REVIEW: Features and Functions of the A-Minor Motif, the Most Common Motif in RNA Structure
Eugene F. Baulin1,2
1Institute of Mathematical Problems of Biology RAS – the Branch of Keldysh Institute of Applied Mathematics of the Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russia2Moscow Institute of Physics and Technology, 141701 Dolgoprudny, Moscow Region, Russia
* To whom correspondence should be addressed.
Received May 5, 2021; Revised June 13, 2021; Accepted June 13, 2021
A-minor motifs are RNA tertiary structure motifs that generally involve a canonical base pair and an adenine base forming hydrogen bonds with the minor groove of the base pair. Such motifs are among the most common tertiary interactions in known RNA structures, comparable in number with the non-canonical base pairs. They are often found in functionally important regions of non-coding RNAs and, in particular, play a central role in protein synthesis. Here, we review local variations of the A-minor geometry and discuss difficulties associated with their annotation, as well as various structural contexts and common A-minor co-motifs, and diverse functions of A-minors in various processes in a living cell.
KEY WORDS: RNA structure, tertiary motif, A-minor interaction, A-minor motif, A-patchDOI: 10.1134/S000629792108006X
INTRODUCTION
It is now widely accepted that non-coding RNAs are the major players in a variety of molecular processes in living organisms [1]. The function of an RNA molecule strongly depends on its spatial structure formed by the secondary structure elements, such as stems (short helical regions of consecutive canonical base pairs) and loops, as well as their tertiary interactions [2]. These interactions often form recurrent modules termed tertiary motifs, that retain their configuration in various structural contexts [3]. Among the most widespread and functionally important RNA tertiary motifs are A-minor motifs [4].
The A-minor motifs have been originally identified in 1994-1996 in the crystal structures of the group I intron [5, 6] and hammerhead ribozyme [7]. The authors of these early works reported the presence of highly conserved adenines forming hydrogen bonds with the minor grooves of distant canonical base pairs.
In 2001, with the newly resolved structures of ribosomal RNAs, the volume of data on the RNA structures increased nearly ten times, which has led to the identification of numerous recurrent interactions [8]. Ribosomal RNA structures have been shown to involve unexpectedly high number of unpaired adenine bases, a notable portion of which participated in the tertiary interactions and were conserved among all kingdoms of living organisms [4, 8]. Nissen et al. [4] coined the term “A-minor motif” and listed as many as 186 A-minors found in the structure of the 50S ribosomal subunit. They demonstrated that A-minors of the 23S rRNA mediate all possible combinations of helices and loops, such as loop-loop, helix-helix, and loop-helix interactions [4]. In the same year, Doherty et al. [9] suggested that A-minor interactions represent a universal mode of helix packing shared by the hepatitis delta virus ribozyme, 23S rRNA, group I intron, and the hammerhead ribozyme.
In 2002, biochemical methods for the identification of A-minor interactions using adenosine analogs were introduced [10]. In particular, this approach has allowed to demonstrate that the group I intron employs two A-minors for positioning the 5′-exon within the catalytic core [10].
Later, A-minor interactions have been found in a variety of non-coding RNA molecules, including ribonuclease P [11, 12], signal recognition particle RNA [13], viral RNA [14, 15], Pistol ribozyme [16], precursor transfer RNA (pre-tRNA) [17], various riboswitches [18-20], and metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) long non-coding RNA [21].
A-minors are regularly identified as constituents of larger motifs, such as kink-turns [22, 23], GNRA-tetraloop/tetraloop-receptors [24-26], pseudoknots [14, 27, 28], coaxial stacking [29-33], ribose zippers [30, 34], lonepair triloops [35], and adenosine wedges [36].
In this review, we discuss different definitions of the A-minor interaction and A-minor motif and list their geometric types. Next, we present A-minor structural features, as well as databases and annotation software. We also explore the most common and functionally important A-minors with a special focus on the A-minors involved in the ribosome functioning. In the discussion, we review general A-minor attributes and current difficulties in the annotation of A-minors.
WHAT IS AN A-MINOR?
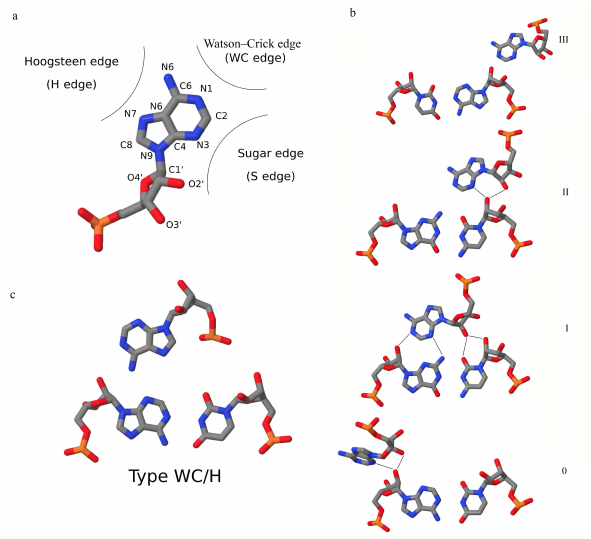
Before we discuss definitions of an A-minor, we have to mention a commonly accepted nomenclature of RNA base pairs known as the Leontis-Westhof (LW) classification [37]. According to this classification, each nucleobase can form hydrogen bonds via one of its three edges: the Hoogsteen edge (H edge), the Watson–Crick edge (WC edge), and the Sugar edge (S edge) (Fig. 1a). The S edge, along with the base atoms, also involves the ribose O2′ atom. Each base pair can be attributed to the three-letter LW geometric type, where two letters denote interacting base edges (H/S/W) and the third letter (c/t) denotes relative orientation of the glycosidic bonds (cis or trans, respectively). For example, canonical Watson–Crick (WC) A-U and G-C pairs, as well as the wobble G-U pairs, are attributed to the cWW type, i.e., the bases interact via their WC edges with the glycosidic bonds being in the cis orientation. In RNA helical regions consisting of consecutive canonical base pairs, the S edges form the minor groove of the helix, and the major groove is formed by the H edges of the paired bases.
Fig. 1. a) Adenine and its three base edges: Hoogsteen edge, Watson–Crick edge, and Sugar edge. The Sugar edge also involves the ribose O2′ atom. b) Four types of A-minor interactions introduced in [4]. Types I and II are considered canonical A-minors. c) Example of a non-canonical WC/H A-minor (PDB 1LPW); residues U4 and A6 of chain A and A6 residue of chain B. Adenines of the WC/H A-minors interact with the base pair via their Watson–Crick or Hoogsteen edges.
