MINI-REVIEW: Informosomes Travel in Time: An Early mRNA Concept in the Current mRNP Landscape
Pavel Ivanov1,2,3
1Division of Rheumatology, Inflammation, and Immunity, Brigham and Women’s Hospital, Boston, Massachusetts, 02115 USA2Department of Medicine, Harvard Medical School, Boston, Massachusetts, 02115 USA
3Harvard Medical School Initiative for RNA Medicine, Harvard University, Boston, Massachusetts, 02115 USA
Received May 27, 2021; Revised July 9, 2021; Accepted July 12, 2021
Messenger RNA is complexed with proteins throughout its life cycle. The first mRNA-containing particles of non-ribosomal nature, named informosomes, were discovered in cytoplasmic extracts of fish embryos by the laboratory of Alexander Spirin, and later described in live cells. Over time, various other nuclear and cytoplasmic mRNA-containing ribonucleoproteins (mRNPs) have been found and characterized. Although these mRNPs are very diverse in their subcellular localization, structure and functions, they share many common characteristics with informosomes. In this mini-review, I will discuss the discovery of informosomes, their characteristics and proposed functions, and their potential relationship to other mRNPs.
KEY WORDS: informosomes, mRNA, ribonucleoproteins, mRNP, RNA granules, protein biosynthesisDOI: 10.1134/S0006297921090029
Abbreviations: mRNPs, messenger RNA nucleoprotein particles; PBs, Processing Bodies; RBPs, RNA-binding proteins; SGs, Stress Granules.
INTRODUCTION: A TYPICAL mRNA LIFE CYCLE
Eukaryotic messenger RNAs (mRNAs) are transcribed in the nucleus by RNA polymerase II. During transcription, these nascent transcripts become decorated with a diverse set of RNA-binding proteins (RBPs), which actively assist in the conversion of mRNA precursors (pre-mRNAs) into mature mRNAs. Such conversion includes three major processing events, namely RNA splicing, 5′-capping, and 3′-cleavage/polyadenylation (reviewed in [1]). Upon completion of these processes, a typical mRNA contains an Open Reading Frame (ORF), which encodes protein, and is flanked by untranslated regions (UTRs), named respectively as 5′- and 3′-UTRs. In turn, the UTRs contain characteristic features such as presence of 7-methylguanosine at the 5′-end of the mRNA (mRNA cap structure), and a string of adenine (A) residues at the 3′-end that form a poly(A) tail. Maturation into mRNA requires coordinated action of multiple enzymes and associated nuclear proteins, many of which are RBPs [2]. In addition, other RBPs, which are not required for the nuclear mRNA maturation, recognize sequences and/or structures in the UTRs (cis-elements) and participate in additional processes that determine the mRNA’s fate [3]. As a result, a diverse set of nuclear messenger RNA nucleoprotein particles (mRNPs) is formed that differ in their protein composition and overall structure [4].
Upon mRNP formation, composition determines how quickly these mRNPs can be exported into the cytoplasm from the nucleus, where they can be immediately available for mRNA translation or stored as “silenced” mRNA-containing particles [5]. Some mRNPs are transported to specific regions of the cell for the purpose of the localized translation, e.g., trafficking to the synapses of neurons for the production of neuromediators. These processes are associated with further active remodeling of mRNPs via association with cytoplasmic RBPs [6]. It should be also noted, that actively growing cells are associated with more active protein translation than some specialized cells (such as oocytes) that typically store mRNPs and engage them in mRNA translation only at specific stages of their development. Both actively translated and untranslated mRNAs are also eventually subject to RNA degradation, which completes their life cycle [7]. One unifying feature of mRNA metabolism is that any step in an mRNA’s life is unthinkable without interaction with its protein partners.
DISCOVERY OF INFORMOSOMES
While it is currently very well accepted that mRNAs do not exist in the “naked” form and instead are associated with proteins as parts of various mRNPs at any stage of their life cycle, the idea that newly synthesized mRNAs are protein-bound and can be found in a ribosome-free state was quite provocative for the early 1960s. As the sole function of mRNAs was understood to be their participation in protein synthesis via association with ribosomes, this idea seemed irrelevant.
In 1964, Spirin and colleagues proposed that eukaryotic messenger RNAs, which are localized in the cytoplasm and temporarily untranslated, are present in association with unknown proteins of non-ribosomal nature [8, 9]. Such conclusions were based on the studies of Misgurnus fossils (loach) embryos in the late blastula stages that were pre-incubated with [14C]-adenine or [14C]-uridine, which can be efficiently incorporated into the newly synthesized transcripts [9]. Using sucrose gradient centrifugation of cytoplasmic embryonic extracts, they found that the newly synthesized transcripts sedimented after the 80S ribosomes (the region between 20S and 75S). Surprisingly, the same approach using cytoplasmic extracts from embryos pre-incubated with radioactive amino acids revealed very similar sedimentation distribution. Taking into consideration that M. fossils embryos do not produce new ribosomes (or ribosomal RNAs) until the very end of gastrulation [8], it was hypothesized that the newly synthesized RNAs sedimenting in the post-ribosomal zone represented mRNAs in complexes with proteins [9].
