3 mutant ribosome: illustration of the
importance of ribosomal proteins for translation efficiency, J. Mol.
Biol., 428, 2195-2202, doi: 10.1016/j.jmb.2016.02.013.
102.Shulman, E., Belakhov, V., Wei, G., Kendall,
A., Meyron-Holtz, E. G., et al. (2014) Designer aminoglycosides that
selectively inhibit cytoplasmic rather than mitochondrial ribosomes
show decreased ototoxicity: a strategy for the treatment of genetic
diseases, J. Biol. Chem., 289, 2318-2330, doi:
10.1074/jbc.M113.533588.
103.Prokhorova, I., Altman, R. B., Djumagulov, M.,
Shrestha, J. P., Urzhumtsev, A., et al. (2017) Aminoglycoside
interactions and impacts on the eukaryotic ribosome, Proc. Natl.
Acad. Sci. USA, 114, E10899-E10908, doi:
10.1073/pnas.1715501114.
104.Dolz, H., Vazquez, D., and Jimenez, A. (1982)
Quantitation of the specific interaction of [14a-3H]cryptopleurine with
80S and 40S ribosomal species from the yeast Saccharomyces
cerevisiae, Biochemistry, 21, 3181-3187, doi:
10.1021/bi00256a023.
105.Khatter, H., Myasnikov, A. G., Natchiar, S. K.,
and Klaholz, B. P. (2015) Structure of the human 80S ribosome,
Nature, 520, 640-645, doi: 10.1038/nature14427.