In 2001, Nissen et al. introduced the term “A-minor motif” to describe recurrent interactions in the ribosomal RNA that involve an insertion of the smooth N1-C2-N3 face of adenine into the minor groove of the Watson–Crick helices, preferentially at the C-G base pairs (Fig. 1b) [4]. Later, other definitions for the A-minor motif or A-minor interaction have been suggested that we will discuss below. Hereinafter, we use the term “A-minor interaction”, or “A-minor”, for the three-nucleotide interaction, and the term “A-minor motif” – for a group of stacked A-minors.
A-minor interaction. Nissen et al. [4] used the following criteria to annotate A-minor interactions: (i) adenine C2 atom have to face the minor groove of the base pair, (ii) at least one of the base pair atoms has to lie within 3.7 Å of the adenine C2 atom, and (iii) the atom of the base pair closest to the adenine C2 atom has to lie within 45° of the adenine plane. The four geometric types of A-minors introduced in [4] are defined by the positions of the O2′ and N3 atoms of the adenosine base relative to the interacting base pair (Fig. 1b). In type I, both N3 and O2′ atoms of adenine lie between the base pair O2′ atoms. In type II, the O2′ atom of adenine lies outside the base pair minor groove, while the N3 atom lies inside it. In type III, entire adenosine base lies outside the minor groove of the base pair with the N3 atom facing the O2′ atom of the closest strand. In type 0 A-minor, only the adenine ribose atoms are located between the O2′ atoms of the base pair. Nissen et al. [4] also stated that the strands of the interacting adenine and the closest base of the receptor base pair are always antiparallel. Type I and type II A-minors are specific to adenine due to its smooth Sugar edge with the N1, N3, and O2′ atoms available for the hydrogen bond formation. Type 0 and III A-minors are less specific, but still prefer adenines [4].
In recent work [38], Torabi et al. introduced the WC/H subclass of A-minor interactions, in which adenines face the minor groove of the receptor base pairs with their WC or H edges instead of the S edge (Fig. 1c). Representatives of this novel subclass had been found before in different RNA molecules [16, 39], but had not been attributed to a separate subclass.
A-minor motif. The A-minor interactions tend to stack forming the so-called A-patches [4]. An A-patch rarely exceeds three A-minors in length, and the A-minors commonly decrease in type order in the 5′-to-3′ direction, generating types II,I or III,II,I A-patches. Larger A-patches are usually formed with two strands of adenines and involve a cross-strand adenine stack; their A-minors’ types increase to the ends (e.g., II,I,I,II). A-patches consisting of two consecutive adenines and two consecutive WC base pairs have been designated in [40] as sextuples, RNA tertiary motifs involving six bases that form a network of hydrogen bonds.
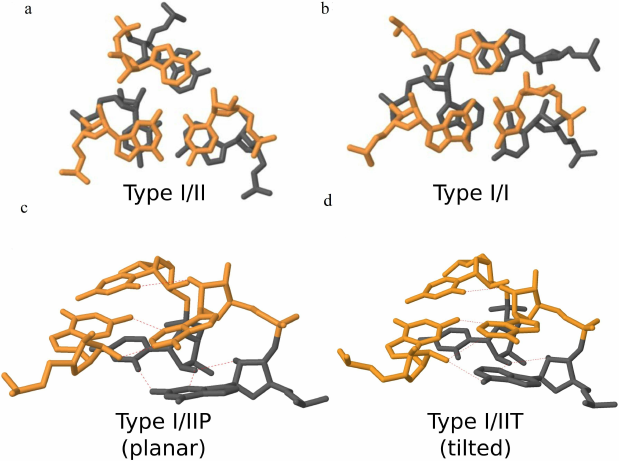
Lescoute et al. [41] defined the type I/II A-minor motif as an assembly of two consecutive adenines forming type II and type I A-minor interactions with two consecutive WC base pairs (Fig. 2a). The authors emphasized that the type II A-minor interaction is formed by the 5′-closest A residue, whereas the 3′-closest A residue interacts with both strands of the receptor helix, forming type I A-minor interaction [41]. Less frequent type I/I A-minor motif [42] is formed by two type I A-minors whose adenines form a cross-strand adenine stack (Fig. 2b).
Fig. 2. a) Type I/II A-minor motif formed by a stack of consecutive adenines (PDB 6QZP); residues G673, C674, A996, A997, (A2M)1031, C1032 of chain S2. b) Type I/I A-minor motif formed by a cross-strand adenine stack (PDB 5TBW); residues U2629, C2630, G2648, A2649, A2696, A2758 of chain 1. c) Type I/IIP (planar) A-minor motif (PDB 2GCV); residues G31, G116, A117 of chain B and residues C10, U11 of chain A. d) Type I/IIT (tilted) A-minor motif (PDB 1HR2); residues A152, A153, C223, U224, G250 of chain B.
Geary et al. [24, 33] distinguished two subclasses of type I/II A-minor motif, namely type I/IIP (planar, Fig. 2c) and type I/IIT (tilted, Fig. 2d). Type I/IIP predominantly includes loop/helix interactions, and type I/IIT usually represents loop/receptor interactions, in which the receptor provides additional stabilizing interactions.
A-MINOR ANNOTATION AND FEATURES
In 2008, Xin et al. [30] showed that A-minors are the most abundant type of RNA tertiary motifs (37%), followed by coaxial stacking (32%) and ribose zippers (20%). Furthermore, 73% of ribose zippers involve at least one adenine forming an A-minor interaction. The analyzed dataset of 229 A-minors consisted of interactions annotated with FR3D [43] in 54 high-resolution crystal structures of non-coding RNAs and included 52% type I interactions, 31% type II interactions, 10% type 0 interactions, and 7% type III interactions. Out of 229 A-minors, 18% receptor base pairs were WC G-C base pairs, 62% – WC C-G base pairs, 5% – WC A-U base pairs, and 9% – WC U-A base pairs. Xin et al. [30] demonstrated that the receptor base pairs in A-minor interactions tend to be located at the end of the helical stems, near the interface with an adjacent hairpin, internal loop, or junction loop. The dataset also contained a few unusual examples of A-minors, e.g., a U base forming type I interaction with a G-C pair, and an A base forming type I interaction with a noncanonical cWW A-A base pair.