To complement these findings, Spirin et al. employed a method relying on fixation of RNPs with formaldehyde, which efficiently crosslinks RNA-protein complexes [10]. Following this stabilization strategy, it was possible to analyze density distribution of the crosslinked components in the CsCl gradient [11]. It was clear that these formaldehyde-fixed post-ribosomal complexes contained both radioactively labeled amino acids and RNA components that coincided in the analyzed density distribution. These RNPs also clearly differed from both the ribosome/ribosomal subunits and free RNA as they corresponded specifically to a low buoyant density of ~1.40-1.45 g/cm3, which is between that of a free protein and RNA [12]. Moreover, their characteristic buoyant density values also pointed to the predominant presence of protein over RNA at a ratio ~3 : 1. Seven such RNA–protein complexes with sedimentation coefficients equal to 20S, 30S, 40S, 50S, 55S, 65S, and 75S were found in the post-ribosomal zone, indicating reproducibility of the phenomenon [12]. Additionally, these complexes could be quantitatively absorbed on nitrocellulose membranes, while free RNA or proteins could not. These complexes were also degraded by pronase (a non-specific protease from Streptomyces griseus) and ribonuclease, which excluded their association with DNA [12]. In addition, when the radiolabeled RNA components were released from the post-ribosomal RNPs by deproteinization and centrifuged in a sucrose gradient, the resulting sedimentation coefficients were clearly distinct from that of 18S and 28S ribosomal RNAs [12], suggesting that RNA components of these RNPs were not of ribosomal RNA origin [13].
Importantly, RNPs of similar nature were also discovered in the sea urchin embryos [14] and in the vaccinia virus infected HeLa cells [15]. In the sea urchin embryo, radiolabeled RNA isolated from the post-ribosomal RNPs was quantitatively hybridizing with DNA, thus indicating an mRNA-like nature [14, 16]. In the vaccinia virus infected HeLa cells, where only one transcript is highly expressed, namely viral mRNA, the post-ribosomal RNPs were also detected [17, 18], and contained the newly synthesized viral mRNA [15].
Altogether, the above-described data suggest that these post-ribosomal ribonucleoproteins with a characteristically low buoyant density, consist of mRNAs and proteins, and do not contain ribosomes or ribosomal subunit. These mRNPs were then collectively called informosomes, the term both stating that the RNA component of these particles contains information (not just a message for the production of an encoded protein) and also suggesting that the protein component actively participates in the regulation of mRNA fate.
DEVELOPMENT OF THE INFORMOSOME CONCEPT
Soon after informosomes were discovered, they were also found in several other experimental models such as uninfected HeLa cells [19], developing epidermis cells of giant silkworms [20], rat liver cells [21], cultivated mouse fibroblasts (L cells) [22], L cells infected with Mengo virus [23], and Ehrlich ascites tumor cells infected with Sendai virus [24]. All the discovered informosomes were heterogeneous in size but retained the core characteristics of post-ribosomal informosomes. Interestingly, in two early reports, analysis of mRNPs in the post-ribosomal zone of normal HeLa cells [19] and rat liver cells [21] indicated formation of 45S particles that could be either informosomes or mRNA complexes with 40S ribosomal subunits [21, 25]. Importantly, analysis of the virus-infected cells allowed for identification of specific types of informosomes loaded with viral mRNA. Such viral-specific 45S informosomes were isolated and characterized [23, 24].
Another important observation was that informosomes are not limited to the post-ribosomal zone but can also be found in the pre-ribosomal zone [12, 26]. According to the results of sucrose gradient centrifugation of loach embryo cytoplasmic extracts, the newly synthesized radiolabeled mRNAs could be found ahead of the 80S ribosomal peak, within RNPs with sedimentation coefficients of 90S, 100S, and 110S [12]. Since even a dimer of 80S ribosomes would have a sedimentation coefficient of ~120S, these mRNAs could not be of polysomal nature. Based on the buoyant density of pre-ribosomal 90S and 110S components, which were similar to buoyant density of the post-ribosomal 50S to 75S components (~1.39 g/cm3), it was concluded that the post-ribosomal mRNPs are bona fide informosomes [12]. Thus, two broad subpopulations of informosomes can be defined: post-ribosomal (20S to 75S) and pre-ribosomal (90S to 110S). Since the discovered informosomes were heterogeneous in size, it was also proposed that they are heterogeneous in their protein and mRNA content [12].
One largely overlooked aspect of these studies was that there are some, although not as abundant as described above, pre-ribosomal informosomes that have high sedimentation coefficients. This may suggest that they are either single large particles or aggregates of many smaller informosomes into a larger mRNP [13]. Biological significance of such high molecular weight particles is questionable but may be connected to the processes of RNA granule assembly (see below).
Once informosomes were found in the cytoplasm, it was also predicted that nuclear counterparts should also exist. Indeed, several research groups observed mRNPs in nuclear extracts analogous to cytoplasmic informosomes [27-32], e.g., they were heterogeneous in size yet similar in buoyant density coefficients (~1.4 g/cm3) [30, 31]. Such nuclear informosomes are decorated by proteins different than the ones associated with informosomes in the cytoplasm, suggesting that during mRNA export mRNAs may change their protein binding partners.
An important question remained regarding whether informosomes actually exist in vivo or their formation is a mere artifact of cell destruction and/or homogenization. It was first found that the addition of free non-protein bound RNAs into cytoplasmic extracts prepared from a number of different cellular models resulted in formation of artificial informosome-like particles, as judged from their behavior in CsCl gradients [33, 34]. Such particles were homogeneous; their formation was independent of the nature of the added free RNAs (both RNA length and origin, e.g., E. coli rRNA was commonly used). The only limitation for formation of such complexes was the amount of RNA to be added to extract: a relatively small RNA amount was allowed, while excessive RNA was not incorporated into RNP [34]. This suggested that a titratable and limited “loading factor” exists in cellular extracts, which is available for association with the added free RNA [34]. Based on several other experiments, it was concluded that this “loading factor” is of proteinaceous nature (single protein or a group of proteins), has a high molecular weight, constitutes approximately 0.3% of the soluble total fraction of the extract and readily interacts with an exogenously introduced RNA. As indicated by these experiments, it was still plausible that free endogenous mRNAs interacted with released “loading factors” and formed artificial informosomes during cell destruction/homogenization, thus suggesting that the pre-existing natural informosomes might not exist.