Sheth et al. [44] introduced 11 geometric parameters that can be used to annotate A-minors of types I,II,0, and III. The parameters have been derived using the combined dataset of 260 A-minors analyzed in [30, 45].
At present, there are several databases with the information on the A-minor interactions annotated in known RNA 3D structures from the Protein Data Bank (PDB [46]). The CaRNAval database [47] stores long-range interaction modules called RINs (RNA interaction networks), including type I A-minor interactions (194 occurrences) and type I/II A-minor motifs (102 occurrences). One of the advantages of the CaRNAval pipeline is sequence independence, as RINs are defined solely based on the geometry of interacting base pairs. Thus, type I A-minors should contain a base involved in two cSS/tSS base pairs with the bases of the receptor cWW base pair, while type II A-minor of the type I/II A-minor motif has to be formed with a single cSS base pair. This approach allowed Reinharz et al. [47] to annotate one type I G-minor, 21 type II G-minors, and one type II U-minor. Another database on A-minors is the InterRNA database [48], which lists A-minors annotated by the NASSAM and COGNAC programs [49, 50] and includes type I and II A-minors, as well as type II G-minors.
Among the most popular programs used for the annotation of A-minors are FR3D [43] and DSSR [51]. The FR3D program [43] is able to find instances of a given motif template or a symbolic pattern of interactions in RNA 3D structures. In particular, it was successfully used to annotate type I,II,0, and III A-minors [30] and type I and II A-minors [47]. FR3D allows to identify A-minors formed by bases other than adenine, but is limited to unmodified nucleotides with no missing atoms. The DSSR program [51] annotates type I,II, and X A-minors, among various RNA tertiary motifs and interactions. Type X (eXtended) is defined as a noncanonical type, in which adenine forms hydrogen bonds with the minor groove of the receptor base pair via its WC or H edges, without involving the O2′ atom. Although A-minors annotated with DSSR are limited to adenosine bases by default, DSSR provides an additional option “--a-minor=BASE” where BASE can be any character in ACGTURYN (Xiang-Jun Lu, personal communication). Some A-minors can be annotated using the base pair annotation programs [52-54] followed by subsequent data processing.
LOCAL A-MINORS
A-minors can be divided into two large groups, namely: local and long-range interactions. We consider an A-minor to be a local interaction if either its three nucleotides lie within a loop of any type (hairpin/internal loop/bulge/junction loop) or its adenine lies within a loop and the receptor base pair is a part of the stem adjacent to this loop. If the adenine and the base pair are from distant secondary structure elements, A-minor belongs to the group of long-range interactions. Among the A-minors found in known RNA spatial structures, 40% are local A-minors and 60% are long-range A-minors [55].
A-minors and kink-turns. More than a half of A-minors located within internal loops in the known RNA 3D structures are elements of the kink-turn motifs [55], which are asymmetric internal loops with a sharp kink in the phosphodiester bond [22]. A kink-turn involves a type I A-minor interaction formed by adenine from one side of the kink and inner C-G base pair of the stem from the other side [22]. There is a strong preference for the inner base pair to be C-G to allow the optimal formation of the A-minor [22, 23]. Rázga et al. [56] showed that the angle between the stems of a kink-turn is regulated by the local geometry of the type I A-minor interaction. The angle is regulated via insertion of a water molecule between the adenine and its cSS partner, forming a water-inserted variation of the A-minor [56, 57].
A-minor junctions. Another example of frequent local A-minor co-motifs is an A-minor junction [29, 31, 33]. A-minor junctions are the three- or four-way junctions involving coaxial stacking of two stems stabilized by at least one A-minor interaction. The strand with the interacting adenines may form a U-turn closed by at least 1 base pair [29], or form more complex and specific motifs to gain a higher stability [33]. Higher-order junctions, confining five or more helical stems, also involve structural modules characteristic of the three- and four-way junctions, including A-minor interactions [45]. The presence of possible A-minor interactions is successfully used as one of the features for predicting coaxial stacking in RNA junctions [58]. Interestingly, Beššeová et al. [59] showed that A-minor junctions are subjects to the hinge-like fluctuations, similar to the kink-turn motif. However, unlike the kink-turns, the authors found no apparent local variations providing junction flexibility.
Furthermore, A-patches have been found in the junctional cores of the A- and G-riboswitches, with bound purines forming one of the stacked A-minors [18, 60]. Klein et al. [22] mentioned a rare three-stranded variant of the kink-turn motif found in the 23S rRNA of Haloarcula marismortui, which was a part of the junction loop and also includes an A-minor interaction.
A-minors and pseudoknots. ABAB-pseudoknots, also known as H-type knots, are usually stabilized by the A-minor interactions formed by adenines from the 3′-closest strand of the pseudoknot and its AA-stem [14, 27, 28]. Aalberts and Hodas [27] explain such asymmetry of the H-type knots by the fact that the A-form RNA helix is right-handed, which brings the 3′-closest strand of the H-knot close to the minor groove of the AA-stem and brings the 5′-closest strand close to the major groove of the BB-stem. A-minors have been also found in other pseudoknot types, e.g., kissing loops (ABACBC-pseudoknots) [55, 61].
A-minors and bulges. A-minors in bulges are less common, but represent a number of functionally important cases. One example is the type WC/H A-minor identified in a single-adenine bulge formed by the precursor mRNA intron and U2 small nuclear (sn)RNA [39]. The bulge is called the spliceosomal branch site recognition motif, as it ensures the nucleophilic attack at the pre-mRNA 5′ splice site at the first stage of splicing [39]. Another example is two symmetric type I A-minors (a15-G9-c12 and A15-g9-C12) identified in the crystal structure of the bulge–helix–bulge (BHB) motif complex with the splicing endonuclease [17]. The BHB motif is found in precursor tRNA structures; it is recognized by the splicing endonuclease, an enzyme responsible for the excision of introns from nuclear tRNAs and all archaeal RNAs [62]. Interestingly, both A-minors are facilitated by stacking with the histidine residues of the endonuclease [17].
A rare example of the long-range A-minor formed with adenines of the bulge is the type I/IIP A-minor motif formed by the conserved A-rich bulge and the P4 helix of the group I intron [5, 6, 63]. The A-minor motif involves A183 and A184 adenines of the bulge interacting with G110-C211 and C109-G212 base pairs. It was suggested that this A-motif is responsible for the ribozyme activation [9, 64].