To address this question, Spirin and colleagues ran several experiments on loach embryo homogenization in the presence of excessive amounts of exogenous free RNA [35]. If informosomes are formed as a result of association of free endogenous mRNAs with a “loading factor” during homogenization, it should be expected that exogenous RNA would serve as an efficient competitor and would decrease the informosome formation. Such competition experiments, however, clearly demonstrated that informosomes exist in live cells prior to their destruction, homogenization, and fractionation, since the addition of an excess of exogenous RNA did not change the typical sedimentation/distribution profiles of informosomes as well as their characteristic buoyant density [35].
The ability of any exogenous RNA to induce informosome-like particle formation raised other important questions. What is identity of the “loading factor”? Is this one or group of proteins that cooperatively bind RNAs? Does the “loading factor” lack any RNA sequence/RNA structure-binding specificity? While the answers to these questions are still largely lacking, one important outcome during attempts to identify/purify the “loading factor” unexpectedly hinted at the differences in stabilities of natural informosomes and artificial ones. Fractionation analysis of these complexes in sucrose gradients with or without formaldehyde crosslinking followed by density analysis with CsCl gradient centrifugation has unambiguously shown this difference: unlike artificial informosome-like RNPs that are reversible and unstable, natural informosomes are inheritably stable and irreversible under any conditions tested [36]. Another interesting finding was that if the endogenously isolated mRNAs from loach embryos were used in the loach cytoplasmic extracts, such informosome-like particles were much more stable than those made with the exogenous RNAs [36].
The search for the “loading factor” has had important consequences for the development of RNA biology and protein synthesis/translational control, in particular. Analysis of different cytoplasmic informosomes (e.g., “masked” and free cytoplasmic) from dormant and actively dividing cells showed that two major proteins and a large number of minor protein species are present in these mRNA complexes. In contrast to ribosome particles, they had characteristic resistance to removal of Mg2+ (e.g., by adding EDTA) yet could be sequentially removed from the preparation of informosomes by increasing salt concentrations [37]. The identified major proteins had molecular mass of 50-55 kDa (p50) and 70-80 kDa (p70) determined from electrophoretic mobility in denaturing gels [38-43]. Subsequently, biochemical analysis suggested that p70 protein exhibited a predominant affinity to poly(A) sequences, and was later termed as a PABP (poly(A)-binding protein) [39, 43]. The p50 proteins were subsequently characterized as DNA-binding transcription factors that stimulate mRNA synthesis from Y-box promoters with Y-box-binding protein 1 (YB-1 or YBX1) being the best characterized and universal among them [44]. This protein has high content of prolines and alanines, demonstrates anomalous electrophoretic mobility (36 kDa) compared to the predicted, has high isoelectric point (pI > 9) and ubiquitously binds to various heterogeneous mRNA sequences [45]. Both YB-1 and PABP are very abundant proteins and have characteristics suitable for the “loading factor” such as the ability of PABP to bind universal poly(A) tails, and of YB-1 to bind great number of RNA sequences and structures.
It should also be noted that during the search of the “loading factor” several other important discoveries were made. It was shown that many of RBPs found in free informosomes were only present as minor proteins in the polysome-associated informosomes (see below, and figure) suggesting that active remodeling takes place during association of informosomes with polysomes [37]. Later, many of these RBPs were characterized as translation initiation and elongation factors. Similarly, a number of new RBPs were identified that specifically bind 5′- or 3′-UTRs via recognition of specific cis-elements such as TOP motifs (5′-terminal oligopyrimidine motifs [46]) or ARE (AU-rich elements [47]). Consequently, physiological relevance of such RBP : cis-element interactions was shown in the context of the translational control regulation, turnover and localization of transcripts, and their relation to the etiology of pathological conditions.
RELATIONSHIP BETWEEN INFORMOSOMES AND PROTEIN SYNTHESIS
Many of the initial studies on informosomes were done in fish and sea urchin embryos, which have significant differences from the actively dividing cells, where protein synthesis and its control are strongly influenced by exogenous stimuli such as nutrient or oxygen availability. In contrast, embryonic development is controlled by intrinsic factors that are collectively called “clock” mechanisms. One of the features of translational control in early embryogenesis is that the unique patterns of development are mirrored by spatiotemporal regulation of mRNA translation [48, 49]. Following transcription, mRNAs are exported from the nucleus into the cytoplasm, where they are “stored” in a silenced (“masked”) form until a specific time point of the “developmental clock”. In this way, these masked mRNAs are available to be translationally activated in a timely manner. Timewise, there is a significant gap between their export into the cytoplasm and actual engagement into protein biosynthesis. For example, such gap is evident in the loach embryo development, where nuclear information transcribed in the late gastrula is only realized in post-gastrulation stages [50].
Spirin and colleagues hypothesized that the existence of “masked” forms of mRNAs could be explained by the existence of informosomes (discussed in details in [13]). Namely that it is the protein component of these mRNPs playing a regulatory role in the decision of whether mRNAs are accumulated and “silenced” or available for association with ribosomes. Using loach embryos pre-incubated with either radiolabeled amino acids (to detect de novo translated polypeptides) or [14C]-uridine (to detect newly synthesized RNAs), cytoplasmic extracts were prepared from the same developmental stage (late gastrula). Both extracts were mixed and fractionated by sucrose gradient centrifugation revealing differential distribution of labeled mRNAs and polypeptides between the post-ribosomal fractions, monosomes, and polysomes [25, 51]. This analysis unambiguously demonstrated that polysomes are engaged in protein synthesis with the mRNAs synthesized earlier, while the newly synthesized mRNAs are localized in the translationally inactive fractions. It was clear from the results of CsCl density gradient centrifugation that almost all newly synthesized mRNAs (>80%) were present in the informosomes (~1.40 g/cm3 density), none – in the 80S monosomes (~1.55 g/cm3), and a small fraction – in the polysomes (~1.51 g/cm3) [25]. These biochemical results logically agree with the observed delay in translation of the newly synthesized mRNAs in embryos via formation of “masked” mRNPs [13].