LONG-RANGE A-MINORS
A-minors are the most abundant type of long-range interactions in known RNA structures, more common even than the non-stem WC base pairs [24]. Along with other tertiary interactions, putative long-range A-minors, facilitate compact packing of helical elements of large structured RNAs [30, 65]. In [66, 67], Sponer et al. demonstrated that A-minor interactions are more hydrophobic than canonical base pairs and thus particularly suitable for helix packing.
A-minors and hairpins. The GNRA-tetraloop/tetraloop-receptor (GNRA/receptor) motif is the most prevalent co-motif of long-range A-minor interactions [55] found in various non-coding RNAs [24-26]. The motif involves one or two A-minor interactions formed between the hairpin of the GNRA sequence (where N is any nucleotide and R is purine) and a distant helix or a specific receptor. The most common and most stable GNRA/receptor motif is the GAAA/11nt motif, whose receptor is a conserved internal loop of 11 residues (CCUAAG-UAUGG) [68]. The type I/II A-minor motif is formed by purines at the third and fourth positions of the GNRA sequence. As a rule, GNGA sequences form the type I/IIP motif with the GA residues, while GNAA sequences favor the type I/IIT motif, which does not tolerate G at the third position of the GNRA sequence [24].
A-minors and internal loops. The second most populated secondary structure context of the long-range A-minor interactions is internal loops with the cross-strand adenine stacks, in particular, the UAA/GAN motif found in the 23S rRNA, RNase P RNA, and group I and II introns [69]. The cross-strand adenine stack in the UAA/GAN motif involves three adenines: two adenines of the UAA strand and the middle adenine of the GAN strand. The AAA stack tends to interact with distant RNA elements forming an A-patch consisting of the type I – type II – type I A-minors. A characteristic feature of this A-patch is that it is formed with three adenines and only two consecutive base pairs [69], whereas the number of interacting base pairs in A-patches is usually equal to the number of interacting adenines [4, 55].
Mitton-Fry et al. [15] identified a type III,II,I A-patch adjacent to the major groove triple helix formed by the poly(A)-tail of the viral polyadenylated nuclear (PAN) RNA and the U-rich internal loop of its expression and nuclear retention element (ENE). In [21], formation of the type I/II A-minor motif has been observed between the ENE element and the poly(A)-tail of the nuclear MALAT1 long non-coding RNA. Recently, Torabi et al. [38] identified lone A-minor interactions of the WC/H type, also close to the triple helices formed by the poly(A) sequence and the U-rich internal loops of the double-domain ENE (dENE), in the crystal structure of the dENE complexed with a 28-mer poly(A). Hence, A-minor interactions participate in the protection of the poly(A)-tail as found in many functional cellular and viral non-coding RNAs.
One of the most fascinating examples of the functional long-range A-minor motif is the interaction of the 16S rRNA A-site with the codon-anticodon helix formed between the mRNA and its cognate tRNA [41]. This interaction has been first predicted in biochemical experiments [70] and then confirmed for the crystal structure of the 30S ribosomal subunit [71]. The type I/II A-minor motif is formed by the universally conserved A1492 and A1493 residues from the A-site internal loop [71]. The adenines either lie within the loop or bulge out for the interaction. A1493 interacts with both the first residue of the codon and the third residue of the anticodon forming a type I A-minor interaction, whereas A1492 interacts only with the second base of the codon forming a type II A-minor. In many type I/II A-minor motifs, adenine of the type II interaction forms hydrogen bonds with water molecules that act as a bridge to the second strand, but in the A-minor formed by A1492 and A1493, it is G530 that fills a part of the minor groove instead of water [72]. It was shown that A1492 and A1493 discriminate between the canonical WC base pairs and mismatches, providing recognition of cognate tRNAs [41, 73]. Of note, there is no specific adenine interacting with the third base pair of the codon-anticodon minihelix, which explains the degeneracy of the genetic code that allows the third base pair to be a wobble G-U pair in addition to the WC base pair [71, 72]. It should be also mentioned that A1492 and A1493 of the bacterial 16S rRNA correspond to A1755 and A1756, respectively, of the eukaryotic 18S rRNA [74].
A-MINORS AND RIBOSOME FUNCTION
The central role of the A-minor motif in the decoding process during protein synthesis favors the RNA world hypothesis, not only by leading to the conclusion that “ribosome is a ribozyme” [4, 41, 75], but also by suggesting that ribosomes have evolved solely from the RNA-based assembly [41]. Noller [76] speculates that such an elegant application of A-minor interactions to discriminate between WC base pairs and mismatches could be a relic of some RNA replication mechanism from the RNA world.
Besides the A-minors monitoring the codon-anticodon helix formation, ribosomal RNA involves many other functionally important A-minor interactions. Here, we list a few of them.
During the translation initiation, the initiator formyl-methionine tRNA binds directly to the P-site. This extra stable binding, at least in part, can be explained by the type I/II A-minor motif formed by the universally conserved G1338 and A1339 residues of the 16S rRNA and G-C base pairs 30-40 and 29-41 of the tRNA anticodon stem [77, 78]. Other tRNAs may contain A-U pairs instead of G-C pairs and are unable to form these interactions. Furthermore, G1338 and A1339 are located between the P- and E-site tRNAs and may prevent inappropriate translocation from the P-site to the E-site [78].
A-minors are also present in the peptidyl transferase center, as the universally conserved A76 residues of both the A-site and P-site tRNAs form type I A-minors with the base pairs in the 23S rRNA domain V [4, 79, 80].
The only conserved tertiary contact between the 5S rRNA and 23S rRNA is the symmetric A-patch formed by three stacked adenines of 5S rRNA loop E and three stacked adenines of helix 38 of the 23S rRNA [4, 81]. This A-patch is a good example of a common tendency of internal loops involving cross-strand adenine stacks to interact with distant secondary structure elements.
rRNA of the 50S subunit includes at least 7 kink-turn motifs [22] fluctuating in a hinge-like manner due to the local variations of their type I A-minor interactions. Such elbow-like properties of the kink-turn motifs can provide significant functional changes in the positions of distant parts of RNA elements, e.g., the A-site finger (helix 38) and the factor-binding L11 site (helices 42-44) [56].