One unexpected aspect of this informosome-80S ribosome-polysome mRNA distribution analysis is that the buoyant density of the polysome fraction is less than that of the monosome fraction, suggesting the presence of components that are less dense than RNA in polyribosomes, such as additional bound proteins [25]. Moreover, forced disassociation of the purified polysomes (with EDTA) into monosomes is accompanied by the release of heterogeneous in sedimentation coefficients mRNPs, and not of free mRNAs, also suggesting that informosomes are stable even in the presence of EDTA [21, 25]. Since these complexes have similar buoyant density (~1.4 g/cm3), these results indicate that the released mRNPs are informosomes [25]. Thus, informosomes are able to associate with translating ribosomes, and mRNAs in live cell do not exist in the “naked” form whether being translated or not.
INFORMOSOMES IN THE COMPLEX WORLD OF mRNPs
Based on the available experimental data, Spirin and colleagues formulated the following concept on the relationship between mRNAs and informosomes [52]:
1.Eukaryotic mRNAs do not exist in free form but are always complexed with proteins in vivo;
2.mRNPs of non-ribosomal nature are informosomes;
3.Many eukaryotic proteins involved in RNA metabolism commonly possess RNA binding activities;
4.Some protein components of informosomes (and other RNPs) are RBPs.
As early as 1969, a few predictions were made based on the assumptions derived from this concept [13]. Firstly, it was hypothesized that the export of mRNAs from the nucleus to the cytoplasm happens in the form of informosomes, where they can be immediately transported to the translating polysomes or stored as “masked” mRNPs. Some informosome proteins could assist in such transport, and whether the protein content of informosomes changes during the nucleo-cytoplasmic transport was not clear. “Attachment” of informosomes to the translating ribosomes may involve some modifications of informosomes or their proteins [13]. Secondly, informosomes are a form of mRNP that protect/stabilize mRNA from further processing, e.g., from enzymatic modifications such as nuclease-mediated degradation. Indeed, informosomes are much more resistant than free RNA to ribonuclease activities, and protein constituents of informosome serve this “protector role” [13]. Thirdly, informosomes are an integral part of protein synthesis regulation. In this respect, protein components of informosomes can serve as modulators of translation, e.g., as a translational repressor [13]. Even more attractive, proteins within informosomes can play direct regulatory roles in the dynamics of association/disassociation of informosomes with the translating ribosomes.
Such projections also echoed earlier conceptual developments, where the term “informational” RNA has been used to describe RNA intermediates of different sizes of non-ribosomal and non-transfer RNA nature, which possess ability to hybridize with DNA (“complementary” RNA) [53, 54]. One important implication of such concept is that every “complementary” RNA still contains a sequence-specific information necessary for hybridization with DNA. Consequently, while every mRNA would also be considered as type of “informational” RNA (in conjunction with its role in protein synthesis), it was predicted that some other types of “informational” RNAs exist that do not serve strictly messenger functions but could also be, for example, of regulatory nature. The discovery of informosomes has further extended these developments by showing that protein component of various non-ribosome containing ribonucleoprotein particles (viral RNPs/mRNPs, nuclear RNPs/mRNPs, cytoplasmic “masked” mRNPs) can actively contribute to their cellular fates such as their subcellular localization, stability or association with ribosomes. In this way, the term “informosomes” is much broader than mRNPs and rather unites different RNPs that may or may not contain mRNA (e.g., pre-mRNA complexes or components of RNA granules).
Tight control of mRNA metabolism such as mRNA transport, decay, and translation are crucial for the regulation of gene expression. All these processes are modulated by interaction with RBPs, which bind the mRNA within mRNPs. The fact that RBPs contribute to the fate and functions of RNPs, as based on the informosomes studies, is a fundamental basis for understanding functional and structural features of ribonucleoproteins [55]. Early work on informosomes have predicted a few principles that are applicable to other mRNPs, many of which are well studied (figure). For example, mRNPs are often localized in the specific cellular and subcellular compartments [56]. Such compartmentalization may facilitate certain aspects of mRNA biogenesis (mRNA processing, etc.), to be a part of specific developmental program (classic “masked” informosomes in loach embryos), or part of response to changing conditions (such as stress) [57]. Many mRNPs are dynamic, meaning that they can associate with other RNPs (such as reversible interaction of informosomes with translating ribosomes) and also change their composition depending on the changes of their microenvironment (conversion of nuclear to cytoplasmic informosomes during mRNA export).