It was found [82] that helix 68 of the 23S rRNA maintains a contact with helix 75 through four conserved adenines (A1853, A1854, A1889, A1890) forming the II,I,I,II A-patch, and this contact is involved in the L1 stalk movement and intersubunit rotation.
Also, A-minors form the protein-binding sites of rRNA; for example, A-patches from helices 95 and 97 of the 23S rRNA mediate interactions with the ribosomal protein L6 [4].
DISCUSSION
In this review, we discussed different definitions of A-minors, listed examples of functional A-minor motifs, and surveyed the most common types of their structural context.
Overall, A-minor motifs have evolved to discriminate WC base pairs against mismatches. For example, interactions between P4 domain helix and A-rich bulge of the group I intron P6 domain, as well as the A-site/codon-anticodon interactions, involve type I/IIP A-minor motifs specific to adenines [24, 64]. At the same time, motifs that distinguish between AU and GC WC base pairs, such as the A-minor interactions formed by the GA residues of the 16S rRNA and the anticodon stem of the initiator P-site tRNA or the GAAA/11nt motif, either tolerate G instead of A or prefer the type I/IIT conformation that requires additional interactions [33, 78].
Another A-minor feature that should be mentioned is the dynamics of its constituents that considerably differs among various A-minor co-motifs [41]. The GNRA/helix interaction occurs between stable preformed motifs, whereas the GAAA/11nt motif occurs between partially preformed motifs. The A-site A1492 and A1493 residues demonstrate the highest structural differences between the bound and unbound states, which allows them to behave as a molecular switch [41].
A-minor interactions are among the most common types of interactions in RNA-RNA interfaces. Beside rRNAs with numerous intermolecular A-minors, such interactions were also found to stabilize the interdomain interface in the crystal structure of Vibrio cholerae glycine aptamer homodimer [20, 83]. Furthermore, A-minor interactions have been utilized in the formation of synthetic tectoRNA homo- and heterodimers [24].
Since type I and type II A-minors are more frequent than the other types, the definition of an A-minor is now limited to a base triple containing a cWW base pair and a base (usually adenine) forming the cSS and/or tSS base pairs with the cWW base pair [84]. Unfortunately, such approach misses many non-canonical A-minors, e.g., the WC/H subclass. Furthermore, annotation of intermediate cases of A-minors with geometric parameters slightly exceeding the thresholds is also a problem. It was suggested that many of such intermediate A-minors found in the ribosome allow the overall flexibility of the ribosomal structure and, in particular, intersubunit translocating relative to each other [80, 85]. As proposed by A. S. Spirin in 1968 [86] the existence of such ribosomal mobility is a necessary element of translation. A good example is the intermolecular A-patch formed between the 5S rRNA and 23S rRNA. This A-patch (PDB 4V9F; adenines A80, A103, A104 of the 5S rRNA and A955, A1013, A1014 of the 23S rRNA) is not annotated by DSSR, which identifies only constituent hydrogen bonds and ribose zippers, whereas FR3D recognizes only some of the base-base and base-ribose interactions of the “near” geometry (i.e., not passing the default threshold but passing the extended threshold).
There is no doubt that with the exponentially growing number of resolved 3D structures of various RNA molecules, the list of functionally important A-minor interactions will also grow rapidly. Therefore, there is an urgent need to develop enhanced programs for the annotation of diverse A-minor types and to map A-minor interactions with other RNA tertiary motifs in order to improve their functional annotation.
Ethics declarations. The authors declare no conflict of interest. This article does not contain any studies involving human participants or animals performed by any of the authors.
REFERENCES
1.Lekka, E., and Hall, J. (2018) Noncoding RNAs in
disease, FEBS Lett., 592, 2884-2900, doi:
10.1002/1873-3468.13182.
2.Novikova, I. V., Hennelly, S. P., Tung, C. S., and
Sanbonmatsu, K. Y. (2013) Rise of the RNA machines: exploring the
structure of long non-coding RNAs, J. Mol. Biol., 425,
3731-3746, doi: 10.1016/j.jmb.2013.02.030.
3.Leontis, N. B., Lescoute, A., and Westhof, E.
(2006) The building blocks and motifs of RNA architecture, Curr.
Opin. Struct. Biol., 16, 279-287, doi:
10.1016/j.sbi.2006.05.009.
4.Nissen, P., Ippolito, J. A., Ban, N., Moore, P. B.,
and Steitz, T. A. (2001) RNA tertiary interactions in the large
ribosomal subunit: the A-minor motif, Proc. Natl. Acad. Sci.
USA, 98, 4899-4903, doi: 10.1073/pnas.081082398.
5.Murphy, F. L., and Cech, T. R. (1994) GAAA
tetraloop and conserved bulge stabilize tertiary structure of a group I
intron domain, J. Mol. Biol., 236, 49-63, doi:
10.1006/jmbi.1994.1117.
6.Cate, J. H., Gooding, A. R., Podell, E., Zhou, K.,
Golden, B. L., et al. (1996) Crystal structure of a group I ribozyme
domain: principles of RNA packing, Science, 273,
1678-1685, doi: 10.1126/science.273.5282.1678.
7.Scott, W. G., Finch, J. T., and Klug, A. (1995) The
crystal structure of an all-RNA hammerhead ribozyme: a proposed
mechanism for RNA catalytic cleavage, Cell, 81, 991-1002,
doi: 10.1016/S0092-8674(05)80004-2.
8.Nissen, P. (2020) The a-minor motif, in
Structural Insights into Gene Expression and Protein Synthesis,
pp. 461-463, doi: 10.1142/9789811215865_0055.
9.Doherty, E. A., Batey, R. T., Masquida, B., and
Doudna, J. A. (2001) A universal mode of helix packing in RNA, Nat.
Struct. Biol., 8, 339-343, doi: 10.1038/86221.
10.Strobel, S. A. (2002) Biochemical identification
of A-minor motifs within RNA tertiary structure by interference
analysis, Biochem. Soc. Transact., 30, 1126-1131, doi:
10.1042/bst0301126.
11.Krasilnikov, A. S., Yang, X., Pan, T., and
Mondragón, A. (2003) Crystal structure of the specificity domain
of ribonuclease P, Nature, 421, 760-764, doi:
10.1038/nature01386.
12.Krasilnikov, A. S., Xiao, Y., Pan, T., and
Mondragón, A. (2004) Basis for structural diversity in
homologous RNAs, Science, 306, 104-107, doi:
10.1126/science.1101489.