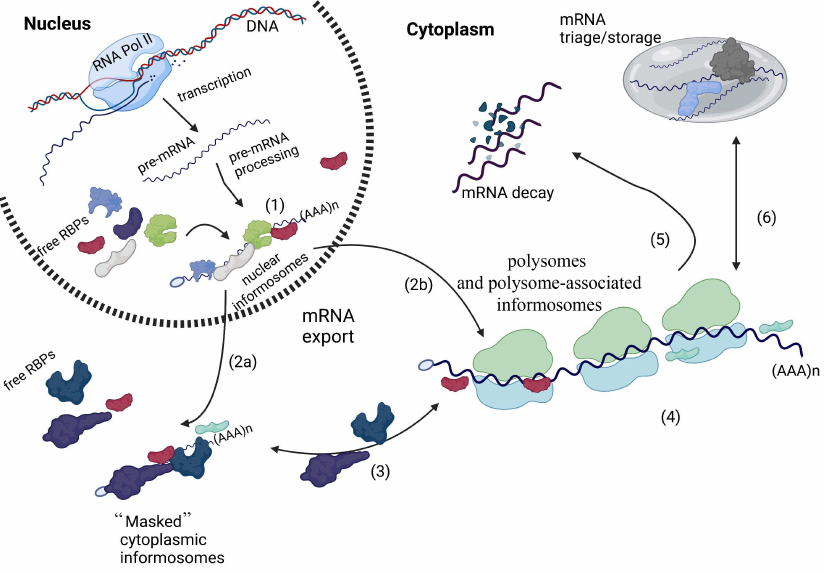
Informosomes and mRNA metabolism. RNA polymerase II transcribes pre-mRNAs in the nucleus, which then undergo several processing steps leading to mature mRNA formation. Processed messenger mRNAs are then decorated with RNA-binding proteins to assemble nuclear informosomes (1). These nuclear complexes are exported into the cytoplasm, where there are either stored in untranslated form (“masked’ cytoplasmic informosomes) (2a) or actively associate with the translating polysomes in the form of polysome-associated informosomes (2b). During mRNA export, some nuclear RBPs are exchanged for cytoplasmic counterparts. In the cytoplasm, “masked” informosomes can be “activated” by external or internal stimuli and then associate with ribosomes as polysome-associated informosomes (3). During mRNA translation (4), polysome-associated informosomes can also dissociate from the translating ribosomes and get back to the “masked” form (3), be directed for mRNA decay (5), or be sorted for mRNA triage or storage within RNA granules such as Stress Granules or Processing Bodies (6). This illustration was created in BioReneder.com.
An important consequence of the informosome concept is coupling of the different mRNA-related processes via interaction of mRNPs with other RBPs (figure). It is now generally accepted that upon transcription and processing, mature mRNAs bind numerous proteins [e.g., family heterogeneous nuclear ribonucleoproteins (hnRNPs) that contribute to several aspects of mRNA metabolism (transport, localization, stability)] [55]. At least some of these nuclear mRNPs are classic nuclear informosomes (figure). After mRNA export, some of the nuclear RBPs are exchanged for other proteins that regulate cytoplasmic mRNA metabolism. Some of these cytoplasmic proteins have multiple functions, e.g., in mRNA localization, stability or/and translation [58]. Multiple mRNPs found in specialized cells (such oocytes) resemble behavior of “masked” informosomes of loach embryos as they contain proteins that contribute to their subcellular localization and stability [57] (figure).
The observation that informosomes interact with translating ribosomes has paved the road to study molecular mechanisms of translational control. That additional change of buoyant density observed in the translating polysomes versus 80S ribosomes indicates that some additional proteins are bound to polyribosomes [51]. It is now very clear that a subset of translation factors, especially translation initiation factors, is first engaged in the interaction with mRNA and only after that with the ribosome [59]. These initiation factors are organized around the universal cap structure and help the small 40S ribosomal subunit both to interact with an mRNA and facilitate proper positioning on the start AUG codon. In this respect, it is worth noting that some reports assumed that informosomes could also be found in the form of mRNA complexes with 40S ribosomal subunits (45S informosomes), reminiscent of the ribosomal pre-initiation complexes. Once such start codon recognition is established, the large 60S ribosomal subunit joins the so-called 48S pre-initiation complex to establish a functional 80S ribosome [59]. The 80S ribosome is then engaged in translation elongation and synthesis of nascent polypeptides encoded in the mRNA’s ORF. Upon translation termination, 80S ribosomes are split again into 60S and 40S subunits. It should, however, be noted that efficiently translated mRNAs simultaneously engage more than one ribosome for translation, and after termination 40S subunits are recycled again for the next round of translation initiation on the same mRNA [60]. This is facilitated by mRNA circularization, where the scaffolding translation initiation factor eIF4G bound to the mRNA cap structure (as a part of the eIF4F complex) interacts with poly(A)-binding protein (PABP) situated on the poly(A) tail [61]. It is tempting to speculate that the observed polysome-associated informosomes or informosomes disassociating from polyribosomes are at least one form of the translation factor-bound mRNPs.
Recent data show that self-organization of mRNPs into diverse membraneless organelles, termed RNA granules, is an evolutionary conserved phenomenon underlying different aspects of mRNA metabolism [62, 63]. High molecular weight informosomes have been identified, and these “gigantic” informosomes may be a product of self-aggregation. Thus, we can speculate that informosomes themselves can be a part of other mRNP-containing RNA granules. Two classes of such cytoplasmic RNA granules are best studied in terms of mRNA metabolism [64]. First are Processing Bodies (PBs), RNA granules enriched with deadenylated mRNAs and specific mRNA decay factors, which are postulated to function in mRNA decay and translation [65]. The second class of mRNA-containing granules is Stress Granules (SGs), which contain translationally stalled mRNAs arrested at the level of translation initiation [66, 67]. SGs are induced by various stresses, and thought to affect cell survival via translational reprogramming [68]. Since both these granules contain multiple mRNAs bound to a diverse set of RBPs, classic informosomes may be constituents of these RNA granules.
CONCLUSIONS
The hypothesis that mRNAs are always associated with proteins and exist in the form of informosomes was revolutionary more than a half century ago. Informosomes are dynamic and dependent on both mRNAs and proteins for their assembly. The informosome model proposed that interaction of proteins with mRNA not only plays a structural role in building of these mRNPs, but also functionally contributes to metabolism of the mRNA. It should be noted that the principle described by Spirin as “omnia mea mecum porto” (“all that is mine I carry with me”) [52], is not limited to informosomes/mRNPs but also applies to other RNPs. It proposes that during evolution, RNPs have acquired an optimal stoichiometric and structural match for RNA and protein partners pointing towards their function as a ribonucleoprotein particle [69]. Such biophysical principles of assembly of RNA-containing complexes are currently under intensive investigation since many of these complexes contribute to human health and disease.