13.Nagai, K., Oubridge, C., Kuglstatter, A.,
Menichelli, E., Isel, C., and Jovine, L. (2003) Structure, function and
evolution of the signal recognition particle, EMBO J.,
22, 3479-3485, doi: 10.1093/emboj/cdg337.
14.Schüler, M., Connell, S. R., Lescoute, A.,
Giesebrecht, J., Dabrowski, M., et al. (2006) Structure of the
ribosome-bound cricket paralysis virus IRES RNA, Nat. Struct. Mol.
Biol., 13, 1092-1096, doi: 10.1038/nsmb1177.
15.Mitton-Fry, R. M., DeGregorio, S. J., Wang, J.,
Steitz, T. A., and Steitz, J. A. (2010) Poly (A) tail recognition by a
viral RNA element through assembly of a triple helix, Science,
330, 1244-1247, doi: 10.1126/science.1195858.
16.Nguyen, L. A., Wang, J., and Steitz, T. A. (2017)
Crystal structure of Pistol, a class of self-cleaving ribozyme,
Proc. Natl. Acad. Sci. USA, 114, 1021-1026, doi:
10.1073/pnas.1611191114.
17.Xue, S., Calvin, K., and Li, H. (2006) RNA
recognition and cleavage by a splicing endonuclease, Science,
312, 906-910, doi: 10.1126/science.1126629.
18.Serganov, A., Yuan, Y. R., Pikovskaya, O.,
Polonskaia, A., Malinina, L., et al. (2004) Structural basis for
discriminative regulation of gene expression by adenine-and
guanine-sensing mRNAs, Chem. Biol., 11, 1729-1741, doi:
10.1016/j.chembiol.2004.11.018.
19.Dann, C. E. 3rd, Wakeman, C. A., Sieling, C. L.,
Baker, S. C., Irnov, I., and Winkler, W. C. (2007) Structure and
mechanism of a metal-sensing regulatory RNA, Cell, 130,
878-892, doi: 10.1016/j.cell.2007.06.051.
20.Jones, C. P., and
Ferré-D’Amaré, A. R. (2015) RNA quaternary
structure and global symmetry, Trends Biochem. Sci., 40,
211-220, doi: 10.1016/j.tibs.2015.02.004.
21.Brown, J. A., Bulkley, D., Wang, J., Valenstein,
M. L., Yario, T. A., et al. (2014) Structural insights into the
stabilization of MALAT1 noncoding RNA by a bipartite triple helix,
Nat. Struct. Mol. Biol., 21, 633, doi:
10.1038/nsmb.2844.
22.Klein, D. J., Schmeing, T. M., Moore, P. B., and
Steitz, T. A. (2001) The kink‐turn: a new RNA secondary
structure motif, EMBO J., 20, 4214-4221, doi:
10.1093/emboj/20.15.4214.
23.Réblová, K., Šponer, J. E.,
Špačková, N., Beššeová, I.,
and Šponer, J. (2011) A-minor tertiary interactions in RNA
kink-turns. Molecular dynamics and quantum chemical analysis, J.
Phys. Chem. B, 115, 13897-13910, doi: 10.1021/jp2065584.
24.Geary, C., Baudrey, S., and Jaeger, L. (2008)
Comprehensive features of natural and in vitro selected GNRA
tetraloop-binding receptors, Nucleic Acids Res., 36,
1138-1152, doi: 10.1093/nar/gkm1048.
25.Wu, L., Chai, D., Fraser, M. E., and Zimmerly, S.
(2012) Structural variation and uniformity among tetraloop-receptor
interactions and other loop-helix interactions in RNA crystal
structures, PLoS One, 7, e49225, doi:
10.1371/journal.pone.0049225.
26.Fiore, J. L., and Nesbitt, D. J. (2013) An RNA
folding motif: GNRA tetraloop–receptor interactions, Quart.
Rev. Biophys., 46, 223-264, doi:
10.1017/S0033583513000048.
27.Aalberts, D. P., and Hodas, N. O. (2005)
Asymmetry in RNA pseudoknots: observation and theory, Nucleic Acids
Res., 33, 2210-2214, doi: 10.1093/nar/gki508.
28.Giedroc, D. P., and Cornish, P. V. (2009)
Frameshifting RNA pseudoknots: structure and mechanism, Virus
Res., 139, 193-208, doi: 10.1016/j.virusres.2008.06.008.
29.Lescoute, A., and Westhof, E. (2006) Topology of
three-way junctions in folded RNAs, RNA, 12, 83-93, doi:
10.1261/rna.2208106.
30.Xin, Y., Laing, C., Leontis, N. B., and Schlick,
T. (2008) Annotation of tertiary interactions in RNA structures reveals
variations and correlations, RNA, 14, 2465-2477, doi:
10.1261/rna.1249208.
31.Laing, C., and Schlick, T. (2009) Analysis of
four-way junctions in RNA structures, J. Mol. Biol., 390,
547-559, doi: 10.1016/j.jmb.2009.04.084.
32.Cruz, J. A., and Westhof, E. (2009) The dynamic
landscapes of RNA architecture, Cell, 136, 604-609, doi:
10.1016/j.cell.2009.02.003.
33.Geary, C., Chworos, A., and Jaeger, L. (2011)
Promoting RNA helical stacking via A-minor junctions, Nucleic Acids
Res., 39, 1066-1080, doi: 10.1093/nar/gkq748.
34.Tamura, M., and Holbrook, S. R. (2002) Sequence
and structural conservation in RNA ribose zippers, J. Mol.
Biol., 320, 455-474, doi: 10.1016/S0022-2836(02)00515-6.
35.Lee, J. C., Cannone, J. J., and Gutell, R. R.
(2003) The lonepair triloop: a new motif in RNA structure, J. Mol.
Biol., 325, 65-83, doi: 10.1016/S0022-2836(02)01106-3.
36.Gagnon, M. G., and Steinberg, S. V. (2010) The
adenosine wedge: A new structural motif in ribosomal RNA, RNA,
16, 375-381, doi: 10.1261/rna.1550310.
37.Leontis, N. B., and Westhof, E. (2001) Geometric
nomenclature and classification of RNA base pairs, RNA,
7, 499-512, doi: 10.1017/s1355838201002515.