Acknowledgments. Pavel Ivanov thanks members of his lab and Dr. Claire Riggs for help in editing.
Ethics declarations. The author declares no conflict of interest in financial or any other sphere. This article does not contain any studies with of human participants or animals performed by the author.
REFERENCES
1.Bentley, D. L. (2014) Coupling mRNA processing with
transcription in time and space, Nat. Rev. Genet., 15,
163-175, doi: 10.1038/nrg3662.
2.Luna, R., Gaillard, H., Gonzalez-Aguilera, C., and
Aguilera, A. (2008) Biogenesis of mRNPs: integrating different
processes in the eukaryotic nucleus, Chromosoma, 117,
319-331, doi: 10.1007/s00412-008-0158-4.
3.Corley, M., Burns, M. C., and Yeo, G. W. (2020) How
RNA-binding proteins interact with RNA: molecules and mechanisms,
Mol. Cell, 78, 9-29, doi:
10.1016/j.molcel.2020.03.011.
4.Khong, A., and Parker, R. (2020) The landscape of
eukaryotic mRNPs, RNA, 26, 229-239, doi:
10.1261/rna.073601.119.
5.Hershey, J. W. B., Sonenberg, N., and Mathews, M.
B. (2019) Principles of translational control, Cold Spring Harb.
Perspect. Biol., 11, doi: 10.1101/cshperspect.a032607.
6.Mateu-Regue, A., Nielsen, F. C., and Christiansen,
J. (2020) Cytoplasmic mRNPs revisited: singletons and condensates,
Bioessays, 42, e2000097, doi: 10.1002/bies.202000097.
7.Keene, J. D. (2007) RNA regulons: coordination of
post-transcriptional events, Nat. Rev. Genet., 8,
533-543, doi: 10.1038/nrg2111.
8.Aitkhozhin, M. A., Belitsina, N. V., and Spirin, A.
S. (1964) Nucleic Acids in the early stages of development of fish
embryos (based on the loach Misgurnus Fossilis), Biokhimiia,
29, 169-175.
9.Belitsina, N. V., Aitkhozhin, M. A., Gavrilova, L.
P., and Spirin, A. S. (1964) The messenger ribonucleic acids of
differentiating animal cells, Biokhimiia, 29,
363-374.
10.Spirin, A. S., Belitsina, N. V., and Lerman, M.
I. (1965) Use of formaldehyde fixation for studies of ribonucleoprotein
particles by caesium chloride density-gradient centrifugation, J.
Mol. Biol., 14, 611-615, doi:
10.1016/s0022-2836(65)80213-3.
11.Spirin, A. S., Belitsina, N. V., and Aitkhozhin,
M. A. (1964) Messenger RNA in early embryogenesis, Zhurn. Obshch.
Biol., 25, 321-338.
12.Ovchinnikov, L. P., Ajtkhozhin, M. A., Bystrova,
T. F., and Spirin, A. S. (1969) Newt embryo informosomes: 1.
Sedimentation and density parameters, Mol. Biol. (USSR),
3, 449-464.
13.Spirin, A. S. (1969) The Second Sir Hans Krebs
Lecture. Informosomes, Eur. J. Biochem., 10, 20-35.
14.Spirin, A. S., and Nemer, M. (1965) Messenger RNA
in early sea-urchin embryos: cytoplasmic particles, Science,
150, 214-217, doi: 10.1126/science.150.3693.214.
15.Belitsina, N. V., Ovchinnikov, L. P., Spirin, A.
S.,Gendon, Yu. Z., and Cheros, V. I. (1968) Informosomes of HeLa cells
infected with vaccinia virus, Mol. Biol. (U.S.S.R.),
2.
16.Infante, A. A., and Nemer, M. (1968)
Heterogeneous ribonucleoprotein particles in the cytoplasm of sea
urchin embryos, J. Mol. Biol., 32, 543-565, doi:
10.1016/0022-2836(68)90342-2.
17.Joklik, W. K., and Becker, Y. (1965) Studies on
the genesis of polyribosomes. II. The association of nascent messenger
RNA with the 40 S subribosomal particle, J. Mol. Biol.,
13, 511-520, doi: 10.1016/s0022-2836(65)80113-9.
18.Shatkin, A. J., Sebring, E. D., and Salzman, N.
P. (1965) Vaccinia virus directed RNA: its fate in the presence of
actinomycin, Science, 148, 87-90, doi:
10.1126/science.148.3666.87.
19.McConkey, E. H., and Hopkins, J. W. (1965)
Subribosomal particles and the transport of messenger RNA in HeLa
cells, J. Mol. Biol., 14, 257-270, doi:
10.1016/s0022-2836(65)80245-5.
20.Kafatos, F. C. (1968) Cytoplasmic particles
carrying rapidly labeled RNA in developing insect epidermis, Proc.
Natl. Acad. Sci. USA, 59, 1251-1258, doi:
10.1073/pnas.59.4.1251.
21.Henshaw, E. C. (1968) Messenger RNA in rat liver
polyribosomes: evidence that it exists as ribonucleoprotein particles,
J. Mol. Biol., 36, 401-411, doi:
10.1016/0022-2836(68)90164-2.
22.Perry, R. P., and Kelley, D. E. (1968) Messenger
RNA-protein complexes and newly synthesized ribosomal subunits:
analysis of free particles and components of polyribosomes, J. Mol.
Biol., 35, 37-59, doi: 10.1016/s0022-2836(68)80035-x.
23.Levy, H. B., and Carter, W. A. (1968) Molecular
basis of the action of interferon, J. Mol. Biol., 31,
561-577, doi: 10.1016/0022-2836(68)90428-2.
24.Volkova, M. Y., Zaides, V. M., and Zaslavsky, V.
G. (1969) Slowly sedimenting particles present in cytoplasmic extract
of Ehrlich ascites cells infected by Sendai virus, Mol. Biol.
(U.S.S.R.), 3, 4-9.
25.Ovchinnikov, L. P., Belitsina, N. V., Avanesov,
A., and Spirin, A. S. (1969) Postribosomal RNA-containing particles of
cytoplasm of animal cells according to CsCl density gradient
centrifugation data, Dokl. Akad. Nauk SSSR, 186,
1202-1205.
26.Neifakh, A. A. (1959) Method of inactivation of
nuclei by radiation and its possible applications for the investigation
of nuclei functions during early development of fish, Zhurn. Obshch.
Biol. (Russian), 20, 202-207.
27.Samarina, O. P., Asriian, I. S., and Georgiev, G.
P. (1965) Isolation of nuclear nucleoproteins containing informational
ribonucleic acid, Dokl. Akad. Nauk SSSR, 163,
1510-1513.
28.Samarina, O. P., Krichevskaya, A. A., and
Georgiev, G. P. (1966) Nuclear ribonucleoprotein particles containing
messenger ribonucleic acid, Nature, 210, 1319-1322, doi:
10.1038/2101319a0.
29.Samarina, O. P., Lerman, M. I., Tumanian, V. D.,
Anan’eva, L. N., and Georgiev, G. P. (1965) Characteristics of
chromosomal information RNA, Biokhimiia, 30, 880-893.
30.Samarina, O. P., Lukanidin, E. M., and Georgiev,
G. P. (1967) On the structural organization of the nuclear complexes
containing messenger RNA, Biochim. Biophys. Acta, 142,
561-564, doi: 10.1016/0005-2787(67)90642-9.
31.Samarina, O. P., Lukanidin, E. M., Molnar, J.,
and Georgiev, G. P. (1968) Structural organization of nuclear complexes
containing DNA-like RNA, J. Mol. Biol., 33, 251-263, doi:
10.1016/0022-2836(68)90292-1.
32.Samarina, O. P., Molnar, J., Lukanidin, E. M.,
Bruskov, V. I., Krichevskaya, A. A., and Georgiev, G. P. (1967)
Reversible dissociation of nuclear ribonucleoprotein particle
containing mRNA into RNA and protein, J. Mol. Biol., 27,
187-191, doi: 10.1016/0022-2836(67)90359-2.
33.Girard, M., and Baltimore, D. (1966) The effect
of HeLa cell cytoplasm on the rate of sedimentation of RNA, Proc.
Natl. Acad. Sci. USA, 56, 999-1002, doi:
10.1073/pnas.56.3.999.
34.Ovchinnikov, L. P., Voronina, A. S., Stepanov, A.
S., Belitsina, N. V., and Spirin, A. S. (1968) Informosome-like
complexes were formed by RNA adding to animal cell homogenates, Mol.
Biol. (U.S.S.R.), 2, 752-761.
35.Ovchinnikov, L. P., Avanesov, A. C., and Spirin,
A. S. (1969) Informosomes from loach embryos, Molek. Biol.
(U.S.S.R.), 3, 465471.
36.Ovchinnikov, L. P., and Avanesov, A. C. (1969)
Informosomes of loach embryos. 3. Specificity of interaction of
“informosome-forming” protein with RNA, Mol. Biol.
(U.S.S.R.), 3, 5-12.
37.Spirin, A. S. (1994) Storage of messenger RNA in
eukaryotes: envelopment with protein, translational barrier at 5′
side, or conformational masking by 3′ side? Mol. Reprod.
Dev., 38, 107-117, doi: 10.1002/mrd.1080380117.
38.Blobel, G. (1972) Protein tightly bound to globin
mRNA, Biochem. Biophys. Res. Commun., 47, 88-95, doi:
10.1016/s0006-291x(72)80014-7.
39.Blobel, G. (1973) A protein of molecular weight
78,000 bound to the polyadenylate region of eukaryotic messenger RNAs,
Proc. Natl. Acad. Sci. USA, 70, 924-928, doi:
10.1073/pnas.70.3.924.
40.Jain, S. K., Pluskal, M. G., and Sarkar, S.
(1979) Thermal chromatography of eukaryotic messenger ribonucleoprotein
particles on oligo (dT)-cellulose. Evidence for common mRNA-associated
proteins in various cell types, FEBS Lett., 97, 84-90,
doi: 10.1016/0014-5793(79)80058-7.
41.Kumar, A., and Pederson, T. (1975) Comparison of
proteins bound to heterogeneous nuclear RNA and messenger RNA in HeLa
cells, J. Mol. Biol., 96, 353-365, doi:
10.1016/0022-2836(75)90165-5.
42.Morel, C., Kayibanda, B., and Scherrer, K. (1971)
Proteins associated with globin messenger RNA in avian erythroblasts:
isolation and comparison with the proteins bound to nuclear
messenger-likie RNA, FEBS Lett., 18, 84-88, doi:
10.1016/0014-5793(71)80413-1.
43.van Venrooij, W. J., van Eekelen, C. A., Jansen,
R. T., and Princen, J. M. (1977) Specific poly-A-binding protein of
76,000 molecular weight in polyribosomes is not present on poly A of
free cytoplasmic mRNP, Nature, 270, 189-191, doi:
10.1038/270189a0.
44.Minich, W. B., Maidebura, I. P., and Ovchinnikov,
L. P. (1993) Purification and characterization of the major 50-kDa
repressor protein from cytoplasmic mRNP of rabbit reticulocytes,
Eur. J. Biochem., 212, 633-638, doi:
10.1111/j.1432-1033.1993.tb17701.x.