38.Torabi, S. F., Vaidya, A. T., Tycowski, K. T.,
DeGregorio, S. J., Wang, J., et al. (2021) RNA stabilization by a poly
(A) tail 3′-end binding pocket and other modes of poly (A)-RNA
interaction, Science, 371, eabe6523, doi:
10.1126/science.abe6523.
39.Newby, M. I., and Greenbaum, N. L. (2002)
Sculpting of the spliceosomal branch site recognition motif by a
conserved pseudouridine, Nat. Struct. Biol., 9, 958-965,
doi: 10.1038/nsb873.
40.Hamdani, H. Y., and Firdaus-Raih, M. (2019)
Identification of structural motifs using networks of hydrogen-bonded
base interactions in RNA crystallographic structures, Crystals,
9, 550, doi: 10.3390/cryst9110550.
41.Lescoute, A., and Westhof, E. (2006) The A-minor
motifs in the decoding recognition process, Biochimie,
88, 993-999, doi: 10.1016/j.biochi.2006.05.018.
42.Lescoute, A., and Westhof, E. (2006) The
interaction networks of structured RNAs, Nucleic Acids Res.,
34, 6587-6604, doi: 10.1093/nar/gkl963.
43.Petrov, A. I., Zirbel, C. L., and Leontis, N. B.
(2011) WebFR3D – a server for finding, aligning and analyzing
recurrent RNA 3D motifs, Nucleic Acids Res., 39, W50-W55,
doi: 10.1093/nar/gkr249.
44.Sheth, P., Cervantes-Cervantes, M., Nagula, A.,
Laing, C., and Wang, J. T. (2013) Novel features for identifying
A-minors in three-dimensional RNA molecules, Computat. Biol.
Chem., 47, 240-245, doi:
10.1016/j.compbiolchem.2013.10.004.
45.Laing, C., Jung, S., Iqbal, A., and Schlick, T.
(2009) Tertiary motifs revealed in analyses of higher-order RNA
junctions, J. Mol. Biol., 393, 67-82, doi:
10.1016/j.jmb.2009.07.089.
46.Burley, S. K., Berman, H. M., Kleywegt, G. J.,
Markley, J. L., Nakamura, H., and Velankar, S. (2017) Protein Data Bank
(PDB): the single global macromolecular structure archive, Protein
Crystallogr., 1607, 627-641, doi:
10.1007/978-1-4939-7000-1_26.
47.Reinharz, V., Soulé, A., Westhof, E.,
Waldispühl, J., and Denise, A. (2018) Mining for recurrent
long-range interactions in RNA structures reveals embedded hierarchies
in network families, Nucleic Acids Res., 46, 3841-3851,
doi: 10.1093/nar/gky197.
48.Appasamy, S. D., Hamdani, H. Y., Ramlan, E. I.,
and Firdaus-Raih, M. (2016) InterRNA: a database of base interactions
in RNA structures, Nucleic Acids Res., 44, D266-D271,
doi: 10.1093/nar/gkv1186.
49.Hamdani, H. Y., Appasamy, S. D., Willett, P.,
Artymiuk, P. J., and Firdaus-Raih, M. (2012) NASSAM: a server to search
for and annotate tertiary interactions and motifs in three-dimensional
structures of complex RNA molecules, Nucleic Acids Res.,
40, W35-W41, doi: 10.1093/nar/gks513.
50.Firdaus-Raih, M., Hamdani, H. Y., Nadzirin, N.,
Ramlan, E. I., Willett, P., and Artymiuk, P. J. (2014) COGNAC: a web
server for searching and annotating hydrogen-bonded base interactions
in RNA three-dimensional structures, Nucleic Acids Res.,
42, W382-W388, doi: 10.1093/nar/gku438.
51.Lu, X. J., Bussemaker, H. J., and Olson, W. K.
(2015) DSSR: an integrated software tool for dissecting the spatial
structure of RNA, Nucleic Acids Res., 43, e142-e142, doi:
10.1093/nar/gkv716.
52.Yang, H., Jossinet, F., Leontis, N., Chen, L.,
Westbrook, J., et al. (2003) Tools for the automatic identification and
classification of RNA base pairs, Nucleic Acids Res., 31,
3450-3460, doi: 10.1093/nar/gkg529.
53.Gendron, P., Lemieux, S., and Major, F. (2001)
Quantitative analysis of nucleic acid three-dimensional structures,
J. Mol. Biol., 308, 919-936, doi:
10.1006/jmbi.2001.4626.
54.Lu, X. J., and Olson, W. K. (2008) 3DNA: a
versatile, integrated software system for the analysis, rebuilding and
visualization of three-dimensional nucleic-acid structures, Nat.
Protoc., 3, 1213, doi: 10.1038/nprot.2008.104.
55.Shalybkova, A. A., Mikhailova, D. S.,
Kulakovskiy, I. V., Fakhranurova, L. I., and Baulin, E. F. (2021)
Annotation of the local context of the RNA secondary structure improves
the classification and prediction of A-minors, RNA, rna-078535,
doi: 10.1261/rna.078535.120.
56.Rázga, F., Koča, J., Šponer,
J., and Leontis, N. B. (2005) Hinge-like motions in RNA kink-turns: the
role of the second A-minor motif and nominally unpaired bases,
Biophys. J., 88, 3466-3485, doi:
10.1529/biophysj.104.054916.
57.Sponer, J., Bussi, G., Krepl, M.,
Banáš, P., Bottaro, S., et al. (2018) RNA structural
dynamics as captured by molecular simulations: a comprehensive
overview, Chem. Rev., 118, 4177-4338, doi:
10.1021/acs.chemrev.7b00427.
58.Laing, C., Wen, D., Wang, J. T., and Schlick, T.
(2012) Predicting coaxial helical stacking in RNA junctions, Nucleic
Acids Res., 40, 487-498, doi: 10.1093/nar/gkr629.
59.Beššeová, I., Reblova, K.,
Leontis, N. B., and Šponer, J. (2010) Molecular dynamics
simulations suggest that RNA three-way junctions can act as flexible
RNA structural elements in the ribosome, Nucleic Acids Res.,
38, 6247-6264, doi: 10.1093/nar/gkq414.
60.Lescoute, A., and Westhof, E. (2005) Riboswitch
structures: purine ligands replace tertiary contacts, Chem.
Biol., 12, 10-13, doi: 10.1016/j.chembiol.2005.01.002.