45.Mordovkina, D., Lyabin, D. N., Smolin, E. A.,
Sogorina, E. M., Ovchinnikov, L. P., and Eliseeva, I. (2020) Y-box
binding proteins in mRNP assembly, translation, and stability control,
Biomolecules, 10, doi: 10.3390/biom10040591.
46.Cockman, E., Anderson, P., and Ivanov, P. (2020)
TOP mRNPs: molecular mechanisms and principles of regulation,
Biomolecules, 10, doi: 10.3390/biom10070969.
47.Ivanov, P., and Anderson, P. (2013)
Post-transcriptional regulatory networks in immunity, Immunol.
Rev., 253, 253-272, doi: 10.1111/imr.12051.
48.Stebbins-Boaz, B., and Richter, J. D. (1997)
Translational control during early development, Crit. Rev. Eukaryot.
Gene Expr., 7, 73-94, doi:
10.1615/critreveukargeneexpr.v7.i1-2.50.
49.Teixeira, F. K., and Lehmann, R. (2019)
Translational control during developmental transitions, Cold Spring
Harb. Perspect. Biol., 11, doi:
10.1101/cshperspect.a032987.
50.Neyfakh, A. A. (1964) Radiation investigation of
nucleo-cytoplasmic interrelations in morphogenesis and biochemical
differentiation, Nature, 201, 880-884, doi:
10.1038/201880a0.
51.Ovchinnikov, L. P., Bystrova, T. F., and Spirin,
A. S. (1969) Sedimentation and density characteristics of ribosomes and
their subunits from embryonic groundlings, Dokl. Akad. Nauk
SSSR, 185, 210-213.
52.Spirin, A. S. (1978) Eukaryotic messenger RNA and
informosomes. Omnia mea mecum porto, FEBS Lett., 88,
15-17, doi: 10.1016/0014-5793(78)80596-1.
53.Spiegelman, S. (1961) The relation of
informational RNA to DNA, Cold Spring Harb. Symp. Quant. Biol.,
26, 75-90, doi: 10.1101/sqb.1961.026.01.013.
54.Spiegelman, S., Hall, B. D., and Storck, R.
(1961) The occurrence of natural DNA–RNA complexes in E.
coli infected with T2, Proc. Natl. Acad. Sci. USA,
47, 1135-1141, doi: 10.1073/pnas.47.8.1135.
55.Bjork, P., and Wieslander, L. (2017) Integration
of mRNP formation and export, Cell. Mol. Life Sci., 74,
2875-2897, doi: 10.1007/s00018-017-2503-3.
56.Fazal, F. M., Han, S., Parker, K. R., Kaewsapsak,
P., Xu, J., et al. (2019) Atlas of subcellular RNA localization
revealed by APEX-Seq, Cell, 178, 473-490.e426, doi:
10.1016/j.cell.2019.05.027.
57.Kong, J., and Lasko, P. (2012) Translational
control in cellular and developmental processes, Nat. Rev.
Genet., 13, 383-394, doi: 10.1038/nrg3184.
58.Singh, G., Pratt, G., Yeo, G. W., and Moore, M.
J. (2015) The clothes make the mRNA: past and present trends in mRNP
fashion, Annu. Rev. Biochem., 84, 325-354, doi:
10.1146/annurev-biochem-080111-092106.
59.Jackson, R. J., Hellen, C. U., and Pestova, T. V.
(2010) The mechanism of eukaryotic translation initiation and
principles of its regulation, Nat. Rev. Mol. Cell Biol.,
11, 113-127, doi: 10.1038/nrm2838.
60.Hellen, C. U. T. (2018) Translation termination
and ribosome recycling in eukaryotes, Cold Spring Harb. Perspect.
Biol., 10, doi: 10.1101/cshperspect.a032656.
61.Wells, S. E., Hillner, P. E., Vale, R. D., and
Sachs, A. B. (1998) Circularization of mRNA by eukaryotic translation
initiation factors, Mol. Cell, 2, 135-140, doi:
10.1016/s1097-2765(00)80122-7.
62.Zhang, H., Ji, X., Li, P., Liu, C., Lou, J., et
al. (2020) Liquid–liquid phase separation in biology: mechanisms,
physiological functions and human diseases, Sci. China Life
Sci., 63, 953-985, doi: 10.1007/s11427-020-1702-x.
63.Hyman, A. A., Weber, C. A., and Julicher, F.
(2014) Liquid-liquid phase separation in biology, Annu. Rev. Cell
Dev. Biol., 30, 39-58, doi:
10.1146/annurev-cellbio-100913-013325.
64.Ivanov, P., Kedersha, N., and Anderson, P. (2019)
Stress granules and processing bodies in translational control, Cold
Spring Harb. Perspect. Biol., 11, doi:
10.1101/cshperspect.a032813.
65.Luo, Y., Na, Z., and Slavoff, S. A. (2018)
P-Bodies: composition, properties, and functions, Biochemistry,
57, 2424-2431, doi: 10.1021/acs.biochem.7b01162.
66.Hofmann, S., Kedersha, N., Anderson, P., and
Ivanov, P. (2021) Molecular mechanisms of stress granule assembly and
disassembly, Biochim. Biophys. Acta Mol. Cell Res., 1868,
118876, doi: 10.1016/j.bbamcr.2020.118876.
67.Riggs, C. L., Kedersha, N., Ivanov, P., and
Anderson, P. (2020) Mammalian stress granules and P bodies at a glance,
J. Cell Sci., 133, doi: 10.1242/jcs.242487.
68.Advani, V. M., and Ivanov, P. (2019)
Translational control under stress: reshaping the translatome,
Bioessays, 41, e1900009, doi: 10.1002/bies.201900009.
69.Mitchell, S. F., and Parker, R. (2014) Principles
and properties of eukaryotic mRNPs, Mol. Cell, 54,
547-558, doi: 10.1016/j.molcel.2014.04.033.