61.Baulin, E., Yacovlev, V., Khachko, D., Spirin,
S., and Roytberg, M. (2016) URS DataBase: universe of RNA structures
and their motifs, Database, 2016, baw085, doi:
10.1093/database/baw085.
62.Calvin, K., and Li, H. (2008) RNA-splicing
endonuclease structure and function, Cell. Mol. Life Sci.,
65, 1176-1185, doi: 10.1007/s00018-008-7393-y.
63.Ikawa, Y., Yoshimura, T., Hara, H., Shiraishi,
H., and Inoue, T. (2002) Two conserved structural components,
A‐rich bulge and P4 XJ6/7 base‐triples, in activating the
group I ribozymes, Genes Cells, 7, 1205-1215, doi:
10.1046/j.1365-2443.2002.00601.x.
64.Battle, D. J., and Doudna, J. A. (2002)
Specificity of RNA–RNA helix recognition, Proc. Natl. Acad.
Sci. USA, 99, 11676-11681, doi: 10.1073/pnas.182221799.
65.Schwalbe, H., Buck, J., Fürtig, B., Noeske,
J., and Wöhnert, J. (2007) Structures of RNA switches: insight
into molecular recognition and tertiary structure, Angewandte Chemie
Int. Edn., 46, 1212-1219, doi: 10.1002/anie.200604163.
66.Šponer, J. E., Leszczynski, J.,
Sychrovský, V., and Šponer, J. (2005) Sugar edge/sugar
edge base pairs in RNA: stabilities and structures from quantum
chemical calculations, J. Phys. Chem. B, 109,
18680-18689, doi: 10.1021/jp053379q.
67.Šponer, J. E., Reblova, K., Mokdad, A.,
Sychrovský, V., Leszczynski, J., and Šponer, J. (2007)
Leading RNA tertiary interactions: structures, energies, and water
insertion of A-minor and P-interactions. A quantum chemical view, J.
Phys. Chem. B, 111, 9153-9164, doi: 10.1021/jp0704261.
68.Costa, M., and Michel, F. (1995) Frequent use of
the same tertiary motif by self‐folding RNAs, EMBO J.,
14, 1276-1285, doi: 10.1002/j.1460-2075.1995.tb07111.x.
69.Lee, J. C., Gutell, R. R., and Russell, R. (2006)
The UAA/GAN internal loop motif: a new RNA structural element that
forms a cross-strand AAA stack and long-range tertiary interactions,
J. Mol. Biol., 360, 978-988, doi:
10.1016/j.jmb.2006.05.066.
70.Yoshizawa, S., Fourmy, D., and Puglisi, J. D.
(1999) Recognition of the codon-anticodon helix by ribosomal RNA,
Science, 285, 1722-1725, doi:
10.1126/science.285.5434.1722.
71.Ogle, J. M., Brodersen, D. E., Clemons, W. M.,
Tarry, M. J., Carter, A. P., and Ramakrishnan, V. (2001) Recognition of
cognate transfer RNA by the 30S ribosomal subunit, Science,
292, 897-902, doi: 10.1126/science.1060612.
72.Ogle, J. M., and Ramakrishnan, V. (2005)
Structural insights into translational fidelity, Annu. Rev.
Biochem., 74, 129-177, doi:
10.1146/annurev.biochem.74.061903.155440.
73.Gromadski, K. B., Daviter, T., and Rodnina, M. V.
(2006) A uniform response to mismatches in codon-anticodon complexes
ensures ribosomal fidelity, Mol. Cell, 21, 369-377, doi:
10.1016/j.molcel.2005.12.018.
74.Prokhorova, I., Altman, R. B., Djumagulov, M.,
Shrestha, J. P., Urzhumtsev, A., et al. (2017) Aminoglycoside
interactions and impacts on the eukaryotic ribosome, Proc. Natl.
Acad. Sci. USA, 114, E10899-E10908, doi:
10.1073/pnas.1715501114.
75.Steitz, T. A., and Moore, P. B. (2003) RNA, the
first macromolecular catalyst: the ribosome is a ribozyme, Trends
Biochem. Sci., 28, 411-418, doi:
10.1016/S0968-0004(03)00169-5.
76.Noller, H. F. (2012) Evolution of protein
synthesis from an RNA world, Cold Spring Harb. Perspect. Biol.,
4, a003681, doi: 10.1101/cshperspect.a003681.
77.Lancaster, L., and Noller, H. F. (2005)
Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination
of initiator tRNA, Mol. Cell, 20, 623-632, doi:
10.1016/j.molcel.2005.10.006.
78.Steitz, T. A. (2008) A structural understanding
of the dynamic ribosome machine, Nat. Rev. Mol. Cell
Biol., 9, 242-253, doi: 10.1038/nrm2352.
79.Hansen, J. L., Schmeing, T. M., Moore, P. B., and
Steitz, T. A. (2002) Structural insights into peptide bond formation,
Proc. Natl. Acad. Sci. USA, 99, 11670-11675, doi:
10.1073/pnas.172404099.
80.Noller, H. F. (2005) RNA structure: reading the
ribosome, Science, 309, 1508-1514, doi:
10.1126/science.1111771.
81.Szymański, M., Barciszewska, M. Z.,
Erdmann, V. A., and Barciszewski, J. (2003) 5S rRNA: structure and
interactions, Biochem. J., 371, 641-651, doi:
10.1042/bj20020872.
82.Mohan, S., and Noller, H. F. (2017) Recurring RNA
structural motifs underlie the mechanics of L1 stalk movement, Nat.
Commun., 8, 1-11, doi: 10.1038/ncomms14285.
83.Bou-Nader, C., and Zhang, J. (2020) Structural
insights into RNA dimerization: Motifs, interfaces and functions,
Molecules, 25, 2881, doi: 10.3390/molecules25122881.
84.Leontis, N. B., and Westhof, E. (2003) Analysis
of RNA motifs, Curr. Opin. Struct. Biol., 13, 300-308,
doi: 10.1016/S0959-440X(03)00076-9.
85.Frank, J., Gao, H., Sengupta, J., Gao, N., and
Taylor, D. J. (2007) The process of mRNA–tRNA translocation,
Proc. Natl. Acad. Sci. USA, 104, 19671-19678, doi:
10.1073/pnas.0708517104.
86.Spirin, A. S. (1968) How does the ribosome work?
A hypothesis based on the two subunit construction of the ribosome,
Curr. Mod. Biol., 2, 115-127, doi:
10.1016/0303-2647(68)90017-8.